Abstract
Chronic idiopathic colitis is a common clinical entity in young captive rhesus monkeys. Eight isolates, cultured from five monkeys in colony 1 with endemic diarrhea and three from colony 2 without diarrhea, were grown under microaerobic conditions on selective agar and were classified by full 16S rRNA sequence, biochemical, and phenotypic analysis as a novel helicobacter, “Helicobacter macacae” (proposed name). All eight strains of H. macacae had 99.5% identical 16S rRNA sequences.
Chronic idiopathic colitis in young rhesus monkeys (Macaca mulatta) is a common clinical entity in animals maintained in captivity (1). Monkeys with this disease have progressive weight loss and dehydration as a result of chronic diarrhea. The affected colons in these monkeys have severe inflammation characterized by lymphoplasmacytic infiltration, crypt epithelial hyperplasia, goblet cell depletion, the presence of multifocal crypt abscesses, and mucosal erosion and ulceration.
In a previous study, we described the presence of two novel Helicobacter spp. Helicobacter sp. strain rhesus monkey 1 was isolated from two monkeys with mild colitis (6). These animals were part of a cohort of monkeys in which chronic idiopathic colitis was endemic (1, 6). To further examine the prevalence of enteric helicobacters in this group of monkeys with endemic chronic diarrhea, as well as in a second established colony of adult macaques used in neurophysiology studies that were without diarrheal disease, we tabulated the prevalence of Helicobacter spp. by Helicobacter genus-specific PCR (5) and the specific presence of a novel (rhesus monkey 1) helicobacter by culturing colonic tissue collected at necropsy or during endoscopic biopsy (colony 1) or by culturing the feces (colony 2). Twenty-six juvenile rhesus monkeys from a colony with endemic idiopathic colitis (colony 1) were necropsied or underwent endoscopic biopsy and had their colonic tissue cultured under microaerobic conditions as previously described (6). A second group of 35 adult rhesus monkeys (colony 2) without a clinical history of endemic diarrhea had their feces cultured for Helicobacter spp.
Helicobacter genus-specific primers C97 (5′-GCT ATG ACG GGT ATC C-3′) and C05 (5′-ACT TCA CCC CAG TCG CTG-3′) were used to amplify a 1,200-base-pair PCR product from the DNA of bacterial isolates suspected based on colony morphology and Gram stain to be Helicobacter spp. (5, 6). In colony 1, Helicobacter spp. in the colonic contents of 21/26 (81%) of the monkeys were identified by colony morphology and Gram stain and confirmed by the amplification of the 1,200-base-pair Helicobacter genus-specific PCR product (5). Using the same criteria, culture results on fecal samples in colony 2 yielded helicobacter isolates in 20/35 (57%) of the animals.
Eight isolates classified as rhesus monkey type 1 by 16S rRNA analysis, five from four monkeys without diarrhea and one monkey with diarrhea from colony 1 and three from three nondiarrheic monkeys from colony 2, were subjected to a detailed biochemical characterization as previously described (11, 12). The bacteria grew under microaerobic conditions at 37°C and 42°C, but not 25°C. These Helicobacter sp. strains were oxidase and catalase positive, urease negative, grew in 1% glycine, did not reduce nitrate nor hydrolyze alkaline phosphatase or indoxyl acetate, and did not have γ-glutamyl transpeptidase activity. They were also resistant to nalidixic acid and cephalothin (Table 1). As per the previously published description, the cells had a spiral appearance, possessed bipolar, sheathed flagella, and measured approximately 0.2 μm by 2 to 3 μm (6).
TABLE 1.
