Abstract
A sensitive chemiluminescence enzyme immunoassay has been developed for hepatitis B virus (HBV) core-related antigen (HBcrAg) detection. We aimed to investigate the usefulness of HBcrAg measurement for monitoring chronic hepatitis B disease. HBcrAg levels were measured by a chemiluminescence enzyme immunoassay in 54 untreated patients and 39 patients treated with either entecavir or lamivudine. The HBcrAg concentration correlated positively with the levels of serum HBV DNA (r = 0.820), intrahepatic total HBV DNA (r = 0.700), and covalently closed circular DNA (cccDNA) (r = 0.664; for all, P values were <0.001). A higher HBcrAg concentration was associated with a greater proportion of hepatitis B core antigen immunostaining. Although the differences were not statistically significant, patients with higher Knodell necroinflammation and fibrosis scores tended to have higher serum HBcrAg concentration levels. In the treated patients, the logarithmic reduction in HBcrAg at week 48 correlated positively with the logarithmic reduction of serum HBV DNA, intrahepatic total HBV DNA, and cccDNA. Of the 31 patients with undetectable serum HBV DNA (<300 copies/ml) at the end of treatment, 20 (65%) still had detectable HBcrAg. A greater reduction in posttreatment HBcrAg concentration was associated with histological improvement and a decrease in hepatitis B core antigen immunostaining. HBcrAg concentrations of <40,000 kU/ml at baseline and <200 kU/ml at week 24 were associated with a higher chance of having undetectable HBV DNA at week 48. In conclusion, serum HBcrAg levels correlated with HBV virological markers and reflected the chronic hepatitis B disease activity in the liver.
Chronic hepatitis B virus (HBV) infection is a global health problem, affecting approximately 400 million people worldwide (7). Serological markers are important in chronic HBV infection for the diagnosis, prognosis, and monitoring of untreated and treated patients.
With the advance of PCR technology, sensitive assays for the detection of HBV DNA are commonly employed to monitor viral replication. However, these assays for HBV DNA measurement are often expensive and cumbersome. Several assays have been developed for the measurement of serum hepatitis B e antigen (HBeAg) or hepatitis B core antigen (HBcAg) concentrations (1, 3, 16). However, these assays are of limited use because of their relatively low sensitivities and the complex procedures. Recently, a relatively simple and sensitive chemiluminescence enzyme immunoassay (CLEIA) has been developed for the simultaneous detection of both HBeAg and HBcAg (5, 13). HBeAg, possibly an immune-modulating protein, is a nonstructural secreted protein translated from HBV e mRNA. HBcAg, the building block of the HBV viral capsids, is translated from the HBV pregenomic mRNA. Since HBeAg and HBcAg share a 149-amino-acid sequence identity, they are collectively called hepatitis B core-related antigens (HBcrAg) (5, 13). The analytical lower detection limit of the HBcrAg assay is 4 × 102 U/ml, which is equivalent to the immunoreactivity of 4 pg/ml of ProHBeAg, a recombinant protein containing HBeAg amino acid sequence from −10 to 183 (5). The dynamic range of the assay ranges from 4 × 102 to 1 × 107 U/ml (5). In this assay, the specimens were pretreated so that HBeAg and HBcAg were released from the patients’ own anti-HBe and anti-HBc antibody-antigen complexes, respectively. Thus, both HBeAg and HBcAg can be accurately detected even in anti-HBc- or anti-HBe-positive specimens. Although the levels of HBcrAg are higher in HBeAg-positive patients than in HBeAg-negative patients, the accuracy of HBcrAg measurement is shown to be unaffected by the HBeAg status and the emergence of HBeAg-negative precore mutations (13). The level of serum HBcrAg concentration has also been shown to reflect the patient's HBV viral load (5) and to decrease during lamivudine treatment (10, 13, 14). However, correlations of serum HBcrAg concentration with other markers of disease progression, such as serum alanine aminotransferase (ALT), intrahepatic HBV DNA, intrahepatic HBcAg staining, and the degree of necroinflammation and fibrosis, have not been thoroughly studied. In addition, its usefulness for the prediction of treatment response after 48 weeks of antiviral therapy has not been explored. The aims of this study were to evaluate the usefulness of HBcrAg measurement in terms of correlation with disease activity (i) in untreated chronic hepatitis B patients and (ii) in patients under treatment with nucleoside analogues.
MATERIALS AND METHODS
Sample collection from untreated patients.
