Abstract
Multiple-locus variable-number tandem-repeat analysis (MLVA), multiplex PCR, and PCR-restriction fragment length polymorphism analysis were compared for typing Brucella suis isolates. A perfect concordance was obtained among these molecular assays. However, MLVA was the only method to demonstrate brucellosis outbreaks and to confirm that wildlife is a reservoir for zoonotic brucellosis.
Brucella suis, the causative agent of swine brucellosis, is classified in five biovars that preferentially infect different animal hosts: biovars 1 and 3 affect domestic pigs and wild boars, biovar 2 also affects hares, biovar 4 infects reindeer and caribou, and biovar 5 infects only rodents. In contrast to biovars 1 and 3, biovar 2 has been isolated rarely from humans, and its zoonotic role is questioned (9). The increasing number of brucellosis outbreaks in pig farms is becoming a serious problem all over Europe (9, 13). Hares and wild boars are widely infected with B. suis, and spillover from wildlife to domestic pigs and cattle has been reported (3, 10, 11, 13). Accurate diagnostic and typing procedures are essential for epidemiological studies aimed at B. suis control or eradication.
The identification of the B. suis biovars is currently performed by standard bacteriological methods (2). However, these tests lack specificity and are not straightforward particularly for the identification of biovars 1, 2, and 3 (7). With the objective of improving the typing of these biovars, different PCR-based assays have been proposed. The most widely used are the PCR-restriction fragment length polymorphism (RFLP) analysis of genes omp2a and omp2b, which can differentiate between reference biovars 1, 2, and 3 (6), and of gene omp31, which is able to differentiate biovars 1 and 3 from biovar 2 (15). In addition, a modification of the original AMOS-PCR multiplex assay (4), including a new pair of primers derived from the ery locus (AMOS-ery-PCR [14]), can differentiate biovar 1 from others. Recently, a new “fingerprinting” approach based on a PCR method for multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) has been developed for molecular typing of Brucella (5, 12, 16). The high variability of VNTR loci has already been applied in a large panel of animal and human Brucella isolates, proving a high discriminatory power for typing purposes (1, 8, 12). However, the concordance of VNTR analysis with other molecular typing assays has not been directly investigated yet. To do this, a representative collection of B. suis field isolates was tested; 35 strains were isolated from pigs, 13 from wild boars, 8 from hares, and 2 from horses, and the strains were isolated from different regions of Europe during more than 20 years to ensure adequate diversity (Fig. 1). All isolates were typed according to standard bacteriological procedures (2), and 38 strains were identified as biovar 2, and 20 were identified as biovar 1 (6 of these strains showed atypical fuchsin resistance). Growth and harvesting of Brucella cells and bacterial DNA were performed as described elsewhere (2, 8, 12). All isolates were subjected to five different PCR-based typing techniques: PCR-RFLP analysis for omp2a, omp2b, and omp31 genes (6, 15), multiplex AMOS-ery-PCR (14), and Brucella MLVA using 15 genetic markers (8, 12).
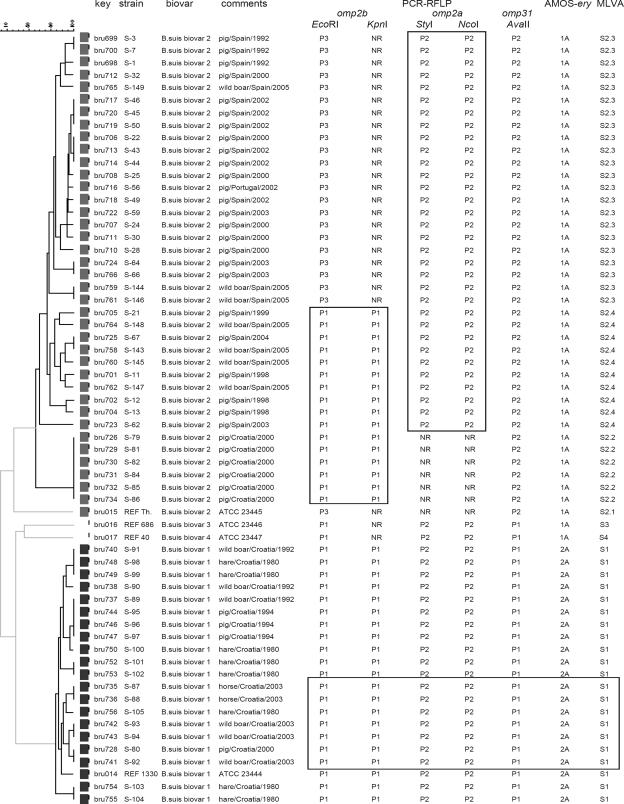
FIG. 1.
