Abstract
Studies on the molecular epidemiologic characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains have demonstrated their genetic and geographical diversity. In addition, it has been reported that there are genetic differences between community-associated (CA) and health care-associated (HA) MRSA strains. Therefore, we investigated the major epidemiologic characteristics of CA MRSA isolates in South Korea and compared them with those of HA MRSA strains. Distributions of staphylococcal chromosome cassette mec (SCCmec) types and other molecular features, including the Panton-Valentine leukocidin (PVL) gene, were studied in 138 invasive MRSA isolates. Multiplex type IVA SCCmec was identified as the major CA MRSA infection type (53.1%), with a significantly higher prevalence than in HA MRSA (P < 0.001). One major group of type IVA strains carried a larger atypical class B mec element and new subtypes of ccrA2 (96% amino acid homology). The PVL gene was detected in one USA300-like isolate only. Seven major clone types determined by combinational grouping (genetic background SCCmec typing) showed representative patterns of antimicrobial susceptibilities. We concluded that less multi-drug-resistant strains of clone types B-I and D-1 (genetic background, B and D complexes; type IVA SCCmec) predominate in CA MRSA and that international PVL-positive strains have not spread in South Korea as yet.
Rates of methicillin-resistant Staphylococcus aureus (MRSA) infection have continuously increased in both communities and hospitals. The staphylococcal cassette chromosome mec element (SCCmec) has contributed to this phenomenon as an important epidemiologic factor and also as a determinant of antibiotic resistance patterns (16, 25). In parallel with the identification of various types and subtypes of SCCmec (14, 26, 30), a new nomenclature for the SCCmec element was proposed (5). In addition to antimicrobial resistance factors, a wide variety of virulent factors are also important for understanding S. aureus infection, which varies from mild to severe (2, 20). In particular, Panton-Valentine leukocidin (PVL) is a major concern with respect to MRSA infection, and community-associated MRSA (CA MRSA) has been isolated recently (13, 31, 32). There are some differences in the genetic and epidemiologic backgrounds of CA MRSA and health care-associated MRSA (HA MRSA). Some studies have suggested that the smaller SCCmec (type IV or V) and PVL are strongly associated with CA MRSA infection (21, 23, 32). Their features correspond to a non-multi-drug-resistant character, and such infections exhibit toxin-like risk factors, unlike HA MRSA infections. However, it has also been reported that type IV SCCmec isolates are rare in South Korea compared with type II and III SCCmec strains, which are isolated mainly from HA MRSA infections (4, 18). We noted that the number of SCCmec variants has increased recently in South Korea, and thus, we thought that the epidemiology of CA MRSA in South Korea could be different from that in other countries, because typical CA MRSA isolates characterized by PVL and type IV SCCmec have been rarely found in South Korea. Initially, we investigated the distribution of SCCmec types, genetic variations among invasive isolates, and other molecular epidemiologic characteristics. We then characterized the major epidemiologic features of CA MRSA isolates and compared them with HA MRSA by studying relationships between SCCmec diversity and molecular features or antimicrobial susceptibilities.
MATERIALS AND METHODS
Bacterial isolates and definitions.
From April 2004 to October 2005, we collected a total of 138 invasive nonduplicate MRSA isolates from patients with bacteremia (n = 74) and skin and soft tissue infections (n = 64) at a tertiary-care hospital and five community hospitals in four regions of South Korea (two hospitals in Seoul, two in Incheon, one in Gyeongki, and one in Gyeongnam). Most of these isolates were recovered from blood, wounds, or pus. Possible colonizations and contaminants were excluded. Using medical records, CA MRSA was defined as described by Fridkin et al. (9) (CA isolates, n = 81; HA, n = 57). Briefly, CA MRSA isolates were recovered from a patient who had none of the following established risk factors; isolation of MRSA 48 h or more after hospitalization; a history of hospitalization, surgery, dialysis, or residence in a nursing home within the past year; the presence of a permanent indwelling catheter or percutaneous medical device at the time of culture; previous isolation of MRSA.
SCCmec typing and genetic variation studies.