Liver biopsy and serum specimens were collected from 54 chronic hepatitis B patients (17 HBeAg positive and 37 anti-HBe positive) who were regularly followed up in the hepatitis clinic, Queen Mary Hospital, Hong Kong. The study was approved by the institutional review board, The University of Hong Kong, Hong Kong. Written informed consent was obtained, and serum HBcrAg was measured in these patients.
Sample collection from patients treated with nucleoside analogues.
Thirty-nine patients (14 HBeAg positive and 25 anti-HBe positive) were enrolled in this part of the study. These patients were part of two phase III international multicenter clinical trials for HBeAg-positive (2) and anti-HBe-positive patients (8), evaluating the efficacy of 0.5 mg entecavir daily versus that of 100 mg lamivudine daily. Baseline and week-48 liver biopsies and serum samples, as well as serum samples at week 24, were taken. The serum HBcrAg concentration was measured at baseline and week 24 and week 48 of treatment.
Serum HBcrAg concentration measurement.
HBcrAg was measured in serum using the CLEIA described previously (5). Briefly, 150 μl of serum was incubated with 150 μl pretreatment solution containing 15% sodium dodecyl sulfate at 60°C for 30 min. The pretreated serum was added to a well coated with three monoclonal antibodies (HB44, HB61, and HB114) against denatured HBcAg and HBeAg. After the wells were washed, two other alkaline phosphatase-labeled monoclonal antibodies against denatured HBcAg and HBeAg (HB91 and HB110) were added as secondary antibodies. The epitope specificity of these monoclonal antibodies has been described previously (5). Two hundred microliters of substrate [AMPPD; 3-(2′-spiroadamantan)-4-methoxy-4-(3"-phosphoryloxy) phenyl-1,2-dioxetane disodium salt] (Applied Biosystems, Bedford, MA) solution was added, and the assay tube was incubated for 5 min at 37°C. The relative chemiluminescence intensity was measured with a fully automated Lumipulse f CLEIA analyzer (Fujirebio Inc., Tokyo, Japan), and the HBcrAg concentration was estimated by comparison to a standard curve generated using recombinant ProHBeAg (amino acids −10 to 183) expressed as a TrpE-fusion protein in Escherichia coli and purified as described previously (5). In the present study, the cutoff value of HBcrAg concentration was 1 kU/ml, which is equivalent to the immunoreactivity of recombinant ProHBeAg at 10 pg/ml as previously determined (5).
Measurement of serum and intrahepatic HBV DNA.
Intrahepatic DNA was isolated from needle liver biopsies by using a QIAamp DNA mini kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. Intrahepatic total HBV DNA and covalently closed circular DNA (cccDNA) were measured by using the Invader assay, described previously, which has been fully validated and shown to be specific for cccDNA measurement (18). Briefly, the extracted total liver DNA was heat denatured, followed by hybridization detection by using different sets of oligonucleotides specific for either HBV DNA minus strand (hence, total HBV DNA detection), uninterrupted HBV DNA plus strand at the direct-repeat-2 junction (for cccDNA detection), or human insulin-like growth factor I gene (for human genomic DNA detection). Based on an estimated 6.667 pg of human genomic DNA per cell, the intrahepatic total HBV DNA and cccDNA levels were expressed as the number of HBV DNA copies/cell. Serum viral loads were measured by using a COBAS Amplicor HBV monitor test (lower limit of detection, 300 copies/ml; Roche Diagnostics, Branchburg, NJ).
Histological evaluation and immunostaining of HBcAg.
The liver tissues available for histological evaluation were at least 1 cm in length, with more than six portal tracts. Immunostaining for HBcAg was performed on 5-μm sections from the formalin-fixed, paraffin-embedded liver tissue as previously described (19). Histological activity index scores were assessed according to the criteria of Knodell et al. (6). Histologic improvement was defined as a reduction by more than 2 of the histological activity score.
Statistical analyses.
Statistical analyses were performed by using the Statistical Program for Social Sciences (SPSS 12.0 for Windows; SPSS, Chicago, IL). Related and unrelated continuous variables were tested by using the Wilcoxon signed-rank test and Mann-Whitney U test, respectively. The correlation between two variables was tested by using Spearman's rank correlation analysis. Categorical variables were tested by the chi-square test or the Fisher exact test.
RESULTS
Use of HBcrAg measurement in monitoring disease activity.
