Abstract
The Escherichia coli adhesin involved in diffuse adherence (AIDA-I) is one of the few glycosylated proteins found in Escherichia coli. Glycosylation is mediated by a specific heptosyltransferase encoded by the aah gene, but little is known about the role of this modification and the mechanism involved. In this study, we identified several peptides of AIDA-I modified by the addition of heptoses by use of mass spectrometry and N-terminal sequencing of proteolytic fragments of AIDA-I. One threonine and 15 serine residues were identified as bearing heptoses, thus demonstrating for the first time that AIDA-I is O-glycosylated. We observed that unglycosylated AIDA-I is expressed in smaller amounts than its glycosylated counterpart and shows extensive signs of degradation upon heat extraction. We also observed that unglycosylated AIDA-I is more sensitive to proteases and induces important extracytoplasmic stress. Lastly, as was previously shown, we noted that glycosylation is required for AIDA-I to mediate adhesion to cultured epithelial cells, but purified mature AIDA-I fused to GST was found to bind in vitro to cells whether or not it was glycosylated. Taken together, our results suggest that glycosylation is required to ensure a normal conformation of AIDA-I and may be only indirectly necessary for its cell-binding function.
Protein glycosylation is widespread in prokaryotes, with more than 70 bacterial glycoproteins reported so far (38). Most of these are surface or secreted proteins that affect how bacteria interact with their environment, for instance, by influencing cell-cell interactions, surface adhesion, or evasion of immune response (34, 38).
In several bacterial species, complex O- and N-glycosylation pathways are encoded by multiple genes clustered in “glycosylation islands” (38, 39). In other cases, highly specific single glycosyltransferases are responsible for the modification of target proteins (2, 20, 35). However, in most cases the exact mechanism of glycosylation remains to be elucidated. How glycosylation exerts its role is also unclear. The addition of sugar moieties can define (9, 18) or mask (21, 31) interaction sites. Carbohydrates have also been shown to influence the stability (7) and protease sensitivity (14) of individual proteins or macromolecular assembly of several polypeptides (21, 33).
Only a few glycoproteins have been identified in Escherichia coli. Among them are the adhesin involved in diffuse adherence (AIDA-I) (2), the TibA adhesion-invasion protein of enterotoxigenic E. coli (20), and the autoaggregation factor antigen 43 (Ag43) (35). Specific glycosyltransferases of AIDA-I and TibA have been identified, but, by contrast, no Ag43-specific glycosyltransferases are known, and Ag43 glycosylation was performed heterologously with the AIDA-I or TibA-specific enzymes. These glycoproteins present several similarities: all three (i) are secreted as autotransporters, which represent a branch of the type V secretion pathway; (ii) have nearly identical N-terminal 19-amino-acid repeats; (iii) are glycosylated by the addition of heptoses mediated by single glycosyltransferases that are functionally interchangeable; and (iv) are versatile virulence factors mediating bacterial autoaggregation and biofilm formation as well as adhesion to and invasion of mammalian cells. Because of these similarities, AIDA-I, TibA, and Ag43 have been named self-associating autotransporters (SAAT) (17).
AIDA-I was originally identified as a plasmid-encoded protein able to confer a pattern of diffuse adherence on the surface of cultured epithelial cells (1). It is associated with a high percentage of the pathogenic E. coli strains involved in neonatal and postweaning diarrhea in piglets, which causes major economic losses in farms worldwide (10, 11, 22, 26, 27). AIDA-I is synthesized as a 132-kDa preproprotein (37). A cleavable N-terminal signal sequence of 49 amino acids allows secretion across the inner membrane via the general sec secretion machinery. After crossing the periplasm, the proprotein is inserted in the outer membrane and is cleaved, probably by an autocatalytic mechanism (3, 37). The cleavage separates an N-terminal extracellular fragment, the mature AIDA-I adhesin, from a C-terminal membrane-embedded fragment, AIDAc. Despite the cleavage, the mature adhesin and AIDAc remain strongly associated at the bacterial surface (3, 37).
The autotransporter adhesin heptosyltransferase (Aah) is responsible for the glycosylation of AIDA-I with heptoses. Aah uses precursors recruited from the lipopolysaccharide biosynthetic pathway, but the modified residues have not been identified (2). The glycosylation of AIDA-I has been shown to be essential for adhesion, since deletion of the aah gene abolishes adherence to cultured epithelial cells, but dispensable for autoaggregation and biofilm formation (2, 36). These observations suggested that heptose residues are involved in the interaction between AIDA-I and a receptor on the surface of epithelial cells. There is, however, no formal proof for this hypothesis, and glycosylation could also affect the conformation or the stability of the protein. It has been shown, for instance, that glycosylation is required for the stability of the HMW1 adhesin of Haemophilus influenzae and its tethering to the bacterial surface (7).
In the present study, we characterized the glycosylation of AIDA-I. We used mass spectrometry (MS) and N-terminal sequencing of AIDA-I peptides to show that the heptoses are O-linked to the 15 serine and 1 threonine residues. We also show that glycosylation provides increased resistance to degradation and does not alter the binding efficiency of purified mature AIDA-I. Our results therefore strongly suggest that glycosylation is mainly necessary for the stability of the protein at the cell surface rather than being required to engage a cellular receptor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli K12 strain C600 (thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21; obtained from New England Biolabs) and the pathogenic strain 2787 (1) were used in this study. Plasmid pAg containing the whole aidA operon (aah and aidA) under the control of ptrc, a promoter inducible with isopropyl-β-d-thiogalactopyranoside (IPTG), has been described before (3). pAg allows the expression of glycosylated AIDA-I. Plasmid pAgH, also previously described (3), is derived from pAg and allows expression of glycosylated AIDA-I tagged at the N terminus of the proprotein with six histidine amino acids and a glycine (HisG). To construct plasmid pAng, which allows the expression of nonglycosylated AIDA-I, we amplified by PCR the aidA gene from E. coli strain 2787 by use of primers introducing NcoI and XbaI restriction sites. The resulting fragment was cloned in the vector pTRC99a (Pharmacia Biotech), resulting in plasmid pAng, which bears the aidA gene alone under the control of the ptrc promoter. Plasmid pAngH, allowing the expression of the HisG-tagged AIDA-I, was obtained by introducing an oligonucleotide coding for the HisG tag in the pAng plasmid by site-directed mutagenesis using mutagenic primers, as described previously for pAg (3). To construct the plasmid pAah, we subcloned the aah gene with its ptrc promoter from the pTRC-Aah plasmid described before (3a) into the plasmid pACYC184 (Pharmacia Biotech) by use of SphI and EcoRI restriction sites. The pAah plasmid has an origin of replication compatible with those of pAng and pAngH. A plasmid allowing the expression of a fusion between glutathione S-transferase (GST) and mature AIDA (encompassing residues 50 to 847) was constructed by PCR amplification and cloning into the pGex-4T-1 vector (Amersham Biosciences), as described elsewhere (M.-È. Charbonneau and M. Mourez, unpublished data). All constructions were verified by restriction mapping and sequencing.