Table of biochemical characteristics of enterohepatic helicobacters isolated from the intestine of humans and nonhuman primatesa
| Source | CAT | UR | Nitrate reduction | Alkaline phosphatase hydrolysis | Indoxyl acetate hydrolysis | γ-Glutamyl transpeptidase activity | Growth at 42°C | Growth with 1% glycine | Susceptibility to NA | Susceptibility to CE |
|---|---|---|---|---|---|---|---|---|---|---|
| H. macacaeb | W | − | − | − | − | − | + | + | R | R |
| Rhesus monkey no. 2 | + | − | + | − | + | − | + | W | R | S |
| Cotton-top tamarin | + | − | − | − | − | − | + | + | R | R |
| H. canis | − | − | − | + | + | + | + | − | S | I |
| H. canadensis | + | − | +/− | − | + | − | + | + | R | R |
| H. cinaedi | + | − | + | − | − | − | − | + | S | I |
| H. fennelliae | + | − | − | + | + | − | − | + | S | S |
| H. pullorum | + | − | + | − | − | ND | + | − | R | S |
| H. rappini | +/− | + | − | − | − | + | + | − | R | R |
| H. winghamensis | − | − | − | − | + | ND | − | + | R | R |
CAT, catalase production; UR, urease activity; NA, nalidixic acid (30-μg disc); CE, cephalothin (30-μg disc); ND, not determined; W, weakly positive.
The results of all tests were the same for eight of eight strains tested.
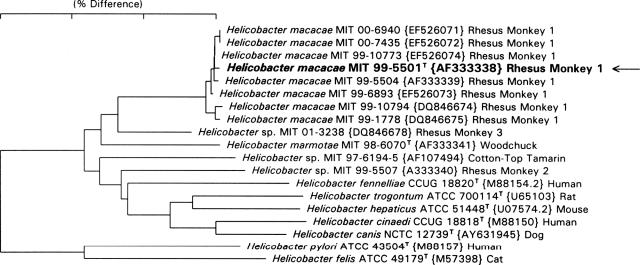
The 16S rRNA sequences for the eight novel strains listed in Fig. 1 were determined by using previously described methods (2). The 23S rRNA sequence for strain 99-5504 was determined in a previous study (3). Sequences were entered in 16S rRNA and 23S rRNA databases and aligned based on secondary structure (3, 9). Phylogenetic trees were constructed using the neighbor-joining method (10). All “Helicobacter macacae” (proposed name) strains except MIT 99-5504 contained an intervening sequence (IVS) of approximately 164 bases located in helix 188 to 219 (E. coli numbering). The phylogenetic tree, based on 16S rRNA sequence analysis, is shown in Fig. 1. Excluding the IVS helix region, the H. macacae strains were more than 99.5% identical. By 16S rRNA analysis, the H. macacae strains were closest to Helicobacter sp. strain MIT 01-3238 rhesus monkey 3 (2.4% difference); Helicobacter marmotae, isolated from woodchucks and cats (3.2%); and Helicobacter sp. strain MIT 97-6194-5 isolated from cotton-top tamarin (4.0%). The relationship of H. macacae to other helicobacters by 23S rRNA analysis was shown in a previous publication (3). By 23S rRNA analysis, H. macacae strains were most closely related to Helicobacter sp. strain CLO-3, Helicobacter canis, and Helicobacter sp. strain MIT 01-5529B that was isolated from a seal.
FIG. 1.
Neighbor-joining tree depicting phylogenetic relationships of H. macacae based on 16S rRNA analysis. Arrow indicates type strain.
Phylogenetic analyses of both 16S and 23S rRNA sequences and phenotypic analyses indicate that H. macacae strains represent a novel species. By 16S rRNA analysis, Helicobacter macacae differed from other Helicobacter species by at least 2.4%, and by 23S rRNA analysis, it differed from other species by at least 6%. As suggested in our previous paper on discordant 16S and 23S rRNA trees, the 23S rRNA analysis is generally more accurate because of the more-than-twofold-greater information content of the 23S rRNA sequences (3). The 16S rRNA IVSs of H. macacae strain MIT 99-5501 are in fact about 60% similar to those from a Helicobacter canis strain (CCUG 29176; GenBank accession number L13634) and strain MIT 01-5529B from a harp seal, representative of two taxa that are the closest relatives by 23S rRNA analysis.
As is the case in nonhuman primates, enteric Helicobacter spp. are also isolated from the feces of diarrheic humans. In selected cases, some of these species are also cultured from the blood or extraintestinal sites in patients with and without immune defects (4, 13). Enteric helicobacters have also been identified in the stools of patients with inflammatory bowel disease (14). Children with diarrhea were transiently positive for H. pylori by an ELISA of antigens in their stool samples; they not only had H. pylori DNA amplified by PCR and identified by sequence, but in addition, a significant percentage of their antigen-positive stool samples had enteric Helicobacter species identified, including, for the first time, MIT 99-5504, a strain of H. macacae (8). We also have recently cultured H. macacae from the liver and intestine of a baboon with islet cell amyloidosis, focal hepatitis, and colitis (7).