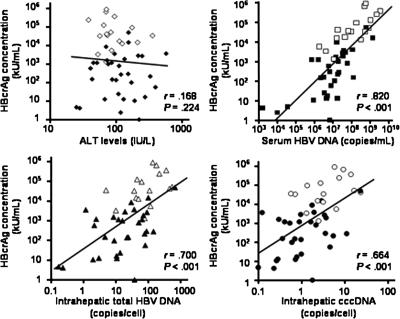
HBcrAg was detectable in the sera of all 17 HBeAg-positive patients and 32/37 (86.5%) anti-HBe-positive patients. As shown in Table 1, there was no significant difference in HBcrAg concentrations between patients of different genders (P = 0.130). There was also no strong correlation between HBcrAg concentrations and patients’ ages (r = −0.25; P = 0.068). As expected, the HBeAg-positive patients had a higher median HBcrAg concentration than the anti-HBe-positive patients (35,700 versus 153 kU/ml; P < 0.001). The correlation between serum ALT levels and HBcrAg concentration was insignificant (r = 0.168; P = 0.224) (Fig. 1). Patients with serum ALT levels that were less than or equal to the upper limit of normal (ULN) had levels of HBcrAg concentration that were comparable to the levels in patients with serum ALT levels of more than two times the ULN (P = 0.621).
TABLE 1.
Correlation of HBcrAg concentration with clinical parameters
| Patient (n = 54) characteristic | Valuea | kU/ml HBcrAg [median (range)] | P value |
|---|---|---|---|
| Males/females | 44:10 | 1,070 (<1.0-899,000) vs 7,660 (12.8-356,000) | 0.130 |
| Age (yrs) | 42 ± 10 | ||
| HBeAg/anti-HBe | 17:37 | 35,700 (1,320-899,000) vs 153 (<1.0-49,600) | <0.001 |
| ALT level (IU/liter) | 86 (21-576) | ||
| ≤2 × ULN (n = 29) vs >2 × ULN (n = 25) | 1,180 (<1.0-899,000) vs 1,860 (1.0-366,000) | 0.621 | |
| Serum HBV DNA (copies/ml) | 1.7 × 107 (<300-3.6 × 109) | ||
| Intrahepatic total HBV DNA (copies/cell) | 33.3 (<0.002-670) | ||
| Intrahepatic cccDNA (copies/cell) | 1.3 (<0.002-23.3) | ||
| HBcrAg concn (kU/ml) | 1,180 (<1.0-9.0 × 105) |
Mean ratio, mean ± standard deviation, or median (range).
FIG. 1.
Correlation between serum HBcrAg concentration and ALT, serum HBV DNA, intrahepatic total HBV DNA, and cccDNA levels. Open squares, circles, and triangles indicate samples which are HBeAg-positive, whereas closed squares, circles, and triangles indicate samples lacking HBeAg.
Serum HBV DNA, intrahepatic total HBV DNA, and intrahepatic cccDNA were detectable in 53 (98%), 51 (94%), and 50 (93%) patients, respectively. There were strong positive correlations between serum HBcrAg concentrations and levels of serum HBV DNA (r = 0.820; P < 0.001), intrahepatic total HBV DNA (r = 0.700; P < 0.001), and intrahepatic cccDNA (r = 0.664; P < 0.001), respectively (Fig. 1).
The correlation between serum HBcrAg concentration, histology data, and hepatocyte HBcAg staining is shown in Table 2. Patients with higher necroinflammation (≥6) and higher fibrosis (≥3) scores tended to have higher serum HBcrAg concentration levels (P = 0.056 and P = 0.079, respectively). Liver tissues from 38 patients were available for immunostaining of cytoplasmic and nuclear HBcAg. As shown in Table 2, 63% and 61% of patients had positive immunostaining results for cytoplasmic and nuclear HBcAg, respectively. There were strong positive correlations between the serum HBcrAg concentrations and the percentages of cytoplasmic and nuclear HBcAg-positive hepatocytes (P = 0.004 and P < 0.001, respectively).
TABLE 2.