Bacterial and cell culture growth conditions.
Bacteria containing the different plasmids were grown on Luria-Bertani (LB) agar plate or in liquid LB (rich) or M9 (minimal) media containing 100 μg ml−1 of ampicillin and, in addition, 50 μg ml−1 chloramphenicol when the plasmid pAah was used. Bacterial cultures were grown at 30°C and induced overnight with 10 μM IPTG when they reached an optical density at 600 nm (OD600) of 0.8 unless indicated otherwise. Strain 2787 was grown on I-medium (1) at 37°C. Hep-2 cells (ATCC CCL-23) were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle medium (Gibco) containing 10 mM sodium pyruvate (Sigma), bovine growth serum (HyClone), 2.5 μg ml−1 of fungizone, and 100 μg ml−1 of penicillin-streptomycin (Gibco).
SDS-PAGE and immunoblotting.
Protein-containing samples were diluted in twice-concentrated sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing β-mercaptoethanol and denatured by heating at 100°C for 10 min. The samples were separated by SDS-PAGE on 10% acrylamide gels. The gels were either stained with Coomassie blue or transferred to polyvinylidene fluoride membranes (Millipore). Immunodetection was performed with an anti-HisG horseradish peroxidase (HRP)-coupled antibody (Invitrogen) diluted 1:5,000 in blocking buffer (5% skim milk, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Triton X-100). Immune complexes were revealed using a 3,3′,5,5′-tetramethylbenzidine solution for membranes (Sigma). For quantitative comparison the membranes were digitally scanned using ImageJ software (National Institutes of Health).
Whole-cell and membrane extract preparations.
Overnight cultures from E. coli C600 harboring pTRC99A, pAngH (nonglycosylated AIDA-I), or pAngH plus pAah (glycosylated AIDA-I) normalized to the same OD600. To obtain whole-cell extracts, the cultures were normalized and pelleted and the bacteria were resuspended in phosphate-buffered saline (PBS). To obtain membrane fractions, the cultures were lysed and fractionated as described previously (3).
Heat extraction and purification of mature or whole AIDA-I.
Heat extraction was performed as described before (3). Briefly, overnight cultures of C600 harboring empty vector, pAngH, or pAngH plus pAah were normalized and bacteria were harvested, resuspended in 10 mM sodium phosphate buffer (pH 7), and heated at 60°C for 20 min. The treated samples were centrifuged for 5 min at 12,000 × g to recover the heat extracts. The HisG-tagged mature AIDA-I was purified from heat extracts, and the HisG-tagged whole AIDA-I was purified from solubilized membrane extracts as described before (3).
Protease accessibility assay.
Normalized cultures of strain C600 bearing empty vector, pAngH, or pAngH plus pAah were pelleted and resuspended in PBS in the presence or absence of trypsin (Sigma) or proteinase K (Invitrogen) at a final concentration of 0.3 μg ml−1 or 3 μg ml−1. After 30 min of incubation on ice, the proteases were neutralized by addition of proteases inhibitor cocktail (Complete Mini; Roche) for 5 min. The samples were pelleted, and whole-cell extracts were obtained as described above.
Purification of the GST fusion proteins.
One liter of E. coli BL21 harboring pGex-4T-1 or pGex-AIDA plasmid with or without pAah was grown until an OD of 0.4 was obtained and was induced with 10 μM IPTG for 3 h. Bacteria were harvested and resuspended in 40 ml of Tris-buffered saline (50 mM Tris-HCl [pH 8], 150 mM NaCl) containing lysozyme (0.4 mg ml−1 final concentration) and EDTA (pH 8; 10 mM final concentration) and lysed with a French press and an ultrasonic processor. The soluble fraction was isolated by 30 min of centrifugation at 16,000 × g. The GST and the GST-AIDA fusion proteins were purified using an ÄKTA purifier system with a 5-ml glutathione Sepharose column (Amersham Biosciences) according to the instructions of the manufacturer. The purity of the purified proteins was confirmed by SDS-PAGE and staining with Coomassie blue. Glycan detection was performed using a digoxigenin glycan detection kit (Roche) according to the instructions of the manufacturer.
Cell enzyme-linked immunosorbent assay.
Hep-2 cells were grown in 96-well plates and fixed for 15 min with PBS containing final concentrations of 2.5% paraformaldehyde and 0.2% glutaraldehyde. After one wash with PBS, the plate was blocked with PBS-bovine serum albumin (3%) for 2 h at 37°C. Purified proteins (GST, a fusion of GST to mature AIDA-I, or whole AIDA-I) were added to the cells at concentrations between 30 nM and 1 μM and incubated overnight at 4°C. After extensive washes with PBS, bound proteins were detected with an anti-GST antibody coupled to HRP diluted 1:10,000 in PBS (Amersham Biosciences) or with a custom polyclonal rabbit anti-AIDA serum (QCB, Hopkinton, MA) and a secondary goat anti-rabbit antibody coupled to HRP. Immune complexes were revealed using a 3,3′,5,5′-tetramethylbenzidine solution for an enzyme-linked immunosorbent assay (Sigma). Background values were subtracted, and the absorption intensities were normalized by dividing the value of intensity determined for each well by the maximum intensity value measured on the plate. Experiments were conducted in duplicate at least twice. Binding curve and dissociation constant data were obtained by nonlinear regression fitting to a one-binding-site hyperbola by use of Prism 4.0 (GraphPad software).
Functional assays.