We screened SCCmec isolates by multiplex PCR, as previously described (26). Single SCCmec PCR typing was then performed to further analyze variations in class mec complex and J region, as previously described (12, 23). SCCmec type was assigned using multiplex type nomenclature and the nomenclature recently proposed by Chongtrakool et al. (5). Atypical elements of the class B mec complex and of the ccrA2 gene were sequenced with the primers listed in Table 1 in order to analyze genetic variations. Sequences homologous with class B mec complex and in the new ccrA2 gene subtype were searched for with BLAST (BLASTP version 2.2.16; http://www.ncbi.nlm.nih.gov/BLAST).
TABLE 1.
PCR primers used for sequencing analysis of class B mec element and ccrA2
| Target | Primer | Sequence (5′→3′) | Location (nt)a | Reference |
|---|---|---|---|---|
| IS1272-mecA | IS5 | AACGCCACTCATAACATATGGAA | 11882-11904 | 23 |
| mA6 | TATACCAAACCCGACAAC | 13860-13877 | 17 | |
| mecA | mAnew1 | TGGAATTAACGTGGAGACGA | 13569-13588 | This study |
| mAnew2 | AACGTTGTAACCACCCCAAG | 15127-15146 | This study | |
| mecA-IS431mec | mA1 | TGCTATCCACCCTCAAACAGG | 14861-14881 | 11 |
| IS2 | TGAGGTTATTCAGATATTTCGATGT | 18911-18935 | 17 | |
| ccrA2 | ccrA2-F | GGATAGGCCCTTCAGGAGTT | 5785-5804 | This study |
| ccrA2-R | TGTGCTTTGCATTTCTGTTGA | 7476-7496 | This study |
Locus of AB245470.
MLST, spa and other molecular characterizations.
Multilocus sequence typing (MLST) was performed as previously described (7). Alleles of each locus were compared, and sequence types (STs) were assigned based on the S. aureus MLST database (http://saureus.mlst.net/).
The typing of the polymorphic region of the protein A gene (spa) was performed as previously described (1, 29). The product was amplified using spa-1113f (5′-TAAAGACGATCCTTCGGTGAGC-3′) and spa-1514r primers (5′-CAGCAGTAGTGCCGTTTGCTT-3′), as previously described (1). Purified spa PCR products were sequenced, and short sequence repeats (SSRs) were assigned using the spa database web site (http://www.ridom.de/spaserver). The spa complex was defined by visual analysis whereby spa types with similar SSRs were clustered into the complexes previously described by Ruppitsch et al. (28).
The accessory gene regulator locus (agr) gene was amplified, and agr group was determined by DraI restriction fragment length polymorphism analysis, as previously described (27). The PVL, hlg, hlg-2, and lukE-lukD genes were detected, as previously described (15). nuc PCR was performed as the control for validation purposes, as previously described (3).
Antimicrobial susceptibilities.
Antimicrobial susceptibilities were determined using the disc diffusion method, as recommended by the CLSI (6). BBL Sensi-Discs (Becton Dickinson, Sparks, MD) of arbekacin, ciprofloxacin, clindamycin, erythromycin, gentamicin, oxacillin, penicillin, rifampin, tetracycline, tobramycin, trimethoprim-sulfamethoxazole, and vancomycin were used. Multidrug resistance was defined as resistance to three or more classes of antimicrobials. S. aureus ATCC 29213 was used as a control strain.
Clone type definitions based on molecular characteristics and antimicrobial susceptibilities.
MRSA isolates were clustered into representative groups based on genetic background as previously described by Oliveira et al. (24, 25), with some modifications. Briefly, genetic backgrounds were determined by MLST profile with one or two allelic variants corresponding to the spa complex. Based on genetic backgrounds, clone types were redefined according to SCCmec type and antimicrobial susceptibilities.
Statistical analysis.
Comparisons were made using the χ2 test or Fisher's exact test in Sigma Stat version 3.10 (Systat Software, San Jose, CA). All hypotheses were two-tailed and were considered significant at the P < 0.05 level.
Nucleotide sequence accession numbers.
