Abstract
Sensitive and accurate quantification of hepatitis B virus (HBV) DNA is necessary for monitoring patients with chronic hepatitis receiving antiviral therapy in order to determine treatment response and to adapt therapy in case of inadequate virologic control. The development of quantitative PCR assays has been crucial in meeting these needs. The objective of this study was to compare the performance of a new real-time PCR assay (Abbott RealTime) for HBV DNA with that of three other commercial assays for the detection of HBV DNA. These were the Versant 3.0 branched-chain DNA assay, the Cobas Amplicor HBV Monitor test, and the Cobas AmpliPrep-Cobas TaqMan hepatitis B virus assay (CAP-CTM). HBV DNA was measured in blood samples taken from two cohorts of patients with chronic hepatitis. HBV DNA levels measured with the Abbott RealTime assay were highly correlated with those measured with the other three tests over their respective dynamic ranges (r, 0.88 to 0.96). The sensitivity (detection limit, 10 IU/ml) and dynamic range of the Abbott RealTime assay (101 to 109 IU/ml) was superior to that of the Versant assay. The RealTime assay recognized both HBV strains belonging to genotypes A to G and those bearing polymerase gene mutations equivalently. In conclusion, this study demonstrates the utility of the Abbott RealTime assay for monitoring HBV DNA levels in patients with chronic hepatitis B. Its sensitivity and wide dynamic range should allow optimal monitoring of antiviral therapy and timely treatment adaptation.
Chronic hepatitis B virus (HBV) infections represent an important public health issue, being a major risk factor for hepatocellular carcinoma and leading to potentially fatal liver damage (19). Globally, these infections are the 10th most common cause of death (13). Prevalence rates vary across the world, with the highest rates being observed in eastern Asia, where perinatal vertical transmission of infection from mother to child represents an important mechanism for sustaining high rates of infection (19). North America and northwestern Europe are areas with low prevalence rates for chronic hepatitis. In the United States, an overall prevalence rate of 5.6% was reported in the NHANES study (23), and in France, the prevalence of chronic HBV infections has been recently estimated at 0.65% (24).
Hepatitis B is caused by infection of the liver by HBV, a small circular, partially double-stranded DNA virus of approximately 3,200 base pairs (6), followed by an immune reaction mounted by the host to eliminate infected hepatocytes. During the viral replication cycle, supercoiled covalently closed circular viral DNA (cccDNA) is generated, and this DNA persists in host cells as a viral minichromosome. This cccDNA can be a source of renewed virus production once the immune response to the acute infection is over and thus constitutes a reservoir of infectious viral particles, thereby leading to the development of chronic hepatitis in certain individuals (37). The probability of developing chronic disease declines with age (19).
Systematic vaccination programs have been recommended by the World Health Organization since 1991 and have been successful in reducing de novo HBV infections in many countries (13). Recent studies have demonstrated a direct link between HBV viral load and the risk of developing disease complications, such as cirrhosis and hepatocellular carcinoma (2). In infected individuals, antiviral drugs, such as lamivudine (9), adefovir (3, 30), and entecavir (22), are effective in reducing viral load, normalizing liver function, and consequently reducing the incidence of long-term complications (16). Nonetheless, these treatments do not generally allow complete eradication of HBV from the organism, and continuous long-term therapy is required to maintain effective viral suppression and symptom control (32, 35, 37). In addition, the long-term effectiveness of these drugs is compromised by the emergence of antiviral resistance which allows viral breakthrough and recovery of disease activity (14, 36). Several studies have shown that the evaluation of viral load levels after 24 to 48 weeks of therapy can predict the subsequent occurrence of antiviral drug resistance (5, 7, 34). Furthermore, the current standard of care is to adapt antiviral therapy in patients with drug resistance as early as possible, namely, at the time of viral breakthrough, defined by an increase in viremia levels by 1 log10 unit of copies/ml compared to the nadir value (11), or even before viral breakthrough if the viral load is not adequately suppressed.
For these reasons, current consensus guidelines recommend that viral load be measured at the first consultation of a chronic HBV carrier to determine the necessity of treatment and then regularly in patients treated with antiviral drugs in order to monitor response to therapy (4, 15, 18, 20). This allows the initial antiviral response to therapy to be defined and primary treatment failure and virologic breakthrough to be detected (10). Measurement of viral load is most simply accomplished by detection of HBV DNA in serum or plasma using nucleic acid amplification or signal amplification technologies (25). A number of assay systems have been developed for quantification of HBV DNA which differ in their sensitivity, dynamic range, and specificity towards genomic variants of the virus (26, 38).