All assays were performed as described before (3). Briefly, the autoaggregation assay was performed using overnight cultures of C600 harboring empty vector, pAng, or pAg or cultures of strain 2787. All cultures were normalized, vortexed for 10 s, and left at 4°C. The OD600 was measured at the top of the culture at the beginning of the assay and after 3 h, and pictures were taken after overnight incubation. For biofilm formation, normalized cultures of C600 harboring empty vector, pAng, or pAg were grown without agitation for 24 h at 30°C in minimal medium in 96-well polyvinyl chloride plates (Falcon). Biofilms were stained for 15 min with 1% crystal violet, and the fixed dye was solubilized by the addition of ethanol-acetone (80:20). The absorbance of the dye solution was measured at 595 nm. For unknown reasons strain 2787 grows extremely slowly in minimal medium, and AIDA-I expression is minimal in those conditions; the biofilm assay was therefore not performed with this strain. The adhesion assay was performed with Hep-2 cells grown in 24-well plates and inoculated with 106 CFU per well of C600 harboring empty vector, pAng, or pAg. Cells were washed with PBS. Bacteria adhering to cells were recovered with 100 μl of 1% Triton X-100 and plated for numbering. Adherence was calculated by dividing the number of adherent bacteria by the number of bacteria found in the inoculum after 3 h of incubation.
β-Galactosidase reporter assay.
The β-galactosidase activity of strains SR1458 (30) and SR1364 (25) transformed with an empty vector, plasmid pAng, or plasmid pAg was assessed as described previously (25), and the results were expressed in Miller units. Statistical comparisons were performed by analysis of variance using Prism 4.0 (GraphPad software).
Mass spectrometry.
Glycosylated AIDA-I was purified from heat extracts as described above and run on an SDS-PAGE 10% acrylamide gel. The protein band corresponding to AIDA-I was cut from the gel and destained with water-sodium bicarbonate buffer and acetonitrile. The protein was reduced with dithiothreitol and alkylated with iodoacetamide prior to in-gel digestion with trypsin or chymotrypsin. The tryptic peptides were eluted from the gel with acetonitrile containing 0.1% trifluoroacetic acid. The tryptic peptides were then separated on an Agilent Nanopump system using a C18 ZORBAX trap and a SB-C18 ZORBAX 300 reversed-phase column (Agilent Technologies, Inc.) (150 mm by 75 μm; 3.5 μm particle size). All mass spectra were recorded on a hybrid linear ion trap-triple quadrupole mass spectrometer (Q-Trap; AB Applied Biosystems) equipped with a nanoelectrospray ionization source. The accumulation of MS-MS data was performed with Analyst software, version 1.4 (AB Applied Biosystems). MASCOT software (Matrix Science, London, United Kingdom) was used to create peak lists from MS and MS-MS raw data.
N-terminal sequencing.
The mature AIDA-I protein was isolated from an SDS gel stained with Coomassie blue as described above. The band corresponding to mature AIDA-I was cut and transferred in a 1.5 ml screw-cap microcentrifuge tube, reduced, alkylated, and digested with trypsin (Promega sequencing grade) as described previously (12). The peptides were extracted with 60% acetonitrile-1% trifluoroacetic acid at 60°C and separated using reverse-phase high-pressure liquid chromatography, a Vydac microbore C18 column (1 mm internal diameter by 50 mm), and an Applied Biosystems 130A separation system. The peptides were detected by absorbance at 220 nm, and fraction peak data were collected manually. Each fraction was applied to a precycled glass fiber filter treated with trifluoroacetic acid and coated with Biobrene Plus (0.5 mg of Polybrene and 0.03 mg of NaCl). The fractions were subjected to automatic Edman degradation (ED) using a model 494 CLC Procise sequencer and a general protocol (15). The phenylthiohydantoin amino acid derivatives were analyzed on line using a capillary separation system (Applied Biosystems model 140 D) and a UV detector (Applied Biosystems model 785A) set at 269 nm.
RESULTS
AIDA-I is O-glycosylated.
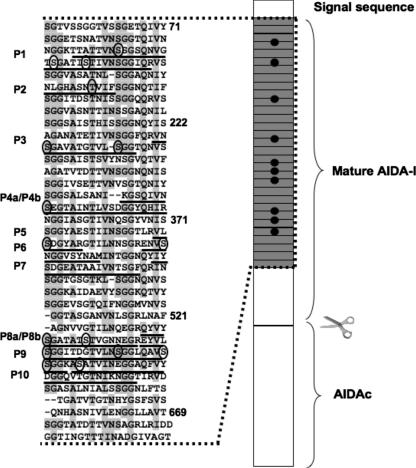
We purified glycosylated mature AIDA-I (corresponding to amino acids 50 to 847 in the preproprotein) by affinity chromatography from heat extracts of the E. coli strain C600 expressing a histidine-tagged protein as described before (3). After SDS-PAGE, the band corresponding to purified mature glycosylated AIDA-I was digested in the gel by trypsin or chymotrypsin and the resulting peptides were submitted to MS-MS with collision-induced dissociation (CID). Some of the parent ions were identified as AIDA-I peptides presenting between one and four incremental additions of 192 Da compared with the theoretical mass of the unsubstituted peptides. This 192-Da addition is consistent with the grafting of a heptose molecule. Thus, we concluded that these peptides were glycosylated. In most cases we could compare the MS spectra of modified and unmodified peptides. With this MS approach, nine peptides were found to be glycosylated with one to four heptose residues whereas five peptides were found not to be glycosylated (Fig. 1 and Table 1). In two instances, there was extensive overlap between two glycosylated peptides; therefore, we in fact identified seven different glycosylated peptides by use of MS. We noted that the glycosylation was heterogeneous, as five out of the nine peptides were found without carbohydrate or with various amounts of heptose residues. When multiple glycosylations were observed, it was not possible to determine whether the observation was the result of multiple sites being glycosylated or the result of a single site being glycosylated by multiple heptose residues. Similarly, we could not ascertain the conformation of the grafted heptose. The glycosylated peptides are found at several different positions in mature AIDA-I but exclusively in a region containing 35 imperfect repeats of a 19-amino-acid sequence (Fig. 1). In most cases, CID of peptides does not allow the identification of the amino acid residue bearing the carbohydrate since the carbohydrate-polypeptide bond is usually preferentially cleaved compared to the peptide bond. However, in one instance, the CID spectrum of a chymotryptic peptide showed peptide fragment ions still bearing the carbohydrate moiety (data not shown). Analysis of these fragment ions revealed that the threonine at position 154 was modified with one heptose.
FIG. 1.
