Abstract
Klebsiella spp. have become an important cause of clinical mastitis in dairy cows in New York State. We describe the occurrence of two Klebsiella mastitis outbreaks on a single dairy farm. Klebsiella isolates from milk, feces, and environmental sources were compared using random amplified polymorphic DNA (RAPD)-PCR typing. The first mastitis outbreak was caused by a single strain of Klebsiella pneumoniae, RAPD type A, which was detected in milk from eight cows. RAPD type A was also isolated from the rubber liners of milking machine units after milking of infected cows and from bedding in the outbreak pen. Predominance of a single strain could indicate contagious transmission of the organism or exposure of multiple cows to an environmental point source. No new cases with RAPD type A were observed after implementation of intervention measures that targeted the prevention of transmission via the milking machine as well as improvement of environmental hygiene. A second outbreak of Klebsiella mastitis that occurred several weeks later was caused by multiple RAPD types, which rules out contagious transmission and indicates opportunistic infections originating from the environment. The diversity of Klebsiella strains as quantified with Simpson's index of discrimination was significantly higher for isolates from fecal, feed, and water samples than for isolates from milk samples. Several isolates from bedding material that had the phenotypic appearance of Klebsiella spp. were identified as being Raoultella planticola and Raoultella terrigena based on rpoB sequencing.
Klebsiella pneumoniae has been reported to be an important cause of clinical mastitis (CM) in the northern United States (20, 25). CM due to Klebsiella infection results in high milk losses and mortality of the affected cows (13). Antimicrobial therapy and vaccination have limited effects against Klebsiella CM (15, 27). Hence, prevention of infections through the reduction of exposure of cows to Klebsiella has been the cornerstone of Klebsiella mastitis control on dairy farms. Sources of Klebsiella spp. in dairy operations include organic bedding material such as wood by-products (16). In addition, fecal shedding by cows contributes to the presence of a large variety of K. pneumoniae strains in dairy herds (20, 21). Despite the increasing prevalence and incidence of Klebsiella infections in dairy herds and the impact of infections on cow health and productivity, information on the molecular epidemiology of Klebsiella mastitis is very limited (25). In this paper, we present a case study with the molecular epidemiology of two Klebsiella mastitis outbreaks on a single dairy farm.
Outbreak history.
An unusual increase in the number of severe cases of CM in a New York State dairy herd occurred in April 2006. The herd, a 410-cow Holstein Friesian herd in free stall facilities bedded with sawdust, had an average daily milk production of 35.5 kg, a 305-day mature equivalent (i.e., age, season, and fat-corrected milk production) of 11,318 kg, and a rolling bulk milk somatic cell count of 244,000 cells/ml. Milk samples (n = 24) from 20 cows with CM were submitted to Quality Milk Production Services (QMPS), Cornell University, Ithaca, NY, for bacteriological culture. Klebsiella pneumoniae was detected in 10 of 17 (59%) of the culture-positive samples. To identify possible sources of K. pneumoniae in the herd so that targeted measures could be taken to prevent new infections, bedding samples and animal feces were collected on 27 April 2006. Klebsiella isolates from milk samples, feces, and the environment were analyzed using random amplified polymorphic DNA (RAPD) typing. Nine of 10 isolates from milk showed indistinguishable DNA banding patterns. The nine isolates with indistinguishable banding patterns were from CM cases that occurred in the management group that was housed in pen 3. The isolate with the second banding pattern originated from a CM case in a different management group, which was housed in pen 4. Based on typing results, the observation that almost all clinical Klebsiella cases had occurred in one group of animals that were housed and milked together and analysis of isolates from the milking equipment, contagious transmission from cow to cow or exposure to an environmental point source of the pathogen was suspected. Preventive measures aimed at the limitation of cow-to-cow transmission were implemented. Briefly, Klebsiella-infected animals were segregated from the herd or marked and milked with a dedicated unit that was sanitized using an acidified iodine solution (Dyne; West Agro, Kansas City, MO) after each use. Antimicrobial therapy of confirmed Klebsiella mastitis cases was not attempted, because treatment is rarely effective. The rubber liners of the milking units were replaced, and the milking vacuum was increased. Using weekly cow cleanliness scores as a guideline, the frequencies of bedding replacement and alley scraping were increased to improve environmental hygiene.
After intervention measures were fully implemented, no new cases of CM due to Klebsiella were reported for 3 weeks. Approximately 6 weeks later, starting in mid-June 2006, a second outbreak of Klebsiella CM was observed. This outbreak lasted through July and affected cows in multiple management groups. In addition to milk samples from cows with CM, milk samples from a whole-herd sampling were available. The whole-herd survey was performed by QMPS personnel in the first week of July 2006 at the request of the producer. Using RAPD-PCR, many different strains were identified among isolates from CM cases that occurred during the second outbreak. The second outbreak exemplifies Klebsiella mastitis cases due to exposure to the large variety of Klebsiella strains that is commonly present in the dairy farm environment. To identify specific environmental sources of Klebsiella strains that caused mastitis, additional environmental samples, including samples from feed and drinking water, were collected and analyzed towards the end of the second outbreak in August 2006.
MATERIALS AND METHODS
Samples from milk and the milking machine.