Nucleotide sequences determined during the present study were submitted to GenBank (http://www.ncbi.nlm.nih.gov/GenBank) under accession numbers EF584543 and EF596937.
RESULTS
Genetic diversities of SCCmec types from CA and HA MRSA isolates.
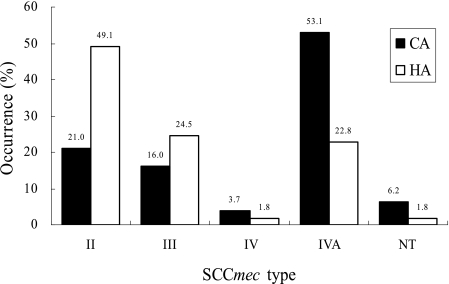
Screening by SCCmec multiplex- and single-PCR typing resulted in the typing of 132 of the 138 (95.7%) isolates; the remaining 6 (4.3%) could not be typed (Table 2). Multiplex type II (including variants) accounted for 32.6% (45/138), III for 19.6% (27/138), IV for 3.8% (4/138), and IVA for 40.6% (56/138). Investigations of the distribution of each SCCmec type in CA and HA MRSA showed that multiplex type IVA was significantly prevalent in CA MRSA (P < 0.001) (Fig. 1). On the other hand, type II was more prevalent in HA MRSA (P < 0.001) (Fig. 1). Also, the occurrence of type III was a little higher in HA than in CA MRSA, although the difference was statistically insignificant (P = 0.276) (Fig. 1).
TABLE 2.
Genetic diversities of SCCmec types classified by multiplex and single PCR
| SCCmec multiplex type1 | Amplified locus or locia | MLST sequence type(s) (n) | Proposed SCCmec typeb | mec classc | ccr typec | J regiond | No. of isolates (%) |
|---|---|---|---|---|---|---|---|
| II | B, C, D, G | ST5 | II.1.1 | A | 2 | 9 (6.5) | |
| II variant | B, C, D | ST5 (25), ST5SLV (1) | II.1.2 | A | 2 | Does not carry pUB110 | 26 (18.9) |
| II NT1 | C, D, G | ST89 (4), ST5 (1) | NDe | A | 2 | Does not carry kdp operon | 5 (3.6) |
| II NT2 | C, D | ST5 | ND | A | 2 | Does not carry kdp and pUB110 | 5 (3.6) |
| III | C, E, F, H | ST239 | III.1 | A | 3 | 13 (9.4) | |
| IIIA | C, E, F | ST239 | III.1.2 | A | 3 | Does not carry pT181 | 12 (8.7) |
| III NT1 | C, H | ST239 | ND | A | 3 | Does not carry RIF | 2 (1.5) |
| IV | D | ST8 (1), ST1 (2) | IV.1 | B | 2 | IVA (1), IVC (2) | 3 (2.2) |
| IV NT1 | D, F | ST254 | ND | B | 2 | Carries RIF5 | 1 (0.7) |
| IVA | D, G | ST72 | IV.N.2 | (B)f | (2)g | IVC, carries pUB110 | 32 (23.2) |
| ST1 (19),h ST493 (1),h ST573 (1)h | ND | B | 2 | IVC, carries pUB110 | 21 (15.2) | ||
| ST89 | ND | (A)i | 2 | ND | 3 (2.2) | ||
| NT | None amplified | ST188, ST72, ST1, ST89, ST30, ST239 | ND | ND | ND | ND | 6 (4.3) |
Multiplex PCR results described by Oliveira and de Lencastre (26).
Proposed SCCmec type described by Chongtrakool et al. (5).
Single PCR results described by Okuma et al. (23).
Subtyping of J region was performed as described by Hisata et al. (12); other characteristics were determined from multiplex PCR results.
ND, not determined.
The increased size of class B mec was due to the presence of IS1272-tnp20-ΔmecR1-mecA-IS431 (GenBank no. EF596937).
ccrA2 (GenBank no. EF584543) shared 96% homology with other ccrA2 genes.
Belongs to CC1 (ST1, 1-1-1-1-1-1-1; ST493, 62-1-1-1-1-1-1; ST573, 1-1-1-1-12-1-1).
