Abstract
Helicobacter pylori cagA and vacA genotypes have been used for almost a decade as stable entities to link the severity of gastritis and ulcer disease. We describe here microevolution of the two genomic islands, cag pathogenicity island (cagPAI; 40 kb) and tfs3 (16 kb) from isolates obtained at inclusion (one subclone) and after a 10-year period (two subclones) from a duodenal ulcer patient. Our results indicate microevolution in cagA, cagE, and cag7 genes of the cagPAI and open reading frames G, P, and L in tfs3, which possibly leads to inactivation or pseudogenization of these genes. Interestingly, no significant reduction in the severity of gastroduodenal pathology was found. These results point to an obvious difficulty in correlating the continuously evolving virulence factors such as the cagPAI genes with disease characteristics that appear to remain stable.
Helicobacter pylori displays genetic variability (8, 17) due to frequent mutations and genetic recombination (5, 11, 22). It has also been shown that during mixed infections recombinant strains emerge with different combinations of parental genotypes (19). However, there has been a long-standing dogma with infection biologists that H. pylori virulence-encoding genes such as cagA and vacA are stable entities and that these could be robustly correlated with disease progression or outcome. Observations contrary to this assumption have been frequently found; hence, in order to adapt to a new environment the selection process concerning virulence encoding genes should take place. Genomic rearrangements aimed at host-specific adaptation include spontaneous mutations as shown via serial passages in gnotobiotic piglets (1) or through allelic recombination leading to differential inactivation of important virulence genes such as the vacA of the subclones in a single patient (23). Another important mechanism establishing adaptive evolution operates through deletion of either individual gene loci such as cagA (3, 10) or the entire genomic island, e.g., the cag pathogenicity island (cagPAI) (19). In addition, various genotyping methods applied to serial isolates obtained from the same patient revealed similar fingerprints with minor differences (2, 13). This may be possible due to the fact that two or more isolates recovered from a patient might share a common ancestor but experience independent genomic alterations (14). This phenomenon has been termed “microevolution” (7, 11). However, sequence data are necessary to verify microevolution (19), and phenotypic confirmation is required to ascertain whether this process leads to optimization of virulence in response to changing gastric environment. Genetic changes within the entire cagPAI sequences obtained from different individuals (6) have been recently described (5). Similarly, a novel genomic island, tfs3 was analyzed from clones of strain J99 derived from patients at different time points (18). Despite these important studies, the nature and extent of genetic polymorphisms during the colonization of different gastric niches on a wide time scale are not known. The forces driving such evolution in vivo and the selective advantage conferred to H. pylori by such microevolution of virulence-encoding genes are also not understood. While some studies have explored strain diversity through family studies (19), no concept of chronological evolution was reported until the study of Israel et al., based on microevolution in H. pylori J99 across 6 years (13). These authors compared several isolates of the reference strain J99 taken 6 years apart from the same patient and, despite the fact that the RAPD [random(ly) amplified polymorphic DNA] profiles were identical, microarray analysis revealed acquisition and deletion of several genes (13). However, although microarray analysis on a genomewide scale does offer a complete picture of acquisition and loss of a whole gene, the nature and extent of DNA sequence divergence should also be understood. The best strategy therefore appears to perform pan-island, large-scale sequencing of regions such as the cagPAI. We report whole-PAI sequence analysis of H. pylori and highlight changes occurring at a wide time scale in a single host. This is an extension of our previous study (21), wherein we showed by various genome-profiling techniques that isolates recovered 10 years apart from the same patient corresponded to a single strain without any evidence of mixed colonization. In the present study, using the same set of isolates (21) we studied molecular evolution of cagPAI and tfs3 to examine whether microevolution can occur in the absence of mixed infection. Several hotspots concerning the microevolution of these genomic islands were identified that could be related to adaptation mechanisms.
Gastric biopsy samples were collected from an African patient living in France after written informed consent were obtained; this patient had an endoscopically proven duodenal ulcer in 1994 (21). H. pylori was isolated from individual biopsy samples and maintained frozen at −70°C after limited passages. The patient was successfully treated with a 10-day triple therapy (amoxicillin, clarithromycin, and lansoprazole). Posttreatment histology followed by a negative culture report proved the absence of H. pylori, and a complete recovery was registered. In January 2003 the patient presented again with the symptoms of duodenal ulcer. Biopsy samples were collected from the antral and corpus mucosa for H. pylori isolation, and histopathological examination was performed.