Milk samples from CM cases were aseptically collected using standard procedures (22), frozen, and sent in cooler boxes to the QMPS laboratory by the farm manager. Additional milk samples were collected by the authors at the time of sampling of the milking machine (see below) and by QMPS personnel during the whole-herd sampling during the first week of July 2006. Milk samples from CM cases and the whole-herd sampling were processed using routine methods for the isolation and identification of mastitis pathogens (22). Briefly, approximately 0.01 ml of each milk sample was swabbed onto trypticase soy agar with 5% sheep blood and 0.1% esculin (TSA-BE; PML Microbiologicals, Mississauga, Ontario, Canada). Growth was evaluated after 24 h and 48 h of incubation at 37°C. For samples from cows with CM, a 1:1 mixture with Todd-Hewitt broth (THB) (CM0189; Oxoid Ltd., Basingstoke, United Kingdom) was prepared, and after 4 h of enrichment at 37°C, additional TSA-BE plates were streaked and incubated in the same manner as the rest of the milk samples. Coliform isolates were identified based on colony morphology, subcultured onto MacConkey agar no. 3 (CM0115; Oxoid Ltd., Basingstoke, United Kingdom), and incubated overnight at 37°C. Pink-yellow, mucoid, lactose-positive colonies were considered to be Klebsiella spp. and identified to the species level based on motility and biochemistry (i.e., citrate utilization and indole production) (22). One phenotypically confirmed Klebsiella isolate per milk sample was selected for storage and molecular typing.
To explore the possibility of transmission of Klebsiella via the milking machine, swabs were taken from the teat cup liners of milking units in May 2006. Following the approach described previously by Zadoks et al. (36), three cows that had CM caused by K. pneumoniae in the first outbreak were selected. Because of the chronic nature of K. pneumoniae infections, the likelihood that these cows would still shed the organism was considered to be high. Quarter milk samples from each animal were collected aseptically to determine the actual infection status with respect to Klebsiella. Following milking of each of the selected cows, two additional cows were milked with the same unit. Quarter milk samples were also collected from those cows. Before and after each of the three selected cows and their two subsequent cows were milked, the inside surfaces of the rubber liners of the milking units were sampled with saline-moisturized cotton wool swabs (BioMérieux, Marcy l'Étoile, France). Swabs were inserted into the liner and withdrawn in a spiraling motion while rotating the swab at the same time. Swabs were put into sterile tubes containing 1.5 ml of THB. Milk samples and swabs were transported to the laboratory in cooler boxes with ice packs. Processing of milk samples was performed as described above for samples from the herd survey. Liner swabs in THB tubes were incubated overnight at 37°C, and approximately 0.01 ml of the enrichment was swabbed onto MacConkey agar containing 10 mg/liter of ampicillin (MacA), a Klebsiella-selective medium (20). MacA plates were incubated for 18 to 24 h at 37°C, and putative Klebsiella isolates were identified as described above for milk samples. Four colonies per liner swab were selected from each plate, i.e., one from the center of the plate and three from the periphery at 120° angles. Phenotypic confirmation of Klebsiella isolates was performed as described above for milk samples, and four isolates per sample were used for further processing.
Bedding.
Bedding samples were collected using procedures described elsewhere (20) and transported to the QMPS laboratory in cooler boxes. Briefly, samples were collected from the back one-third of 10% of stalls in a pen. Bedding samples were collected from two pens that housed lactating animals, pen 1 and pen 3. Pen 3 housed multiparous cows, i.e., animals that have calved more than once, including the majority of cows that had CM due to Klebsiella during the first outbreak. Pen 1 housed heifers, i.e., animals in their first lactation. No clinical Klebsiella mastitis had occurred in animals in pen 1. Pen 3 and pen 1 could be considered to be case and control pens, respectively. Bedding samples were also collected from the stalls in the dry cow pen, from the bedded pack in the maternity pen, and from two piles of stored bedding, that is, an old pile of sawdust and a pile of sawdust that had been purchased recently. Bedding samples were processed as described previously, with slight modifications (20). Bedding (10 g) was mixed with 90 g of sterile water and homogenized in a stomacher. Supernatant was decanted, and 50 μl was plated onto MacConkey agar so that the lower limit of detection for Klebsiella spp. was 200 CFU/g. Plates were incubated for 24 h at 37°C. The goal of the initial culture was to determine whether Klebsiella could be detected. If Klebsiella was detected in a bedding sample, the sample was processed again after 1 week of storage at 4°C to obtain additional Klebsiella isolates for assessment of strain diversity within samples.
Feces, feed, and water.
Fecal samples were collected using individual palpation sleeves and transported in cooler boxes to the QMPS laboratory. Fecal samples were collected from the rectum of a convenience sample of five animals in pen 1 and five animals in pen 3 and processed as described previously (20). Briefly, a 1:10 dilution of fecal matter in saline was prepared and incubated for 4 h at 37°C. Ten microliters of the enrichment was swabbed onto MacA, and plates were incubated for 24 h at 37°C before reading. If Klebsiella was detected in a fecal sample, up to four isolates were selected for assessment of strain diversity within samples.
Feed samples included the total mixed ration as fed to the cows and feed refusals that were collected from the feeding alley in front of pen 1 and pen 2. Feed samples and commingled manure samples were collected in Ziploc bags. Commingled manure samples were collected from the alleyways in pen 2 and pen 3. Pen 2 housed mature, lactating cows that had not shown CM during the first outbreak. Feed and commingled manure samples were processed in the same manner as that used for individual fecal samples. Water samples were collected from troughs in pen 2 and pen 3 and from the water source that was used to fill the troughs. Drinking water was collected in sterile snap-cap sample vials (Capitol Vial, Inc., Fultonville, NY). In the laboratory, water samples were filtered using a 150-ml bottle top with a 0.22-μm cellulose acetate filter (Corning Inc., Corning, NY) and a vacuum pump. Filters were submerged in 12 ml THB in a 200-ml plastic vial (Capitol Vial) and incubated at 37°C for 4 h. The enrichment broth was subsequently processed in the same manner as that described above for enrichments of feces, feed, and commingled manure.
Molecular typing.
