Abstract
Burkholderia pseudomallei is the etiologic agent of melioidosis. Many disease manifestations are associated with melioidosis, and the mechanisms causing this variation are unknown; genomic differences among strains offer one explanation. We compared the genome sequences of two strains of B. pseudomallei: the original reference strain K96243 from Thailand and strain MSHR305 from Australia. We identified a variable homologous region between the two strains. This region was previously identified in comparisons of the genome of B. pseudomallei strain K96243 with the genome of strain E264 from the closely related B. thailandensis. In that comparison, K96243 was shown to possess a horizontally acquired Yersinia-like fimbrial (YLF) gene cluster. Here, we show that the homologous genomic region in B. pseudomallei strain 305 is similar to that previously identified in B. thailandensis strain E264. We have named this region in B. pseudomallei strain 305 the B. thailandensis-like flagellum and chemotaxis (BTFC) gene cluster. We screened for these different genomic components across additional genome sequences and 571 B. pseudomallei DNA extracts obtained from regions of endemicity. These alternate genomic states define two distinct groups within B. pseudomallei: all strains contained either the BTFC gene cluster (group BTFC) or the YLF gene cluster (group YLF). These two groups have distinct geographic distributions: group BTFC is dominant in Australia, and group YLF is dominant in Thailand and elsewhere. In addition, clinical isolates are more likely to belong to group YLF, whereas environmental isolates are more likely to belong to group BTFC. These groups should be further characterized in an animal model.
A gram-negative, soil-dwelling bacterium, Burkholderia pseudomallei is the causative agent of melioidosis, a tropical disease that is a significant cause of mortality and morbidity for people in Southeast Asia and northern Australia (3, 26). Many disease manifestations are associated with melioidosis (27, 28), which may be due to variation in host immune response, different modes of acquisition, or genomic differences among strains (3). B. pseudomallei is also a potential biological threat agent and is classified as a category B select agent (23) in the United States. The genome of B. pseudomallei shares a high degree of similarity with the genomes of B. thailandensis and B. mallei (15, 19, 29), although the former species is nonpathogenic and is thought to occur only in Thailand, and the latter species is believed to be restricted to specific host animals and is not isolated from the environment.
Several recent studies have compared the whole-genome sequences of B. pseudomallei strains to those of sequenced strains of B. thailandensis and/or another closely related species, B. mallei (16, 19, 29). Genomic comparison of B. pseudomallei strain K96243 (15) and B. thailandensis strain E264 revealed high similarity between the two syntenic chromosomes, with most of the differences attributed to the presence of several virulence-related genes in B. pseudomallei that are absent in B. thailandensis (29). These genes include a capsular polysaccharide gene cluster and a type III secretion system. Genomic comparisons of B. pseudomallei strain K96243 (15) and B. mallei strain ATCC 23344 (20) revealed that the genome of B. pseudomallei is 1.31 Mb larger than the genome of B. mallei but that most of the homologous regions are very similar.
Several horizontal gene transfer events are believed to be the mechanisms by which B. pseudomallei obtained some of the virulence-related genes that are absent in B. thailandensis. These events include the replacement of a distinct set of capsular polysaccharide synthesis genes with the polysaccharide cluster and the horizontal acquisition of a Yersinia-like fimbrial (YLF) cluster by B. pseudomallei, resulting in the replacement of a flagellum biosynthesis cluster (16, 29). Here, we show that the acquisition of the YLF cluster is not universal for B. pseudomallei strains. Indeed, it appears to be mutually exclusive with the flagellum biosynthesis gene cluster also found in B. thailandensis. This genomic difference defines two distinct groups within B. pseudomallei populations.
MATERIALS AND METHODS
Sequencing of strain MSHR305.