The Abbott RealTime HBV test is an in vitro real-time PCR assay for the quantification of HBV DNA in human plasma or serum from HBV-infected individuals. The target sequence in the HBV DNA consists of a highly conserved region in the S gene, which is essential for the assembly and secretion of subviral particles, and tolerates only minor structural changes (27). The choice of this target region means that all HBV genotypes (genotypes A to H) can be adequately detected and that assay sensitivity is not compromised by YMDD mutations conferring antiviral drug resistance, HBsAg escape mutations, or the HBe phenotype.
The objective of this study was to compare the performance of the Abbott RealTime HBV assay with that of three other commercial assays for the detection of HBV DNA in clinical samples obtained from infected patients. These three assays were the Versant HBV DNA 3.0 assay (Bayer Healthcare), a branched-chain DNA (bDNA) signal amplification assay (33), the Cobas Amplicor HBV Monitor test (CAM; Roche Molecular Diagnostics), an end point PCR assay, and the Cobas AmpliPrep-Cobas TaqMan hepatitis B virus assay (CAP-CTM; Roche Molecular Diagnostics), a real-time PCR-based assay (8, 21).
MATERIALS AND METHODS
Study samples.
Samples of HBV DNA were acquired from two patient cohorts monitored in two HBV reference centers in France. The first set of 93 serum samples was obtained from a group of 68 patients (cohort A) infected with HBeAg-negative HBV, treated with lamivudine, and monitored as part of a resistance surveillance program (39). All these serum samples were prospectively quantified using the Versant HBV bDNA 3.0 assay (Bayer, Eragny, France) according to the supplier's instructions. The genotypic characteristics of the HBV strains isolated from these samples are provided in Table 1.
TABLE 1.
Genotypic characterization of HBeAg-negative HBV strains isolated from 93 samples obtained from cohort A
| HBV strain and relevant genotype | No. of samples with genotype |
|---|---|
| Surface gene variants | |
| A | 31 |
| A/G | 2 |
| B | 9 |
| C | 7 |
| D | 35 |
| E | 6 |
| F | 2 |
| G | 1 |
| Polymerase gene variants | |
| Wild-type | 34 |
| L180M | 1 |
| M204I | 14 |
| M204V | 1 |
| L180M + M204I | 3 |
| L180M + M204V | 39 |
| L180M + M204I + M204V | 1 |
Cohort B consisted of 106 plasma samples from 104 patients addressed to La Pitié Hospital Virology laboratory for measurement of HBV viral load. All these samples were prospectively quantified using the CAP-CTM (Roche, Meylan, France) according to the supplier's instructions. Among those, 79 samples were also tested in parallel with CAM (Roche, Meylan, France) as recommended by the manufacturer. Residual plasma samples were stored at −80°C until they were shipped to Abbott central facilities for RealTime HBV testing. Samples were dispatched with a code so that the HBV DNA levels determined with the Roche systems were unknown to the evaluator.
Sample preparation.
The purpose of sample preparation is to extract and concentrate target DNA, to make the target accessible for amplification, and to remove potential inhibitors of amplification from the sample. This process is accomplished by the Abbott m Sample Preparation SystemDNA (4 × 24 Preps) (m2000sp), an automated sample preparation system designed to use magnetic microparticle processes for the purification of nucleic acids from samples. A sample volume of 500 μl is processed in the system for HBV DNA quantification. The m2000sp reagents lyse the virion, capture the nucleic acids, and wash the particles to remove unbound sample components. Proteinase K is included in the lysis step to digest proteins associated with the nucleic acids (12, 13). The bound nucleic acids are eluted and transferred to a 96-deep well plate. The nucleic acids are then ready for amplification.
Internal control.
A DNA sequence that is unrelated to the HBV target sequence is introduced into each specimen at the beginning of sample preparation. This unrelated DNA sequence is simultaneously amplified by PCR and serves as an internal control to demonstrate that the process has proceeded correctly for each sample. The target sequence of the internal control is derived from the hydroxypyruvate reductase gene from the pumpkin plant Cucurbita pepo and is provided as a DNA plasmid in a buffer solution.
Reagent preparation and reaction plate assembly.
