Abstract
With the availability of more potent nucleotide/nucleoside analogues, the early detection of drug-resistant mutants of hepatitis B virus (HBV) is important for the strategic treatment of chronic hepatitis B. We studied 336 serum samples from 80 patients chronically infected with HBV who were receiving lamivudine treatment for the presence of lamivudine resistance mutations at codons 80, 173, 180, and 204 of the HBV polymerase. The sequencing data were compared with the results generated with the INNO-LiPA HBV DR (drug resistance) v2 strip, a line probe assay (LiPA) covering wild-type and mutant motifs, for resistance mutations to lamivudine and adefovir dipivoxil. This method provided at least the same information as sequencing for 99.1% of all codons analyzed. On the basis of the LiPA results, 20 of 80 patients developed a lamivudine resistance mutation after 1 year. In all 20 patients, the mutation occurred in the YMDD motif at reverse transcriptase position 204 (rt204; M204V/I) either with or without the compensatory mutation at position rt180 (L180M). A compensatory mutation at position rt80 (L80V/I) was detected in half of these patients. After 36 months, a compensatory mutation was seen at position rt173 (V173L) in 3/15 patients. Time-to-event survival analysis indicated a 2.8 times greater chance for LiPA to detect a given mutation than sequencing at any moment in time (hazard ratio, 2.8, 95% confidence interval, 1.79, 4.41; P < 0.0001). These results demonstrate that a highly sensitive and specific assay such as the INNO-LiPA HBV DR v2 can precociously detect and monitor the emergence of primary and compensatory lamivudine resistance mutations in patients chronically infected with HBV and is more sensitive than sequencing.
Chronic infection with hepatitis B virus (HBV) occurs in at least 5% of the world's population. Given the increasing use and extending repertoire of nucleotide/nucleoside analogues for the treatment of this ubiquitous disease, drug resistance mutations are becoming increasingly problematic. The beneficial effects of antiviral agents in halting disease progression are blunted by the occurrence of these mutations (5, 18). Furthermore, severe hepatitis reactivation flares due to drug-resistant virus, resulting in hepatic decompensation and even mortality, have been reported (4, 17). Such unsatisfactory outcomes of drug resistance could be prevented by the early detection of its emergence, thereby permitting the opportune alteration of treatment with appropriate alternatives.
The INNO-LiPA HBV DR (drug resistance) v2 strip, an update of the INNO-LiPA HBV DR strip (for research use only, not for use in diagnostic procedures; Innogenetics NV, Ghent, Belgium), is an in vitro, reverse hybridization line probe assay (LiPA) used to detect the presence of different genetic variants of HBV in human serum or plasma samples. As reviewed recently (6), this update includes new and clinically relevant wild-type and mutant motifs for codons L80V/I, V/G173L, L180M, A181T/V, M204V/I/S, and N236T, located in the HBV polymerase protein, that confer resistance to lamivudine and adefovir dipivoxil. The differences between the previous and updated strips and the clinical relevance of the drug resistance mutations are summarized in Table 1.
TABLE 1.
Differences between the previous INNO-LiPA HBV DR strip and the updated INNO-LiPA HBV DR v2 strip and the clinical relevance of the mutations
| Codon | Isolate typea | Mutation detected
|
Clinical relevance of the mutation | |
|---|---|---|---|---|
| Previous strip (INNO-LiPA HBV DR) | Updated strip (INNO-LiPA HBV DR v2) | |||
| 80 | WT | L80 | Possibly relevant for lamivudine (compensatory mutation) | |
| MUT | V80 | |||
| MUT | I80 | |||
| 173 | WT | V173 | Possibly relevant for lamivudine (compensatory mutation) | |
| WT | G173 | |||
| MUT | L173 | |||
| 180 | WT | L180 | L180 | Relevant for lamivudine |
| MUT | M180 | M180 | ||
| 181 | WT | A181 | Possibly relevant for lamivudine and adefovir | |
| MUT | T181 | |||
| MUT | V181 | |||
| 204 | WT | M204 | M204 | Relevant for lamivudine |
| MUT | V204 | V204 | ||
| MUT | I204 | I204 | ||
| MUT | S204 | |||
| 207 | WT | V207 | No longer relevant (famciclovir) | |
| WT | L207 | |||
| WT | M207 | |||
| MUT | I207 | |||
| 236 | WT | N236 | Relevant for adefovir | |
| MUT | T236 | |||
WT, wild type; MUT, mutant.
Our primary objective was to confirm the ability of this new test to detect the emergence of lamivudine resistance primary and compensatory mutations in a Chinese population of HBV-infected patients who were receiving continuous lamivudine treatment. Secondarily, we compared the results of this assay with those of direct sequencing for concordance and determined whether the emergence of lamivudine resistance mutations could be detected earlier with the INNO-LiPA HBV DR v2 strip than by sequencing.
MATERIALS AND METHODS
Sample collection.