Molecular methods were used for strain typing and to confirm the species identity of isolates that had been classified as Klebsiella spp. based on phenotypic characteristics. Strain typing was performed using RAPD-PCR (21). Briefly, crude DNA extracts from Klebsiella isolates were obtained by 10 min boil preparation and used as templates for RAPD-PCR with primer set ERIC-2/ERIC-1026 (5′-AAGTAAGTGACTGGGGTGAGCG-3′ and 5′-TACATTCGAGGACCCCTAAGTG-3′, respectively) and cycling conditions described previously (34). Electrophoresis of amplified products was carried out using 1.5% agarose gels, with 20 5-mm-wide wells, run in 0.5× Tris-borate-EDTA buffer for 1.5 h in a horizontal electrophoresis system at ∼95 V. Gels were stained with ethidium bromide and visualized through UV transillumination with the Molecular Imager Gel Doc XR system and Quantity One software, version 4.4.1 (Bio-Rad, Hercules, CA). For evaluation of the within-sample heterogeneity of Klebsiella strains, isolates from a sample were run in adjacent lanes. If necessary, isolates were included in the lysate preparation, RAPD-PCR, and electrophoresis repeatedly to allow comparisons of banding patterns from isolates that were initially not processed in the same batch or on the same gel. The positive control strain, K. pneumoniae ATCC 13883, was included in all lysate preparations, PCR runs, and corresponding gels. Banding patterns from isolates with the same numbers and sizes of DNA fragments were considered to belong to the same strain, regardless of band intensity. Two observers independently read the banding pattern from each Klebsiella isolate. When discrepancies in interpretation occurred, lysate preparation, PCR, and gel electrophoresis were repeated to resolve disagreement. One isolate per Klebsiella RAPD type per sample was used for genotypic confirmation of species identity and stored at −80°C using the Microbank preservation system (Pro-Lab Diagnostic, Austin, TX). Data on the origin of isolates are publicly available online in Cornell University's searchable isolate database PathogenTracker 2.0 (http://www.pathogentracker.net).
Species confirmation was carried using sequence data for the housekeeping gene rpoB (beta subunit of RNA polymerase). PCR was performed using primers described previously by Diancourt and coworkers (7). PCR amplicons were treated with ExoSAP-IT (USB Corporation, Cleveland, OH) to remove excess primers and nucleotides. Sequencing of treated PCR products was performed in two directions using PCR primers and Big Dye Terminator chemistry on an ABI PRISM 3700 DNA analyzer (Applied Biosystems, Foster City, CA). Sequence data were proofread using SeqMan (version 5.08, Lasergene; DNASTAR Inc., Madison, WI), and compared with publicly available sequence data using nucleotide-nucleotide BLAST (1).
Statistical analysis.
To compare the strain diversities between isolate populations from milk and from other sources, Simpson's index of discrimination (SID) and its 95% confidence interval (CI) were calculated (14). In addition, the proportions of isolates belonging to the strain that caused the first outbreak were compared between milk samples and samples from feces, feed, and water using the chi-square statistic (Statistix, version 8.0; Analytical Software, La Jolla, CA). For both analyses, each combination of strain and sample was included once, regardless of how many isolates belonging to the same RAPD type were found in a sample. Furthermore, if a strain was obtained from the udder quarter of a cow on multiple occasions, e.g., during an outbreak and during the analysis of the milking machine, only one of the observations was included in the statistical analysis. Liner samples were excluded from the analysis because isolates from liners can originate from milk or from the environment. Bedding samples were excluded because of the low number of samples and RAPD types from bedding. Statistical significance was considered to be a P value of <0.05.
RESULTS
Milk and milking machine samples.
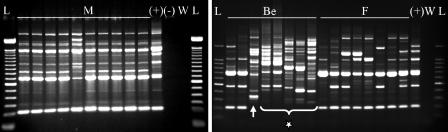
Of 24 samples from CM cases submitted in April 2006, 17 were culture positive and 11 isolates were identified as being Klebsiella based on colony morphology. Ten of 11 isolates were confirmed to be K. pneumoniae based on citrate, motility, and indole testing and rpoB sequencing. One isolate was identified as being an Enterobacter sp. isolate based on citrate and motility testing and was excluded from further analyses. The remaining six isolates were identified as being Staphylococcus spp. (n = 3), Escherichia coli (n = 2), and Staphylococcus aureus (n = 1). RAPD typing showed indistinguishable patterns (RAPD type A) for 9 of 10 K. pneumoniae isolates (Fig. 1, left), all of which originated from eight cows in pen 3. The first sample that tested positive for RAPD type A was obtained on 2 April 2006, and the cow from which this sample originated was considered to be the index case for the outbreak. Approximately 2 weeks later, the same cow developed a second case of CM in a different quarter. RAPD type A was also obtained from this quarter. The 10th K. pneumoniae isolate that was obtained during the first outbreak originated from a cow in pen 4 and showed a different banding pattern (Fig. 1).
FIG. 1.
(Left) RAPD-PCR banding patterns of isolates from milk of all cows involved in the first clinical mastitis outbreak. (Right) Examples of all RAPD-PCR banding patterns of isolates found in bedding and feces shortly after the first clinical mastitis outbreak. Isolates were identified as being K. pneumoniae based on phenotypic methods and rpoB sequencing, with the exception of six strains from bedding, which were identified as being Klebsiella spp. with phenotypic methods but were identified as being Raoultella spp. (star) based on rpoB sequencing (n = 5) or as being K. oxytoca (arrow) with all methods (n = 1). L, DNA ladder; M, isolates from milk; Be, isolates from bedding; F, isolates from feces; +, positive control lysate (ATCC 13883); −, negative control lysate; W, water.