The Abbott m2000sp combines the Abbott RealTime HBV amplification reagent components (HBV oligonucleotide reagent, Amplitaq Gold enzyme, and activation reagent). The Abbott m2000sp dispenses the resulting master mix to the Abbott 96-well optical reaction plate along with aliquots of the nucleic acid samples prepared by the Abbott m2000sp. The plate is ready, after manual application of the Abbott optical adhesive cover, for transfer to the Abbott m2000rt.
Amplification.
During the amplification/detection reaction on the Abbott m2000rt instrument, the target DNA is amplified by Amplitaq Gold enzyme in the presence of deoxynucleotide triphosphates and magnesium. In the initial step, forward and reverse primers, designed to hybridize to the target sequence in the S gene of the HBV genome, anneal to the target sequence and are extended by the polymerase. After a denaturation step in which the temperature of the reaction is raised above the melting point of the double-stranded DNA product, the newly created DNA strand is denatured from the target DNA.
During each round of thermal cycling, amplification products dissociate to single strands at a high temperature, allowing primer annealing and extension as the temperature is lowered. Exponential amplification of the product is achieved through repeated cycling between high and lower temperatures, resulting in a billionfold or greater amplification of target sequences. Amplification of both targets (HBV sequence and the internal control) takes place simultaneously in the same reaction.
Detection.
The amount of HBV target sequence that is present at each amplification cycle is quantified by measuring the fluorescence of the HBV probe that binds to the target during the extension/annealing step. This probe is a single-stranded DNA oligonucleotide consisting of a probe sequence with a fluorescent moiety covalently linked to its 5′ end and a quenching moiety covalently linked to its 3′ end. In the absence of the target sequence, probe fluorescence is quenched. In contrast, in the presence of target sequence, the probe specifically binds to the target. During the extension/annealing step, the exonuclease activity of the DNA polymerase cleaves the bound probe as it moves along the template strand. This separates the fluorophore from the quencher, allowing fluorescence emission and detection.
An analogous probe is used to detect the presence of amplification products of the internal control. Each probe is labeled with a different fluorophore, thus allowing simultaneous detection of both amplified products at each cycle. The amplification cycle at which a fluorescent signal is detected by the Abbott m2000rt is proportional to the log of the HBV DNA concentration present in the original sample.
The Abbott RealTime HBV PCR assay is standardized against the World Health Organization International Standard for Hepatitis B Virus DNA (NIBSC Code 97/746) and has recently been approved in the European Union (29). Results are reported in international units per milliliter or copies/ml.
Other quantification assays.
The three other assays, Versant, CAM, and CAP-CTM, were performed using standard published protocols on 50 μl, 100 μl, and 850 μl of sample, respectively, as recommended by each manufacturer (8, 21, 33). These three assays are also approved for use for viral load monitoring in the European Union.
RESULTS
Dynamic range.
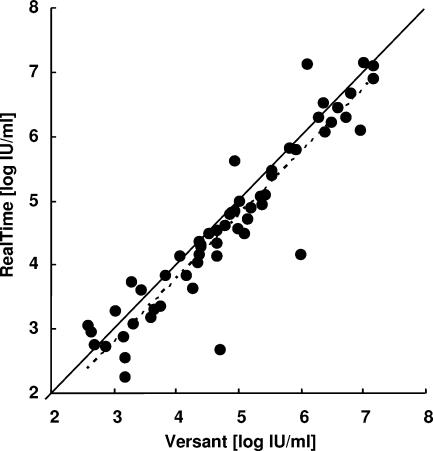
The dynamic range of the Abbott RealTime assay was compared with that of the Versant bDNA assay. Using the latter assay, HBV DNA could be quantified in 59 of the 93 samples obtained from cohort A (63.4%). Four samples were below the limit of detection of this assay (<357 IU/ml), and 30 samples were above the limit of quantification (7.25 log of IU/ml). Using the Abbott RealTime assay, HBV DNA was quantifiable in 92 samples (98.9%), including 3 of the 4 samples in which viral DNA could not be detected with the Versant assay (Table 2). HBV DNA concentrations in these three samples were found to be 0.44, 1.28, and 2.20 log IU/ml. In the 59 samples in which viral DNA could be quantified by both assays, the correlation between the two measures was high (r = 0.9298; Fig. 1).
TABLE 2.