Serum samples from 80 patients who were chronically infected with HBV and who were receiving continuous lamivudine treatment in Hong Kong (Department of Medicine, The University of Hong Kong, Queen Mary Hospital, Pokfulam Road, Hong Kong, China) were sent to Innogenetics for analysis and were stored at −20°C. The patients had been positive for HBsAg and HBeAg for at least 6 and 3 months, respectively, before they entered the trial. All except eight patients received lamivudine treatment (100 mg daily) until week 156; the eight patients who were the exceptions defaulted on follow-up visits at various times after the first year of lamivudine treatment. Patients were monitored every 2 weeks for the first 4 weeks, every 4 weeks thereafter until week 52, and then every 8 weeks to 104 weeks and subsequently every 16 weeks (19).
Patients and samples.
A total of 336 samples from the 80 patients were analyzed, with a range of 2 to 5 samples obtained from each patient at different time points: the baseline and weeks 12, 24, 52 (year 1), and 156 (year 3). All patients were Chinese, and 20 patients infected with genotype B HBV and 60 patients infected with genotype C HBV were included.
Serologic tests and HBeAg seroconversion.
HBsAg, HBeAg, and antibodies to HBeAg (anti-HBe) were tested by a microparticle enzyme immunoassay (Abbott Laboratories, Chicago, IL). HBeAg seroconversion was defined as a loss of HBeAg with the development of anti-HBe on at least two consecutive follow-up visits.
HBV DNA measurement.
The levels of HBV DNA were measured in all patients by the Cobas Amplicor HBV Monitor test (Roche Diagnostics, Branchburg, NJ; lower limit of detection, 300 copies/ml) at the baseline, week 24, and week 52 (year 1). Serum drawn from 15 patients at week 156 (year 3) was available for HBV DNA level measurement.
HBV DNA purification and amplification with the INNO-LiPA HBV DR v2 strip.
HBV DNA was isolated from the serum samples by using the commercially available QIAamp DNA blood mini kit (Qiagen, Venlo, The Netherlands). Purified DNA was amplified in a single-round PCR with the INNO-LiPA HBV DR v2 primers (Innogenetics): sense primer HBPr950 (HBV nucleotides 255 to 278) and antisense primer HBPr952 (HBV nucleotides 1121 to 1099). These primers are specific for the reverse transcriptase region of the HBV polymerase gene (which contains polymerase domains A to F) and generate an amplicon of 867 bp that covers codons 42 to 330. The reference sequence used for numbering purposes was the sequence with GenBank accession number X70185.
Amplification was performed according to the manufacturer's instructions (Innogenetics). Briefly, 20 μl of extracted DNA was added to 22.2 μl autoclaved distilled water, 5 μl of 10× PCR buffer [containing Tris-HCl, KCl, (NH4)2SO4, 15 mM MgCl2, pH 8.7; Qiagen], 1 μl of 25 mM MgCl2, 1 μl primer mix, 0.4 μl deoxynucleoside triphosphate mix (25 mM), and 0.4 μl HotStarTaq DNA polymerase (5 U/μl; Qiagen). The reactions were performed in a PE-9700 thermal cycler (Perkin-Elmer, Wellesley, MA), with an initial denaturation step at 95°C for 15 min and 50 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 40 s, with a final elongation step at 72°C for 10 min. The presence of the amplified product was checked by adding 5 μl of the PCR product to a 2% agarose gel. The amplicon, with a band length of 867 bp, was visualized with ethidium bromide.
Sequencing.
HBV DNA was amplified, after extraction, for HBV polymerase domains A to F, as described above. Sequence analysis was performed with an AB377 sequencer (Applied Biosystems, Foster City, CA); the samples obtained at 3 years, however, were sequenced by AGOWA (Berlin, Germany) by using four-color fluorescent sequencing technology (with the ABI 3730 xl and ABI 3700 systems). A wild-type, mutated, or mixed status was scored for each of the six codons. If any of the mutations mentioned above were detected by sequencing at any time point, sera from all time points for that particular patient were retested with the INNO LiPA HBV DR v2 strip.
INNO-LiPA HBV DR v2 strip preparation.