H. pylori was isolated from antral biopsy samples at the initial endoscopy (isolate 908) and from both antrum and corpus biopsy samples (isolates 2017 and 2018) at the follow-up endoscopy. All of the three bacterial isolates corresponding to different niches of the stomach, isolated at different time periods, were considered as identical based on (i) RAPD fingerprinting, (ii) analyses of enterobacterial repetitive intergenic consensus, and (iii) repetitive extragenic palindromes and multilocus sequence typing (21). Any possibility of mixed infection was thus ruled out. Biopsy specimens were embedded in paraffin and 4-μm-thick sections were stained with hematoxylin and eosin. The pathologist was blinded to the patient's clinical condition. Chronic gastritis was graded according to the updated Sydney system (9). The whole cagPAI of the three isolates was subjected to PCR and sequencing as described by Blomstergren et al. (6) and analyzed through the CLUSTAL X program (15), together with other known cagPAI sequences. The resulting alignment was used to construct a neighbor-joining tree. The complete sequence of the tfs3 cluster of the three isolates was also determined essentially as described by Kersulyte et al. (18). Comparative genomic analysis based on the tfs3 sequences was performed as for the cagPAI. The cagPAI sequences of isolates 908, 2017, and 2018 were deposited in GenBank under accession numbers EF195721, EF195722, and EF195723, respectively. The accession numbers for tfs3 were EF195724, EF195725, and EF195726 for isolates 908, 2017, and 2018, respectively.
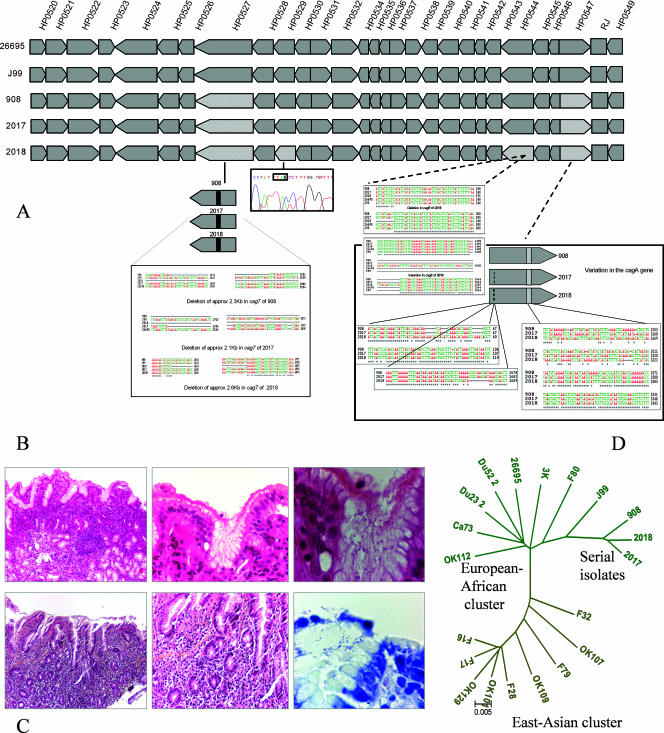
The details of the nature and extent of the DNA sequence rearrangements occurring in the cagPAI of the three serial isolates are depicted schematically in Fig. 1A. Briefly, we noticed extensive rearrangements in the cag7 gene, the cagE gene, and the cagA gene of the three isolates. Compared to the H. pylori 26695 genome, extensive deletions were found in the cag7 gene: (i) deletions at the 5′ end of 357 bp (positions 564 to 920), 423 bp (positions 563 to 985), and 438 bp (positions 557 to 994) in isolates 908, 2017, and 2018, respectively, and (ii) large deletions of 2,049 bp (positions 2007 to 4055), 1,834 bp (positions 2127 to 3960), and 2,258 bp (positions 1899 to 4156) in isolates 908, 2017, and 2018, respectively. The HP0529 gene was found to be pseudogenized through the generation of a stop codon in isolate 2018, which originated in the corpus region of the stomach. A frameshift mutation caused by both independent insertion and deletion events each was found in the cagE gene (HP0544). Similarly, the cagA gene was also found to have undergone multiple rearrangements due to various insertion, deletion, and substitution events. Although the cagA gene was apparently nonfunctional even in the first isolate (isolate 908), it was subjected to further rearrangement in isolates 2017 and 2018. The extent of evolution of the cagPAI genes was dramatic and significant in isolate 2018 compared to isolates 2017 and 908, possibly due to its location in a different niche, i.e., the corpus of the stomach.