Correlation of HBcrAg concentration with histology and hepatocyte HBcAg immunostaining data
| Histology data (n = 54) | No. (%) of patients | kU/ml HBcrAg [median (range)] | P value |
|---|---|---|---|
| Knodell necroinflammation score | |||
| 0-5 | 27 (50) | 23.8 (<1.0-899,000)} | 0.056 |
| 6-9 | 23 (42)} | 2,850 (55.6-51,900) | |
| >9 | 4 (8) | ||
| Knodell fibrosis score | |||
| 0 | 18 (33)} | 193 (<1.0-899,000)} | 0.079 |
| 1 | 12 (22) | ||
| 3 | 21 (39)} | 2,990 (55.6-92,600) | |
| 4 | 3 (6) | ||
| % Staining (n = 38) | |||
| Cytoplasmic HBcAg | |||
| 0 | 14 (37) | 124 (2.4-8,700) | <0.001 |
| ≤5 | 13 (34) | 2,670 (23.8-92,600) | |
| 6-25 | 5 (13) | 49,600 (17,200-126,000) | |
| 26-50 | 4 (11) | 129,000 (1,860-899,000) | |
| 51-75 | 2 (5) | 204,000 (41,700-366,000)} | |
| Nuclear HBcAg | |||
| 0 | 15 (39) | 1,150 (2.4-51,900) | 0.004 |
| ≤5 | 14 (37) | 2,210 (23.8-92,600) | |
| 6-25 | 6 (16) | 10,500 (1,860-244,000) | |
| 26-50 | 3 (8) | 366,000 (41,700-899,000)} |
Role of HBcrAg measurement in disease monitoring in patients receiving nucleoside analogue treatment.
At baseline, all 39 patients on either entecavir (n = 20) or lamivudine (n = 19) therapy had detectable serum HBcrAg (median, 1,860 kU/ml; range, 2.4 to 899,000 kU/ml), serum HBV DNA (median, 1.9 × 107 copies/ml; range, 11,100 to 3.6 × 109 copies/ml), intrahepatic total HBV DNA (median, 42 copies/cell; range, 1.2 to 670 copies/cell), and cccDNA (median, 1.8 copies/cell; range, 0.12 to 23.3 copies/cell). There were no differences in the reduction of viral markers between patients receiving entecavir and lamivudine because of the small number of patients in each group (data not shown). The patients were pooled together in the subsequent analysis of HBcrAg in this study.
Nine (23%) and 11 (28%) patients had undetectable serum HBcrAg levels, and 22 (56%) and 31 (79%) patients had serum HBV DNA levels that were undetectable by the COBAS Amplicor HBV monitor test after 24 weeks and 48 weeks of nucleoside analogue therapy, respectively. Of the 31 patients who had undetectable serum HBV DNA after 48 weeks of therapy, 20 (65%) still had measurable levels of HBcrAg. At week 24, the median serum HBcrAg concentration was 77.5 kU/ml (range, 1.0 to 617,000 kU/ml), and there was a median logarithmic reduction of 1.23 log10 kU/ml (range, 0.13 to 3.57 log10 kU/ml) from baseline. The median logarithmic reduction of serum HBV DNA at week 24 was 4.63 log10 copies/ml (range, −3.98 to 6.02 log10 copies/ml). At week 48, the median serum HBcrAg concentration was 49.9 kU/ml (range, 1.0 to 283,000 kU/ml), and the median logarithmic reductions of HBcrAg concentration and serum HBV DNA were 1.38 log10 kU/ml (range, −1.19 to 3.57 log10 kU/ml) and 4.78 log10 copies/ml (range, −1.24 to 6.56 log10 copies/ml), respectively. The logarithmic reduction of HBcrAg concentration correlated positively with the logarithmic reduction in serum HBV DNA at week 24 (r = 0.378; P = 0.018) and week 48 (r = 0.402; P = 0.011). The median logarithmic reductions of intrahepatic total HBV DNA and cccDNA were 1.90 log10 copies/cell (range, −0.3 to 3.73 log10 copies/cell) and 1.00 log10 copies/cell (range, −0.11 to 2.34 log10 copies/cell), respectively. The logarithmic reduction in HBcrAg concentration at week 48 also correlated positively with the logarithmic reductions of intrahepatic total HBV DNA (r = 0.736; P < 0.001) and cccDNA (r = 0.378; P = 0.027).
Of the 14 HBeAg-positive patients, after 48 weeks of therapy, 1 had undergone HBeAg seroconversion, 1 lost HBeAg, and 1 had concomitant detectable HBeAg and anti-HBe. These 3 patients had undetectable serum HBV DNA and lower serum HBcrAg concentrations at week 48 (for all, <200 kU/ml) than the 11 patients without HBeAg seroconversion (for all, >200 kU/ml).