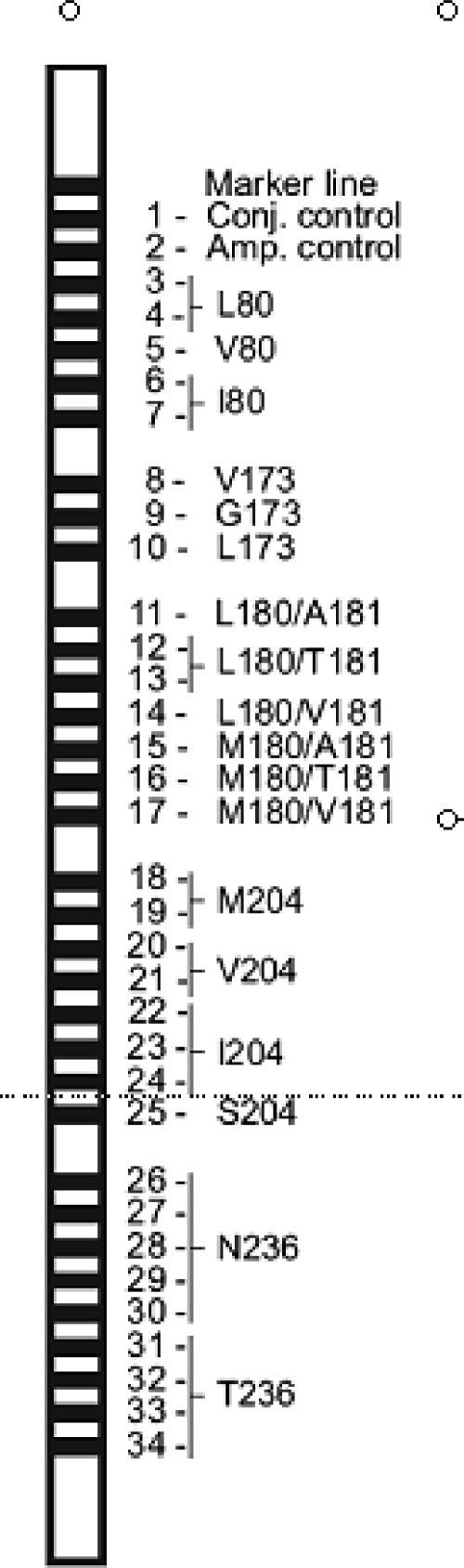
A set of 62 highly specific oligonucleotide probes was designed on the basis of known viral resistance-specific sequence motifs in the amplified region. The following parameters were also taken into consideration: percent G+C content, probe length, ionic strength of the hybridization buffer, and temperature of incubation. After optimization, these specific probes were applied as 32 different parallel probe lines on a membrane strip. Control lines for amplification and conjugate incubation (biotinylated DNA) were also applied. A schematic representation of the INNO-LiPA HBV DR v2 strip is shown in Fig. 1.
FIG. 1.
Locations of the marker line, conjugate control line, amplification line, and the 32 probe lines on the INNO-LiPA HBV DR v2 strip.
The INNO-LiPA HBV DR v2 strip covers the most important amino acid variations at six different codon positions: at codon 80, wild-type leucine and mutant valine and isoleucine; at codon 173, wild-type valine and glycine and mutant leucine; at codon 180, wild-type leucine and mutant methionine; at codon 181, wild-type alanine and mutant valine and threonine; at codon 204, wild-type methionine and mutant valine, isoleucine, and serine; and at codon 236, wild-type asparagine and mutant threonine.
INNO LiPA HBV DR v2 strip performance.
For all patients, the samples obtained at week 52 were subjected to reverse hybridization with the INNO-LiPA HBV DR v2 strip according to the manufacturer's instructions (Innogenetics). Briefly, 10 μl of biotinylated amplified product was denatured and hybridized to specific oligonucleotide probes coated on the reaction strip. Hybridization and color development were performed as described previously (12, 13). The reaction strips were then aligned against a plastic reading card, and the results were interpreted according to the manufacturer's instructions. If any of the mutations mentioned above were detected, sera from all time points for that particular patient were also tested with the INNO LiPA HBV DR v2 strip. Samples obtained at week 156 were tested only when a mutation appeared at week 52 or if a patient displayed an unconfirmed mutation on LiPA at any time point, in order to check for the presence of this mutation. All these samples were also subjected to direct sequencing.
Genotype determination.
Genotypes were determined by using Kodon software (Applied Maths, Sint-Martens-Latem, Belgium). The sequences of all samples were aligned together with reference sequences of all genotypes.
Statistical analysis.
The data were analyzed post hoc to determine whether the chances of detecting a given mutation were greater by LiPA or sequencing. For this purpose, a time-to-event survival analysis was performed by using a Cox proportional hazard regression, with correction for clustering within the data. The results are given as hazard ratios with 95% confidence intervals.
RESULTS
Distribution of mutations.
According to sequence analysis, the frequency of mutations at the baseline and weeks 12, 24, 52, and 156 totaled 1, 2, 6, 12, and 11, respectively, while the corresponding frequencies according to LiPA totaled 3, 10, 17, 25, and 14 mutations. M204I mutations in combination with L80I were often detected by sequencing during weeks 24 and 52, while LiPA frequently found the former combination as well as A181T mutations and combinations at weeks 12, 24, and 52.
Serum samples from only 15 patients were sequenced at year 3 (on the basis of the LiPA results). Mutant virus was detected in 11 of these patients by sequencing and in 14 by LiPA. The different mutations and their frequencies are listed in Table 2, according to the results of sequencing and LiPA analysis.
TABLE 2.