Localization of heptose modifications. The sequences of the glycosylated peptides identified in this study are underlined on a schematic representation of the preproprotein showing the N-terminal signal sequence required for inner membrane translocation, a region encompassing 35 imperfect 19-amino-acid repeats (gray boxes) and the autoproteolytic cleavage site separating mature AIDA-I from the membrane-embedded AIDAc. The shaded residues correspond to the consensus sequence of the imperfect repeats. Numbering corresponds to the positions of the amino acids in the preproprotein. In several peptides, a heptose residue could be identified on threonine or serine residues. These residues are circled in the sequence.
TABLE 1.
Glycosylation of peptides identified in AIDA-Ia
| Peptide (designation) | No. of heptoses | Means of identification |
|---|---|---|
| Glycosylated peptides | ||
| K↓T95TATTVNSSGSQNVGTSGATISTIVNSGGIQR126↓ (P1) | 2, 3, or 4 | MS, ED |
| Y↓N147LGHASNTVIF157↓ (P2) | 0 or 1 | MS |
| R↓V240NSGAVATGTVLSGG- (P3) | NDb | ED |
| K↓G328SQIVNSEGTAINTLVSDGGYQHIR352↓ (P4a) | 0, 1, or 2 | MS, ED |
| L↓S323ANIKGSQIVNSEGTAINTLVSDGGY348↓ (P4b) | 2 | MS |
| R↓V389LSDGYAR396↓ (P5) | ND | ED |
| R↓E406NVSNGGVSYNAM- (P6) | ND | ED |
| Y↓I427YSDGEATAAIVNTSGF443↓ (P7) | 1 | MS |
| R↓Q536YVYSGATATSTVGNNEGR554↓ (P8a) | 0, 1, or 2 | MS, ED |
| Y↓V538YSGATATSTVGNNEGREY556↓ (P8b) | 1, 2 | MS |
| R↓E555YVLSGGITDGTVLNSGGLQAVSSGGK581↓ (P9) | 4 | MS, ED |
| K↓A582SATVINEGGAQFVYDGGQVTGTNIK607↓ (P10) | 0 or 1 | MS, ED |
| Unglycosylated peptides | ||
| Y↓Q349HIRNGGIASGTIVNQSGY367↓ | MS | |
| R↓G397TILNNSGR405↓ | ED | |
| K↓A470IDAEVYSGGK480↓ | ED | |
| Y↓S485GGEVSGTQIF495↓ | MS | |
| R↓L518NAFAGNVVGTILNQEGR535↓ | MS, ED | |
| K↓D739NTGIMTYAGTLTQAQGVNK759↓ | MS, ED | |
| K↓L824LLSATVNGSLVNNK837↓ | MS, ED | |
| K↓N838NIILNPTK845↓ | ED |
Glycosylated mature AIDA-I (corresponding to amino acids 50 to 847 in the preproprotein) was processed by digestion with trypsin or chymotrypsin, and the resulting peptides were identified by MS or ED. In MS, glycosylation of a peptide was identified as an excess of mass corresponding to a multiple of 192 Da (the mass of one heptose residue). In ED, glycosylated residues are identified when a signal corresponding to the modified residue is lacking or reduced during a degradation cycle. Two residues (S577, S578) showed an only 80% reduction in signal, suggesting that these residues were not modified in all peptides. The sequences of peptides P1, P3, and P6 could not be completely obtained by ED. Numbering corresponds to the positions of the amino acids in the preproprotein. MS yielded the number of heptose molecules bound per peptide. However, except in one case (T154), MS did not permit to identification of the modified residues. The number of heptoses cannot be determined using ED, but several residues (S102, S111, S116, S242, S252, S334, S391, S409, S540, S546, S559, S570, S577, S578, and S583) were identified as modified. Modified residues are underlined.
ND, not determined.
In a parallel approach, purified tryptic peptides of mature AIDA-I were sequenced by N-terminal ED. Six peptides had the expected sequences of AIDA-I peptides, with a perfect signal for each residue of the peptides. However, for eight other peptides, no signal was observed at positions corresponding to some of the serine residues they comprised (Table 1). This suggested that these residues had been modified and thus could not be identified during the degradation cycle. Five of the eight peptides identified as potentially modified corresponded to peptides identified as glycosylated by MS, suggesting that all the peptides identified by ED in this manner were indeed glycosylated. The signal corresponding to the serine residues at positions 577 and 578 was sharply reduced but not absent, suggesting that these residues were heterogeneously modified. This result is in agreement with the heterogeneity observed by MS. Also in agreement with the previous results, the modified peptides identified by ED were located in the 19-amino-acid repeats.
Together, our approaches revealed 10 different glycosylated peptides, and 15 serine and 1 threonine residues were found to be modified. Adding the maximum number of heptoses bound to each of the peptide identified by MS and the number of modified serine residues in the peptides identified solely by ED, we estimate that up to 19 heptoses could be present on AIDA-I (Table 1). All attempts to obtain the mass of the whole mature AIDA-I by MS were unsuccessful. This was most likely due to the high molecular weight of the protein and the heterogeneity conferred by the glycosylation.
Glycosylation influences the abundance of AIDA-I.
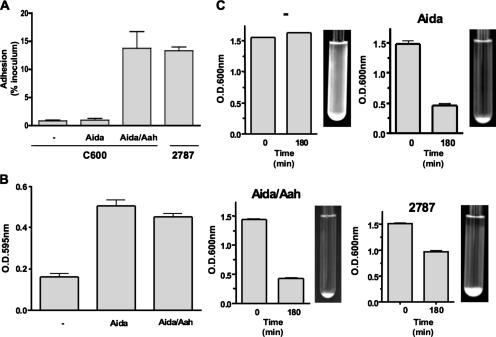
We compared the abilities of bacteria expressing the glycosylated or the nonglycosylated forms of AIDA-I to autoaggregate, form a biofilm, or adhere to cultured epithelial cells. Glycosylation seems dispensable for autoaggregation and biofilm formation, but the nonglycosylated form of AIDA-I cannot mediate adhesion to Hep-2 cells (Fig. 2), as previously observed (2, 36). This observation could have been the result of the fact that unglycosylated AIDA-I is unable to mediate adhesion, that there is not enough unglycosylated AIDA-I present at the bacterial surface to mediate adhesion, or that the conformation of unglycosylated AIDA-I is abnormal and cannot mediate adhesion.
FIG. 2.