The emergence of drug-resistant mutations was assessed in all the week-48 samples by using an INNO-LiPA line-probe assay (Innogenetics, Ghent, Belgium). A reverse transcriptase M204I mutation was detected in one lamivudine-treated patient. The HBV DNA levels for this patient at baseline, week 24, and week 48 of treatment were 6.39 × 108, 1.53 × 103, and 1.10 × 1010 copies/ml, respectively. The HBcrAg concentrations at baseline, week 24, and week 48 of treatment were 49,590, 400, and 126,200 kU/ml, respectively. At week 48, this patient had an increase in serum HBV DNA of 1.2 log and an increase in serum HBcrAg concentration of 0.41 log above the baseline values.
Thirty-eight patients had paired liver tissues for the assessment of histological improvement. However, changes in the Knodell histological index were not assessed in two patients, as they had baseline Knodell histological indices of less than 2. Thirty-four patients had sufficient liver tissues for intrahepatic HBV DNA measurement. The relationships between histological improvement and the changes in serum and intrahepatic markers are shown in Table 3. Twenty-six patients had improved histology, while 10 had no or little improvement at week 48. Patients with an improvement in histology at week 48 had a significantly greater reduction in HBV cccDNA and HBcrAg levels than patients with no or little improvement (cccDNA, 1.09 versus 0.75 log10 copies/cell, respectively [P = 0.022], and HBcrAg, 1.72 versus 0.83 log10 kU/ml, respectively [P = 0.031]). Patients with improved histology tended to have greater median logarithmic reductions in intrahepatic total HBV DNA (2.07 versus 1.83 log10 copies/cell; P = 0.071) and serum HBV DNA (4.84 versus 4.42 log10 copies/ml; P = 0.094) than patients with no histological improvement.
TABLE 3.
Correlation between histological improvement and reduction in levels of intrahepatic and serum HBV DNA and serum HBcrAg during treatment
| Marker | Median log reduction at wk 48 with improvement in Knodell histological index of:
|
P value | |
|---|---|---|---|
| >2 | ≤2 | ||
| Intrahepatic total HBV DNA (copies/cell) | 2.07 | 1.83 | 0.071 |
| cccDNA (copies/cell) | 1.09 | 0.75 | 0.022 |
| HBcrAg (kU/ml) | 1.72 | 0.83 | 0.031 |
| Serum HBV DNA (copies/ml) | 4.84 | 4.42 | 0.094 |
The reduction in HBcrAg at week 48 of therapy also correlated with the degree of cytoplasmic and nuclear HBcAg staining at week 48 (Table 4). Patients with greater reductions in HBcrAg concentration at week 48 had smaller percentages of hepatocytes stained positive for cytoplasmic and nuclear HBcAg (P = 0.017 and P = 0.007, respectively).
TABLE 4.
Correlation between the degree of cytoplasmic and nuclear HBcAg staining and the logarithmic reduction in level of HBcrAg at week 48 of therapy
| Location | % of cells that stained pos at wk 48 for HBcAg | No. of patients | Median reduction in HBcrAg (log10 kU/ml) | P value |
|---|---|---|---|---|
| Cytoplasm | 0 | 26 | 1.87 | 0.017 |
| ≤5 | 8 | 1.36 | ||
| 6-25 | 3 | 0.5 | ||
| 26-50 | 1 | 0.3 | ||
| Nucleus | 0 | 27 | 1.75 | |
| ≤5 | 7 | 1.34 | 0.007 | |
| 6-25 | 4 | 0.4 |
Analyses were performed to see whether there was a pretreatment value for HBcrAg concentration, intrahepatic total HBV DNA, cccDNA, and cytoplasmic and nuclear HBcAg immunostaining which could predict PCR nondetectability of serum HBV DNA at week 48 of therapy (Table 5). Patients with a baseline serum HBcrAg concentration lower than 40,000 had a higher chance of having HBV DNA undetectable by PCR at week 48. It was also found that baseline intrahepatic total HBV DNA and cccDNA levels of <50 and <5 copies/cell, respectively, were associated with a greater chance of HBV DNA being nondetectable by PCR. Since we have shown that the HBV DNA level at week 24 of lamivudine treatment predicts the subsequent treatment response (20), different levels of HBcrAg, starting from 100 kU/ml and rising to 1,000 kU/ml at increments of 100 kU/ml, were assessed for their predictive values for undetectable HBV DNA at week 48. We found that an HBcrAg level of 200 kU/ml (Table 5) was the best cutoff level (data not shown).
TABLE 5.