Comparison of the dynamic ranges of three HBV DNA quantification assaysa
| Test and result | No. of samples with the following result by Abbott RealTime assay:
|
||
|---|---|---|---|
| Below detection threshold | Quantifiable | Above quantification limit | |
| Versant assay (n = 93) | |||
| Below detection threshold | 1 | 3 | 0 |
| Quantifiable | 0 | 59 | 0 |
| Above quantification limit | 0 | 30 | 0 |
| Amplicor assay (n = 79) | |||
| Below detection threshold | 0 | 0 | 0 |
| Quantifiable | 0 | 66 | 0 |
| Above quantification limit | 0 | 13 | 0 |
| TaqMan assay (n = 106) | |||
| Below detection threshold | 0 | 2 | 0 |
| Quantifiable | 0 | 101 | 0 |
| Above quantification limit | 0 | 3 | 0 |
The dynamic range of the Abbott RealTime assay was compared with those of the Versant, Amplicor, and TaqMan assays for detecting HBV DNA in 93 samples isolated from cohort A (Versant) and 106 samples isolated from cohort B (TaqMan). The cohort tested with Amplicor is a subgroup of 79 samples from cohort B.
FIG. 1.
Correlation between HBV DNA determinations within dynamic ranges using the Abbott RealTime and Versant assays for samples from 59 HBV-infected, HBeAg-negative patients for which a quantitative result was obtained with both assays. The identity line (solid line) as well as the best-fit correlation (broken line) is shown (y = 0.9863x − 0.154, R2 = 0.8646).
Genotype specificity.
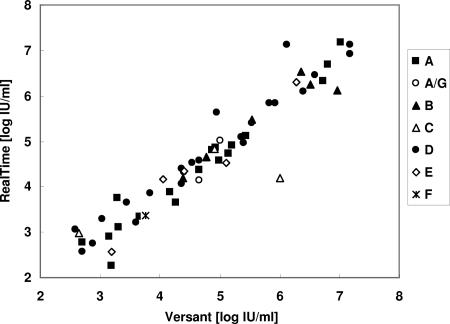
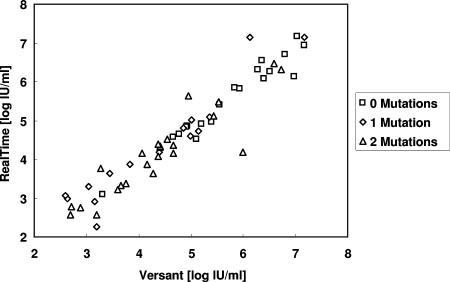
The ability of the Abbott RealTime assay to detect accurately HBV DNA irrespective of viral genotype was also investigated in comparison with the Versant bDNA assay. Concerning the S-gene variation across viral genotypes A to G, no pattern of preferential detection emerged (Fig. 2). Similarly, over the dynamic range of the tests, no differences were observed in the capacity of the two assays to detect HBV DNA from viral strains bearing multiple polymerase gene mutations known to confer antiviral drug resistance (Fig. 3). For the total population of viral isolates from cohort A, a systematic bias could be attributed to higher values with the Versant assay. The bias between the two methods was estimated at −0.22 log IU/ml (95% confidence limits, −0.35 to −0.09).
FIG. 2.
Correlation between HBV DNA determinations within dynamic ranges using the RealTime and Versant assays for samples from 59 HBV-infected, HBeAg-negative patients according to surface gene variants (genotypes are indicated).
FIG. 3.
Correlation between HBV DNA determinations within dynamic ranges using the RealTime and Versant assays for samples from 59 HBV-infected, HBeAg-negative patients according to the number of polymerase gene mutations.
Comparison with other PCR methods.