FIG. 1.
(A) Genomic alignments based on pan-island sequencing of cagPAIs from isolates 908, 2017, and 2018 and their comparison with sequences from the two sequenced strains, namely, J99 and 26695. For details of sequencing strategy and alignment methods, see the text. (B) Histopathology of biopsy sections corresponding to isolate 908, originally obtained in 1994 from three antropyloric biopsy samples that revealed duodenal ulcer (inflammatory infiltrate [3+] with polymorphs [3+] and numerous H. pylori on the surface [3+]). (C) Endoscopic diagnosis revealed duodenal ulcer, while histopathological examination of two gastric biopsy samples—one each from antrum (inflammatory infiltrate [2+] with eosinophils, atrophy [+], no metaplasia, no lymphoid follicle, and the presence of H. pylori [2+]) and corpus (slight superficial gastritis)—revealed relatively low-grade pathology in 2003. Isolates 2017 and 2018 corresponding to antrum and corpus, respectively, were successfully cultured from this source. (D) Bifurcating neighbor-joining tree based on whole cagPAI sequence comparison. The clonal association among isolates 908, 2017, and 2018 clearly reveals that they belonged to the same founder strain that shares genogroup affinity with J99 strain (HspSAfrica).
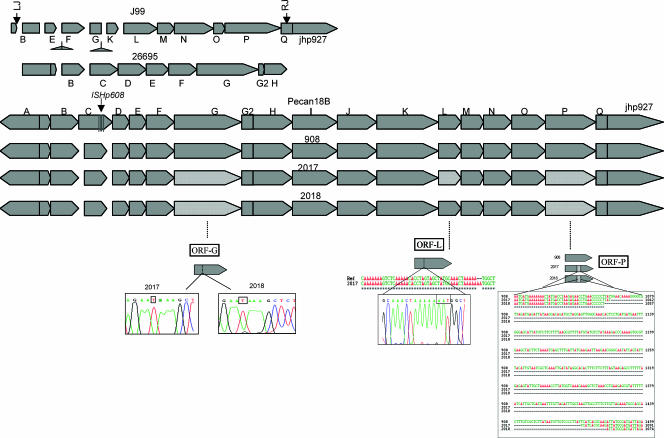
An intact tfs3 cluster was observed in isolate 908, similar to that of the Peruvian cancer patient isolate PeCan18B, within which the tfs3 was originally described (18). However, substantial rearrangements were found to accumulate in the tfs3 cluster of both of the successive isolates, where 3 of the 19 tfs3 open reading frames (ORFs) were rearranged and rendered nonfunctional during the follow-up period. Single base substitutions and deletions leading to pseudogenization of ORF G and ORF P, respectively, were observed. In addition, isolate 2017 also revealed a single base change leading to a stop codon in ORF L, thus leading to its pseudogenization. A more important change, however, was the loss of DNA fragments of 406 and 421 bp at positions 1066 to 1471 and positions 1063 to 1483 (compared to H. pylori 26695) in ORF P of isolates 2017 and 2018, respectively. Genetic changes, however, in the cluster tfs3 compared to the cagPAI were not seemingly correlated to the change in niche; for example, isolate 2018 compared to 2017 did not appear to undergo different rearrangements despite the fact that the former was isolated from the corpus region. Hence, both of the isolates evolved similarly at both the niches (Fig. 2).
FIG. 2.
Genomic alignments based on pan-island sequencing of tfs3 islands from isolates 908, 2017, and 2018 and their comparison with sequences from the two sequenced strains, namely, J99 and 26695 and the Peruvian cancer strain Pecan18B. For details of the sequencing strategy and alignment methods, see the text.
Figure 1B and C present the histopathological lesions observed in the patient at different time points after H. pylori infection. At the time H. pylori isolate 908 was isolated, the patient suffered from duodenal ulcer. Antropyloric mucosa revealed a large-scale inflammatory infiltrate with a high number of polymorphonuclear cells and numerous H. pylori organisms visible on the surface. Ten years later, the histological preparation showed a limited infiltration of polymorphonuclear cells and lymphocytes, the presence of H. pylori, and the absence of lymphoid follicles.