Effect of glycosylation on the function of AIDA-I. (A) Adhesion assay. Bacteria bearing an empty vector (−) or expressing unglycosylated AIDA-I (Aida) or glycosylated AIDA-I (Aida/Aah) were inoculated onto a monolayer of confluent Hep-2 cells, and adhering bacteria were plated and counted. E. coli strain 2787, from which AIDA-I was originally identified (1), was used as a control. The adhesion values were calculated by dividing the level of CFU of adhering bacteria recovered by the level of CFU found in the inoculum after 3 h of incubation. (B) Biofilm formation. The same strains were grown in minimal medium for 24 h at 30°C in microtiter plates, and biofilms were stained with crystal violet. (C) Autoaggregation assay. Cultures of the same strains were left standing at 4°C, and the turbidity at the top of the culture was measured. Pictures of the culture tubes were taken after an overnight incubation at 4°C. Experiments were performed at least three times in duplicate (A and C) or quadruplicate (B), and the values represent means ± standard errors of the means.
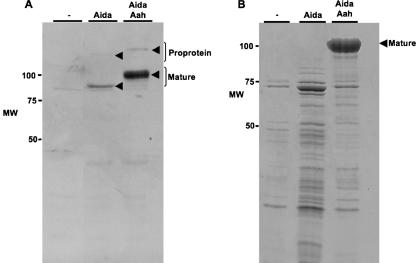
To distinguish between these possibilities, we compared the amounts of the glycosylated and nonglycosylated forms of AIDA-I present at the cell surface. As previously suggested, we observed that antibodies directed against the glycosylated mature adhesin do not efficiently recognize nonglycosylated AIDA-I (2), hindering efforts to compare expression levels. To remedy this situation, we used a plasmid allowing the expression of nonglycosylated AIDA-I with a HisG tag localized at the N terminus of the mature adhesin. In order to express the glycosylated protein, we cotransformed bacteria with this plasmid and a compatible plasmid containing the aah gene. With those constructs, we could prepare whole-cell extracts of normalized overnight cultures expressing glycosylated or unglycosylated forms of the histidine-tagged protein. Proteins of 100 and 132 kDa for glycosylated AIDA-I and 80 and 120 kDa for nonglycosylated AIDA-I were detected with an anti-HisG monoclonal antibody, as is consistent with the presence of the mature protein and the proprotein precursor forms of AIDA-I (Fig. 3A). We observed that there was dramatically less of the nonglycosylated form of AIDA-I than of the glycosylated form. We observed the same decrease with untagged versions of AIDA-I by separating membrane fractions of bacteria expressing glycosylated or unglycosylated forms of AIDA-I and staining with Coomassie blue (data not shown).
FIG. 3.
Effect of glycosylation on the expression level and on mild heat extraction of AIDA-I. (A) Whole-cell extracts of bacteria bearing an empty vector (−) or expressing unglycosylated AIDA-I (Aida) or glycosylated AIDA-I (Aida/Aah) were separated by SDS-PAGE and probed by immunoblotting with antiserum against the HisG tag fused to mature AIDA-I. (B) Bacteria harboring the same plasmids were heat extracted and the extracts recovered by centrifugation prior to separation by SDS-PAGE and Coomassie blue staining. MW, molecular weight.
Altered expression levels might be due to the fact that glycosylated AIDA-I is expressed in bacteria bearing two plasmids whereas unglycosylated AIDA-I is expressed in bacteria bearing only one plasmid. To eliminate the possibility that our results were affected by these expression characteristics, we expressed unglycosylated AIDA-I in bacteria bearing one plasmid by engineering a point mutation in the aah gene, resulting in the expression of an inactive glycosyltransferase, as previously observed (24). As reported above, we observed dramatically less nonglycosylated AIDA-I compared to its glycosylated form in heat extracts (data not shown).
Glycosylation confers partial resistance to proteases.
We observed that when the mature unglycosylated and glycosylated adhesins were heat extracted, the glycosylated polypeptide ran as a clear band of approximately 100 kDa, whereas the nonglycosylated protein appeared as a less intense band of approximately 80 kDa along with a degradation profile absent from extracts of bacteria expressing glycosylated AIDA-I (Fig. 3B). Additionally, the heat-extracted unglycosylated polypeptides were undetectable with antibodies directed against the HisG tag (data not shown). This result suggests that unglycosylated AIDA-I is more sensitive to proteolytic degradation. To determine whether and where unglycosylated AIDA-I is degraded in vivo, we compared the expression levels of glycosylated and unglycosylated AIDA-I in a degP-negative background, but there was no visible effect of the presence of the periplasmic protease DegP on the relative amounts of unglycosylated and glycosylated AIDA-I (data not shown). We also tested whether the outer membrane protease OmpT could have an influence on the levels of AIDA-I, but the presence of that protease also seemed not to have an effect (data not shown). Since AIDA-I autoproteolytically matures itself (3, 37), it could be that unglycosylated AIDA-I is responsible for its own degradation when it is in an abnormal conformation.
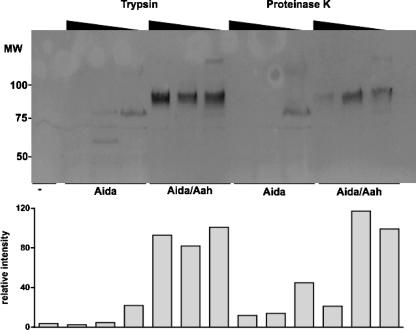
To confirm the hypothesis that unglycosylated AIDA-I is more sensitive to proteolytic degradation, we performed a limited digestion of bacterial surface proteins by use of trypsin, a protease cleaving after lysine and arginine residues. We observed that unglycosylated AIDA-I was degraded at the lowest concentration of trypsin whereas the glycosylated protein was resistant at all concentrations tested (Fig. 4).
FIG. 4.
Effect of glycosylation on the resistance to proteases. Bacteria bearing an empty vector (−) or expressing unglycosylated AIDA-I (Aida) or glycosylated AIDA-I (Aida/Aah) were pelleted and resuspended in PBS in the presence or absence of trypsin or proteinase K (0.3 μg/ml−1 and 3 μg/ml−1). The amount of AIDA-I was revealed by immunoblotting with an anti-HisG antibody (upper panel), and the intensity of the bands was quantitated by densitometry and normalized (lower panel). MW, molecular weight.