Class A mec complex variant, ΔmecI-mecR1-mecA.
FIG. 1.
Distribution of SCCmec types in invasive CA and HA MRSA isolates. Each type includes variants.
Diverse genetic variants or subtypes were observed within each multiplex type (Table 2). In particular, multiplex type IVA was subdivided into two major groups and one minor group carrying a class A mec complex variant (ΔmecI-mecR1-mecA) (Table 2). The class B mec complex of the first group was different from that of type IV due to increased size of the element and new subtypes of ccrA2 (GenBank no. EF584543; 96% homology), while the other major group carried typical class B mec and ccrA2 (Table 2). Atypical class B mec was composed of IS1272-tnp20 (pfam02371)-ΔmecR1-mecA-IS431 (GenBank no. EF596937).
MLST, spa complex, and other molecular epidemiologic factors.
MLST analysis showed that some STs (ST1, -5, -72, and -239) were prevalent in MRSA infections and also that there should be an epidemiological relationship among STs, SCCmec types, and spa types (Tables 2 and 3). A total 22 of spa types were analyzed in 137 of the 138 MRSA isolates, and 5 of them (t2457 to t2461) were assigned as new types in the spa database (http://www.ridom.de/spaserver). The spa gene of isolate ST254 (SSR profile, 3-32-1-1-4-4-3) could not be amplified by PCR.
TABLE 3.
The spa complex of MRSA isolates as determined by visual analysis
| MLST sequence type | spa complex (n)a | spa type | SSR profile | No. of Isolates | Other characteristics | Genetic backgroundb (n)a |
|---|---|---|---|---|---|---|
| ST5 (1-4-1-4-12-1-10)c | spa class A (41) | t002 | 26-23-17-34-17-20-17-12-17-16 | 15 | CA (13/81, 16.0%) | A (41) |
| t601 | 26-23-17-34-34-17-20-17-12-17-16 | 13 | HA (28/57, 49.1%) | |||
| t2458d | 26-16-34-34-17-20-17-12-17-16 | 8 | agr group II, hlg-2 | |||
| t2460d | 26-17-34-34-17-20-17-17-17-16 | 3 | ||||
| t010 | 26-17-34-17-20-17-12-17-16 | 1 | ||||
| t306 | 26-23-17-34-17-20-17-12-17-17-16 | 1 | ||||
| ST72 (1-4-1-8-4-4-3) | spa class B (33) | t324 | 07-23-12-12-17-20-17-12-12-17 | 25 | CA (22, 27.2%), | B (33) |
| t664 | 07-23-12-12-17-20-17-12-17 | 4 | HA (11, 19.3%) | |||
| t148 | 07-23-12-21-12-17-20-17-12-12-17 | 2 | agr group I, hlg-2 | |||
| t901 | 07-23-12-17-20-17-12-12-17 | 1 | ||||
| t2461d | 07-23-12-12-12-17-20-17-12-12-17 | 1 | ||||
| ST239 (2-3-1-1-4-4-3) | spa class C (29) | t037 | 15-12-16-02-25-17-24 | 27 | CA (15, 18.5%) | C (27)e |
| ST580 (3-35-48-19-20-26-39) | t021 | 15-12-16-02-16-02-25-17-24 | 1 | HA (14, 24.6%) | ||
| ST30 (2-2-2-2-6-3-2) | t138 | 08-16-02-25-17-24 | 1 | hlg-2, agr group I (1) | ||
| ST1 (1-1-1-1-1-1-1) | spa class D (24) | t286 | 07-23-13-34-16-34-33-13 | 21 | CA (22, 27.2%) | D (24) |
| ST493 (62-1-1-1-1-1-1) | t1533 | 07-23-13-13-16-34-33-13 | 1 | HA (2, 3.5%) | ||
| ST573 (1-1-1-1-12-1-1) | t2457d | 07-23-13-34-34-16-34-33-13 | 1 | agr group III, hlg-2 | ||
| t2459d | 07-23-34-34-16-34-33-13 | 1 | ||||
| ST89 (1-26-28-18-18-33-50) | spa class E (8) | t375 | 49-13-23-05-17-34-33-34 | 7 | CA (8, 9.9%) | E (8) |
| t1728 | 49-20-13-23-05-17-34-33-34 | 1 | agr group III, hlg | |||
| ST8 (3-3-1-1-4-4-3) | Singleton | t008 | 11-19-12-21-17-34-24-34-22-25 | 1 | ST8 (PVL, agr group I) | |
| ST188 (3-1-1-8-1-1-1) | Singleton | t189 | 07-23-12-21-17-34 | 1 | ST188 (agr group I) |
Defined by visual analysis as described by Ruppitsch et al. (28); 137 of 138 isolates were analyzed, as 1 HA isolate was not amplified by spa PCR.