It is generally understood that the disease developing in people with H. pylori infection is determined by the pattern of colonization in the individual stomach. For example, antral gastritis predisposes patients to duodenal ulcers, whereas body gastritis and pangastritis predispose patients to gastric ulcers and gastric carcinoma. If we assume that specific strain characteristics resulting from in vivo microevolution allow colonization of different niches of stomach, it is possible that rapid evolution of a parent strain culminating in a variety of subclones will eventually allow colonization of the whole stomach, thus causing pangastritis and carcinoma. However, due to the rapid evolution of the cagPAI genes in all of the three subclones we studied, it is tempting to speculate that these clones are limiting their aggressiveness via rapid pseudogenization of essential genes such as cagA and cagE (Fig. 1). If this scenario holds true, we must uncover the as-yet-unknown selective advantages conferred upon H. pylori by such differently evolved forms of virulence-encoding genes and different selective forces driving this type of evolution.
We suggest that isolate 2018 recovered from the corpus of our patient might have undergone distinctive microevolution compared to isolates 908 and 2017 obtained from the antrum of the stomach. This probably suggests the effect of a niche on the genotype or the adaptation of an evolved substrain to a defined niche in the stomach.
The tfs3 cluster also showed evidence of microevolution (Fig. 2), although not as extensive as that observed by Kersulyte et al. (18), in different subclones of J99 strain obtained in serial endoscopies across 6 years (13). Our aim in studying tfs3 was mainly to (i) validate the stability of this new type IV secretory apparatus through a decade-long colonization and (ii) use it as a seemingly”benign“ control against the cagPAI, which is under greater selection pressure than tfs3, because of its association with virulence (16). Despite the fact that our serial isolates belonged to the H. pylori subpopulation HspWAfrica (whose well-known member J99 revealed extensive deletions of tfs3 in its serial isolates [18]), tfs3 largely remained intact. Comparing, therefore, our tfs3 results to those obtained with J99 (18), we assumed that the stability or fragility of this element may be strain specific and largely determined by the environmental drivers and in vitro conditions (such as repeated subculture that has happened in the case of strain J99). The niche-specific“adaptive evolution” as seen for cagPAI was not observed for the tfs3 cluster of our isolates. This is because the loss of the fragments due to deletion has been minimal and limited to only one gene, ORF P. The other two ORFs (ORFs G and L) have been pseudogenized due to specific nonsense substitutions, a more subtle and transient phenomenon that might be reversed.
In our previous study, we initially assumed isolate 908 to be the parent strain and 2017 as its “surviving” descendant posteradication. Reinfection with an altogether new strain was also ruled out in that study by various fingerprinting and genotyping approaches (21). The pan-island sequencing performed for all of the available strains from this patient reconfirmed this. We understand that the isolates of H. pylori responsible for a severe pathological outcome as seen with our patient in 1994 (Fig. 1B) could possibly be harboring functional cagA and vacA genes. In contrast to this, we found that the subclone of H. pylori that we cultured in 1994 (isolate 908) from the antropyloric mucosal biopsy samples could be an evolved version of the parent strain. This evolved version has been suggested to be deficient in cagA function due to a frameshift deletion (Fig. 1A) and nontoxigenic due to the deletion of the middle region of the vacA gene (21). Therefore, it appears that isolate 908 might have undergone rapid and dramatic evolution of the cagA gene after the pathology developed. This scenario supports our hypothesis that virulence factor types are not the fixed characteristic of H. pylori and that they may evolve during colonization, hence the difficulty in precisely correlating these genotypes with disease. Although, the comparatively lower severity of the ulcer disease recurring after 10 years (Fig. 1C) might correlate with evolved cagPAI composition (evolved cagA, cagE, and cag7), it is possible that cagPAI alone may not explain everything.
Finally, it appears that H. pylori continuously evolves (11, 13, 19, 23) and spreads within the niches of the stomach. However, a majority of its “evolved” subclones might just be the beneficiaries of the inflammation and ulceration processes, feeding on the exudate and debris with a minimal role in pathology. Nonetheless, few highly aggressive substrains with “conserved” cag functions might be active as drivers of gastroduodenal pathology in different niches. Alternatively, recombination might be occurring frequently among all of the subclones wherein some loci become nonfunctional for a while but regain functional alleles later in the life cycle. The acquisition of a functional vacA mid-region fragment by isolate 2017 as described by us previously (21) demonstrates this potential hypothesis. In any case, it will be necessary to study this microevolution process especially in the context of the development of pan-gastritis, gastric ulcers, and gastric adenocarcinoma. A large number of paired isolates (12, 20) from different niches of the stomach obtained at different time intervals may be analyzed to explore transient loss or gain of function at important virulence loci such as cagA and vacA. Also, other potential virulence factors may be investigated for their association with development and progression of pathology as juxtaposed to microevolutionary changes acquired in a single host. Determining therefore the in vivo evolutionary status (of these three important strains in hand) concerning the “nonclassical” virulence factors such as several outer-membrane proteins, members of the plasticity region cluster, porins, and flagellins, etc., may be undertaken in the future. The functional status of these subclones evolving through a variety of insertion, deletion, and substitution events needs to be checked via in vitro phenotyping methods (4) for rapid loss or gain of virulence. Also, environmental switches responsible for such a fast intragenic (and intergenic) recombination may be identified in a suitable animal model of H. pylori colonization.