The genome of strain MSHR305 (referred to herein as 305) was sequenced at the Joint Genome Institute using small (2- to 3-kb) and medium (6- to 8-kb) insert plasmid libraries. Draft assemblies were based on 13× average genome coverage. The Phred/Phrap/Consed software package was used for sequence assembly and quality assessment (13). After shotgun sequencing, reads were assembled with parallel phrap (High Performance Software, LLC). Two rounds of finishing were performed, resulting in 36 contigs and 17 scaffolds. During finishing, many possible misassemblies were corrected with Dupfinisher (14). Gaps between contigs were closed with custom primer walk reactions.
Bioinformatics.
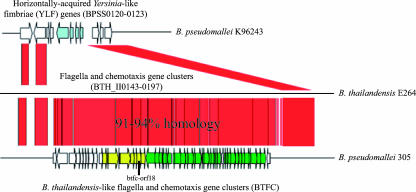
We first identified the B. thailandensis-like flagellum and chemotaxis biosynthesis (BTFC) gene cluster in the genome sequence of B. pseudomallei strain 305 when it was compared to the genome sequences of the original B. pseudomallei reference strain, K96243, and B. thailandensis strain E264 (16) (Fig. 1). These comparisons were made using the software packages BioEdit (Ibis Therapeutics) and Artemis Comparison Tool (2). Strain K96243 was originally isolated in Thailand (15), whereas strain 305 was isolated from an autopsy sample from the brain of a fatal melioidosis encephalomyelitis case at the Royal Darwin Hospital in the Northern Territory of Australia as part of the Darwin prospective melioidosis study (6).
FIG. 1.
Genomic diversity between B. pseudomallei strains K96243 and 305 compared to the related region in the genome of B. thailandensis E264. A horizontal gene transfer event caused the ancestral BTFC gene cluster to be replaced by the acquired YLF genes (BPSS0120 to -0123), as observed in strain K96243. The BTFC cluster remains in strain 305 and has 91 to 94% nucleotide similarity to B. thailandensis E264. This diverse region was used to design markers that differentiate B. pseudomallei into two distinct groups: group YLF and group BTFC. Yellow, chemotaxis protein genes; green, flagellum biosynthesis genes; blue, YLF genes; red, matched areas.
To annotate this diverse region in the genome sequence of strain 305, we used the software packages Glimmer3 (7) and GeneMark (17) to predict open reading frames (ORFs) and then aligned predicted ORFs against NCBI's “nonredundant” protein database using BLASTX. This annotation identified 55 predicted ORFs that were clustered into two main groups, including genes for flagellum biosynthesis and genes for chemotaxis biosynthesis proteins (Fig. 1). We named this region the BTFC gene cluster. Genes in the BTFC cluster were compared to other flagellum and chemotaxis genes in the B. pseudomallei genome using BLAST to determine if they were paralogs of these other genes or distinct. Sequences for predicted ORFs were extracted and checked against NCBI's nonredundant database using the BLASTN algorithm.
To determine if the BTFC cluster was present in other B. pseudomallei strains, we examined the region in nine other whole-genome sequences (Table 1). The other genome sequences were compared to the sequences for K96243 and 305 using BioEdit and the Artemis Comparison Tool. Table 1 provides information on the country and laboratory of origin for these sequenced B. pseudomallei strains, as well as the sequencing centers that produced the genome sequences.
TABLE 1.
Eleven whole-genome sequences of B. pseudomallei examined in this study
| Strain | Group | Country of origin | Laboratory of origina | Sequencing centerb | GenBank Accession no. |
|---|---|---|---|---|---|
| 305 | BTFC | Australia | MSHR | JGI | NZ_AAYX00000000 |
| 668 | BTFC | Australia | MSHR | TIGR | NC_009074 and NC_009075 |
| 1655 | BTFC | Australia | MSHR | TIGR | NZ_AAHR00000000 |
| 406e | BTFC | Thailand | WTMU | TIGR | NZ_AAMM00000000 |
| 1106a | YLF | Thailand | WTMU | TIGR | NC_009076 and NC_009078 |
| 1106b | YLF | Thailand | WTMU | TIGR | NZ_AAMB00000000 |
| 1710a | YLF | Thailand | WTMU | TIGR | NZ_AAHS00000000 |
| 1710b | YLF | Thailand | WTMU | TIGR | NC_007434 and NC_007435 |
| K96243 | YLF | Thailand | WTMU | SANG | NC_006350 and NC_006351 |
| Pasteur | YLF | Viet Nam | PAV | TIGR | NZ_AAHV00000000 |
| S13 | YLF | Singapore | UNK | TIGR | NZ_AAHW00000000 |
MSHR, Menzies School of Health Research; WTMU, Wellcome Trust Unit at Mahidol University; PAV, Pasteur Institute—Viet Nam; UNK, unknown.