Determined by MLST profile with one or two allelic variants corresponding to the spa complex.
Includes the ST5 single locus variant.
Newly reported in this study.
ST580 and ST30 were excluded because of an MLST profile with three or more allelic variants.
Visual analysis of the SSR profile was used to group spa types with similar repeat profiles into five spa complexes with two singletons (Table 3). spa class A (type II SCCmec including variants, agr group II) and C (type III and IIIA SCCmec, agr group I) complexes were associated mainly with HA MRSA infection, and their major STs were, respectively, ST5 and ST239 (Table 3). CA MRSA strains were strongly associated with spa class B and D complexes, which mainly exhibited ST72 and CC1 (ST1, ST493, and ST573) (Table 3). ST72 isolates exhibited type IVA SCCmec (IV.N.2) and agr group I, and isolates of the spa class D complex were associated with SCCmec type IVA with typical class B mec complexes and agr group III. Isolates of the spa class E complex (ST89) were infrequently found in CA MRSA infections (type II; NT1 and IVA SCCmec carrying a class A mec variant). The PVL gene was detected in only one USA300-like strain exhibiting ST8, spa t008, type IVA SCCmec, and agr group I. Most strains carried hlg-2, except for isolates of the spa class E complex and an ST30 isolate, which carried hlg (Table 3).
Clone types of MRSA isolates by molecular characteristics and antimicrobial susceptibilities.
All isolates showed resistance against oxacillin, penicillin, and tobramycin but no resistance against vancomycin. Antimicrobial resistance patterns were dependent mainly on genetic background and SCCmec type. Thus, we could group 131 of the 138 isolates into clone types, which are expected to represent antimicrobial susceptibility patterns (Table 4) In the same spa complex, isolates with three more allelic variants in the MLST profile were considered to have a different genetic background and were classified as singletons (ST580, 3-35-48-19-20-26-39, compared to ST239 in the spa class C complex). Also, a single isolate with no spa type (ST254 isolate) and six with nonamplified SCCmec were excluded from the analysis. Isolates with genetic background A were divided into two clone types (A-I and A-II), and these exhibited different antimicrobial susceptibilities, especially with respect to tetracycline (P < 0.001) (Table 4). Isolates with genetic background C were also subgrouped into two clone types (C-I and C-II), and there were some differences in antimicrobial susceptibility patterns between them, especially with respect to trimethoprim-sulfamethoxazole (P < 0.001) (Table 4). Major CA MRSA isolates (clone types B-I, D-I, and E-I) were less multi-drug resistant than HA MRSA isolates (A-I, II, C-I, and II). Isolates of clone type B-I were much less resistant than those of clone type D-I, especially against erythromycin (P = 0.0014), gentamicin (P < 0.001), and tetracycline (P < 0.001).
TABLE 4.