A description of H. macacae (ma.ca.cae. N.L. gen. fem. n. macacae, from Macaca, the taxonomic genus name for rhesus monkeys, from which the bacterium was isolated) follows.
Cells are slender and slightly curved (0.2 by 2 to 3 μm). The bacterium is gram negative and nonsporulating. The organism is motile, having a single, sheathed flagellum at each end. Cultures grow slowly on solid agar and appear as small, pinpoint colonies on the surface. The bacterium grows at 37° and 42°C under microaerobic but not aerobic or anaerobic conditions. The bacterium is catalase and oxidase positive but urease, alkaline phosphatase, and γ-glutamyl transpeptidase negative. The bacterium hydrolyzes indoxyl acetate and grows in 1% glycine but does not reduce nitrate to nitrite. It is resistant to both nalidixic acid and cephalothin. The type strain is MIT 99-5501. The 16S rRNA sequence accession number in GenBank for the type strain is AF333338.
Acknowledgments
We thank Hans G. Truper for providing taxonomic expertise in the proper naming of this novel helicobacter.
This work was supported by NIH grants T32 RR07036, P30 ES02109, and R01CA67529 to J.G.F.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Adler, R. R., P. F. Moor, D. L. Schmucker, and L. J. Lowenstine. 1993. Chronic colitis, juvenile Macaca mulatta, p. 81-87. In T. Jones, U. Moher, and R. Hunt (ed.), Nonhuman primates. Springer-Verlag, New York, NY.
- 2.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewhirst, F. E., Z. Shen, M. S. Scimeca, L. N. Stokes, T. Boumenna, T. Chen, B. J. Paster, and J. G. Fox. 2005. Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J. Bacteriol. 187:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox, J. G. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox, J. G., F. E. Dewhirst, Z. Shen, Y. Feng, N. S. Taylor, B. J. Paster, R. L. Ericson, C. N. Lau, P. Correa, J. C. Araya, and I. Roa. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755-763. [DOI] [PubMed] [Google Scholar]
- 6.Fox, J. G., L. Handt, S. Xu, Z. Shen, F. E. Dewhirst, B. J. Paster, C. A. Dangler, K. Lodge, S. Motzel, and H. Klein. 2001. Novel Helicobacter species isolated from rhesus monkeys with chronic idiopathic colitis. J. Med. Microbiol. 50:421-429. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, A., S. Xu, F. E. Dewhirst, P. R. Nambiar, and J. G. Fox. 2006. Enterohepatic Helicobacter species isolated from the ileum, liver and colon of a baboon with pancreatic islet amyloidosis. J. Med. Microbiol. 55:1591-1595. [DOI] [PubMed] [Google Scholar]
- 8.Haggerty, T. D., S. Perry, L. Sanchez, G. Perez-Perez, and J. Parsonnet. 2005. Significance of transiently positive enzyme-linked immunosorbent assay results in detection of Helicobacter pylori in stool samples from children. J. Clin. Microbiol. 43:2220-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 10.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 11.Saunders, K. E., Z. Shen, F. E. Dewhirst, B. J. Paster, C. A. Dangler, and J. G. Fox. 1999. Novel intestinal Helicobacter species isolated from cotton-top tamarins (Saguinus oedipus) with chronic colitis. J. Clin. Microbiol. 37:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen, Z., J. G. Fox, F. E. Dewhirst, B. J. Paster, C. J. Foltz, L. Yan, B. Shames, and L. Perry. 1997. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int. J. Syst. Bacteriol. 47:627-634. [DOI] [PubMed] [Google Scholar]
- 13.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14:59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, L., A. Day, G. McKenzie, and H. Mitchell. 2006. Nongastric Helicobacter species detected in the intestinal tract of children. J. Clin. Microbiol. 44:2276-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]