Abstract
We describe the development and validation of an agar dilution method for the detection of inducible clindamycin resistance by using 227 previously characterized erythromycin-resistant, clindamycin-susceptible Staphylococcus sp. isolates. Mueller-Hinton agar with defibrinated horse blood containing a range of erythromycin concentrations (1 to 8 mg/liter) combined with clindamycin at 0.5 mg/liter was used to determine the optimal concentration that produced growth of inducible isolates while inhibiting that of isolates without the inducible phenotype. A concentration of clindamycin of 0.5 mg/liter with erythromycin at 1 mg/liter was the optimal combination for detection of inducible resistance and resulted in a sensitivity of 100% (95% confidence interval [CI], 97.9 to 100) and a specificity of 100% (95% CI, 93.0 to 100). Attention must be paid to ensuring that a sufficient inoculum has been used, since an inoculum below the standard 107 bacteria/ml may result in false-negative results. This method has been incorporated into routine use in our laboratory.
Macrolide resistance in Staphylococcus spp. is generally attributable to one of two mechanisms (7). One is active efflux due to a pump encoded by the msrA gene, which confers resistance to macrolides and streptogramin antibiotics but not lincosamides such as clindamycin (MS phenotype). The second is ribosomal methylation mediated by enzymes encoded by one of a variety of erm genes. This mechanism results in resistance to macrolides, lincosamides, and streptogramin B antibiotics (MLSB phenotype). The MLSB phenotype is inducible (iMLSB) or constitutive (cMLSB). Induction of resistance occurs in the presence of macrolide antibiotics but not lincosamides. Thus, isolates exhibiting iMLSB resistance appear susceptible to clindamycin by routine methods such as broth microdilution, disk diffusion, and agar dilution. However, treatment failures have occurred with such isolates when clindamycin is used, because of selection of mutations during therapy, leading to cMLSB resistance (4, 8, 12).
It is therefore important for microbiology laboratories to show that an isolate that is erythromycin resistant but apparently clindamycin susceptible on routine testing is of the MS phenotype before reporting it as clindamycin susceptible. This is most commonly done by using the disk diffusion test, in which a 2-μg clindamycin disk is placed next to a 15-μg erythromycin disk (D-zone test). Flattening of the clindamycin zone adjacent to the erythromycin disk (positive D-zone test) indicates the presence of iMLSB resistance, while a circular zone (negative D-zone test) indicates MS resistance. This method has been shown to have high sensitivity and specificity compared with genotypic analysis and has been incorporated into the Clinical and Laboratory Standards Institute (CLSI) method (3), but false-negative tests may occur if the disk separation distance is too wide (5, 9).
For laboratories that use agar dilution for routine antimicrobial susceptibility testing, disk diffusion methods for detection of inducible clindamycin resistance are inconvenient. An agar dilution method has the potential benefit of allowing large numbers of isolates to be simultaneously tested. We describe the development and validation of an agar dilution method for detection of inducible clindamycin resistance that has been incorporated into routine use at our laboratory.
MATERIALS AND METHODS
Collection and characterization of isolates.
Consecutive, nonduplicate Staphylococcus sp. isolates which were clindamycin susceptible (MIC, ≤0.5 mg/liter) but erythromycin intermediate (MIC, 1 to 4 mg/liter) or resistant (MIC, ≥8 mg/liter) by broth microdilution were collected, identified, and stored as outlined in a previous study (9). The D-zone test was performed according to the CLSI method with Mueller-Hinton agar as described elsewhere (3, 9), and genes conferring macrolide resistance, namely, ermA, ermB, ermC, ermTR, and msrA, were identified with a multiplex PCR-based reverse line blot assay as previously described (9). Three isolates used in the previous study were nonviable, and a fourth, with iMLSB resistance by D-test and no erythromycin resistance gene detected, was excluded from the analysis. Of the 227 remaining isolates, 176 had the iMLSB phenotype (100 methicillin-resistant Staphylococcus aureus, 56 methicillin-sensitive S. aureus [MSSA], and 20 coagulase-negative Staphylococcus sp. strains) while 51 had the MS phenotype (3 methicillin-resistant S. aureus, 3 MSSA, and 45 coagulase-negative Staphylococcus sp. strains). All isolates with the MS phenotype harbored msrA alone, while all those with the iMLSB phenotype harbored either ermA (92 isolates) or ermC (84 isolates). Two with ermA also harbored msrA and had the iMLSB phenotype. All isolates were erythromycin resistant by broth microdilution except one MSSA strain which was D-zone test positive and carried the ermA gene.
Agar dilution.