Dendrogram constructed from MLVA testing and molecular typing results of representative B. suis isolates. The dendrogram was generated using a distance matrix calculated with the categorical coefficient and the unweighted-pair group method using average linkages as previously described (12). An identical weight was given to each marker. The 58 animal B. suis isolates investigated clustered (indicated by boxes) in 35 different MLVA genotypes. In the columns the following data are presented: DNA batch (key); strain identification; biovar, according to standard bacteriological procedures; animal host, geographic origin, and year of isolation (comments); omp2b, omp2a, and omp31 PCR-RFLP patterns, as defined originally in references 5 and 14; AMOS-ery-PCR patterns, as defined originally in references 4 and 13; and MLVA patterns, see text for details. B. suis biovar 1, 2, 3 and 4 ATCC reference strains were included as controls. Based on detailed background information collected from the veterinary records, strains S-3 and S-7, strains S-45, S-46, and S-50, strains S-64 and S-66, strains S-87 and S-88, strains S-95, S-96, and S-97, and strains S-144 and S-146 corresponded to specific outbreaks.
Figure 1 summarizes the molecular typing results and represents a dendrogram construct from MLVA genotyping assay data on the B. suis isolates. Strains of the reference biovars 1, 2, and 3 could be differentiated clearly according to the RFLP identified in omp genes and to MLVA results. By contrast, the AMOS-ery-PCR assay was unable to differentiate between biovars 2 and 3. Twenty field strains typed as biovar 1 were also grouped together using the different PCR-based techniques (S1 pattern). All these strains gave the same results that the reference biovar 1 strain, suggesting a very high genetic homogeneity for this biovar. Interestingly, this is the first evidence showing that B. suis biovar 1 is a cause of brucellosis in hares. Neither PCR-RFLP nor AMOS-ery-PCR assays were able to differentiate the six biovar 1 strains showing atypical fuchsin resistance (strains S-80, S-87, S-88, S-92, S-93, and S-94 in Fig. 1). In contrast, these six strains were placed in very closely related MLVA clusters (Fig. 1). A similar correlation between this phenotypic characteristic and VNTR clustering has been recently reported in Brucella melitensis (1). This is also the first evidence showing the existence of fuchsin-resistant B. suis biovar 1 strains in Europe. When testing the 38 strains identified as biovar 2, the omp31 PCR-RFLP and AMOS-ery-PCR assays resulted in patterns identical to those obtained with the reference biovar 2 strain (S2 patterns). However, the RFLP patterns of the omp2a and omp2b PCR-amplified product (Fig. 1) demonstrated additional polymorphism within this biovar, and at least four genetic subgroups were identified (S2.1, S2.2, S2.3, and S2.4), which were also identified in separate clusters by the MLVA assay (Table 1). Subgroup S2.1 included only the reference biovar 2 strain, which was also clearly differentiated from the remaining S2 clusters by MLVA. MLVA results were in perfect agreement with those of standard typing procedures and clearly differentiated these 38 strains from the rest of the patterns (S1, S3, and S4).
TABLE 1.
Repeat copy numbers at each locus in the MLVA assay for 58 B. suis representative isolates
| Markera | No. of repeat copies at each locus in the following patternb:
|
||||||
|---|---|---|---|---|---|---|---|
| S1 | S2.1 | S2.2 | S32.3 | S2.4 | S3 | S4 | |
| bruce04 | 2-7 | 9 | 2 | 2 | 2-7 | 7 | 4 |
| bruce06 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| bruce07 | 0-8 | 9 | 5 | 0-7 | 0-10 | 5 | 5 |
| bruce08 | 5 | 4 | 7 | 7 | 7 | 3 | 3 |
| bruce09 | 0-8 | 18 | 3 | 0-21 | 5-17 | 10 | 9 |
| bruce11 | 6 | 8 | 8 | 8 | 8 | 4 | 9 |
| bruce12 | 10 | 15 | 15 | 9 | 9 | 11 | 11 |
| bruce16 | 0-5 | 2 | 2 | 2 | 2 | 4 | 6 |
| bruce18 | 0-4 | 6 | 4 | 5-6 | 5-6 | 4 | 5 |
| bruce21 | 0-9 | 9 | 9 | 9 | 9 | 9 | 9 |
| bruce30 | 3 | 4 | 8 | 0-8 | 0-8 | 5 | 3 |
| bruce42 | 4 | 6 | 5 | 5 | 5 | 3 | 3 |
| bruce43 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| bruce45 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| bruce55 | 3 | 2 | 8 | 0-5 | 5-6 | 2 | 2 |
Table 1 summarizes the repeat copy numbers at each locus in the MLVA assay. Four markers (bruce06, bruce21, bruce43, and bruce45) proved to be almost identical in all B. suis strains tested. In contrast, markers bruce04, bruce07, bruce09, bruce12, bruce18, bruce30, and bruce55 were highly discriminatory. The four B. suis biovars tested were clearly differentiated with markers bruce11 and bruce16.