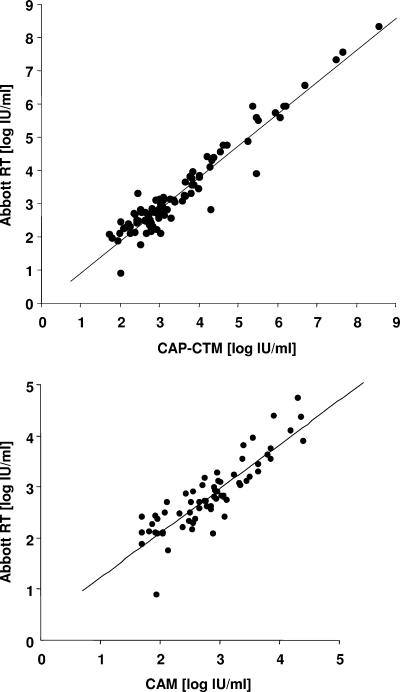
The performance of the Abbott RealTime assay was compared with that of two other quantitative PCR methods, CAM and CAP-CTM. In both cases, equivalence between the assays was observed over the entire dynamic range (Fig. 4). The correlation coefficients determined from regression analysis were 0.8757 for the Abbott RealTime assay versus the CAM assay on 66 samples within the dynamic range of the CAM assay (38 to 38,000 IU/ml) and 0.9566 for the Abbott RealTime assay versus the CAP-CTM on 101 samples within the dynamic range of the CAP-CTM assay (54 to 1.1 × 108 IU/ml). The bias between the techniques was −0.02 (95% confidence interval, −0.105/0.064) and −0.197 (95% confidence interval, −0.268/−0.126) for CAM and CAP-CTM, respectively. When comparing the two real-time PCR based methods, six samples had discordant results falling outside the 95% confidence interval (samples 81, 54, 3, 50, 70, and 35). Retesting of the samples with the Abbott assay confirmed the differences observed on the first run for all but one sample (sample 54). Two samples (samples 81 and 50) had a low viral load below 300 IU/ml, and one explanation for the discrepancy could be the relative imprecision of the techniques at this level. For the remaining three samples, the DNA was sequenced in order to genotype each sample. One (sample 50) could not be sequenced due to a low viral load, and the last two were genotype E viruses.
FIG. 4.
Correlation between HBV DNA determinations using the RealTime and other quantitative PCR assays in HBV DNA samples isolated from patients in cohort B for which a quantitative result was obtained with both assays. (Top) CAM assay (n = 66). (Bottom) CAP-CTM assay (n = 101). The best-fit correlation (solid line) is shown (for the CAM assay, y = 0.8653x + 0.3629 and R2 = 0.7668; for the CAP-CTM assay, y = 0.9586x − 0.0543 and R2 = 0.9276).
DISCUSSION
The objective of this study was to compare four different methods for detection of HBV DNA in serum or plasma samples obtained from individuals with chronic HBV infections and, in particular, to characterize the sensitivity and genomic specificity of the Abbott RealTime PCR assay.
Compared with the Versant 3.0 bDNA assay, the dynamic range and the sensitivity of the Abbott RealTime assay were superior, allowing detection of HBV DNA in 99% of the samples compared with only 63% using the reference method. The Abbott RealTime assay can detect and quantify viral DNA over a range from 10 IU/ml (lower limit of detection) to 109 IU/ml (upper limit of quantification).
The performance of the Abbott RealTime assay was comparable to that of two other quantitative PCR assays, the Cobas Amplicor HBV Monitor test and real-time PCR assay (CAP-CTM). Compared with the former, the Abbott RealTime assay offers the advantage of a much wider dynamic range (38 to 38,000 IU/ml for the Amplicor assay) and improved probe selectivity. The probe of the Cobas Amplicor HBV Monitor test targets a sequence in the viral DNA core region, and the sensitivity of the assay may be compromised with respect to viral strains bearing mutations in this region (17) and genotype F variant strains (12, 31). In contrast, the Abbott RealTime assay probe hybridizes within a highly conserved region of the S gene, allowing accurate detection of all viral strains evaluated. However, it should be noted that in the present comparison, only two samples bearing an F genotype strain were evaluated.
Compared to the CAM assay, an end point PCR method, the technology based on real-time PCR amplification offers many advantages, in particular a wider range of quantification. Indeed, compared to conventional PCR assays, this extended quantification may in some cases eliminate the necessity of performing laborious and potentially inexact dilutions in order to determine an accurate viral load at treatment initiation. The Abbott RealTime assay also has a slightly broader dynamic range than the CAP-CTM, allowing better quantification at the upper end of the range (up to 9 log IU/ml) that should reduce the need to dilute samples from individuals with high replicating HBV. At the lower end, the extended dynamic range of the Abbott RealTime assay reaches the sensitivity limit of the assay of 10 IU/ml, allowing low viral DNA titers to be measured accurately down to this threshold. In contrast, the dynamic range of CAP-CTM (lower limit, 54.5 IU/ml) does not extend down to the sensitivity limit of the assay (12 IU/ml), precluding quantification of DNA in this interval and leaving a gray zone in which the virus is detectable but not quantifiable. Interestingly, all samples from cohort B that had been quantified with CAP-CTM were also quantified using the Abbott RealTime assay. Consistent with previous comparative evaluations of CAP-CTM, this study confirmed that real-time PCR-based assays have an improved sensitivity compared to the Versant bDNA method that should translate into an improved management of chronic HBV carriers (8, 28). Although we did not evaluate the genotypic specificity of the CAP-CTM in our study specifically, this assay has previously been shown to quantify different HBV variants with equivalent precision and specificity (8).