JGI, Department of Energy's Joint Genome Institute; TIGR, The Institute for Genomic Research (now the J. Craig Venter Institute); SANG, Wellcome Trust Sanger Institute.
PCR analyses.
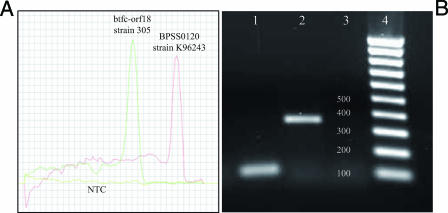
To investigate the global prevalence of BTFC and YLF gene clusters among diverse isolates of B. pseudomallei, we developed a multiplex real-time PCR assay using SYBR green as a fluorescent dye. Gene BPSS0120 (fimbrial usher protein) was utilized as a marker for the horizontal acquisition of YLF, whereas gene btfc-orf18 was utilized as a representative marker for BTFC (Fig. 1). The PCR primers that we used in this assay were as follows: BPSS0120_forward, 5′-TGA CCC ATT CAG GCA AGG GAT TCT-3′; BPSS0120_reverse, 5′-TCC GTC CTG TTC GGT GAT TTC GAT-3′; btfc-orf18_forward, 5′-GTC GAT TTC GGC TGC GAA ACA ACA-3′; and btfc-orf18_reverse, 5′-ATG CCG TCG CAA CCA TTG ATG ATG-3′. We designed primer btfc-orf18_forward so that it differed from the nucleotide sequence of B. thailandensis E264 at 8 nucleotides. The assay was conducted in 10-μl reaction mixtures containing 1× SYBR master mix (Applied Biosystems), 0.3 μM of each PCR primer, and 0.1 to 1.0 ng of DNA template. The reactions were performed on an ABI 7900HT Sequence Detection System (Applied Biosystems) utilizing 40 cycles. Each cycle contained two steps: denaturation at 95°C for 15 s and annealing at 60°C for 30 s. The PCR products were further analyzed by melting them continuously from 60°C to 95°C to generate a dissociation curve. The melting temperatures of PCR amplicons for genes btfc-orf18 and BPSS0120 were constant at 80.0°C and 88.0°C, respectively (Fig. 2A). We used this assay to analyze DNA templates from 571 diverse B. pseudomallei strains isolated from clinical and environmental situations in Australia (n = 231), Thailand (n = 310), and other countries (n = 30), as well as 77 B. thailandensis strains, 2 B. mallei strains, and 3 B. cepacia strains (see Table S1 in the supplemental material). DNA from strains K96243 and 305 were used as positive controls for genes BPSS0120 and btfc-orf18, respectively.
FIG. 2.
Multiplex SYBR green real-time PCR assay targeting genes btfc-orf18 (the BTFC gene cluster target) and BPSS0120 (the YLF gene cluster target). This assay divides B. pseudomallei into two distinct groups. Strain 305 was the positive control strain for group BTFC, whereas strain K96243 was the positive control for group YLF. (A) Derivative dissociation curves of two different PCR amplicons for gene btfc-orf18, melted at 80.0°C, and gene BPSS0120, melted at 88.0°C. NTC, no-template control. (B) PCR amplicons of 115 bp and 350 bp resolved by agarose gel electrophoresis for genes btfc-orf18 and BPSS0120, respectively. Lanes 1 to 4 contain strain 305, strain K96243, NTC, and a 100-bp DNA ladder, respectively.