Mueller-Hinton agar (BBL, Sparks, MD) with 3.3% defibrinated horse blood (MHA-HB), containing 1, 2, 4, or 8 mg/liter erythromycin and 0.5 mg/liter clindamycin (Upjohn Laboratories, Kalamazoo, MI) was prepared. In addition, agar plates with 0.5 mg/liter clindamycin alone or with 1 mg/liter erythromycin alone and agar plates without antibiotics were prepared, the latter two serving as growth controls. Mueller-Hinton agar supplemented with blood is used in our laboratory for all agar dilution susceptibility testing so that all organisms can be tested on the same plates. Additionally, we find that the opaque red background facilitates the detection of scanty growth. MHA-HB was poured into 100-mm-square plates to a depth of 3 mm. Once prepared, plates were stored at 4°C and used within 7 days. Approximately four colonies of an 18- to 20-h-old subculture were inoculated into 4 ml of Trypticase soy broth (BBL, Sparks, MD). After 4 h of incubation at 37°C, this suspension was adjusted to a concentration equivalent to a 0.5 McFarland standard. A 100-μl volume of this suspension was then added to 900 μl of broth contained in a 64-well seed block to produce a concentration of approximately 1 × 107 bacteria/ml. Plates were then inoculated with an automated multipoint inoculator (Denley-Tech, Billingshurst, United Kingdom) which delivers a volume of approximately 1 μl per spot, resulting in an estimated final inoculum of 1 × 104 bacteria. Plates were incubated for 18 h at 35°C under atmospheric conditions. Growth was recorded if at least one colony was evident at the inoculation site after careful visual inspection.
Test interpretation.
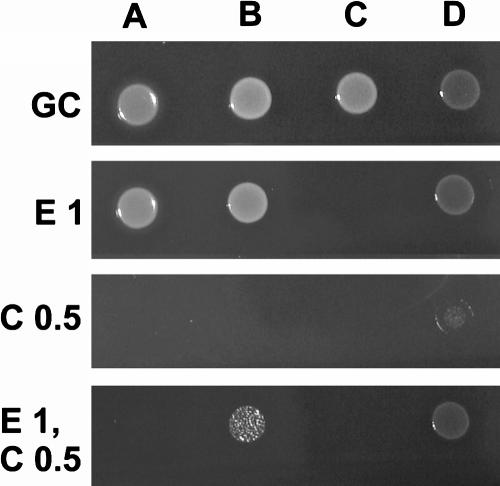
A test was deemed to be positive (i.e., inducible resistance was present) if there was any visible growth on the erythromycin-only and combined plates but not on the clindamycin-only plate. A test was deemed to be negative (i.e., MS resistance phenotype) if growth was found on the erythromycin-only plate but not on the combined or clindamycin-only plate (Fig. 1). Genotyping was used as the reference for comparison, where a negative result was indicated by an msrA genotype and no erm gene detected, while a positive result was an ermA and/or ermC genotype regardless of whether msrA was present.
FIG. 1.
Agar dilution method for detection of inducible clindamycin resistance. Columns: A, MS strain (ATCC BAA-976); B, iMLSB strain (ATCC BAA-976); C, macrolide-sensitive strain; D, cMLSB strain. Rows: GC, growth control (no antibiotics); E 1, erythromycin at 1 mg/liter; C 0.5, clindamycin at 0.5 mg/liter; E1, C 0.5, erythromycin at 1 mg/liter and clindamycin at 0.5 mg/liter.
Quality control.
Negative and positive control strains—ATCC BAA-976 (MS phenotype, msrA gene positive) and BAA-977 (iMLSB phenotype, ermA gene positive)-and two in-house isolates—one erythromycin and clindamycin susceptible and one erythromycin and clindamycin resistant (cMLSB phenotype)—were inoculated onto each plate.
Inoculum study.
To determine the importance of an adequate inoculum, 11 S. aureus isolates and 1 S. epidermidis isolate with inducible clindamycin resistance (comprising 9 ermA-positive and 3 ermC-positive isolates), as well as the four control strains, were tested with dilutions of 108, 106, and 105 bacteria/ml in addition to the standard inoculum concentration of 107 bacteria/ml. These were inoculated onto the same set of MHA-HB plates, incubated, and inspected as described above. In addition, 10-μl loops were used to inoculate blood agar plates for colony counts to ensure the correct inoculum density.
Statistics.
Confidence intervals were calculated by Confidence Interval Analysis for Windows (available at http://www.medschool.soton.ac.uk/cia).
RESULTS
Determination of the optimal erythromycin concentration.
The effects of increasing erythromycin concentrations on the sensitivity and specificity of inducible clindamycin resistance detection are shown in Table 1. The number of false-negative results increased at higher concentrations. All isolates with the MS phenotype were inhibited at all of the erythromycin concentrations tested. A combination of erythromycin at 1 mg/liter and clindamycin at 0.5 mg/liter was determined to be optimal for testing since this was the only combination that gave 100% sensitivity and specificity.
TABLE 1.