Baseline and week-24 parameters which are predictive of undetectable serum HBV DNA at week 48 of therapy
| Parameter | Value | Proportion (%) of patients with undetectable serum HBV DNA | P value |
|---|---|---|---|
| Baseline HBcrAg | <40,000 vs ≥40,000 kU/ml | 29/31 (94) vs 2/8 (25) | <0.001 |
| Baseline intrahepatic total HBV DNA | <100 vs ≥100 copies/cell | 27/29 (93) vs 2/8 (25) | < 0.001 |
| Baseline cccDNA | <5 vs ≥5 copies/cell | 27/29 (93) vs 2/8 (25) | < 0.001 |
| Baseline cytoplasmic HBcAg staining | ≤25% vs >25% | 28/32 (88) vs 2/6 (33) | 0.012 |
| Baseline nuclear HBcAg staining | ≤25% vs >25% | 30/35 (86) vs 0/3 (0) | 0.007 |
| Wk-24 HBcrAg | <200 vs ≥200 kU/ml | 22/23 (96) vs 9/16 (56) | 0.004 |
DISCUSSION
The use of HBcrAg measurement for monitoring chronic hepatitis B disease has been suggested by the results of three previous studies which demonstrate that serum HBcrAg concentrations have a good correlation with HBV DNA levels in Asian patients (5, 12, 13). In the present study, we further investigated the use of HBcrAg measurement for monitoring the disease in untreated and treated patients by examining the correlation of serum HBcrAg concentrations with other disease parameters, specifically, those which reflect the disease activities within the liver.
The results of this study confirmed those of the previous studies. There was a good correlation between serum HBcrAg concentrations and HBV DNA levels, but not with ALT levels. More importantly, the present study showed that HBcrAg concentrations correlated well with intrahepatic parameters, including Knodell necroinflammation and fibrosis scores, the percentage of cells positive for cytoplasmic and nuclear HBcAg, and intrahepatic total HBV DNA and cccDNA levels. Serum HBcrAg measurement is a good quantitative serological marker for measuring chronic hepatitis B disease activity. In a recent study, HBcrAg measurement was shown to be useful in monitoring the disease in patients before and after seroconversion and for providing additional information about patients’ virological characteristics (11).
We also attempted to evaluate the use of HBcrAg measurement in patients treated with either entecavir or lamivudine. Entecavir and lamivudine are potent nucleoside analogues which inhibit HBV reverse transcriptase, reducing the production of HBV DNA-containing virions (9, 17). Since nucleotide analogues have no direct inhibiting action on the transcription and translation activities of viral mRNA, HBcAg- and HBeAg-related proteins would continue to be produced for a certain period of time in spite of the achievement of adequate suppression of the viral DNA synthesis. Moreover, it has been demonstrated that a 22-kDa truncated precore protein, which is also detected by the HBcrAg CLEIA, is found in “empty” HBV DNA-negative Dane particles (4). Therefore, the production of these particles and HBeAg is not dependent on the formation of HBV DNA but reflects the level of transcription and translation of the HBV core/precore gene in the liver. It has been postulated that the levels of HBcrAg may reflect the production of viral proteins within the hepatocytes (13). This hypothesis is indirectly supported by the observation in this study that the serum HBcrAg concentration correlated positively to the proportion of cytoplasmic and nuclear HBcAg-positive hepatocytes.
This study also showed that 1 year of nucleoside analogue therapy could successfully reduce HBcrAg concentration, as well as serum HBV DNA, intrahepatic total HBV DNA, and cccDNA levels. The extent of the reduction of HBcrAg also correlated well with the reduction of the serum and intrahepatic HBV DNA and cccDNA levels. After 48 weeks of therapy, the improvement in histology was found to correlate with greater reductions in serum HBcrAg concentrations and cccDNA levels (Table 3). The magnitude of the reduction of HBcrAg also correlated negatively with the degree of cytoplasmic and nuclear HBcAg immunostaining at week 48 (Table 4). These results suggest that the HBcrAg concentration can serve as a sensitive serum marker to reflect improvement in histology and to monitor disease activity and therapeutic efficacy in patients treated with nucleoside analogues.
In a previous study (13), five out of six patients treated with lamivudine for 6 to 8 months had their levels of HBV DNA become undetectable, while their levels of HBcrAg were still detectable. A similar result was also seen in this study. After 48 weeks of nucleoside analogue therapy, 65% of the patients who had undetectable serum HBV DNA levels (<300 copies/ml) still had measurable HBcrAg levels. In nucleoside analogue-treated patients whose HBV DNA has become undetectable by PCR assays, the use of HBcrAg measurement would be particularly useful for monitoring the decline of viral translational activities. It would also allow for the comparison of the efficacies of potent antiviral agents when a large proportion of patients have HBV DNA levels below PCR detectability.