To determine if the homolog of BTFC found in B. thailandensis E264 is conserved in other B. thailandensis isolates, we designed a second SYBR green real-time PCR assay to target a portion of this region that is distinct from that found in B. pseudomallei strains (Fig. 1). A modified forward primer, Bt-cheB-forward (5′-GCC GTT CTC GAC TGC AAA AAC ACG-3′), was designed and utilized, together with the above-mentioned primer btfc-orf18_reverse, to target the homolog of the BTFC found in the B. thailandensis E264 genome. With the exception of the different forward primer, the PCR reagents and conditions for this assay were the same as those described above for btfc-orf18. We screened this assay across 77 diverse B. thailandensis strains, 3 B. pseudomallei strains, 2 B. mallei strains, and 3 B. cepacia strains (see Table S1 in the supplemental material). DNA from B. thailandensis strain E264 was used as a positive control for the assay.
Analysis of isolate source data.
We used chi-square tests of association to examine the relationship between the presence of the BTFC cluster or the YLF cluster and disease events in humans. These tests were conducted using source data (i.e., clinical versus environmental) that were available for 558 of the 571 B. pseudomallei isolates utilized in this study (see Table S1 in the supplemental material). For the purposes of these analyses, the “environmental” category included isolates obtained directly from the environment (e.g., soil), as well as isolates obtained from nonhuman animals.
Nucleotide sequence accession number.
The BTFC gene cluster has been deposited in GenBank under accession no. EF377328.
RESULTS
The BTFC and YLF gene clusters are mutually exclusive in B. pseudomallei.
The genomic comparison of B. pseudomallei strains K96243 and 305 indicates that these two gene clusters are located at the same location on chromosome 2 and are mutually exclusive (Fig. 1). In addition, all other sequenced strains of B. pseudomallei possessed either the BTFC cluster or the YLF cluster at this location (Table 1). This pattern was confirmed by the results of the multiplex PCR assay. Each of 571 B. pseudomallei DNA templates produced only a single PCR amplicon when screened with the multiplex assay described above: either 350 bp for the BPSS0120 gene target in the YLF cluster or 115 bp for the btfc-orf18 target in the BTFC cluster (Fig. 2B; see Table S1 in the supplemental material). It is important to note that the genomic comparisons compared the entire gene clusters, whereas the PCR assay simply targeted a portion of each cluster.
The genomic differences define two distinct groups within B. pseudomallei.
We used these findings to differentiate B. pseudomallei into two distinct groups. Group BTFC strains contain the BTFC cluster, whereas group YLF strains contain the YLF gene cluster to the exclusion of the BTFC gene cluster. We determined the BTFC gene cluster to be the ancestral genomic state because a similar set of flagellum and chemotaxis genes was present in all of the tested B. thailandensis isolates. This is an appropriate phylogenetic outgroup; a recent rigorous phylogenetic study (12) that employed diverse strain sets for both species, an outgroup species, data from multiple genes, and unbiased markers found that B. pseudomallei and B. thailandensis share a common ancestor. The similar flagellum and chemotaxis biosynthesis gene clusters in B. thailandensis and some strains of B. pseudomallei (Fig. 1) argue that the ancestor would have possessed the flagellum and chemotaxis biosynthesis gene clusters rather than the YLF genes, which would then have been horizontally transferred and evolutionarily derived in B. pseudomallei.
Groups BTFC and YLF have distinct geographic distributions.