Occurrence and frequency of the various drug resistance mutations (per codon) by sequence analysis and LiPA
| Assay | Mutation(s) at codon:
|
Frequency (no. of mutants)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 173 | 180 | 181 | 204 | 236 | Baseline | Wk 12 | Wk 24 | Wk 52 | Wk 156 | |
| Sequencing | M204I | 1 | 1 | 1 | |||||||
| L180M | M204I | 1 | |||||||||
| L180M | M204I, M204V | 2 | |||||||||
| L80I | M204I | 1 | 3 | 7 | 1 | ||||||
| L80I | L180M | M204I | 1 | ||||||||
| L80I | L180M | M204I, M204V | 2 | ||||||||
| L80I | V173L | M204I | 1 | ||||||||
| L80V | M204I | 1 | |||||||||
| L80V | L180M | M204I | 1 | ||||||||
| L80I | L180M | M204V | 1 | ||||||||
| V173L | L180M | M204V | 1 | ||||||||
| L80I | 1 | ||||||||||
| A181T | 1 | 2 | 1 | ||||||||
| N236T | 1 | ||||||||||
| Total | 1 | 2 | 6 | 12 | 11 | ||||||
| LiPA | M204I | 3 | 3 | 3 | |||||||
| L180M | M204I | ||||||||||
| L180M | M204I, M204V | 1 | |||||||||
| L80I | M204I | 1 | 2 | 3 | 1 | ||||||
| L80I | L180M | M204I | 1 | ||||||||
| L80I | L180M | M204I, M204V | 1 | ||||||||
| L80I | V173L | M204I | 1 | ||||||||
| L80V | M204I | ||||||||||
| L80V | L180M | M204I | |||||||||
| L80I | L180M | M204V | |||||||||
| V173L | L180M | M204V | |||||||||
| L80I | |||||||||||
| A181T | 3 | 5 | 4 | 2 | |||||||
| N236T | 1 | ||||||||||
| L80I | M204I, M204V | 1 | |||||||||
| L80I, L80V | M204I | 1 | 1 | 1 | |||||||
| L80I, L80V | A181T | M204I, M204V | N236T | 1 | |||||||
| L80I, L80V | L180M | M204I, M204V | N236T | 1 | 1 | 1 | |||||
| L180M | A181T | M204I | N236T | 1 | |||||||
| A181T | M204I | 1 | 3 | ||||||||
| L80V | L180M | A181T | M204V | 1 | |||||||
| L80V | A181T | 2 | |||||||||
| L80V | M204I, M204V | 1 | |||||||||
| L80V | L180M | M204I, M204V | 1 | 1 | |||||||
| M204I, M204V | 1 | 1 | |||||||||
| V173L | L180M | M204I, M204V | 1 | ||||||||
| L80I | V173L | L180M | M204V | 1 | |||||||
| M204V | 1 | 1 | 2 | ||||||||
| L80I | M204I | N236T | 1 | ||||||||
| L180M | M204V | 1 | |||||||||
| L80I, L80V | L180M | M204I, M204V | 1 | 1 | |||||||
| L80I | A181T | M204I, M204V | 1 | ||||||||
| L80I, L80V | L180M | M204I | 1 | ||||||||
| L80I | L180M | A181T | M204I | 1 | |||||||
| Total | 3 | 10 | 17 | 25 | 14 | ||||||
Primary and compensatory lamivudine resistance mutations.
On the basis of the LiPA results, a lamivudine-resistant strain developed in 20 of 80 patients after 1 year of treatment. In all 20 patients, the mutation occurred in the YMDD motif at reverse transcriptase position 204 (rt204; M204V/I) either with (6 patients) or without (14 patients) the compensatory mutation at position rt180 (L180M). A compensatory mutation at position rt80 (L80V/I) was detected in 10 of these 20 patients: in 5 patients together with a mutation at position rt204 only and in the other 5 patients together with a mutation at positions rt204 and rt180. After 3 years of lamivudine treatment, another common compensatory mutation was seen at position rt173 (V173L) in 3 of the 15 patients tested. In two of these three patients, this mutation was combined with mutations rtM204V and rtL180M, of which one also occurred together with the rtL80I mutation. In the other patient, the rtV173L mutation was combined with rtL80I and rtM204I.
Adefovir dipivoxil resistance mutations.
On the basis of the LiPA results, six patients showed a transient emergence of adefovir-resistant mutants (rtA181T or rtN236T), while nine patients had adefovir-resistant mutants that persisted for several weeks, of whom eight had mutants with the rtA181T mutation. Of the eight latter patients, half also developed a mutation at position rt204 (M204V/I).
Sensitivity of INNO-LiPA HBV DR v2 strip amplification.
Of the 336 samples amplified with INNO-LiPA HBV DR v2 primers, 328 samples were positive for an 867-bp band on gels (97.6%). This included 13 samples that were negative at first but that became positive upon reamplification (i.e., a new PCR was performed with DNA extracts and the same primers). Among the eight samples that remained negative, six had viral loads less than 1,000 copies/ml (below the lower detection limit of the INNO-LiPA HBV DR v2 test), one had a viral load of 1,430 copies/ml, and one had a viral load of less than 2,000 copies/ml. Of the 336 samples, 43 samples had viral loads less than 1,000 copies/ml, and of these 43 samples, 37 were positive on gels (86.0%).
Sequencing versus LiPA.