One patient, whose HBcrAg level at week 24 was 400 kU/ml, developed lamivudine resistance at week 48. This preliminarily agrees with the results of a recent study which show that a week-24 HBcrAg level of less than 4.6 log10 U/ml (equivalent to 40 kU/ml) can be used to identify patients with a low risk of lamivudine resistance (15). A larger sample is required to validate the relationship between HBcrAg levels and risk of lamivudine resistance. We found that HBcrAg levels of <40,000 kU/ml at baseline and HBcrAg levels of <200 kU/ml at week 24 were associated with a more-than-94% chance of achieving nondetectability of HBV DNA at week 48 (Table 5). Although baseline intrahepatic total HBV DNA and cccDNA levels and the baseline proportion of cytoplasmic and nuclear HBcAg immunostaining can also be used as predictive markers for PCR nondetectability of serum HBV DNA at week 48 (Table 5), HBcrAg measurement, unlike these markers, does not require liver biopsies. The usefulness of HBcrAg measurement as a predictive marker was also demonstrated in two recent studies whose results showed that the HBcrAg concentration at the time when lamivudine treatment was stopped could be a prognostic predictor for relapse (10, 14).
Finally, this CLEIA has some practical advantages over an HBV DNA amplification assay. CLEIA is relatively simple and inexpensive compared to PCR-based or signal amplification assays. Chemiluminescence detection is performed with an automatic microplate reader, takes only 1 hour to complete, and does not require a specially trained person to perform the assay. It also has high reproducibility (5). The CLEIA used in this study is more sensitive than common enzyme immunoassays and has a lower risk of contamination and errors than nucleic acid amplification assays.
In summary, the measurement of HBcrAg is a useful additional tool for monitoring chronic hepatitis B infection. The level of serum HBcrAg may reflect the degree of intrahepatic viral translational activity. HBcrAg measurement allows continuous monitoring, especially in patients whose HBV DNA has become undetectable by PCR.
Acknowledgments
This study was supported by the Hepatitis Research Fund, The University of Hong Kong, Hong Kong.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Bredehorst, R., H. von Wulffen, and C. Granato. 1985. Quantitation of hepatitis B virus (HBV) core antigen in serum in the presence of antibodies to HBV core antigen: comparison with assays of serum HBV DNA, DNA polymerase, and HBV e antigen. J. Clin. Microbiol. 21:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, T. T., R. G. Gish, R. de Man, A. Gadano, J. Sollano, Y. C. Chao, A. S. Lok, K. H. Han, Z. Goodman, J. Zhu, A. Cross, D. DeHertogh, R. Wilber, R. Colonno, and D. Apelian. 2006. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 354:1001-1010. [DOI] [PubMed] [Google Scholar]
- 3.Gowans, E. J. 1986. Relationship between HBeAg and HBV DNA in patients with acute and persistent hepatitis B infection. Med. J. Aust. 145:439-441. [DOI] [PubMed] [Google Scholar]
- 4.Kimura, T., N. Ohno, N. Terada, A. Rokuhara, A. Matsumoto, S. Yagi, E. Tanaka, K. Kiyosawa, S. Ohno, and N. Maki. 2005. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280:21713-21719. [DOI] [PubMed] [Google Scholar]
- 5.Kimura, T., A. Rokuhara, Y. Sakamoto, S. Yagi, E. Tanaka, K. Kiyosawa, and N. Maki. 2002. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knodell, R. G., K. G. Ishak, W. C. Black, T. S. Chen, R. Craig, N. Kaplowitz, T. W. Kiernan, and J. Wollman. 1981. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1:431-435. [DOI] [PubMed] [Google Scholar]
- 7.Lai, C. L., V. Ratziu, M. F. Yuen, and T. Poynard. 2003. Viral hepatitis B. Lancet 362:2089-2094. [DOI] [PubMed] [Google Scholar]