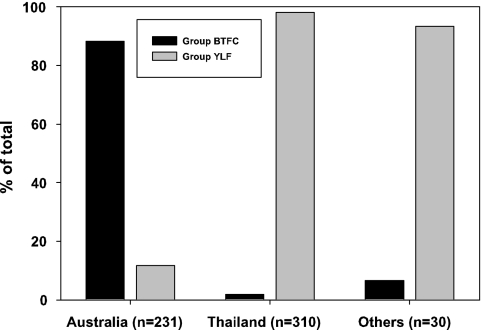
An examination of the countries of origin for the 571 B. pseudomallei strains (see Table S1 in the supplemental material) revealed differences in the geographic distributions of the two types (Fig. 3). Group BTFC is common in Australia (204/231; 88%) but rare in Thailand (6/310; 2%) and other countries (2/30; 7%; Cambodia, n = 1; Ecuador, n = 1). In contrast, group YLF was predominant among the isolates from Thailand (304/310; 98%) and other countries (28/30; 93%) but relatively rare in Australia (27/231; 12%).
FIG. 3.
Countries of origin for isolates in B. pseudomallei groups BTFC and YLF. Group BTFC strains are dominant in Australia, whereas group YLF strains are dominant in Thailand and elsewhere.
Group YLF isolates are associated with clinical settings.
The isolate source (environmental or clinical) was not independent of the group assignment (BTFC or YLF; χ2 = 13.8; df = 1; P < 0.001). A larger percentage of clinical isolates were assigned to group YLF (252 of 347; 73%) than to environmental isolates (95 of 185; 51%). As only seven BTFC isolates with source information originated outside of Australia, we also examined this association using only Australian isolates. Again, the isolate source was not independent of the group assignment (χ2 = 9.6; df = 1; P < 0.01). Although only 17% of Australian clinical isolates (23 of 139) were assigned to group YLF, just 3% of environmental isolates were assigned to this group (3 of 91).
The BTFC cluster contains distinct genes.
The genes found in the BTFC gene cluster of B. pseudomallei strain 305 are distinct from other flagellum and chemotaxis genes in the B. pseudomallei genome. A BLAST search for the individual BTFC genes on chromosome 1 yielded just two weak matches. Gene Btfc-orf20 was similar to the flagellar biosynthesis protein FlhA gene over 34% of the query sequence and with 79% maximum nucleotide identity throughout the region. Gene Btfc-orf31 was similar to the flagellar protein export ATPase FliI gene over 51% of the query sequence with 77% maximum nucleotide identity to the region. No other genes in the BTFC cluster returned matches with other genes on B. pseudomallei chromosome 1 or chromosome 2 using BLASTN.
The BTFC gene cluster is conserved within B. pseudomallei and is similar but distinct from the homologous region in B. thailandensis.
There is some genomic variability between the BTFC gene cluster in B. pseudomallei and the homologous flagellum and chemotaxis genes of B. thailandensis E264. Genomic comparisons revealed approximately 91 to 94% nucleotide similarity between the two species in this region (Fig. 1). In contrast, within B. pseudomallei, the nucleotide sequence of the BTFC region was highly conserved, with as much as 99% similarity among the genome sequences of strains 406e, 1655, and 668 (data not shown). All 77 of the tested B. thailandensis DNA templates failed to amplify with the multiplex assay targeting the btfc-orf18 (BTFC target) and BPSS0120 (YLF target) genes in B. pseudomallei. However, all of these templates produced a 115-bp amplicon when screened with the second assay targeting the cheB gene in the flagellum and chemotaxis gene cluster of B. thailandensis; no PCR amplicons were produced for the three B. pseudomallei strains that were screened with this assay. These findings indicate that at least this region of the flagellum and chemotaxis gene cluster is conserved across diverse B. thailandensis isolates (see Table S1 in the supplemental material) and is distinct from the BTFC found in B. pseudomallei strains. The two B. mallei and three B. cepacia strains did not produce an amplicon with either the multiplex assay targeting the YLF and BTFC clusters in B. pseudomallei or the singleplex assay targeting the flagellum and chemotaxis gene cluster in B. thailandensis (see Table S1 in the supplemental material). This finding was expected, as we did not find either gene cluster in the available genome sequences for B. mallei (strain ATCC2344, accession numbers CP000010 and CP000011; strain NCTC10229, CP000545 and CP000546; strain NTCT10247, CP000547 and CP000548; and strain SAVP1, CP000525 and CP000526) and B. cepacia (strain AMMD, accession numbers CP000440, CP000441, CP000442, and CP000443) (data not shown).