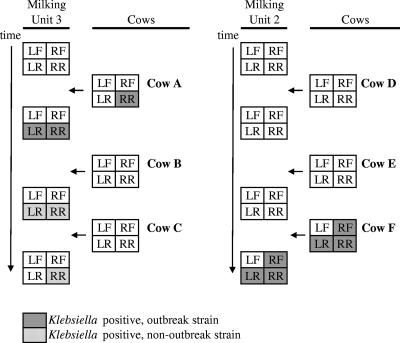
Three cows that had CM due to Klebsiella in April 2006 were selected to evaluate Klebsiella transmission via the milking machine in May. Only one of three cows still shed Klebsiella in milk in May (unit 3, cow A, right rear [RR] quarter) (Fig. 2). During milking of this cow, the RR and left rear (LR) teat cups were detached, and the LR teat cup was reattached to the RR teat. The outbreak strain, RAPD type A, was isolated from milk of the RR udder quarter and from both rear liners immediately after milking of the cow (four isolates per teat cup). After milking of the next cow with unit 3, RAPD types C and H were isolated from the LR teat cup (two isolates per RAPD type), and RAPD type H was isolated from the RR teat cup (four isolates). After milking of the third cow with unit 3, RAPD type H was again isolated from the RR teat cup (four isolates) (Fig. 2). One cow that was milked with unit 2, cow F, shed Klebsiella in milk from three udder quarters (Fig. 2). Cow F had not shown CM, and Klebsiella spp. isolates had not been obtained from this cow before. Isolates from all three udder quarters belonged to RAPD type A. RAPD type A was also isolated from the corresponding teat cup liners after this cow was milked (one to four isolates per teat cup liner). No Klebsiella was isolated from liners in the third milking unit or from cows milked with that unit. All Klebsiella strains that were collected from milk and liners during analysis of the milking machine were identified as being K. pneumoniae by phenotypic and molecular methods.
FIG. 2.
Detection of K. pneumoniae in quarter milk samples from cows and in milking machine liners after milking of cows. Gray blocks show the detection of K. pneumoniae strains belonging to the outbreak strain, RAPD type A. Dotted blocks represent K. pneumoniae isolates belonging to other RAPD types.
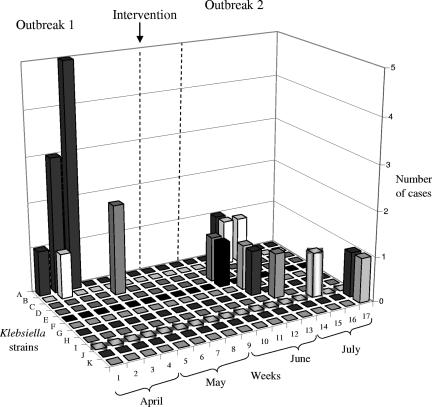
From mid-April to mid-May 2006, when intervention measures were gradually put into place, five presumptive Klebsiella spp. isolates were obtained from milk samples of cows with CM. Two isolates were saved for further characterization. Both isolates were identified as being K. pneumoniae based on rpoB sequencing, and both isolates belonged to RAPD type C (Fig. 3). Intervention measures, as described above, were fully implemented by 15 May 2006. After this time, no new cases of CM attributed to Klebsiella spp. were observed for 3 weeks (Fig. 3).
FIG. 3.
Number of CM cases caused by specific strains of Klebsiella identified by RAPD-PCR during a 17-week period in a dairy herd. All strains were identified as being K. pneumoniae based on rpoB sequencing, with the exception of RAPD type G, which was identified as being a K. oxytoca isolate.
In June and July 2006, a second outbreak of CM was observed. Between 23 June and 26 July, 39 milk samples from 37 cows with CM were submitted to the QMPS laboratory by the herd manager. Sixteen of 39 samples were culture positive, 6 of which were K. pneumoniae and 1 of which was K. oxytoca based on phenotypic and molecular identification. The remaining culture-positive samples contained yeast (n = 2), E. coli (n = 2), S. aureus (n = 1), Staphylococcus spp. (n = 1), Streptococcus spp. (n = 1), Enterobacter spp. (n = 1), and gram-negative bacilli (n = 1). During the whole-herd sampling, Klebsiella was obtained from four additional cows with mild CM. Figure 3 shows the total number of samples with specific RAPD types of K. pneumoniae or K. oxytoca. The figure includes all genotypically confirmed Klebsiella spp. isolates from milk samples from cows with CM during the study period. The strain that caused the first outbreak, RAPD type A, was found once during the second outbreak, in the last week of June (Fig. 3). RAPD type B was found in the last week of June and again in the first week of July. The remaining Klebsiella isolates from CM cases belonged to distinct RAPD types (Fig. 3).
Klebsiella isolates were also obtained from milk samples that were collected from cows that did not show CM during the whole-herd survey. Twenty-one nonclinical samples yielded cultures that were identified as being putative Klebsiella spp. based on colony morphology on MacConkey agar. Nineteen of those isolates were confirmed to be Klebsiella isolates by additional phenotypic testing, and 13 were confirmed by rpoB sequencing. Of six isolates that were identified as being Klebsiella spp. by phenotypic methods but not by rpoB sequencing, five were Enterobacter cloacae and one was Providencia stuartii. Klebsiella isolates from nonclinical samples that were obtained during the whole-herd sampling belonged to RAPD types A, C, B, K, and H and were found in three, four, one, two, and one milk samples, respectively. Two isolates from milk samples collected during the whole-herd sampling had unique RAPD types, i.e., RAPD types that were not identified in any other sample. For four milk samples, multiple isolates (n = 4) per sample were characterized. All isolates within a milk sample showed the same RAPD pattern, as was the case for 10 milk samples from a concurrent study (results not shown). Therefore, testing of multiple isolates per sample was not deemed necessary for the remainder of the milk samples.
Bedding samples.