To ensure that this result was not specific for trypsin, which could simply indicate that most trypsin cleavage sites are protected by the heptoses, we performed the same assay with various concentrations of proteinase K, a protease cleaving after hydrophobic residues. Consistent with our previous results, we observed that the unglycosylated protein was completely degraded at the lowest concentration of proteinase K used (Fig. 4). To further exclude artifacts whose presence might have been due to the use of histidine-tagged proteins, we performed the same assays with untagged proteins revealed by Coomassie blue staining. As described above, in these experiments, the unglycosylated protein was completely degraded by 0.3 μg/ml−1 of proteinase K whereas the glycosylated protein was resistant (data not shown).
The unglycosylated form of AIDA-I induces an extracytoplasmic stress.
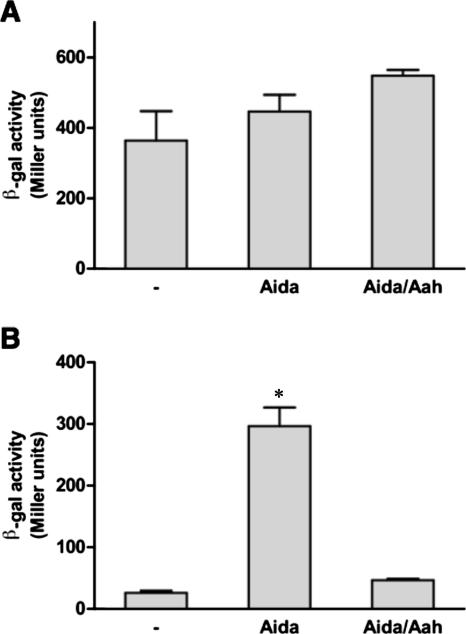
The sensitivity of unglycosylated AIDA-I to proteolytic degradation suggests that it is in an abnormal conformation. Polypeptides with abnormal conformation are usually sensed by specialized stress-sensing systems, which in turn induce the production of folding catalysts and proteolytic enzymes to alleviate the problem (23). We therefore tested whether unglycosylated AIDA-I can induce a stress response. We used two reporter strains: strain SR1458, which bears the β-galactosidase lacZ gene under the control of the degP promoter (30), and strain SR1364, a strain bearing the lacZ gene under the control of the rpoH promoter (25). The former is induced by an extracytoplasmic stress and the latter by a cytoplasmic stress. The strains were transformed with the plasmids bearing the aah-aidA operon or the aidA gene alone or with a control empty plasmid (Fig. 5). We observed that glycosylated and unglycosylated AIDA-I did not induce any cytoplasmic stress (Fig. 5A). The β-galactosidase activity indicated, however, that unglycosylated AIDA-I caused a dramatic extracytoplasmic stress response (Fig. 5B). Note that SR1458 is only a reporter strain and that the induction of the degP promoter does not prove that unglycosylated AIDA-I is degraded by the periplasmic DegP protease. Indeed, as described above, we have observed that DegP does not seem to be involved in the degradation of unglycosylated AIDA-I. Note also that these results were obtained in a context of overexpression; we do not know whether a similar stress response would be induced in an aah mutant of the wild-type strain 2787. Nevertheless, the induction of the stress response upon overexpression is consistent with the enhanced sensitivity to proteolytic degradation and suggests that unglycosylated AIDA-I adopts an abnormal conformation.
FIG. 5.
Induction of an extracytoplasmic stress by the nonglycosylated AIDA-I. An empty vector (−) or plasmids allowing expression of unglycosylated AIDA-I (Aida) or glycosylated AIDA-I (Aida/Aah) were transformed into the SR1364 (A) and SR1458 (B) reporter strains; the β-galactosidase (β-gal) activity levels (presented in Miller units) measured in these strains indicate cytoplasmic and extracytoplasmic stress, respectively. The experiment was performed three times in duplicate. The values represent means ± standard errors of the means and were compared by analysis of variance (*, P < 0.01).
Glycosylation is not required for the binding of a domain of AIDA-I to cultured epithelial cells.
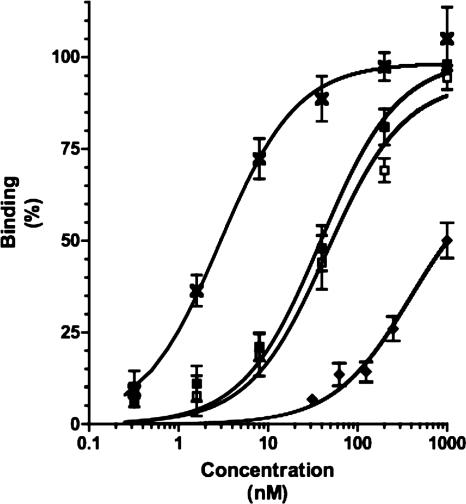
We decided to test directly the role of glycosylation in the binding of AIDA-I to epithelial cells by comparing the binding characteristics of glycosylated and unglycosylated AIDA-I. Whereas we can purify whole glycosylated AIDA-I by solubilizing outer membrane extracts (3), we are unable to purify unglycosylated AIDA-I because it is unstable and degraded. We therefore purified a fusion of mature AIDA-I (corresponding to amino acids 50 to 847) to GST, unglycosylated or glycosylated as a result of coexpression in the presence of Aah. The presence or absence of glycosylation of the GST-AIDA was ascertained using a glycan detection kit (data not shown). Glycosylated GST-AIDA bound specifically to cultured epithelial cells in a saturable manner (Fig. 6). The equilibrium dissociation constant was calculated to be approximately 45 nM. By comparison, we observed that whole glycosylated AIDA-I could bind to cells with a dissociation constant of 4 nM, in close agreement with a previous estimate (19). In a structure-function study we have observed that the N terminus of AIDA-I harbors a cell-binding domain (3a). In this context, when the GST is fused to the N terminus of mature AIDA-I it is likely that it sterically hinders the interaction mediated by the N-terminal cell-binding domain. This might explain the difference between the dissociation constant of our GST fusion protein and that of wild-type AIDA-I. The fusion of mature AIDA to GST, purified in the absence of Aah (and therefore unglycosylated), was also able to result in binding to cultured epithelial cells in a manner similar to that seen with the glycosylated fusion, with an equilibrium dissociation constant calculated at 39 nM. This result suggests that the carbohydrate moieties do not directly participate in the binding to a cellular receptor.
FIG. 6.