DISCUSSION
The BTFC and YLF gene clusters are mutually exclusive in the genome of B. pseudomallei, and their relative frequencies vary among regional populations. BTFC is the ancestral state and is the predominant state found in B. pseudomallei strains in Australia, although group YLF strains are also found there (Fig. 3). However, outside of Australia, group YLF strains are clearly dominant and group BTFC strains, although present, are quite rare. These patterns suggest that a single horizontal-transfer event may have been responsible for replacing the BTFC cluster with the YLF cluster and that this event may have taken place in Australia, with the YLF group subsequently becoming the dominant group in Southeast Asia. Why group YLF is dominant in Southeast Asia is unclear. It may be due to a founder's effect (i.e., B. pseudomallei populations in Southeast Asia originated from group YLF isolates that dispersed from Australia). A previous study found patterns consistent with this scenario, as multilocus sequence typing has shown separation between Australian and Thai strains of B. pseudomallei (4). In that previous study, it was suggested that B. pseudomallei may possibly have originated in Australia and been propagated through animal migration during the Miocene Period around 15 million years ago, when a land bridge joined the Australia-New Guinea continent and Southeast Asia. Another study (18) refuted the distinctiveness of Australian and Thai populations of B. pseudomallei, but this claim is based upon a single sequence type that is shared between the two populations, which might be expected if one population founded the other or because of travel between the two countries. Alternatively, group YLF may be better adapted to environmental conditions in Southeast Asia with its high frequency due to selective forces. The latter hypothesis will be addressed in future studies.
If B. pseudomallei evolved in Australia, as our data suggest, then why are there no similar species present in that country that also possess the ancestral BTFC gene cluster? B. thailandensis is present in Thailand and possesses the BTFC gene cluster, which suggests that a common ancestor of the two species is present in that country currently or was at some time in the past. To date, no closely related species possessing the BTFC gene cluster has been isolated in Australia. However, this does not mean that such a species does not exist in Australia. Indeed, B. thailandensis was first described only in 1998 (1). It is possible that the most recent ancestor of B. pseudomallei in Australia is extinct or undiscovered (i.e., B. thailandensis may not be the most recent common ancestor of B. pseudomallei, just the closest relative we currently know about).
B. pseudomallei strains containing the YLF gene cluster are more commonly observed in clinical cases than might be expected. Across the 571 B. pseudomallei strains that we examined, group YLF isolates were more likely to be isolated in clinical settings than in environmental settings. This pattern could simply be due to the geographical differences described above, as 222 of the 571 isolates that we examined in this study were from clinical settings in Thailand and YLF is the dominant group in that country. However, this pattern remains consistent in Australia, where both groups are found. An alternative explanation is that group YLF strains are more virulent than group BTFC strains. In Thailand, where group YLF is dominant (Fig. 3), mortality rates for primary disease are higher (50%) than in melioidosis regions of endimicity in Australia (15 to 20%) (5, 26). Although these differences have been attributed to more advanced medical technology available in Australia (27), they may also be due in part to the dominance of group YLF in Thailand. Cheng and Currie (3) previously suggested that genomic differences may account for differences in disease manifestation and therefore patient outcomes. To determine if group YLF is truly more virulent than group BTFC, we plan to examine differences between the two groups in an animal model in the near future. It is important to note that group BTFC isolates are clearly capable of causing disease. As noted above, strain 305, which was assigned to group BTFC, caused a fatal case of melioidosis encephalomyelitis.
Other studies of genomic diversity among strains of B. pseudomallei have been performed using suppression subtractive hybridization (8, 10) and comparative genomic hybridization (16, 21). These studies demonstrated variability in genomic islands and prophages but did not analyze these differences across populations or geographic regions. In this study, we have identified genomic differences that are highly correlated with specific geographic regions, findings that are significant and provide potential insights into the evolutionary past of B. pseudomallei. Clearly, other genomic differences need to be analyzed across large strain panels to understand their potential importance to the biology of this pathogen.