Both sequencing and LiPA results were available for 194 samples, including all samples with mutations detected by sequencing. The results obtained for codons 80, 173, 180, 181, 204, and 236 were compared. Taking into consideration that sequencing has heretofore been considered the “gold standard,” the results were divided into four classes: fully concordant, partially concordant, partially discordant, and completely discordant with sequencing. The results were considered fully concordant if both direct sequencing and LiPA showed a wild-type, a mutant, or a mixed sequence. The results were considered partially concordant if LiPA provided additional information compared to that provided by sequencing, meaning that LiPA showed a mixture of wild-type and mutant sequences, while sequencing showed only a wild-type or a mutant sequence. The results were considered partially discordant if sequencing showed a mixture of wild-type and mutant sequences, while LiPA showed a wild-type sequence only. The results were considered completely discordant if one test showed a mutant and the other test showed a wild type. The data are summarized in Table 3. Full concordance between sequence analysis and LiPA was calculated for each codon. The number of samples scored as a mixture of wild type and mutant on LiPA but not on sequencing (or vice versa) was also indicated. Those samples for which LiPA detected a mixed sequence were further subdivided into samples for which sequence analysis revealed a wild type and samples for which sequence analysis showed a mutant. The number of indeterminate results (no information for a specific codon) and completely discordant samples is also indicated.
TABLE 3.
Comparison of sequence data and LiPA results: overview of the results obtained for the six codonsa
| Codon | No. (%) of samples
|
Partial concordance
|
No. (%) of samples
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Full concordance | Full concordance
|
Partial discordance | Complete discordance | IND on LiPA | Total | |||||
| WT | MUT | MIX | No. (%) of samples | No. of samples MIX on LiPA/ no. of samples WT or MUT on sequencing | ||||||
| 80 | 170 (87.6) | 160 | 5 | 5 | 21 (10.8) | 12/9 | 1 (0.5)b | 2c | 194 | |
| 173 | 188 (96.9) | 187 | 1 | 2 (1.0) | 1/1 | 4c | 194 | |||
| 180 | 181 (93.3) | 176 | 5 | 12 (6.2) | 8/4 | 1c | 194 | |||
| 181 | 172 (88.7) | 168 | 4 | 21 (10.8) | 21/0 | 1c | 194 | |||
| 204 | 162 (83.5) | 142 | 17 | 3 | 32 (16.5) | 26/6 | 194 | |||
| 236 | 185 (95.4) | 185 | 7 (3.6) | 7/0 | 1 (0.5)b | 1d | 194 | |||
| Total | 1,058 (90.9) | 1,018 | 23 | 17 | 95 (8.2) | 75/20 | 2 (0.2) | 9 (0.8) | 1,164 | |
WT, wild type; MUT, mutant; MIX, mixture; IND, indeterminate.
A mutation was also detected by LiPA at an earlier or later time point.
Indeterminate results were due to weak reactivities (always in samples with viral loads less than 1,000 copies/ml).
Indeterminate result due to a polymorphism in the probe region.
Full concordance and partial concordance.
Full concordance between LiPA and sequence analysis was observed for 90.9% (n = 1,058) of the codons analyzed. There was partial concordance between LiPA and sequencing for an additional 8.2% (n = 95) of the codon results, meaning that LiPA provided additional information compared to that provided by sequencing. LiPA showed at least the same or more information compared with that provided by sequencing for 98.4% of the results for codon 80, 97.9% for codon 173, 99.5% for codons 180 and 181, 100% for codon 204, and 99.0% for codon 236.
Complete discordance and partial discordance.
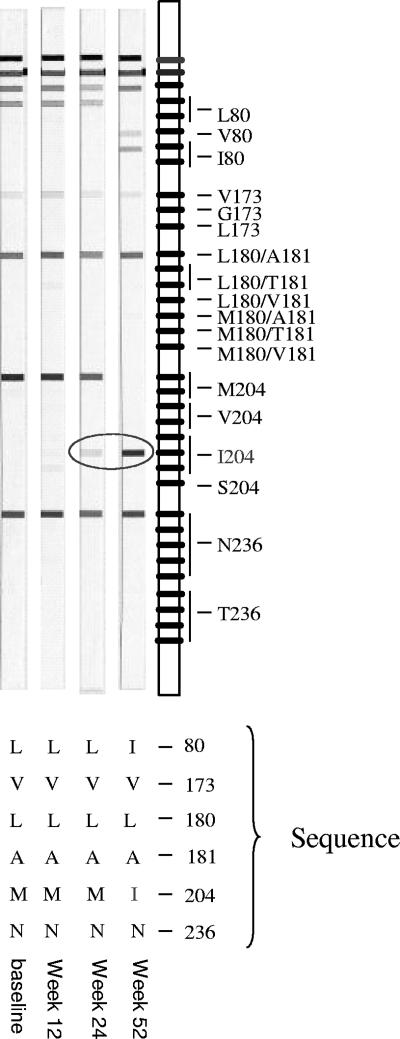
In two cases (0.2% of the codons analyzed), a partial discordant result between LiPA and sequencing was observed, meaning that sequencing showed a mixture of wild-type and mutant sequences, while LiPA showed a wild-type sequence only. However, in both cases, the mutant was observed by LiPA, once at an earlier time point and once at a later time point (both appeared in patient 38) (Fig. 2). No completely discordant results were observed.
FIG. 2.
Comparison of LiPA and sequencing results for patient 38. The mutant virus was detected by LiPA once at an earlier time point (L80I) and once at a later time point (N236T) compared to the results of sequencing.
Indeterminate results.