There was a perfect correlation of classical typing, omp31 PCR-RFLP, AMOS-ery-PCR, and MLVA assays. However, the MLVA was the only assay able to evidence epidemiological relationship between strains. For example, some strains that clustered together after the MLVA analysis had been isolated from different animal hosts. As an example, strain 89 (from a wild boar) and strains 95, 96 and 97 (from domestic pigs) share exactly the same genotype. These results suggest the spread of particular B. suis strains from one animal species to another and that spillover from wildlife to domestic pigs seems to be a frequent event (3, 10, 11, 13). In addition, the MLVA assay demonstrated the same genotype in strains isolated from the same outbreak: strains 45, 46, and 50 from Spain, for instance. In conclusion, the MLVA is the most sensitive assay suitable for simultaneously typing B. suis and epidemiologically tracing the infection.
Acknowledgments
Fellowship support to D. García-Yoldi from the Asociación de Amigos de la Universidad de Navarra and from Short-Term Scientific Mission COST Action 845 (Brucellosis in Animals and Man) are also gratefully acknowledged. This work was partially supported by the Ministerio de Educación y Ciencia, Spain (AGL2005-07401-C03-03). The development of methods improving the accountability of dangerous pathogens is supported by the French “Delegation Generale pour l'Armement” (DGA).
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Al Dahouk, S., P. Le Fleche, K. Nockler, I. Jacques, M. Grayon, H. C. Scholz, H. Tomaso, G. Vergnaud, and H. Neubauer. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69:137-145. [DOI] [PubMed] [Google Scholar]
- 2.Alton, G. G., L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France.
- 3.Andersen, F. M., and K. B. Pedersen. 1995. Brucellosis. A case of natural infection of a cow with Brucella suis biotype 2. Dan. Vet. 78:408. [Google Scholar]
- 4.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker, B. J., D. R. Ewalt, and S. M. Halling. 2003. Brucella ‘HOOF-Prints’: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grépinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 141:2111-2121. [DOI] [PubMed] [Google Scholar]
- 7.Ferrao-Beck, L., R. Cardoso, P. M. Muñoz, M. J. de Miguel, D. Albert, A. C. Ferreira, C. M. Marín, M. Thiebaud, I. Jacques, M. Grayon, M. S. Zygmunt, B. Garín-Bastuji, J. M. Blasco, and M. I. Sa. 2006. Development of a multiplex PCR assay for polymorphism analysis of Brucella suis biovars causing brucellosis in swine. Vet. Microbiol. 115:269-277. [DOI] [PubMed] [Google Scholar]
- 8.García-Yoldi, D., P. Le Fleche, C. M. Marín, M. J. De Miguel, P. M. Muñoz, G. Vergnaud, and I. López-Goñi. 2007. Assessment of genetic stability of Brucella melitensis Rev 1 vaccine strain by multiple-locus variable-number tandem repeat analysis. Vaccine 25:2858-2862. [DOI] [PubMed] [Google Scholar]
- 9.Garin-Bastuji, B., and J. Hars. 2001. Situation epidemiologique de la brucellose a Brucella suis biovar 2 en France. Bull. Epidémiol. L'AFSSA 2:3-4. [Google Scholar]
- 10.Godfroid, J., P. Michel, L. Uytterhaegen, C. De Smedt, F. Rasseneur, F. Boelaert, C. Saegerman, and X. Patigny. 1994. Endemic brucellosis due to Brucella suis biotype 2 in the wild boar (Sus scrofa) in Belgium. Ann. Med. Vet. 138:263-268. [Google Scholar]
- 11.Godfroid, J., C. Saegerman, V. Wellemans, K. Walravens, J. J. Letesson, A. Tibor, A. McMillan, S. Spencer, M. Sanna, D. Bakker, R. Pouillot, and B. Garin-Bastuji. 2002. How to substantiate eradication of bovine brucellosis when aspecific serological reactions occur in the course of brucellosis testing. Vet. Microbiol. 90:461-477. [DOI] [PubMed] [Google Scholar]
- 12.Le Fleche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leuenberger, R., P. Boujon, B. Thur, R. Miserez, B. Garin-Bastuji, J. Rufenacht, and K. D. Stark. 2007. Prevalence of classical swine fever, Aujeszky's disease and brucellosis in a population of wild boar in Switzerland. Vet. Rec. 160:362-368. [DOI] [PubMed] [Google Scholar]
- 14.Ocampo-Sosa, A. A., J. Agüero-Balbin, and J. M. García-Lobo. 2005. Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet. Microbiol. 110:41-51. [DOI] [PubMed] [Google Scholar]
- 15.Vizcaíno, N., J. M. Verger, M. Grayon, M. S. Zygmunt, and A. Cloeckaert. 1997. DNA polymorphism at the omp-31 locus of Brucella spp.: evidence for a large deletion in Brucella abortus, and other species-specific markers. Microbiology 143:2913-2921. [DOI] [PubMed] [Google Scholar]
- 16.Whatmore, A. M., S. J. Shankster, L. L. Perrett, T. J. Murphy, S. D. Brew, R. E. Thirlwall, S. J. Cutler, and A. P. MacMillan. 2006. Identification and characterisation of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 44:1982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]