Our multiplex PCR assay targeting the BTFC and YLF gene clusters of B. pseudomallei can be used to definitively identify this species and distinguish it from closely related species. All of the B. pseudomallei isolates we screened produced an amplicon with this assay, whereas none of the B. mallei, B. thailandensis, or B. cepacia isolates produced an amplicon (see Table S1 in the supplemental material). Although we describe a real-time PCR approach above, this assay can be easily converted to be utilized in a more traditional gel-based approach (Fig. 2B). This, coupled with the relatively low cost of the SYBER green reporter, will allow this assay to be utilized in a variety of research laboratories. A number of other PCR-based assays have been produced for the identification of B. pseudomallei (9, 11, 22, 24, 25). Our assay provides another means of identifying this important public health threat and potential agent of bioterrorism while at the same time providing assignment to either group YLF or group BTFC.
Acknowledgments
Partial funding for this study was provided by grants to D.M.W. and P.K. from the Pacific Southwest Regional Center of Excellence (PSWRCE). S.J.P. was supported by a Wellcome Trust Training Fellowship in Clinical Tropical Medicine. Funding for the sequencing of strain 305 was provided by the Intelligence Technology Innovation Center.
We thank Mark Mayo and Daniel Gal (Menzies School of Health Research, Darwin, Australia), Richard Robison (Brigham Young University, Utah), Jonathan Warawa (NIAID-NIH Rocky Mountain Laboratories, Montana), and Jay Gee and Mindy Glass (Centers for Disease Control and Prevention, Atlanta, Georgia) for providing some of the DNAs used in this study.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 2.Carver, T. J., K. M. Rutherford, M. Berriman, M.-A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, A. C., D. Godoy, M. Mayo, D. Gal, B. G. Spratt, and B. J. Currie. 2004. Isolates of Burkholderia pseudomallei from northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 42:5477-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, A. C., J. N. Hanna, R. Norton, S. L. Hills, J. Davis, V. L. Krause, G. Dowse, T. J. Inglis, and B. J. Currie. 2003. Melioidosis in northern Australia, 2001-02. Commun. Dis. Intell. 27:272. [DOI] [PubMed] [Google Scholar]
- 6.Currie, B. J., D. A. Fisher, D. M. Howard, J. N. Burrow, D. Lo, S. Selva-Nayagam, N. M. Anstey, S. E. Huffam, P. L. Snelling, P. J. Marks, D. P. Stephens, G. D. Lum, S. P. Jacups, and V. L. Krause. 2000. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin. Infect. Dis. 31:981-986. [DOI] [PubMed] [Google Scholar]
- 7.Delcher, A. L., K. A. Bratke, E. C. Powers, and S. L. Salzberg. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeShazer, D. 2004. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 186:3938-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dharakul, T., S. Songsivilai, S. Viriyachitra, V. Luangwedchakarn, B. Tassaneetritap, and W. Chaowagul. 1996. Detection of Burkholderia pseudomallei DNA in patients with septicemic melioidosis. J. Clin. Microbiol. 34:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duangsonk, K., D. Gal, M. Mayo, C. A. Hart, B. J. Currie, and C. Winstanley. 2006. Use of a variable amplicon typing scheme reveals considerable variation in the accessory genomes of isolates of Burkholderia pseudomallei. J. Clin. Microbiol. 44:1323-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gal, D., M. Mayo, E. Spencer, A. C. Cheng, and B. J. Currie. 2005. Application of a polymerase chain reaction to detect Burkholderia pseudomallei in clinical specimens from patients with suspected melioidosis. Am. J. Trop. Med. Hyg. 73:1162-1164. [PubMed] [Google Scholar]
- 12.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 14.Han, C. S., and P. Chain. 2006. Finishing repeat regions automatically with Dupfinisher, p. 141-146. In H. R. Arabnia and H. Valafar (ed.), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology. CSREA Press, Las Vegas, NV.