Binding of unglycosylated mature AIDA-I to cultured epithelial cells. The glycosylated (open squares) or unglycosylated (closed squares) fusion of GST to mature AIDA-I (encompassing amino acids 50 to 847) was purified by affinity chromatography. Various amounts of the proteins were incubated with fixed Hep-2 cells. Pure GST (diamonds) and pure whole AIDA-I (crosses) were used as controls. Bound proteins were revealed with an anti-GST antiserum directly coupled to HRP or an anti-AIDA-I serum and a secondary antibody coupled to HRP. Background values were subtracted, and the absorption was normalized by dividing the absorption of each well by the maximal absorption measured on the plate in order to obtain percentages of maximal binding.
DISCUSSION
In the present study, we identified 10 peptides of AIDA-I bearing heptose residues, and our results suggest that at least up to 19 molecules can be added to AIDA-I, in perfect agreement with a previous evaluation (2). Recently, five different peptides were found to contain heptoses in Ag43 (35). Strikingly, in AIDA-I as well as in Ag43, glycosylation appears to be heterogeneous; i.e., most of the glycosylated peptides could be identified in unglycosylated form and/or with various numbers of heptoses. This fact was not initially appreciated when AIDA-I was shown to be glycosylated (2). The coexistence of modified and unmodified forms of bacterial glycoproteins has previously been reported (31, 41). In some instances it has been shown that this heterogeneity influences the immunogenicity of the protein (31), but the role, if any, of this heterogeneity in AIDA-I is unclear. The mechanism that results in such heterogeneity is also unknown.
The glycosylated peptides were identified in a region of the protein composed of imperfect 19-amino-acid repeats. The same was true with Ag43 (35). It was expected that the specificities of the heptosyltransferases acting on these proteins are similar, since the SAAT glycosyltransferases are functionally exchangeable (24, 35). The role of these repeats is unknown, but it has been postulated that they are involved in the adhesion mediated by these proteins, since it has often been observed that repeated motifs are involved in adhesion. Alternatively, the repeated motifs could provide the backbone for the expected β-helical structure of the proteins, itself a repetitive structure.
In addition to what was reported with respect to Ag43, we unambiguously showed that AIDA-I is modified on serine and threonine residues, proving that Aah mediates O-glycosylation. This was expected based on the similarities of the Aah sequence to those of the known E. coli heptosyltransferases involved in lipopolysaccharide biosynthesis (2). Indeed, these enzymes mediate the transfer of heptose precursors to the hydroxyl group of lipopolysaccharide biosynthetic intermediates (8). We did not identify any glycosylation consensus sequence, but it should be noted that no consensus sequence for O-glycosylation has been established for either eukaryotes or prokaryotes (29, 34, 38, 40). Consequently, the specificity of the Aah glycosyltransferases remains elusive: how does Aah recognize its SAAT substrates, and why does it transfer heptoses in the 19-amino-acid imperfect repeats? It is possible that this region adopts a specific structure that is recognized by the enzyme, but this would contradict the idea that glycosylation occurs in the cytoplasm prior to transport across the inner membrane via the sec machinery, which requires exported polypeptides to be unfolded. Alternatively, the enzyme might recognize a part of the repeated consensus sequence itself. Specificity of O-glycosylation for amino acid sequences has been proposed before, as in the case of proline-rich domains of glycoproteins from Mycobacterium tuberculosis (4) and Clostridium thermocellum (6).
Important biological functions can be affected by glycosylation, such as maintenance of protein conformation, resistance against proteases, and modulation of intermolecular interactions (2, 38). Our results confirmed that glycosylation of AIDA-I is essential for adhesion (2) but not for autoaggregation or biofilm formation (36). Based on these results, it was suggested that the glycans could be involved in receptor recognition. Carbohydrates from glycoproteins have indeed been shown in some cases to mediate the interaction with the host cell receptor, as for the major outer membrane protein of Chlamydia trachomatis (18). All our observations, however, suggest that glycosylation is required for AIDA-I to adopt a normal conformation, which in turn would be responsible for the lack of adhesion. Indeed, we observed that unglycosylated AIDA-I is expressed to a lesser degree and is more sensitive to degradation but that purified mature AIDA-I could specifically bind to cultured epithelial cells even when unglycosylated. There are other examples of bacterial proteins being protected against degradation by glycosylation (14), and the conformation and protease sensitivities of polypeptides have often been shown to be altered by glycosylation (5, 16). The H. influenzae HMW1 adhesin, for instance, is stabilized by glycosylation (7). Unglycosylated HMW1 was prematurely degraded in the cytoplasm and periplasm, and its tethering to the bacterial surface was compromised. Interestingly, HMW1 and SAAT are both secreted by the type V secretion pathway (13), raising the possibility that glycosylation might also exert its role in the context of the secretion of some substrates of this pathway. Such a role could be to prevent premature periplasmic folding, since it was shown that such folding can be incompatible with secretion (32), or to promote extracellular folding, a process which has been proposed to drive secretion (28).
Two observations seem to be at odds with the notion that unglycosylated AIDA-I is in an abnormal conformation. First, Ag43 is usually not glycosylated and there is no indication that it is unstable or that glycosylation increases its stability. Despite their similarities, Ag43 and AIDA-I might have subtle structural differences that make glycosylation more important for AIDA-I, and it should be noted that the latter seems to be more glycosylated than Ag43. Second, it is surprising that glycosylation is dispensable for autoaggregation. One possible explanation is that the domains of the protein involved in autoaggregation do not require the folding and/or the stability provided by glycosylation. The unglycosylated protein could also be degraded into an intermediate that still bears an autoaggregation domain.
Many uncertainties remain about the glycosylation of AIDA-I. The mechanism of glycosylation itself needs to be investigated in order to understand the specificity of Aah toward its substrates and, more precisely, toward specific sites in the latter. How glycosylation affects the conformation of AIDA-I and why glycosylation is not necessary for autoaggregation are also unclear. Further characterization of the glycosylation of AIDA-I, as well as that of other SAAT, is warranted to tackle these puzzling questions.
Acknowledgments
This work was supported by financial contributions from the Groupe de Recherche et d'Etudes sur les Maladies Infectieuses du Porc (GREMIP), the Natural Sciences and Engineering Research Council of Canada (NSERC discovery grant 262746), the Canada Research Chair program, and the Canada Foundation for Innovation (project 201414). M.-È.C. is supported by a graduate fellowship from the Fonds de Recherche sur la Nature et les Technologies du Québec (FQRNT 114663).