For six samples (0.8% of the codons analyzed), the LiPA results for one or two of the six codons were indeterminate. In one case (0.1% of the codons analyzed), sequence analysis revealed a polymorphism at codon 235 that interfered with the annealing of the LiPA probes, thus creating an indeterminate result for codon 236. No polymorphisms were observed for the other cases, but the indeterminate results could be related to the low viral loads of the samples.
Samples showing mixed virus populations.
For 95 codon results (8.2% of the codons analyzed), derived from 26 patients, LiPA revealed the presence of both mutant and wild-type virus, while sequencing detected only either a wild-type or a mutant virus (Table 3). These results were of particular interest because they provided information about the sensitivity of LiPA at detecting an emerging minority population earlier than sequencing or detecting a disappearing virus population for a period of time longer than that observed with sequencing. Therefore, the 75 of 95 codon results (6.4% of the codons analyzed), which represented the results for 25 patients, in which LiPA detected a mixed sequence, while sequencing detected a wild type only, were analyzed in more detail.
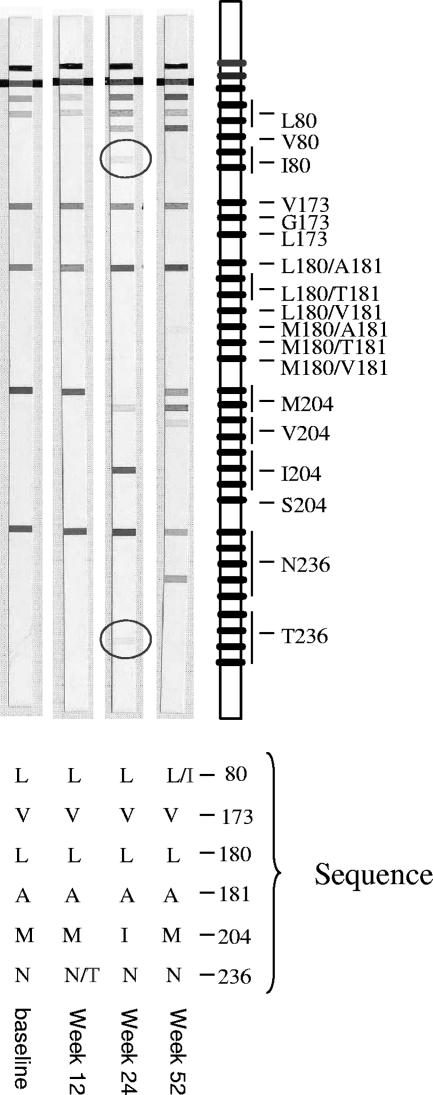
For 28 of these 75 codon results, the mixed or mutant sequence was confirmed by both sequencing and LiPA upon testing of follow-up samples. This clearly indicates that the mutant virus was indeed present in the patient and that LiPA detected this virus population earlier than sequencing. A representative example (for patient 72) is shown in Fig. 3.
FIG. 3.
Comparison of LiPA and sequencing results for patient 72. The mutant virus was detected earlier by LiPA (week 24) and was confirmed by both sequencing and LiPA at week 52.
For 30 of the 75 codon results, the mutant sequence was also detected by LiPA in one or more samples taken at another time point, proving that the mutant virus was indeed present in the patient and that LiPA detected this virus population before sequencing.
For 2 of the 75 codon results, the mutant sequence was confirmed by sequencing in a sample taken once at a later time point and once at a previous time point.
For 15 of the 75 codon results, no definitive conclusion could be made as to whether LiPA detected the mutant strain earlier, because the mutant virus was not detected by LiPA or sequence analysis in samples taken at an earlier time point.
Detection of mutants by LiPA in relation to viral breakthrough.
For 16 of 26 patients (61.5%), the mutant was detected by LiPA before viral breakthrough. A relevant case in which LiPA detected the mutant before viral breakthrough was seen in patient 72. At week 24, the viral load in this patient was low and progressively decreased, and LiPA had already detected a mix of M204I mutant and the wild type. After viral breakthrough, only the mutant virus was found.
Early detection of mutations by LiPA versus sequencing.
Time-to-event survival analysis indicated a 2.8 times greater chance for LiPA to detect a given mutation than sequencing at any moment in time (hazard ratio, 2.8; 95% confidence interval, 1.79, 4.41; P < 0.0001).
DISCUSSION
While new and effective nucleotide/nucleoside analogues are increasingly available for the treatment of chronic HBV infection, lamivudine remains the first-line therapy in many areas of the world with a high prevalence of chronic HBV infection for reasons of efficacy, safety, and cost. Therefore, monitoring of the evolution of primary and compensatory lamivudine resistance mutations during long-term lamivudine treatment remains a clinically relevant issue (10, 16).
In our study, 12 and 25 patients tested by sequencing and with the INNO-LiPA HBV DR v2 strip, respectively, developed a lamivudine resistance mutation after 1 year of treatment. Most displayed the hallmark lamivudine resistance mutation rtM204V/I. In general, the mutations and the mixed combinations that were observed (M204V/I/L180M/V173L, M204V/L180M/V173L/L80I, and M204I/V173L/L80I) agree with those detected in previous studies (2, 9, 11).