- 8.Lai, C. L., D. Shouval, A. S. Lok, T. T. Chang, H. Cheinquer, Z. Goodman, D. DeHertogh, R. Wilber, R. C. Zink, A. Cross, R. Colonno, and L. Fernandes. 2006. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 354:1011-1020. [DOI] [PubMed] [Google Scholar]
- 9.Marion, P. L., F. H. Salazar, M. A. Winters, and R. J. Colonno. 2002. Potent efficacy of entecavir (BMS-200475) in a duck model of hepatitis B virus replication. Antimicrob. Agents Chemother. 46:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto, A., E. Tanaka, M. Minami, T. Okanoue, H. Yatsuhashi, S. Nagaoka, F. Suzuki, M. Kobayashi, K. Chayama, M. Imamura, H. Yotsuyanagi, S. Nakaoka, N. Maki, S. Kawata, H. Kumada, S. Iino, and K. Kiyosawa. 2007. Low serum level of hepatitis B core-related antigen indicates unlikely reactivation of hepatitis after cessation of lamivudine therapy. Hepatol. Res. 37:661-666. [DOI] [PubMed] [Google Scholar]
- 11.Misawa, N., A. Matsumoto, E. Tanaka, A. Rokuhara, K. Yoshizawa, T. Umemura, N. Maki, T. Kimura, and K. Kiyosawa. 2006. Patients with and without loss of hepatitis B virus DNA after hepatitis B e antigen seroconversion have different virological characteristics. J. Med. Virol. 78:68-73. [DOI] [PubMed] [Google Scholar]
- 12.Rokuhara, A., X. Sun, E. Tanaka, T. Kimura, A. Matsumoto, D. Yao, L. Yin, N. Wang, N. Maki, and K. Kiyosawa. 2005. Hepatitis B virus core and core-related antigen quantitation in Chinese patients with chronic genotype B and C hepatitis B virus infection. J. Gastroenterol. Hepatol. 20:1726-1730. [DOI] [PubMed] [Google Scholar]
- 13.Rokuhara, A., E. Tanaka, A. Matsumoto, T. Kimura, T. Yamaura, K. Orii, X. Sun, S. Yagi, N. Maki, and K. Kiyosawa. 2003. Clinical evaluation of a new enzyme immunoassay for hepatitis B virus core-related antigen; a marker distinct from viral DNA for monitoring lamivudine treatment. J. Viral Hepat. 10:324-330. [DOI] [PubMed] [Google Scholar]
- 14.Shinkai, N., Y. Tanaka, E. Orito, K. Ito, T. Ohno, N. Hirashima, I. Hasegawa, F. Sugauchi, R. Ueda, and M. Mizokami. 2006. Measurement of hepatitis B virus core-related antigen as predicting factor for relapse after cessation of lamivudine therapy for chronic hepatitis B virus infection. Hepatol. Res. 36:272-276. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka, E., A. Matsumoto, F. Suzuki, M. Kobayashi, M. Mizokami, Y. Tanaka, T. Okanoue, M. Minami, K. Chayama, M. Imamura, H. Yatsuhashi, S. Nagaoka, H. Yotsuyanagi, S. Kawata, T. Kimura, N. Maki, S. Iino, and K. Kiyosawa. 2006. Measurement of hepatitis B virus core-related antigen is valuable for identifying patients who are at low risk of lamivudine resistance. Liver Int. 26:90-96. [DOI] [PubMed] [Google Scholar]
- 16.Usuda, S., H. Okamoto, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1998. An enzyme-linked immunosorbent assay with monoclonal antibodies for the determination of phosphorylated hepatitis B core protein (p21c) in serum. J. Virol. Methods 72:95-103. [PubMed] [Google Scholar]
- 17.Wolters, L. M., B. E. Hansen, H. G. Niesters, S. Zeuzem, S. W. Schalm, and R. A. De Man. 2002. Viral dynamics in chronic hepatitis B patients during lamivudine therapy. Liver 22:121-126. [DOI] [PubMed] [Google Scholar]
- 18.Wong, D. K. H., M. F. Yuen, H. Yuan, S. S. Sum, C. K. Hui, J. Hall, and C. L. Lai. 2004. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology 40:727-737. [DOI] [PubMed] [Google Scholar]
- 19.Yuan, H. J., M. F. Yuen, D. K. H. Wong, S. M. Sum, E. Sablon, I. O. L. Ng, and C. L. Lai. 2005. Impact of precore and core promoter mutations on hepatic histology in patients with chronic hepatitis B. Aliment. Pharmacol. Ther. 22:301-307. [DOI] [PubMed] [Google Scholar]
- 20.Yuen, M. F., E. Sablon, C. K. Hui, H. J. Yuan, H. Decraemer, and C. L. Lai. 2001. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology 34:785-791. [DOI] [PubMed] [Google Scholar]