Interestingly, the best correlation between all these assays was obtained with the two real-time PCR-based assays (r = 0.96). However, it should be pointed out that there was also very little discrepancy with the two other assays. Analysis of the few samples for which measures of viral load were discrepant between assays did not reveal any particular viral feature which may be associated with the mismatch. Further studies will be needed to determine whether certain viral strains might be incorrectly quantified by a given technique.
As has been recently demonstrated for the CAP-CTM assay (1), the Abbott RealTime HBV assay meets all requirements for use in monitoring viral response to antiviral treatment. Our study also provides good evidence that it performs similarly on serum or plasma samples. Its high sensitivity (10 IU/ml) will be adequate for the detection of viral breakthrough at an early stage and will allow the rapid addition of a salvage therapy before clinical breakthrough. Early introduction of a rescue therapy in patients with viral breakthrough has been demonstrated to provide important benefits in terms of long-term clinical outcome (11). The test allows detection of HBV DNA at levels well below the levels considered necessary for the control of disease progression (4) or associated with an elevated risk of hepatocellular carcinoma (2). Furthermore, since the latest concept of treatment adaptation is to adjust therapy during the first year when viral load is not suppressed below 3 log10 copies/ml (10), this assay is also well suited to this new treatment algorithm. Coupled with genomic sequencing, this assay would also allow the rapid detection of strains bearing resistance mutations. The low bias (−0.197 log IU/ml) and the good correlation observed between the two real-time PCR techniques are important to consider when serial results for the same patient are obtained with different methods. Although it is recommended not to change techniques between samples, one can assume that the use of different real-time PCR assays is likely to generate only minor quantification changes that may be clinically irrelevant if results are reported in IU/ml.
The choice of one assay over another in a routine laboratory should certainly consider all the aforementioned points, but other parameters, such as the number of samples to be processed, the need for other virus viral load measurements, the volume of sample to process, and the cost per assay, will also be crucial for this decision. Our study was not designed to assess all these conditions, and further work should be conducted to evaluate the practical use of these assays in real practice.
In conclusion, this study demonstrates the utility of the Abbott RealTime test for monitoring HBV DNA levels in serum or plasma samples from patients with chronic HBV infections. Its sensitivity and wide detection range should allow optimal monitoring of antiviral therapy and timely treatment adaptation.
Acknowledgments
This work was supported in part by grants from the European Community to F.Z. (ViRgil network of excellence; ViRgil LSHM-CT-2004-503359) and by a grant from the National Agency for Research against AIDS and Hepatitis (ANRS) to V.T. and F.Z.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Allice, T., F. Cerutti, F. Pittaluga, S. Varetto, S. Gabella, A. Marzano, A. Franchello, G. Colucci, and V. Ghisetti. 2007. COBAS AmpliPrep-COBAS TaqMan hepatitis B virus (HBV) test: a novel automated real-time PCR assay for quantification of HBV DNA in plasma. J. Clin. Microbiol. 45:828-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C. J., H. I. Yang, J. Su, C. L. Jen, S. L. You, S. N. Lu, G. T. Huang, and U. H. Iloeje. 2006. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295:65-73. [DOI] [PubMed] [Google Scholar]
- 3.Dando, T., and G. Plosker. 2003. Adefovir dipivoxil: a review of its use in chronic hepatitis B. Drugs 63:2215-2234. [DOI] [PubMed] [Google Scholar]
- 4.de Franchis, R., A. Hadengue, G. Lau, D. Lavanchy, A. Lok, N. McIntyre, A. Mele, G. Paumgartner, A. Pietrangelo, J. Rodes, W. Rosenberg, D. Valla, and EASL Jury. 2003. EASL International Consensus Conference on Hepatitis B. 13 to 14 September, 2002 Geneva, Switzerland. Consensus statement (long version). J. Hepatol. 39(Suppl. 1):S3-S25. [PubMed] [Google Scholar]