Antibiotic susceptibilities of major MRSA clone types according to genetic background and SCCmec typea
| Clone type | Genetic background | SCCmec type | % of isolates resistant tob:
|
No. of CA isolates/total isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | ABK | CIP | CLI | ERY | TET | RIF | SXT | ||||
| A-I | A (ST5-spa A) | II, II NT1, or II NT2 | 86.7 | 0 | 100 | 66.7 | 100 | 100 | 0 | 6.7 | 6/15 |
| A-II | A | II variant | 100 | 0 | 100 | 100 | 100 | 19.2 | 0 | 0 | 7/26 |
| B-I | B (ST72, spa B) | IVA (IV.N.2) | 3.1 | 0 | 0 | 3.1 | 50.0 | 0 | 0 | 0 | 21/32 |
| C-I | C (ST239, spa C) | III or III NT1 | 100 | 0 | 100 | 73.3 | 100 | 100 | 0 | 100 | 7/15 |
| C-II | C | IIIA | 100 | 18.2 | 81.8 | 90.9 | 100 | 81.8 | 18.2 | 0 | 5/11 |
| D-I | D (CC1, spa D) | IVA or IV (IVC) | 100 | 0 | 0 | 0 | 91.3 | 56.5 | 0 | 0 | 21/23 |
| E-I | E (ST89, spa E) | IVA minor or II NT1 | 85.7 | 0 | 0 | 71.4 | 100 | 0 | 0 | 0 | 7/7 |
| Minor 1 | USA300-like (ST8, spa t008) | IVA | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 1/1 |
| Minor 2 | ST580, spa C | IIIA | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 1/1 |
In the same spa complex, the isolate with three more allelic variants of MLST profile was considered to carry another genetic background and was classified as a singleton (ST580, 3-35-48-19-20-26-39, compared to ST239 in the spa C complex). Isolates with no spa type (ST254 isolate) or nonamplified SCCmec (n = 6; CA n = 5) were excluded from the analysis.
All isolates showed resistance against oxacillin, tobramycin, and penicillin, but no isolate showed resistance against vancomycin. Abbreviations: ABK, arbekacin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; RIF, rifampin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; and VAN, vancomycin.
DISCUSSION
It has been reported that ST5 and ST239 strains predominate in HA MRSA infections in South Korea (4, 18). However, few reports on the molecular characteristics of CA MRSA strains in South Korea have been issued. Thus, we tried to characterize CA MRSA strains in South Korea and compare them with HA MRSA strains. Oliveira et al. (25) have described the characterization of MRSA pandemic clones, in which major clones were classified according to genetic background and SCCmec type as an evolutionary marker (25). As spa types were very diverse in isolates of the same ST, we needed to group them into complexes which could represent the diversity as well as common motifs. spa complexes as well as MLST STs were meaningful for analyzing genetic backgrounds and relationships among isolates. In addition, as it could be difficult to define SCCmec types by previous nomenclature (12, 23, 26) due to increasing numbers of variants, we classified our results according to a new nomenclature which is able to represent the variants of each types. Along with genetic background, this nomenclature was very helpful for classifying isolates into clone types corresponding to antibiotic susceptibility as well as molecular epidemiological features of the isolates (Table 4).
In our study, the ST5 strains (clone types A-I and A-II) seemed to belong to the NY/Japan clone, and the ST239 strains (clone types C-I and C-II) seemed to be consistent, respectively, with Hungarian (type III SCCmec) and Brazilian (type IIIA SCCmec) clones, although there were some variants (Table 4) (25). This suggested that HA MRSA clones in South Korea were consistent with a pandemic clone.
While HA MRSA clones in South Korea are expected to have some features in common with pandemic HA clones, genetic features of major CA MRSA strains in South Korea may be unique compared with those of clones that have spread internationally. In particular, type IVA SCCmec is common in South Korea, whereas type IV is not (Fig. 1; Table 2). Moreover, the genetic background of CA MRSA strains in the present study was mainly associated with clone types B-I, D-I, and E-I, which may differ from prevalent CA MRSA strains in other countries (Table 4). Multiplex type IVA SCCmec was firstly mentioned by Oliveira and de Lencastre (26), and Shore et al. (30) reported that multiplex type IVA SCCmec carried the class A mec complex with some variants (30). However, our study showed that the upstream vicinity of type IVA of major groups could have been derived from type IVC and that it carries the class B mec complex, except for a minor group (spa E-type IVA) carrying a class A mec complex variant (Table 2). Chongtrakool et al. (5) proposed that type IVA be described as IV.N.2 (5). But we found three subtypes in type IVA SCCmec, and each subtype was found to represent a different antimicrobial susceptibility pattern (clone types B-I, D-I, and E-I).