Seven bedding samples were analyzed. Five samples were culture negative for Klebsiella spp., i.e., sawdust from pen 1, hay from the maternity pen, sand from the dry cow pen, unused fiberboard bedding, and unused sawdust from a recently delivered pile. The fiberboard and the recently delivered sawdust were used as bedding in pens 1 through 4. Two bedding samples were culture positive for Klebsiella spp., i.e., used bedding from pen 3 and the old pile of unused sawdust. After repeated culture, 14 and 7 isolates were available for pen 3 and the old sawdust, respectively. Based on phenotypic methods and rpoB sequencing, 2 isolates from pen 3 were identified as being K. oxytoca, and the remaining 12 isolates were identified as being K. pneumoniae. The isolates from the unused sawdust had the phenotypic appearance of K. pneumoniae, but they were identified as being members of the closely related genus Raoultella based on rpoB sequencing. Four isolates were identified as being Raoultella planticola, and three were identified as being Raoultella terrigena. The 14 genotypically confirmed Klebsiella isolates from bedding in pen 3 belonged to three RAPD types, i.e., RAPD type A (K. pneumoniae) (n = 10), RAPD type H (K. pneumoniae) (n = 2), and RAPD type N (K. oxytoca) (n = 2). Among the R. planticola isolates from unused bedding, two RAPD types were identified, neither of which was found in any other sample. Each of the R. terrigena isolates from unused bedding had a unique RAPD type.
Fecal, feed, and water samples.
Ten fecal samples were analyzed. Klebsiella pneumoniae was detected in one of five fecal samples from heifers in pen 1 and in five of five fecal samples from cows in pen 3. Species identity was confirmed by phenotypic and genotypic methods. Nineteen isolates were available for strain typing, and eight RAPD types were detected (Fig. 1, right). Between one and three RAPD types per sample were identified. One RAPD type, type M, was found in feces from four of five cows from pen 3. Klebsiella isolates were not detected in the commingled manure samples, fresh feed, feed refusals from pen 3, or tap water. By contrast, feed refusals from pen 1 and the water samples from troughs in pen 2 and pen 3 harbored K. pneumoniae, as confirmed by phenotypic and genotypic methods. In each Klebsiella-positive feed and water sample, multiple RAPD types were identified. Among nine K. pneumoniae isolates from feed refusals, three RAPD types were found. Among 10 K. pneumoniae isolates from the water trough in pen 2, five RAPD types were detected, including the outbreak strain, RAPD type A. Among 9 and 10 K. pneumoniae isolates from the water troughs in pen 3, four and five RAPD types, respectively, were identified. None of the RAPD types from water was found in more than one trough.
Overview of molecular typing results.
In total, 142 isolates that had been identified as being Klebsiella spp. based on morphology on MacConkey agar and citrate and motility testing were characterized by means of RAPD typing, i.e., 39 isolates from milk, 32 isolates from milking machine liners, 14 isolates from bedding, and 57 isolates from feces, feed, and water (Table 1). For each RAPD type/sample combination, i.e., for 97 isolates, species identity was determined using rpoB sequence data. Of 89 isolates that had been identified as being K. pneumoniae using citrate, motility, and indole testing, 83 were confirmed as being K. pneumoniae, 3 were R. planticola, 2 were R. terrigena, and 1 was E. cloacae. Three isolates that had been identified as K. oxytoca based on phenotypic methods were confirmed to be K. oxytoca. Five isolates were citrate positive and motility negative but had uncharacteristic colony morphology on MacConkey agar. These isolates were initially considered to be Klebsiella spp. but were shown to be E. cloacae (n = 4) and P. stuartii (n = 1) based on rpoB sequencing. All Raoultella isolates originated from bedding, and all Enterobacter and Providencia isolates originated from composite milk samples collected during the whole-herd sampling from cows that did not show CM.
TABLE 1.
Klebsiella-positive samples, Klebsiella isolates used for strain typing, and RAPD types by sample categorya
| Sample type | No. of positive samples | No. of isolates | No. of RAPD types detected |
|---|---|---|---|
| Milk | 39 | 39 | 16 |
| Linersb | 8 | 32 | 3 |
| Bedding | 1 | 14 | 3 |
| Feces, feed, waterc | 10 | 57 | 24 |
Numbers are based on the identification of Klebsiella spp. with phenotypic methods.
Rubber liners of milking machine units.
Samples were grouped for calculation of SID. Eight RAPD types were detected among 19 isolates from six fecal samples. Three RAPD types were detected among nine isolates from one feed sample. Thirteen RAPD types were detected among 29 isolates from three water samples.
Among genotypically confirmed Klebsiella spp. isolates from milk, 11 RAPD types were identified, including 10 types for K. pneumoniae and 1 type for K. oxytoca. Genotypically confirmed K. pneumoniae isolates from the milking machine belonged to three RAPD types. Klebsiella spp. isolates from bedding also belonged to three RAPD types. Klebsiella pneumoniae isolates from other sample types, i.e., feces, feed, and water, displayed 24 RAPD types, 20 of which were unique to a single sample (Table 1). Strain diversity of isolates from feces, feed, and water (SID, 0.98; 95% CI, 0.96 to 1.00) was significantly higher than strain diversity of isolates from milk (SID, 0.80; 95% CI, 0.70 to 0.91). RAPD type A was significantly more common among isolates from milk (16 of 39 [41%] samples) than among isolates from feces, feed, and water (2 of 28 [7%] strain-sample combinations) (P < 0.01). RAPD types A and H were found in milk, liners, bedding, and water (Table 2). RAPD type C was found in milk, liners, and feces. All other RAPD types were unique to one sample type (types B, K, and M, found in milk, milk, and feces only, respectively) or to a single sample (unique strains) (Table 2). For seven isolates, lysate preparation, PCR, and gel electrophoresis needed to be repeated before the RAPD type could be determined. For other isolates that were included in lysate preparation, PCR, and gel electrophoresis on more than one occasion, as was done to compare strain typing results across samples, RAPD patterns were generally reproducible, with the exception of weak bands of more than 2,000 bp (results not shown).
TABLE 2.
Breakdown of the number of samples that tested positive for specific RAPD types of genotypically confirmed Klebsiella spp.
| Sample type | No. of positive samples (no. of unique RAPD types) with RAPD type:
|
||||||
|---|---|---|---|---|---|---|---|
| Uniqueb | A | B | C | H | K | M | |
| Milk | 8c | 16 | 4 | 6 | 2 | 3 | 0 |
| Linersa | 0 | 5 | 0 | 1 | 3 | 0 | 0 |
| Bedding | 1 (1)c | 1 | 0 | 0 | 1 | 0 | 0 |
| Feces, feed, water | 8 (20) | 2 | 0 | 1 | 1 | 0 | 4 |
Rubber liners of milking machine units.
