Abstract
We analyzed IS6110-associated polymorphisms in the phospholipase C genes of 107 isolates of Mycobacterium tuberculosis selected to be representative of isolates circulating in central Russia. We found that the majority of Latin American-Mediterranean family strains contained an insertion in a unique position in the plcA gene, suggesting a common ancestor. This insertion can serve as a specific genetic marker for this group, which we designate the LAM-RUS family.
Russia is a country with a high prevalence of tuberculosis (TB), with an estimated 119 new cases per population of 100,000 in 2005 (18). Although the incidence rate has begun to decrease slowly during the past few years, the number of TB-related deaths in Russia remains substantial. It is generally accepted that different Mycobacterium tuberculosis strains have distinctive epidemiological and clinical characteristics: virulence, clinical presentation, and behavior in animals appear to be strain dependent (7). Some M. tuberculosis strains are noted for their wide dissemination and acquisition of drug resistance (4). As a result, there has been increased attention paid to M. tuberculosis strain identification recently.
Phospholipase C proteins have been shown to be involved in M. tuberculosis virulence (9). Four genes, plcA, plcB, plcC, and plcD, encode phospholipase C proteins. It has been shown previously that plc genes in clinical isolates are often interrupted with IS6110 insertion elements (15, 17). The plcD gene was previously suggested to be a hot spot for IS6110 integration; insertions into plcABC are less common (15). In this study, we analyzed the frequency of IS6110 insertions into the phospholipase C locus (plcABC genes) among clinical isolates of M. tuberculosis from central Russia.
A set of strains was selected from a collection of M. tuberculosis clinical isolates maintained at the State Research Center for Applied Microbiology and Biotechnology, Obolensk, Russia. The collection contains more than 1,200 isolates obtained during the period from 1998 to 2006 from patients in Moscow city and in the Moscow, Tula, and Kaluga regions. All strains with a unique spoligotype (accounting for 57 of a total of 72 spoligotypes) were included in the sample. Among the strains with shared spoligotypes, two to eight epidemiologically unlinked strains were chosen for analysis. The resulting panel of 107 strains should be representative of the diversity of M. tuberculosis strains circulating in the region.
IS6110 insertions into the plcABC locus were detected by measuring the sizes of six overlapping PCR amplicons that cover the entire locus. Of the 107 isolates analyzed, 47 contained an IS6110 insertion. In 45 of the 47 isolates, the IS6110 element was inserted at the same position and in the same orientation within the plcA gene (insertion type 2 [Ins2]), as determined by sequencing. One of the remaining two isolates contained an insertion at a different position in plcA (Ins1), and the other contained an insertion in the plcC gene (Ins3) (Table 1).
TABLE 1.
Prevalence of plcABC::IS6110 genotype in different phylogenetic groups
| Phylogenetic group | Total no. of isolates | No. of isolates with plcABC::IS6110 genotype (insertion type) | Position of insertion (nt)a | Adjacent sequenceb |
|---|---|---|---|---|
| PGG1/SCG2 | 10 | 0 | ||
| PGG2/SCG3 | 30 | 0 | ||
| PGG2/SCG5 | 47 | 45 (Ins2, plcA) | 2630571 | CGGGTGTG-GTGGTTTC |
| PGG3/SCG6 | 20 | 1 (Ins1, plcA) | 2630703 | GCAACGGG-GGTGGCTC |
| 1 (Ins3, plcC) | 2628680 | TTAGCCAG-CCAGGAAT |
Positions are numbered according to the M. tuberculosis H37Rv genome. nt, nucleotide.
Duplicated nucleotides are shown in bold.