Acknowledgments
Research in the laboratory of N.A. was funded by CDFD core grants (Department of Biotechnology, Government of India). F.M. and N.A. are the Fellows of the European Helicobacter Study Group.
We are grateful for the guidance of Seyed E. Hasnain. We also thank two anonymous referees whose constructive comments and suggestions helped us improve this study.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Akopyants, N. S., K. A. Eaton, and D. E. Berg. 1995. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect. Immun. 63:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aras, R. A., Y. Lee, S. K. Kim, D. Israel, R. M. Peek, Jr., and M. J. Blaser. 2003. Natural variation in populations of persistently colonizing bacteria affect human host cell phenotype. J. Infect. Dis. 188:486-496. [DOI] [PubMed] [Google Scholar]
- 4.Argent, R. H., M. Kidd, R. J. Owen, R. J. Thomas, M. C. Limb, and J. C. Atherton. 2004. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127:514-523. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkholm, B., M. Sjolund, P. G. Falk, O. G. Berg, L. Engstrand, and D. I. Andersson. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomstergren, A., A. Lundin, C. Nilsson, L. Engstrand, and J. Lundeberg. 2004. Comparative analysis of the complete cag pathogenicity island sequence in four Helicobacter pylori isolates. Gene 328:85-93. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. Morain, C. M. Habibullah, and C. J. Smyth. 2004. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J. Med. Microbiol. 53:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Devi, S. M., I. Ahmed, A. A. Khan, S. A. Rahman, A. Alvi, L. A. Sechi, and N. Ahmed. 2006. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics 7:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon, M. F., R. M. Genta, J. H. Yardley, and P. Correa. 1996. Classification and grading of gastritis: the updated Sydney System. Am. J. Surg. Pathol. 20:1161-1181. [DOI] [PubMed] [Google Scholar]
- 10.Evans, D. L., Jr., and D. G. Evans. 2001. Helicobacter pylori CagA: analysis of sequence diversity in relation to phosphorylation motifs and implications for the role of CagA as a virulence factor. Helicobacter 6:187-198. [DOI] [PubMed] [Google Scholar]
- 11.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustavsson, A., M. Unemo, B. Blomberg, and D. Danielsson. 2005. Genotypic and phenotypic stability of Helicobacter pylori markers in a nine-year follow-up study of patients with noneradicated infection. Dig. Dis. Sci. 50:375-380. [DOI] [PubMed] [Google Scholar]
- 13.Israel, D. A., N. Salama, U. Krishna, M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 15.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with CLUSTAL X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 16.Kauser, F., A. A. Khan, M. A. Hussain, I. M. Carroll, N. Ahmad, S. Tiwari, Y. Shouche, B. Das, M. Alam, S. M. Ali, C. M. Habibullah, R. Sierra, F. Megraud, L. A. Sechi, and N. Ahmed. 2004. The cag pathogenicity island (cag-PAI) of Helicobacter pylori is disrupted in majority of patient isolates from different human populations. J. Clin. Microbiol. 42:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 20.Kuipers, E. J., D. A. Israel, J. G. Kusters, M. M. Gerrits, J. Weel, A. van der Ende, R. W. van der Hulst, H. P. Wirth, J. Hook-Nikanne, S. A. Thompson, and M. J. Blaser. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prouzet-Maule'on, V., M. A. Hussain, H. Lamouliatte, F. Kauser, F. Me'graud, and N. Ahmed. 2005. Pathogen evolution in vivo: genome dynamics of two isolates obtained 9 years apart from a duodenal ulcer patient infected with a single Helicobacter pylori strain. J. Clin. Microbiol. 43:4237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Ende, A., E. A. J. Rauws, M. Feller, C. J. J. Mulder, G. N. J. Tytgat, and J. Dankert. 1996. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology 111:638-647. [DOI] [PubMed] [Google Scholar]