- 5.Di Bisceglie, A., C. Lai, E. Gane, Y.-C. Chen, S. Thongsawat, Y. Wang, et al. 2006. Telbivudine Globe Trial: maximal early HBV suppression is predictive of optimal two-year efficacy in nucleoside-treated hepatitis B patients. Hepatology 44:230A-231A. [Google Scholar]
- 6.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 7.Hadziyannis, S. J., N. C. Tassopoulos, E. J. Heathcote, T. T. Chang, G. Kitis, M. Rizzetto, P. Marcellin, S. G. Lim, Z. Goodman, J. Ma, C. L. Brosgart, K. Borroto-Esoda, S. Arterburn, and S. L. Chuck. 2006. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology 131:1743-1751. [DOI] [PubMed] [Google Scholar]
- 8.Hochberger, S., D. Althof, R. Gallegos de Schrott, N. Nachbaur, H. Rock, and H. Leying. 2006. Fully automated quantitation of hepatitis B virus (HBV) DNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 35:373-380. [DOI] [PubMed] [Google Scholar]
- 9.Jarvis, B., and D. Faulds. 1999. Lamivudine. A review of its therapeutic potential in chronic hepatitis B. Drugs 58:101-141. [DOI] [PubMed] [Google Scholar]
- 10.Keeffe, E., S. Zeuzem, R. S. Koff, D. T. Dieterich, R. Esteban-Mur, E. J. Gane, I. M. Jacobson, S. G. Lim, N. Naoumov, P. Marcellin, T. Piratvisuth, and F. Zoulim. 2007. Report of an international workshop: roadmap for management of patients receiving oral therapy for chronic hepatitis B. Clin. Gastroenterol. Hepatol. 5:890-897. [DOI] [PubMed] [Google Scholar]
- 11.Lampertico, P., M. Vigano, E. Manenti, M. Iavarone, G. Lunghi, and M. Colombo. 2005. Adefovir rapidly suppresses hepatitis B in HBeAg-negative patients developing genotypic resistance to lamivudine. Hepatology 42:1414-1419. [DOI] [PubMed] [Google Scholar]
- 12.Laperche, S., V. Thibault, F. Bouchardeau, S. Alain, S. Castelain, M. Gassin, M. Gueudin, P. Halfon, S. Larrat, F. Lunel, M. Martinot-Peignoux, B. Mercier, J. M. Pawlotsky, B. Pozzetto, A. M. Roque-Afonso, F. Roudot-Thoraval, K. Saune, and J. J. Lefrere. 2006. Expertise of laboratories in viral load quantification, genotyping, and precore mutant determination for hepatitis B virus in a multicenter study. J. Clin. Microbiol. 44:3600-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavanchy, D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 11:97-107. [DOI] [PubMed] [Google Scholar]
- 14.Leung, N. 2003. Viral breakthrough during lamivudine therapy for chronic hepatitis B. Intervirology 46:344-349. [DOI] [PubMed] [Google Scholar]
- 15.Liaw, Y. F., N. Leung, R. Guan, G. K. Lau, I. Merican, G. McCaughan, E. Gane, J. H. Kao, and M. Omata. 2005. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2005 update. Liver Int. 25:472-489. [DOI] [PubMed] [Google Scholar]
- 16.Liaw, Y. F., J. J. Sung, W. C. Chow, G. Farrell, C. Z. Lee, H. Yuen, T. Tanwandee, Q. M. Tao, K. Shue, O. N. Keene, J. S. Dixon, D. F. Gray, and J. Sabbat. 2004. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351:1521-1531. [DOI] [PubMed] [Google Scholar]
- 17.Lindh, M., C. Hannoun, S. Malmstrom, J. Lindberg, and G. Norkrans. 2006. Lamivudine resistance of hepatitis B virus masked by coemergence of mutations in probe region of the COBAS AMPLICOR assay. J. Clin. Microbiol. 44:2587-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lok, A. S., and B. J. McMahon. 2007. Chronic hepatitis B. Hepatology 45:507-539. [DOI] [PubMed] [Google Scholar]
- 19.Lok, A. S., and B. J. McMahon. 2001. Chronic hepatitis B. Hepatology 34:1225-1241. [DOI] [PubMed] [Google Scholar]
- 20.Lok, A. S., and B. J. McMahon. 2004. Chronic hepatitis B: update of recommendations. Hepatology 39:857-861. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, V. A., E. J. Bourne, M. W. Lutz, and L. D. Condreay. 2002. Assessment of the COBAS Amplicor HBV Monitor test for quantitation of serum hepatitis B virus DNA levels. J. Clin. Microbiol. 40:1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews, S. J. 2006. Entecavir for the treatment of chronic hepatitis B virus infection. Clin. Ther. 28:184-203. [DOI] [PubMed] [Google Scholar]
- 23.McQuillan, G. M., D. Kruszon-Moran, B. J. Kottiri, L. R. Curtin, J. W. Lucas, and R. S. Kington. 2004. Racial and ethnic differences in the seroprevalence of 6 infectious diseases in the United States: data from NHANES III, 1988-1994. Am. J. Public Health 94:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meffre, C., and Institut de Veille Sanitaire. 19 December 2006, posting date. Prévalence des hépatites B et C en France en 2004. Institut de Veille Sanitaire, Saint-Maurice, France. http://www.invs.sante.fr/publications/2006/prevalence_b_c/vhb_france_2004.pdf.