For milk samples, one isolate per sample was characterized, and the number of unique RAPD types equals the number of samples. For bedding, feces, feed, and water samples, multiple isolates per sample were characterized, and the number of unique RAPD types (shown in parentheses) is higher than the number of samples. RAPD types were considered to be unique if they were found in one sample only.
Unique RAPD types of K. oxytoca were detected in one milk sample and in one bedding sample. All other isolates were K. pneumoniae.
DISCUSSION
Klebsiella pneumoniae is an opportunistic, environmental organism that can cause intramammary infections in dairy cows (15, 32). The environment can contain a large variety of K. pneumoniae strains with pathogenic potential (26, 30), and mastitis cases within a dairy herd are usually caused by many different strains (3, 23-25). Predominance of a single K. pneumoniae strain, such as RAPD type A in the first outbreak of CM described here, is unusual. Predominance of a single strain can result from several mechanisms: (i) contagious transmission of the strain between cows, (ii) predominance of the strain in the environment, or (iii) an increased ability of an environmental strain to cause udder infections. Mechanisms 1 and 2 are often thought to be mutually exclusive but could occur together if infected cows contaminate the environment with milk with high pathogen loads, resulting in the subsequent exposure of other cows to the same bacterial strain (35). Thus, the relative abundance of RAPD type A observed in bedding during the first outbreak could be the cause of the CM outbreak or a result of it. RAPD type A was the predominant strain in bedding in the outbreak pen, but it was not detected in unused bedding or in any of the other pens. Hence, contamination of bedding with the outbreak strain probably happened after its introduction into pen 3. Because the chronology of events is unknown and because not every possible source of Klebsiella could be tested, it is impossible to determine with certainty where RAPD type A occurred for the first time. Based on the large proportion of cows that shed Klebsiella in feces (20) and the large variety of Klebsiella strains in feces (21), we think that feces are likely to have been the original source of RAPD type A. Infection of the index case, a cow that showed CM in early April 2006, with RAPD type A may subsequently have resulted in the amplification of this strain, followed by dissemination via the milking machine or bedding material. Sawdust bedding is an excellent substrate for growth of Klebsiella, and sawdust that is contaminated with milk supports much higher levels of coliform bacteria than sawdust that is not contaminated with milk (8). In the outbreak that we observed, contaminated sawdust may have been a vehicle for transmission, or it may have become a secondary source due to additional amplification of the outbreak strain after contamination of the bedding with milk. The suggestion that cattle may be the source of Klebsiella in bedding material has been raised previously (24). Humans can be a source of Klebsiella, but the prevalence of carriage on skin or in the nasopharynx is low (1 to 6%) (5, 28). Because carriage of Klebsiella by humans is rare and because Klebsiella is abundant in dairy barns, it was not considered to be necessary to investigate farm personnel as a possible source of the outbreak strain. Humans or nonbovine animals can be a source of pathogens that cause mastitis in dairy cattle, but this has been described mainly for mastitis pathogens that are extremely rare in the farm environment, such as methicillin-resistant S. aureus (6), Streptococcus canis (31), and Streptococcus agalactiae (9).
An outbreak of mastitis caused by a single strain of Klebsiella was described previously. Based on capsular serotyping (23) and plasmid profiling (18), nine CM cases from this outbreak were shown to be caused by the same strain of K. pneumoniae. Because of its sudden occurrence, Kikuchi et al. (18) previously suggested that a common source, such as the milking machine, played a role in this outbreak. Other authors also suggested a role of the milking machine in the transmission of Klebsiella, but strain typing was not used to determine whether contagious transmission might have occurred (11, 17, 33).
The presence of coliform bacteria in the milking machine may result from contact with milk from infected cows or from environmental contamination (17). Jasper and Dellinger (17) previously observed that coliform bacteria from infected quarters were more likely to constitute a risk to uninfected quarters and cows than those from contaminated teat ends. Coliform populations on the teat apex that come from environmental sources were transitory and did not persist throughout a milking (17). In our study, we obtained the strain that was shed by infected cows as well as strains that were not shed by infected cows from liners (Fig. 2). The number of samples collected was not sufficiently high to determine whether strains from cows were more likely to persist than other strains.
In the second outbreak of Klebsiella mastitis described in our case study, almost every case of mastitis was caused by a different strain. High variability of strains is typical of opportunistic, environmental pathogens (19, 25). Opportunistic udder infections in dairy cows can occur when there is a decrease in host resistance or an increase in exposure or by chance. An increase in exposure may result from a failure to persist in the implementation of management measures to improve environmental hygiene or from increases in environmental temperature and humidity. Often, recommendations for mastitis control are implemented by dairy farmers only temporarily, until the acute problem subsides and the perceived need for implementation of control measures diminishes (33, 35). The incidence of Klebsiella mastitis increases under hot and humid weather conditions (15). According to the herd manager of the farm, environmental hygiene was maintained throughout the summer. Hence, an increase in environmental exposure due to the summer weather, possibly combined with decreased host resistance due to heat stress, is likely to have precipitated the second outbreak.
The coexistence of different epidemiologies within a bacterial species on a single dairy, as observed for K. pneumoniae in this case study, was described previously for other mastitis pathogens (29, 36). In the case of gram-positive mastitis pathogens, host adaptation of strains is thought to contribute to strain-specific epidemiology. For K. pneumoniae, there is no indication that host or niche adaptation exists (25, 30). Mastitis outbreaks due to a single strain of Klebsiella may be the result of an unknown mechanism of host or niche adaptation or the result of a chance accumulation of events.