Effects of various concentrations of erythromycin on detection of inducible clindamycin resistance by the agar dilution method
| Erythromycin concn (mg/liter)a | No. of strains with genotype of:
|
Agar dilution result | % Sensitivityb | % Specificityb | |
|---|---|---|---|---|---|
| msrA only (n = 51) | ermA and/or ermC (n = 176) | ||||
| 1 | 51 | 0 | Negative | 100 (97.9-100) | 100 (93.0-100) |
| 0 | 176 | Positive | |||
| 2 | 51 | 3 | Negative | 98.3 (95.1-99.7) | 100 (93.0-100) |
| 0 | 173 | Positive | |||
| 4 | 51 | 5 | Negative | 97.2 (90.5-97.6) | 100 (93.0-100) |
| 0 | 171 | Positive | |||
| 8 | 51 | 9 | Negative | 94.9 (91.6-98.1) | 100 (93.0-100) |
| 0 | 167 | Positive | |||
The clindamycin concentration was 0.5 mg/liter with an inoculum concentration of 107 bacteria/ml.
95% confidence intervals are in parentheses.
Assay validation.
At antibiotic concentrations of 1 mg/liter erythromycin and 0.5 mg/liter clindamycin, all isolates with inducible resistance showed visible growth and all with the MS phenotype failed to grow, resulting in a sensitivity and specificity of 100% (Table 1). However, plates had to be inspected carefully for growth, which was often scanty, with as few as one colony visible on the inoculum for some isolates.
Determination of inoculum effect.
There was a clear relationship between the inoculum density and the amount of growth visible on the plates. With erythromycin at 1 mg/liter and clindamycin at 0.5 mg/liter and a standard inoculum concentration of 1 × 107 bacteria/ml, all 12 isolates with inducible resistance showed visible growth, but in some instances this was as little as one colony. At 1 × 108 bacteria/ml, there was good growth of all of the isolates tested. Lower inoculum densities resulted in false-negative results—1 of 12 at an inoculum concentration of 1 × 106 bacteria/ml and 4 of 12 at 1 × 105 bacteria/ml.
DISCUSSION
Isolates of Staphylococcus spp. that are erythromycin resistant but clindamycin susceptible should not be reported as clindamycin susceptible unless iMLSB resistance has been excluded (4, 7, 11, 12). Currently, the only method routinely recommended for testing of iMLSB resistance is disk approximation testing (1-3). While this accurately distinguishes iMLSB resistance (5, 9), it is relatively labor intensive for laboratories which routinely use agar dilution methods for susceptibility testing or when large numbers of isolates are to be tested.
In this study, we have found that agar dilution testing with erythromycin at 1 mg/liter and clindamycin at 0.5 mg/liter is a sensitive and specific method for detection of iMLSB resistance in Staphylococcus spp. There was an obvious inoculum effect associated with iMLSB resistance, which is similar to the effect seen for some other antibiotic resistance mechanisms such as β-lactamase production (10, 13). As for all of the agar dilutions tests, the importance of the correct inoculum should not be underestimated (6). While using the standard inoculum concentration of 1 × 107 bacteria/ml resulted in accurate test performance, growth was frequently scant (as little as one colony). Thus, it is important to ensure that a sufficiently dense inoculum is used. A higher inoculum concentration (1 × 108 bacteria/ml) could be used to produce more obvious growth, but this would not integrate easily into the standard CLSI susceptibility testing methods.
Acknowledgments
Many thanks to Judy Kelly for laboratory assistance.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Bell, S. M., B. J. Gatus, J. N. Pham, and D. L. Rafferty. 2006. Antibiotic susceptibility testing by the CDS method. [Online.] http://web.med.unsw.edu.au/cdstest. South Eastern Area Laboratory Services, Sydney, New South Wales, Australia.
- 2.British Society For Antimicrobial Chemotherapy. April 2004. Testing for dissociated resistance in staphylococci. [Online.] http://www.bsac.org.uk/_db/_documents/Testing_for_dissocit.pdf. Accessed 24 October 2006.
- 3.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests; approved standard—ninth edition. CLSI document M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 5.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen, J. H., and J. D. Turnidge. 2003. Susceptibility test methods: dilution and disk diffusion methods, p. 1111. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington DC. [Google Scholar]
- 7.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 8.Levin, T. P., B. Suh, P. Axelrod, A. L. Truant, and T. Fekete. 2005. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents Chemother. 49:1222-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Sullivan, M. V. N., Y. Cai, F. Kong, X. Zeng, and G. L. Gilbert. 2006. The influence of disk separation distance on the accuracy of the disk approximation testing for inducible clindamycin resistance in Staphylococcus spp. J. Clin. Microbiol. 44:4072-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Queenan, A. M., B. Foleno, C. Gownley, E. Wira, and K. Bush. 2004. Effects of inoculum and β-lactamase activity in AmpC- and extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae clinical isolates tested by using NCCLS ESBL methodology. J. Clin. Microbiol. 42:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayner, C., and W. J. Munckhof. 2005. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 35(Suppl. 2):S3-S16. [DOI] [PubMed] [Google Scholar]
- 12.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 13.Syriopoulou, V. P., D. W. Scheifele, C. M. Sack, and A. L. Smith. 1979. Effect of inoculum size on the susceptibility of Haemophilus influenzae b to β-lactam antibiotics. Antimicrob. Agents Chemother. 16:510-513. [DOI] [PMC free article] [PubMed] [Google Scholar]