Remarkably, although the patients in our study had never received adefovir treatment, six patients showed the transient emergence of adefovir resistance mutations (rtA181T or rtN236T). Another nine patients developed adefovir-resistant mutants that persisted for several weeks, of whom eight were found to carry the mutant with the rtA181T mutation. Virus from four of these eight patients also showed a mutation at position rt204. A possible explanation for the four patients with the single rtA181T mutation could be that instead of an adefovir dipivoxil resistance mutation, a novel lamivudine resistance mutation emerged. Indeed, recently, Yatsuji et al. (15) described the emergence of a novel lamivudine-resistant strain of HBV with an intact YMDD motif, which included the amino acid substitution rtA181T in the reverse transcriptase domain of HBV polymerase. The finding that a significant number of patients under lamivudine monotherapy also develop adefovir dipivoxil resistance mutations can have consequences for the eventual choice of replacement or add-on therapy following viral breakthrough.
Similarly, even by the less sensitive sequencing method, a transient emergence of mutations at codons 181 and 236 was seen in two patients. However, in all cases these mutations were present as only a small proportion of the total virus population and as such give rise to weakly reactive lines on the LiPA strip. During the development of the LiPA, extreme care was taken to select probes that would not give any cross-reactivity at a considerable margin around the correct incubation temperature. However, since in this study we did not confirm the presence of the minor mutant population by an alternative method, the possibility of such cross-reactivity, even though it is unlikely, cannot be completely ruled out.
For patients on long-term lamivudine treatment, the frequent development of drug resistance and the subsequent loss of a treatment benefit remain overriding clinical concerns. Therefore, HBV treatment optimization necessitates the use of sensitive methods to detect drug resistance as early as possible before HBV isolates with emerging mutations acquire replicative capacities (9).
For our set of patient samples, the sequencing and the LiPA data were compared for 1,164 codon results. For 90.9% (n = 1,058) of these codon results, full concordance between both tests was observed. In an additional 8.2% (n = 95) of the cases, partial concordance between LiPA and sequencing was found, meaning that LiPA provided additional information compared to the information provided by sequencing. These findings are in accord with those of previous studies (1, 3, 7, 8, 14). Such results are of particular clinical interest, as they provide information about the sensitivity of the LiPA in detecting minority populations that are not picked up by sequencing. This implies that the LiPA may detect the emergence of a mutant earlier than would be possible by sequencing. Therefore, the 75 codon results (6.4% of the codons analyzed), representing 25 patients, in which LiPA detected a mixed sequence, while sequencing detected a wild type only, were analyzed in more detail. For 60 of these 75 codon results (80.0%) with a mixed or mutant sequence by LiPA but a wild-type sequence only by sequencing, the mutant sequence could be confirmed.
For the other 15 codon results, no definitive conclusion as to whether LiPA detected the mutant strain earlier could be made, because the mutant virus was not detected by LiPA or sequence analysis in samples taken at another time point. For at least 12 of 24 patients with a YMDD mutation, the mutation was detected earlier by LiPA than by sequencing, which demonstrates the higher sensitivity of LiPA in detecting minor virus populations. Such findings have also been reported in other studies (1, 3, 7).
Of the 336 samples amplified with the INNO-LiPA HBV DR v2 primers, 328 samples gave a positive 867-bp band on gels (97.6%). Forty-three of these 336 samples had viral loads less then 1,000 copies/ml, and of these 43 samples, 37 were positive (86.0%).
These results corroborate the high sensitivity of the INNO-LiPA HBV DR v2 strip, even for samples with very low viral loads.
A possible limitation of our study is that it drew samples from an entirely Asian (Hong Kong) population, which is known for its strong representation of genotypes B and C. However, a recent article by a Canadian group (8), in which all genotypes (except for the rare genotype F) were represented, showed that the new version of the INNO-LiPA HBV DR strip (INNO-LiPA HBV DR v2) works equally well with all common genotypes. The results for the INNO-LiPA HBV DR v2 strip versus those of sequencing in that study (8) were quite similar to those of the present study.
In conclusion, this study showed that for a Hong Kong population consisting of 80 patients chronically infected with HBV and treated exclusively with lamivudine for up to 3 years, the INNO-LiPA HBV DR v2 strip proved to be a sensitive and specific assay that was able to detect the emergence of primary and compensatory lamivudine resistance mutations, as well as the development of adefovir dipivoxil resistance mutations, and was able to provide such clinically relevant information significantly earlier than sequencing.