In previous studies, we showed that cows’ feces are a common source of a large variety of Klebsiella strains (20, 21). In the case study described here, we showed that RAPD type C was isolated from feces and from milk, which strengthens the suggestion that at least some strains from feces have the ability to cause mastitis. RAPD type C was also isolated from a milking machine liner after milking of cows that did not shed K. pneumoniae at detectable levels. The same was true for RAPD type H. In our opinion, contamination of the liner with fecal matter or bedding material, either directly or via contamination on teat skin of the cows that were milked (17, 37), is the most likely explanation for this observation. An oral-fecal transmission cycle has been suggested for K. pneumoniae in dairy herds, with fecal shedding resulting in the contamination of feed and water and subsequent reingestion of the organism, resulting in renewed fecal shedding (21). In the case presented here, K. pneumoniae was not detected in fresh feed and tap water, but feed refusals and water troughs in the barn did contain K. pneumoniae. Klebsiella strains found in water troughs may originate from the mouths of cows (4, 24). In our opinion, the presence of K. pneumoniae in feed refusals, water troughs, and the oral cavities of cows is most likely the result of fecal contamination.
Several isolates that were identified as being K. pneumoniae based on phenotypic methods that are used routinely in laboratories for mastitis diagnosis (22) were shown to belong to the genus Raoultella based on rpoB sequencing. Raoultella terrigena and R. planticola were considered Klebsiella species until they were renamed in 2001 (10). Phenotypic methods that are used in clinical laboratories in human medicine also fail to differentiate Raoultella spp. from Klebsiella spp. (2, 12). To our knowledge, this is the first report of the identification of Raoultella on a dairy farm. In the case reported here, Raoultella spp. were detected only in bedding material. We have since detected R. planticola, but not R. terrigena, in milk and feces from dairy cattle (unpublished data from our laboratory [available at www.pathogentracker.net]). The true prevalence and the importance of Raoultella spp. on dairy farms and in milk are unknown, because routine methods fail to differentiate Raoultella from Klebsiella. In addition to Raoultella species, some E. cloacae isolates and a single Providencia isolate were incorrectly identified as being Klebsiella isolates by phenotypic methods.
Conclusions.
Two outbreaks of clinical mastitis due to K. pneumoniae were observed in one dairy herd. The first outbreak was caused by a predominant RAPD type and may have been caused by contagious transmission via the milking machine or by exposure to a common source, specifically, bedding. By contrast, the second outbreak was caused by a large variety of RAPD types, indicating opportunistic infections from the environment. Without the use of molecular typing methods such as RAPD-PCR, it would not have been possible to distinguish the two outbreaks and to target control measures. The strain diversity of milk isolates was significantly lower than the strain diversity of environmental isolates, including isolates from feces, feed, and water. Strains found in feces, water, and bedding were also found to be the cause of mastitis in dairy cows. Routine phenotypic methods for mastitis diagnosis may result in the misclassification of E. cloacae and Raoultella spp. as Klebsiella.
Acknowledgments
This project was supported in part by the New York State Agricultural Experiment Station with U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service grant NYC 478801.
We thank the participating producer and our colleagues at QMPS, in particular, Christina Ahlström and Carlos Santisteban, for help with collection and processing of samples.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves, M. S., R. C. Dias, A. C. de Castro, L. W. Riley, and B. M. Moreira. 2006. Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. J. Clin. Microbiol. 44:3640-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braman, S. K., R. J. Eberhart, M. A. Asbury, and G. J. Hermann. 1973. Capsular types of Klebsiella pneumoniae associated with bovine mastitis. J. Am. Vet. Med. Assoc. 162:109-111. [PubMed] [Google Scholar]
- 4.Carroll, E. J. 1971. Bactericidal activity of bovine serums against coliform organisms isolated from milk of mastitic udders, udder skin, and environment. Am. J. Vet. Res. 32:689-701. [PubMed] [Google Scholar]
- 5.Davis, T. J., and J. M. Matsen. 1974. Prevalence and characteristics of Klebsiella species: relation to association with a hospital environment. J. Infect. Dis. 130:402-405. [DOI] [PubMed] [Google Scholar]
- 6.Devriese, L. A., and J. Hommez. 1975. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res. Vet. Sci. 19:23-27. [PubMed] [Google Scholar]
- 7.Diancourt, L., V. Passet, J. Verhoef, P. A. Grimont, and S. Brisse. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd, F. H., T. M. Higgs, and A. J. Bramley. 1984. Cubicle management and coliform mastitis. Vet. Rec. 114:522-523. [DOI] [PubMed] [Google Scholar]
- 9.Dogan, B., Y. H. Schukken, C. Santisteban, and K. J. Boor. 2005. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J. Clin. Microbiol. 43:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drancourt, M., C. Bollet, A. Carta, and P. Rousselier. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 51:925-932. [DOI] [PubMed] [Google Scholar]