The genetic relatedness of the isolates in the sample was assessed using five genotyping methods: IS6110-restriction fragment length polymorphism (16), spoligotyping (6), mycobacterial interspersed repetitive unit-variable-number tandem repeat (MIRU-VNTR) typing (14), and single-nucleotide polymorphism (SNP)-based identification of principal genetic groups (PGGs) (13) and SNP clustered groups (SCGs) (3). Genetic distance calculations and phylogenetic tree construction were performed using MathLab 6.5 statistics and bioinformatics toolboxes. The genetic characteristics of the strains are listed in Fig. S1 in the supplemental material.
The distribution of isolates with plcABC::IS6110 insertions between the PGGs and SCGs was uneven (Table 1). Both of the isolates with IS6110 Ins1 or Ins3 were members of SCG6/PGG3. All but two isolates from SCG5/PGG2 contained Ins2 in plcA.
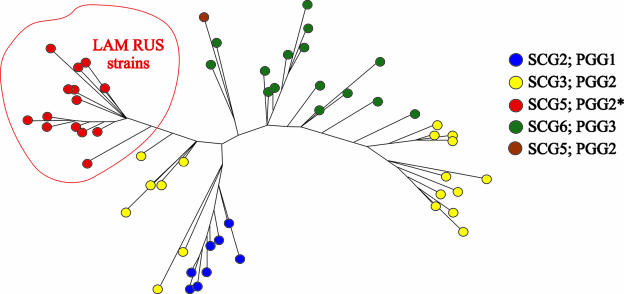
A dendrogram of the 105 unique IS6110 fingerprint patterns found in the 107 isolates was constructed (see Fig. S1 in the supplemental material). Ins2-containing strains showed highly similar IS6110 fingerprint patterns and were grouped together on the dendrogram. An unrooted phylogenetic tree of 58 MIRU-VNTR genotypes was constructed using the neighbor-joining method. The tree is shown in Fig. 1, and the groups are colored according to their SCG/PGG genotype. The Ins2-containing isolates form a compact, discrete group on the dendrogram. In addition, the SCG5 isolates with an uninterrupted plcA gene showed a high degree of similarity to some of the SCG6 strains.
FIG. 1.
Distribution of Ins2-containing strains on a phylogenetic tree based on their MIRU-VNTR patterns. PGGs and SCGs are color coded. LAM-RUS strains are circled. *, strains containing IS6110 Ins2.
The small genetic distance across the Ins2-containing group and the presence of an IS6110 element at a unique site of integration suggest that the strains are derived from a common progenitor. The combination of IS6110-restriction fragment length polymorphism, spoligotyping, and SCG/PGG genotyping methods allowed the recognition of this group as being related to the Latin American-Mediterranean (LAM) family (1). The M. tuberculosis LAM family was initially described as a clade on a neighbor-joining tree of spoligotypes (12). However, as data accumulated, the genetic heterogeneity of this family became evident and suggested the presence of a number of distinct LAM groups in various geographical regions (2, 8, 10). To reflect the high prevalence of this type of strain in central Russia as well as its epidemiological importance, we propose naming the group of Ins2-containing strains the LAM-RUS family and using Ins2 as a specific genetic marker for the group.
To identify members of the LAM-RUS family, a multiplex PCR assay was developed. In brief, two pairs of primers were used in the PCR: U (universal plcA-specific forward primer, 5′GAAGTTGATTCGCGCCCGGTT) and R (universal plcA-specific reverse primer, 5′GCTGGGAGTCCCGCGGACG) and H (hybrid forward primer specific to the IS6110 Ins2 site in the plcA gene, 5′CCAACTCAGGAAACCACACTGAACC [boldface indicates the site of insertion]) and I (reverse IS6110-specific primer, 5′CTCCGAATCGTGCTGACCGC). The amplification reaction was performed in a total volume of 30 μl containing ∼1 ng of DNA template, 200 μM (each) deoxynucleoside triphosphates, 0.3 μM (each) primers, and 2 U of Taq DNA polymerase (Fermentas, Lithuania) in the buffer recommended by the manufacturer. LAM-RUS strains produced two amplicons (163 and 410 bp), while strains without an insertion produced a product of 268 bp. Strains with other insertions and deletions in the analyzed region produced PCR products of various lengths.