- 25.Mommeja-Marin, H., E. Mondou, M. R. Blum, and F. Rousseau. 2003. Serum HBV DNA as a marker of efficacy during therapy for chronic HBV infection: analysis and review of the literature. Hepatology 37:1309-1319. [DOI] [PubMed] [Google Scholar]
- 26.Pawlotsky, J. M., A. Bastie, C. Hezode, I. Lonjon, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J. Virol. Methods 85:11-21. [DOI] [PubMed] [Google Scholar]
- 27.Prange, R., R. Nagel, and R. E. Streeck. 1992. Deletions in the hepatitis B virus small envelope protein: effect on assembly and secretion of surface antigen particles. J. Virol. 66:5832-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronsin, C., A. Pillet, C. Bali, and G. A. Denoyel. 2006. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J. Clin. Microbiol. 44:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saldanha, J., W. Gerlich, N. Lelie, P. Dawson, K. Heermann, and A. Heath. 2001. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 80:63-71. [DOI] [PubMed] [Google Scholar]
- 30.Thibault, V. 2006. Where does adefovir stand amongst newly developed antivirals: from pharmacology to virology. Future Virol. 1:553-565. [Google Scholar]
- 31.Weiss, J., H. Wu, B. Farrenkopf, T. Schultz, G. Song, S. Shah, and J. Siegel. 2004. Real time TaqMan PCR detection and quantitation of HBV genotypes A-G with the use of an internal quantitation standard. J. Clin. Virol. 30:86-93. [DOI] [PubMed] [Google Scholar]
- 32.Werle-Lapostolle, B., S. Bowden, S. Locarnini, K. Wursthorn, J. Petersen, G. Lau, C. Trepo, P. Marcellin, Z. Goodman, W. E. Delaney IV, S. Xiong, C. L. Brosgart, S. S. Chen, C. S. Gibbs, and F. Zoulim. 2004. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 126:1750-1758. [DOI] [PubMed] [Google Scholar]
- 33.Yao, J. D., M. G. Beld, L. L. Oon, C. H. Sherlock, J. Germer, S. Menting, S. Y. Se Thoe, L. Merrick, R. Ziermann, J. Surtihadi, and H. J. Hnatyszyn. 2004. Multicenter evaluation of the VERSANT hepatitis B virus DNA 3.0 assay. J. Clin. Microbiol. 42:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuen, M. F., E. Sablon, C. K. Hui, H. J. Yuan, H. Decraemer, and C. L. Lai. 2001. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology 34:785-791. [DOI] [PubMed] [Google Scholar]
- 35.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoulim, F. 2003. Hepatitis B virus resistance to antivirals: clinical implications and management. J. Hepatol. 39:(Suppl. 1)S133-S138. [DOI] [PubMed] [Google Scholar]
- 37.Zoulim, F. 2005. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J. Hepatol. 42:302-308. [DOI] [PubMed] [Google Scholar]
- 38.Zoulim, F. 2006. New nucleic acid diagnostic tests in viral hepatitis. Semin. Liver Dis. 26:309-317. [DOI] [PubMed] [Google Scholar]
- 39.Zoulim, F., T. Poynard, F. Degos, A. Slama, A. El Hasnaoui, P. Blin, F. Mercier, P. Deny, P. Landais, P. Parvaz, and C. Trepo. 2006. A prospective study of the evolution of lamivudine resistance mutations in patients with chronic hepatitis B treated with lamivudine. J. Viral Hepat. 13:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]