It was interesting to find that representative HA MRSA strains have been spread in CA infections and CA MRSA strains have also been detected in HA infections (Tables 3 and 4). We do not know what epidemiologic factors have contributed to this spread, but these findings emphasize the need for continuous monitoring.
PVL-positive CA MRSA strains have recently spread globally (8, 10, 31, 32). However, no PVL-positive strain had been isolated from humans in South Korea, although it had been isolated from bovine milk (type IVG SCCmec, ST5) (19). In the present study, we detected just one PVL-positive isolate, which seemed to be a USA300-like strain exhibiting PVL, t008, ST8, and SCCmec IVA (22). This strain was recovered from a patient that had recently returned from Hawaii and subsequently developed an invasive MRSA infection.
In summary, we concluded that non-multi-drug-resistant strains of clone types B-I and D-I predominate in CA MRSA in South Korea. Moreover, the prevalence of type IVA SCCmec among CA MRSA strains contrasted with its prevalence in strains found in other countries. In addition, the international PVL-positive CA MRSA clone has not been frequently found in South Korea. Future studies are required to determine other factors that might contribute to the high occurrence of invasive MRSA infection in South Korea.
Acknowledgments
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2005-003-E00119).
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 3.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha, H. Y., D. C. Moon, C. H. Choi, J. Y. Oh, Y. S. Jeong, Y. C. Lee, S. Y. Seol, D. T. Cho, H. H. Chang, S. W. Kim, and J. C. Lee. 2005. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J. Clin. Microbiol. 43:3610-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. Sixteenth informational supplement. Approved standard M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etienne, J. 2005. Panton-Valentine leukocidin: a marker of severity for Staphylococcus aureus infection? Clin. Infect. Dis. 41:591-593. [DOI] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, M. M. Farley, and the Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. 2005. Methicillin resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 10.Gillet, Y., B. Issartel, P. Vnahems, J. Foumet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotizing pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., H. Kihara, and T. Yokota. 1992. Analysis of borderline resistant strains of methicillin-resistant Staphylococcus aureus using polymerase chain reaction. Microbiol. Immunol. 36:445-453. [DOI] [PubMed] [Google Scholar]
- 12.Hisata, K., K. Kuwahara-Arai, M. Yamanoto, T. Ito, Y. Nakatomi, L. Cui, T. Baba, M. Terasawa, C. Sotozono, S. Kinoshita, Y. Yamashiro, and K. Hiramatsu. 2005. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J. Clin. Microbiol. 43:3364-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, P. L., C. W. Tse, G. C. Mak, K. H. Chow, and T. K. Ng. 2004. Community-acquired methicillin-resistant Staphylococcus aureus arrives in Hong Kong. J. Antimicrob. Chemother. 54:845-846. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, N. H., K. T. Park, J. S. Moon, W. K. Jung, S. H. Kim, J. M. Kim, S. K. Hong, H. C. Koo, Y. S. Joo, and Y. H. Park. 2005. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J. Antimicrob. Chemother. 56:624-632. [DOI] [PubMed] [Google Scholar]
- 20.Lina, G., Y. Gillet, F. Vandenesch, M. E. Jones, D. Floret, and J. Etienne. 1997. Toxin involvement in staphylococcal scalded skin syndrome. Clin. Infect. Dis. 25:1369-1373. [DOI] [PubMed] [Google Scholar]
- 21.Lo, W. T., W. J. Lin, M. H. Tseng, S. R. Wang, M. L. Chu, and C. C. Wang. 2006. Community acquired methicillin-resistant Staphylococcus aureus in children, Taiwan. Emerg. Infect. Dis. 12:1267-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin resistant Staphylococcus aureus. Lancet Infect. Dis. 2:280-289. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 28.Ruppitsch, W., A. Indra, A. Stoger, B. Mayer, S. Stadlbauer, G. Wewalka, and F. Allerberger. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and D. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tristan, A., M. Bes, H. Meugnier, G. Lina, B. Bozdogan, P. Courvalin, M. Reverdy, M. Enright, F. Vandenesch, and J. Etienne. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]