Although further study is needed to determine the actual prevalence of the LAM-RUS family, the high degree of clustering of these strains, the high frequency of multidrug resistance in this family, and the increased risk of outbreaks associated with these strains have already been reported. About 45 and 30% of M. tuberculosis isolates collected in prison hospitals in Tula and Moscow regions, respectively, were members of the newly defined LAM-RUS family (5, 11). More than 80% of these isolates were multidrug resistant, and 68% of them were resistant to four or five of the drugs tested (rifampin, isoniazid, streptomycin, and ethambutol or kanamycin). Wide dissemination of these strains in two remote penitentiaries implies that we may face an outbreak of extensively drug-resistant TB in the region before long.
Molecular methods are gradually establishing their role in studies of TB epidemiology. The availability of convenient strain- and lineage-specific PCR markers simplifies strain classification, speeds up the identification of clinically relevant strains, and may give a hint about the nature of the disease before microbiological and drug resistance tests can be completed. Epidemiological studies benefit from efficient tools for unambiguous strain differentiation and tracking. We believe that the description of the specific genetic characteristics of the LAM-RUS M. tuberculosis family will contribute to the TB control programs in the region and worldwide.
Supplementary Material
Acknowledgments
This work was supported by BTEP63/ISTC2628 and CRDF2718.
We are grateful to Thomas M. Shinnick from the CDC, Atlanta, GA, and Richard O'Brien from the Foundation for Innovative New Diagnostics for critical reading of the manuscript.
Footnotes
Published ahead of print on 17 October 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowan, L. S., and J. T. Crawford. 2002. Genotype analysis of Mycobacterium tuberculosis isolates from a sentinel surveillance population. Emerg. Infect. Dis. 8:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatova, A., S. Dubiley, V. Stepanshina, and I. Shemyakin. 2006. Predominance of multi-drug-resistant LAM and Beijing family strains among Mycobacterium tuberculosis isolates recovered from prison inmates in Tula Region, Russia. J. Med. Microbiol. 55:1413-1418. [DOI] [PubMed] [Google Scholar]
- 6.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez, B., D. Aguila, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marais, B. J., T. C. Victor, A. C. Hesseling, M. Barnard, A. Jordaan, W. Brittle, H. Reuter, N. Beyers, P. D. van Helden, R. M. Warren, and H. S. Schaaf. 2006. Beijing and Haarlem genotypes are overrepresented among children with drug-resistant tuberculosis in the Western Cape Province of South Africa. J. Clin. Microbiol. 44:3539-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203-217. [DOI] [PubMed] [Google Scholar]
- 10.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 11.Shemyakin, I. G., V. N. Stepanshina, I. Y. Ivanov, M. Y. Lipin, V. A. Anisimova, A. G. Onasenko, O. V. Korobova, and T. M. Shinnick. 2004. Characterization of drug-resistant isolates of Mycobacterium tuberculosis derived from Russian inmates. Int. J. Tuberc. Lung. Dis. 8:1194-1203. [PubMed] [Google Scholar]
- 12.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 13.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talarico, S., R. Durmaz, and Z. Yang. 2005. Insertion- and deletion-associated genetic diversity of Mycobacterium tuberculosis phospholipase C-encoding genes among 106 clinical isolates from Turkey. J. Clin. Microbiol. 43:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viana-Niero, C., P. E. de Haas, D. van Soolingen, and S. C. Leao. 2004. Analysis of genetic polymorphisms affecting the four phospholipase C (plc) genes in Mycobacterium tuberculosis complex clinical isolates. Microbiology 150:967-978. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. 2007. WHO report 2007, no. WHO/HTM/TB/2007.376. Global tuberculosis control: surveillance, planning, financing, p. 133-136. World Health Organization, Geneva, Switzerland. http://www.who.int/tb/publications/global_report/2007.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.