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 3.Charbonneau, M. E., F. Berthiaume, and M. Mourez. 2006. Proteolytic processing is not essential for multiple functions of the Escherichia coli autotransporter adhesin involved in diffuse adherence (AIDA-I). J. Bacteriol. 188:8504-8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Charbonneau, M.-E., and M. Mourez. 2007. Functional organization of the autotransporter adhesin involved in diffuse adherence. J. Bacteriol. 189:9020-9029. [DOI] [PMC free article] [PubMed]
- 4.Dobos, K. M., K. H. Khoo, K. M. Swiderek, P. J. Brennan, and J. T. Belisle. 1996. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J. Bacteriol. 178:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fares, F. 2006. The role of O-linked and N-linked oligosaccharides on the structure-function of glycoprotein hormones: development of agonists and antagonists. Biochim. Biophys. Acta 1760:560-567. [DOI] [PubMed] [Google Scholar]
- 6.Gerwig, G. J., J. P. Kamerling, J. F. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1993. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J. Biol. Chem. 268:26956-26960. [PubMed] [Google Scholar]
- 7.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 8.Grizot, S., M. Salem, V. Vongsouthi, L. Durand, F. Moreau, H. Dohi, S. Vincent, S. Escaich, and A. Ducruix. 2006. Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. J. Mol. Biol. 363:383-394. [DOI] [PubMed] [Google Scholar]
- 9.Guerry, P., C. P. Ewing, M. Schirm, M. Lorenzo, J. Kelly, D. Pattarini, G. Majam, P. Thibault, and S. Logan. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha, S. K., C. Choi, and C. Chae. 2003. Prevalence of a gene encoding adhesin involved in diffuse adherence among Escherichia coli isolates in pigs with postweaning diarrhea or edema disease. J. Vet. Diagn. Investig. 15:378-381. [DOI] [PubMed] [Google Scholar]
- 11.Ha, S. K., C. Choi, K. Jung, J. Kim, D. U. Han, Y. Ha, S. D. Lee, S. H. Kim, and C. Chae. 2004. Genotypic prevalence of the adhesin involved in diffuse adherence in Escherichia coli isolates in pre-weaned pigs with diarrhoea in Korea. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:166-168. [DOI] [PubMed] [Google Scholar]
- 12.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann, J. L., P. O'Gaora, A. Gallagher, J. E. Thole, and D. B. Young. 1996. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 15:3547-3554. [PMC free article] [PubMed] [Google Scholar]
- 15.Hewick, R. M., M. W. Hunkapiller, L. E. Hood, and W. J. Dreyer. 1981. A gas-liquid solid phase peptide and protein sequenator. J. Biol. Chem. 256:7990-7997. [PubMed] [Google Scholar]
- 16.Imperiali, B., and S. E. O'Connor. 1999. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3:643-649. [DOI] [PubMed] [Google Scholar]
- 17.Klemm, P., R. M. Vejborg, and O. Sherlock. 2006. Self-associating autotransporters, SAATs: functional and structural similarities. Int. J. Med. Microbiol. 296:187-195. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S. Hakomori. 1996. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J. Clin. Investig. 98:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laarmann, S., and M. A. Schmidt. 2003. The Escherichia coli AIDA autotransporter adhesin recognizes an integral membrane glycoprotein as receptor. Microbiology 149:1871-1882. [DOI] [PubMed] [Google Scholar]
- 20.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 22.Mainil, J. G., E. Jacquemin, P. Pohl, A. Kaeckenbeeck, and I. Benz. 2002. DNA sequences coding for the F18 fimbriae and AIDA adhesin are localised on the same plasmid in Escherichia coli isolates from piglets. Vet. Microbiol. 86:303-311. [DOI] [PubMed] [Google Scholar]
- 23.Mogensen, J. E., and D. E. Otzen. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57:326-346. [DOI] [PubMed] [Google Scholar]
- 24.Moormann, C., I. Benz, and M. A. Schmidt. 2002. Functional substitution of the TibC protein of enterotoxigenic Escherichia coli strains for the autotransporter adhesin heptosyltransferase of the AIDA system. Infect. Immun. 70:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mourez, M., S. Skouloubris, J. M. Betton, and E. Dassa. 1997. Heat shock induction by a misassembled cytoplasmic membrane protein complex in Escherichia coli. Mol. Microbiol. 26:821-831. [DOI] [PubMed] [Google Scholar]
- 26.Ngeleka, M., J. Pritchard, G. Appleyard, D. M. Middleton, and J. M. Fairbrother. 2003. Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. J. Vet. Diagn. Investig. 15:242-252. [DOI] [PubMed] [Google Scholar]
- 27.Niewerth, U., A. Frey, T. Voss, C. Le Bouguenec, G. Baljer, S. Franke, and M. A. Schmidt. 2001. The AIDA autotransporter system is associated with F18 and stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, D. C., G. Huang, E. Nodel, S. Pleasance, and R. C. Fernandez. 2003. A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol. Microbiol. 47:1367-1383. [DOI] [PubMed] [Google Scholar]
- 29.Peter-Katalinić, J. 2005. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 405:139-171. [DOI] [PubMed] [Google Scholar]
- 30.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romain, F., C. Horn, P. Pescher, A. Namane, M. Riviere, G. Puzo, O. Barzu, and G. Marchal. 1999. Deglycosylation of the 45/47-kilodalton antigen complex of Mycobacterium tuberculosis decreases its capacity to elicit in vivo or in vitro cellular immune responses. Infect. Immun. 67:5567-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford, N., M. E. Charbonneau, F. Berthiaume, J. M. Betton, and M. Mourez. 2006. The periplasmic folding of a cysteineless autotransporter passenger domain interferes with its outer membrane translocation. J. Bacteriol. 188:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 35.Sherlock, O., U. Dobrindt, J. B. Jensen, R. Munk Vejborg, and P. Klemm. 2006. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 188:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suhr, M., I. Benz, and M. A. Schmidt. 1996. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a beta-barrel structure. Mol. Microbiol. 22:31-42. [DOI] [PubMed] [Google Scholar]
- 38.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 39.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 40.Van den Steen, P., P. M. Rudd, R. A. Dwek, and G. Opdenakker. 1998. Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33:151-208. [DOI] [PubMed] [Google Scholar]
- 41.Young, N. M., J. R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St. Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]