Acknowledgments
We thank Katrien Verscheuren and Geert De Meyer (both of Innogenetics) for their fine efforts in the statistical analysis of the data.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Aberle, S. W., J. Kletzmayr, B. Watschinger, B. Schmied, N. Vetter, and E. Puchhammer-Stockl. 2001. Comparison of sequence analysis and the INNO-LiPA HBV DR line probe assay for detection of lamivudine-resistant hepatitis B virus strains in patients under various clinical conditions. J. Clin. Microbiol. 39:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delaney, W. E., IV, H. Yang, C. E. Westland, K. Das, E. Arnold, C. S. Gibbs, M. D. Miller, and S. Xiong. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77:11833-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain, M., S. Fung, E. Libbrecht, E. Sablon, C. Cursaro, P. Andreone, and A. S. Lok. 2006. Sensitive line probe assay that simultaneously detects mutations conveying resistance to lamivudine and adefovir. J. Clin. Microbiol. 44:1094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liaw, Y. F., R. N. Chien, C. T. Yeh, S. L. Tsai, and C. M. Chu. 1999. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology 30:567-572. [DOI] [PubMed] [Google Scholar]
- 5.Liaw, Y. F., J. J. Sung, W. C. Chow, G. Farrell, C. Z. Lee, H. Yuen, T. Tanwandee, Q. M. Tao, K. Shue, O. N. Keene, J. S. Dixon, D. F. Gray, J. Sabbat, and the Cirrhosis Asian Lamivudine Multicentre Study Group. 2004. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351:1521-1531. [DOI] [PubMed] [Google Scholar]
- 6.Lok, A. S., F. Zoulim, S. Locarnini, A. Bartholomeusz, M. G. Ghany, J. M. Pawlotsky, Y. F. Liaw, M. Mizokami, C. Kuiken, and the Hepatitis B Virus Drug Resistance Working Group. 2007. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 46:254-265. [DOI] [PubMed] [Google Scholar]
- 7.Lok, A. S., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osiowy, C., J. P. Villeneuve, E. J. Heathcote, E. Giles, and J. Borlang. 2006. Detection of rtN236T and rtA181V/T mutations associated with resistance to adefovir dipivoxil in samples from patients with chronic hepatitis B virus infection by the INNO-LiPA HBV DR line probe assay (version 2). J. Clin. Microbiol. 44:1994-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallier, C., L. Castera, A. Soulier, C. Hezode, P. Nordmann, D. Dhumeaux, and J. M. Pawlotsky. 2006. Dynamics of hepatitis B virus resistance to lamivudine. J. Virol. 80:643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pas, S. D., R. A. de Man, E. Fries, A. D. Osterhaus, and H. G. Niesters. 2002. The dynamics of mutations in the YMDD motif of the hepatitis B virus polymerase gene during and after lamivudine treatment as determined by reverse hybridisation. J. Clin. Virol. 25:63-71. [DOI] [PubMed] [Google Scholar]
- 11.Sheldon, J., B. Rodes, F. Zoulim, A. Bartholomeusz, and V. Soriano. 2006. Mutations affecting the replication capacity of the hepatitis B virus. J. Viral Hepat. 13:427-434. [DOI] [PubMed] [Google Scholar]
- 12.Stuyver, L., C. Van Geyt, S. De Gendt, G. Van Reybroeck, F. Zoulim, G. Leroux-Roels, and R. Rossau. 2000. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Virol. 38:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Geyt, C., S. De Gendt, A. Rombaut, A. Wyseur, G. Maertens, R. Rossau, and L. Stuyver. 1998. A line probe assay for hepatitis B virus genotypes, p. 139-145. In R. F. Schinazi, J. P. Sommadossi, and H. Thomas (ed.), Therapies of viral hepatitis. International Medical Press, London, United Kingdom.
- 14.Van Laethem, K., K. Van Vaerenbergh, J.-C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clerq, and A.-M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assay for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 15.Yatsuji, H., C. Noguchi, N. Hiraga, N. Mori, M. Tsuge, M. Imamura, S. Takahashi, E. Iwao, Y. Fujimoto, H. Ochi, H. Abe, T. Maekawa, C. Tateno, K. Yoshizato, F. Suzuki, H. Kumada, and K. Chayama. 2006. Emergence of a novel lamivudine-resistant hepatitis B virus variant with a substitution outside the YMDD motif. Antimicrob. Agents Chemother. 50:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuen, M. F., E. Sablon, C. K. Hui, H. J. Yuan, H. Decraemer, and C. L. Lai. 2001. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology 34:785-791. [DOI] [PubMed] [Google Scholar]
- 17.Yuen, M. F., T. Kato, M. Mizokami, A. O. Chan, J. C. Yuen, H. J. Yuan, D. K. Wong, S. M. Sum, I. O. Ng, S. T. Fan, and C. L. Lai. 2003. Clinical outcome and virologic profiles of severe hepatitis B exacerbation due to YMDD mutations. J. Hepatol. 39:850-855. [DOI] [PubMed] [Google Scholar]
- 18.Yuen, M. F., W. K. Seto, D. H. F. Chow, K. Tsui, D. K. H. Wong, P. Chan, D. But, and C. L. Lai. 2005. Long-term beneficial outcome of Chinese asymptomatic patients with HBeAg-positive chronic hepatitis B on continuous lamivudine therapy: 7-year experience. Hepatology 42(Suppl. 1):583A. [Google Scholar]
- 19.Yuen, M. F., E. Sablon, E. Libbrecht, H. Van De Velde, D. K. H. Wong, J. Fung, B. C. Y. Wong, and C. L. Lai. 2006. Significance of HBV viral load, core promoter/ precore mutations and specific sequences of polymerase gene in HBV-infected patients on 3-year lamivudine treatment. Antivir. Ther. 11:779-786. [PubMed] [Google Scholar]