- 15.Holden, M. T. G., R. W. Titball, S. J. Peacock, A. M. Cerdeno-Tarraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thomson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davis, D. DeShazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Barrell, P. C. F. Oyston, and J. Parkhill. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. S., M. Schell, Y. Yu, R. Ulrich, S. Sarria, W. Nierman, and D. DeShazer. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCombie, R. L., R. A. Finkelstein, and D. E. Woods. 2006. Multilocus sequence typing of historical Burkholderia pseudomallei isolates collected in Southeast Asia from 1964 to 1967 provides insight into the epidemiology of melioidosis. J. Clin. Microbiol. 44:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monastyrskaya, G., A. Fushan, I. Abaev, O. Filyukova, M. Kostina, E. Pecherskih, and E. Sverdlov. 2004. Genome-wide comparison reveals great inter- and intraspecies variability in B. pseudomallei and B. mallei pathogens. Res. Microbiol. 155:781-793. [DOI] [PubMed] [Google Scholar]
- 20.Nierman, W. C., D. DeShazer, H. S. Kim, H. Tettelin, K. E. Nelson, T. Feldblyum, R. L. Ulrich, C. M. Ronning, L. M. Brinkac, S. C. Daugherty, T. D. Davidsen, R. T. Deboy, G. Dimitrov, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, H. Khouri, J. F. Kolonay, R. Madupu, Y. Mohammoud, W. C. Nelson, D. Radune, C. M. Romero, S. Sarria, J. Selengut, C. Shamblin, S. A. Sullivan, O. White, Y. Yu, N. Zafar, L. Zhou, and C. M. Fraser. 2004. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA 101:14246-14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou, K., C. Ong, S. Y. Koh, F. Rodrigues, S. H. Sim, D. Wong, C. H. Ooi, K. C. Ng, H. Jikuya, C. C. Yau, S. Y. Soon, D. Kesuma, M. A. Lee, and P. Tan. 2005. Integrative genomic, transcriptional, and proteomic diversity in natural isolates of the human pathogen Burkholderia pseudomallei. J. Bacteriol. 187:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattanathongkom, A., R. W. Sermswan, and S. Wongratanacheewin. 1997. Detection of Burkholderia pseudomallei in blood samples using polymerase chain reaction. Mol. Cell. Probes 11:25-31. [DOI] [PubMed] [Google Scholar]
- 23.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomaso, H., T. L. Pitt, O. Landt, S. A. Dahouk, H. C. Scholz, E. C. Reisinger, L. D. Sprague, I. Rathmann, and H. Neubauer. 2005. Rapid presumptive identification of Burkholderia pseudomallei with real-time PCR assays using fluorescent hybridization probes. Mol. Cell. Probes 19:9-20. [DOI] [PubMed] [Google Scholar]
- 25.U'Ren, J. M., M. N. Van Ert, J. M. Schupp, W. R. Easterday, T. S. Simonson, R. T. Okinaka, T. Pearson, and P. Keim. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 43:5771-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, N. J. 2003. Melioidosis. Lancet 361:1715. [DOI] [PubMed] [Google Scholar]
- 27.Wiersinga, W. J., T. van der Poll, N. J. White, N. P. Day, and S. J. Peacock. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4:272. [DOI] [PubMed] [Google Scholar]
- 28.Yee, K. C., M. K. Lee, C. T. Chua, and S. D. Puthucheary. 1988. Melioidosis, the great mimicker: a report of 10 cases from Malaysia. J. Trop. Med. Hyg. 91:249-254. [PubMed] [Google Scholar]
- 29.Yu, Y., H. S. Kim, H. Chua, C. Lin, S. Sim, D. Lin, A. Derr, R. Engels, D. DeShazer, B. Birren, W. Nierman, and P. Tan. 2006. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]