- 11.Eberhart, R. J., R. P. Natzke, F. H. S. Newbould, B. J. Nonnecke, and P. Thompson. 1979. Coliform mastitis—a review. J. Dairy Sci. 62:1-22. [DOI] [PubMed] [Google Scholar]
- 12.Granier, S. A., V. Leflon-Guibout, F. W. Goldstein, and M. H. Nicolas-Chanoine. 2003. Enterobacterial repetitive intergenic consensus 1R PCR assay for detection of Raoultella sp. isolates among strains identified as Klebsiella oxytoca in the clinical laboratory. J. Clin. Microbiol. 41:1740-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gröhn, Y. T., D. J. Wilson, R. N. González, J. A. Hertl, H. Schulte, G. Bennett, and Y. H. Schukken. 2004. Effect of pathogen-specific clinical mastitis on milk yield in dairy cows. J. Dairy Sci. 87:3358-3374. [DOI] [PubMed] [Google Scholar]
- 14.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan, J., and K. L. Smith. 2003. Coliform mastitis. Vet. Res. 34:507-519. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, J. S., K. L. Smith, K. H. Hoblet, D. A. Todhunter, P. S. Schoenberger, W. D. Hueston, D. E. Pritchard, G. L. Bowman, L. E. Heider, B. L. Brockett, et al. 1989. Bacterial counts in bedding materials used on nine commercial dairies. J. Dairy Sci. 72:250-258. [DOI] [PubMed] [Google Scholar]
- 17.Jasper, D. E., and J. D. Dellinger. 1975. Teat apex coliform populations and coliform mastitis—a herd study. Cornell Vet. 65:380-392. [PubMed] [Google Scholar]
- 18.Kikuchi, N., C. Kagota, T. Nomura, T. Hiramune, T. Takahashi, and R. Yanagawa. 1995. Plasmid profiles of Klebsiella pneumoniae isolated from bovine mastitis. Vet. Microbiol. 47:9-15. [DOI] [PubMed] [Google Scholar]
- 19.McDougall, S., T. J. Parkinson, M. Leyland, F. M. Anniss, and S. G. Fenwick. 2004. Duration of infection and strain variation in Streptococcus uberis isolated from cows’ milk. J. Dairy Sci. 87:2062-2072. [DOI] [PubMed] [Google Scholar]
- 20.Munoz, M. A., C. Ahlström, B. J. Rauch, and R. N. Zadoks. 2006. Fecal shedding of Klebsiella pneumoniae by dairy cows. J. Dairy Sci. 89:3425-3430. [DOI] [PubMed] [Google Scholar]
- 21.Munoz, M. A., and R. N. Zadoks. 2007. Patterns of fecal shedding of Klebsiella by dairy cows. J. Dairy Sci. 90:1220-1224. [DOI] [PubMed] [Google Scholar]
- 22.National Mastitis Council. 1999. Laboratory handbook on bovine mastitis. National Mastitis Council, Madison, WI.
- 23.Nomura, T., H. Moriya, N. Kikuchi, and T. Hiramune. 1989. Capsular types of Klebsiella associated with bovine mastitis in Japan. Nippon Juigaku Zasshi 51:1287-1289. [DOI] [PubMed] [Google Scholar]
- 24.Nonnecke, B. J., and F. H. Newbould. 1984. Biochemical and serologic characterization of Klebsiella strains from bovine mastitis and the environment of the dairy cow. Am. J. Vet. Res. 45:2451-2454. [PubMed] [Google Scholar]
- 25.Paulin-Curlee, G. G., R. S. Singer, S. Sreevatsan, R. Isaacson, J. Reneau, D. Foster, and R. Bey. 2007. Genetic diversity of mastitis-associated Klebsiella pneumoniae in dairy cows. J. Dairy Sci. 90:3681-3689. [DOI] [PubMed] [Google Scholar]
- 26.Podschun, R., S. Pietsch, C. Holler, and U. Ullmann. 2001. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67:3325-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberson, J. R., L. D. Warnick, and G. Moore. 2004. Mild to moderate clinical mastitis: efficacy of intramammary amoxicillin, frequent milk-out, a combined intramammary amoxicillin, and frequent milk-out treatment versus no treatment. J. Dairy Sci. 87:583-592. [DOI] [PubMed] [Google Scholar]
- 28.Rosenthal, S., and I. B. Tager. 1975. Prevalence of gram-negative rods in the normal pharyngeal flora. Ann. Intern. Med. 83:355-357. [DOI] [PubMed] [Google Scholar]
- 29.Smith, T. H., L. K. Fox, and J. R. Middleton. 1998. Outbreak of mastitis caused by one strain of Staphylococcus aureus in a closed dairy herd. J. Am. Vet. Med. Assoc. 212:553-556. [PubMed] [Google Scholar]
- 30.Struve, C., and K. A. Krogfelt. 2004. Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 6:584-590. [DOI] [PubMed] [Google Scholar]
- 31.Tikofsky, L. L., and R. N. Zadoks. 2005. Cross-infection between cats and cows: origin and control of Streptococcus canis mastitis in a dairy herd. J. Dairy Sci. 88:2707-2713. [DOI] [PubMed] [Google Scholar]
- 32.Todhunter, D. A., K. L. Smith, J. S. Hogan, and P. S. Schoenberger. 1991. Gram-negative bacterial infections of the mammary gland in cows. Am. J. Vet. Res. 52:184-188. [PubMed] [Google Scholar]
- 33.Vecht, U., K. C. Meijers, and H. J. Wisselink. 1987. Klebsiella pneumoniae mastitis as a dairying problem. Tijdschr. Diergeneeskd. 112:653-659. (In Dutch.) [PubMed] [Google Scholar]
- 34.Vogel, L., G. Jones, S. Triep, A. Koek, and L. Dijkshoorn. 1999. RAPD typing of Klebsiella pneumoniae, Klebsiella oxytoca, Serratia marcescens and Pseudomonas aeruginosa isolates using standardized reagents. Clin. Microbiol. Infect. 5:270-276. [DOI] [PubMed] [Google Scholar]
- 35.Zadoks, R. N., H. G. Allore, H. W. Barkema, O. C. Sampimon, Y. T. Grohn, and Y. H. Schukken. 2001. Analysis of an outbreak of Streptococcus uberis mastitis. J. Dairy Sci. 84:590-599. [DOI] [PubMed] [Google Scholar]
- 36.Zadoks, R. N., B. E. Gillespie, H. W. Barkema, O. C. Sampimon, S. P. Oliver, and Y. H. Schukken. 2003. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol. Infect. 130:335-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zdanowicz, M., J. A. Shelford, C. B. Tucker, D. M. Weary, and M. A. von Keyserlingk. 2004. Bacterial populations on teat ends of dairy cows housed in free stalls and bedded with either sand or sawdust. J. Dairy Sci. 87:1694-1701. [DOI] [PubMed] [Google Scholar]