Abstract
YhbO is a member of the DJ-1/ThiJ/Pfp1 superfamily, which includes chaperones, peptidases, and the Parkinson's disease protein DJ-1. A yhbO-disrupted mutant of Escherichia coli is highly sensitive to oxidative, thermal, UV, and pH stresses, and the putative nucleophilic cysteine C104 of YhbO is required for stress resistance. These results suggest that YhbO affects a central process in stress management.
The gene encoding the 19-kDa YhbO protein has homologs in almost every organism and cell that has been examined to date, ranging from Escherichia coli to Homo sapiens. This ubiquity and evolutionary conservation indicate that it may play a fundamental physiological role. YhbO is a member of the DJ-1/ThiJ/Pfp1 superfamily, which includes proteins with diverse functions, such as chaperones/peptidases (11, 20, 22), proteases (3, 6), catalases, and the Parkinson's disease protein DJ-1 (10, 17, 23). Its closest homologs are ThiJ/YajL in E. coli (2, 29), YfkM and YraA in Bacillus subtilis (involved in protection against environmental stresses) (19, 26), the Pyrococcus furiosus protease 1 (3), and the Parkinson's disease protein DJ-1. The crystal structures of several members of the ThiJ superfamily have been solved. They all contain a similar domain with a nucleophilic elbow displaying an important cysteine which, in Php1 and Hsp31, is part of a Cys-His-Glu/Asp catalytic triad responsible for their peptidase activities (6, 20, 32). Several members of this superfamily have been biochemically characterized. Hsp31 has been characterized as a chaperone (11, 22) and a peptidase (12), and Pfp1 has been characterized as a protease/peptidase with activity towards gelatin and the fluorescent substrate Ala-Ala-Phe-7-aminomethyl coumarin (3). The biochemical characterization of DJ-1 led to contradictory results concerning its putative chaperone, peptidase, and redox activities (18, 23, 30). ThiJ was mistakenly believed to be involved in thiamine synthesis, and its function is presently unknown (29). YhbO also possesses a putative catalytic triad, and its three-dimensional (3D) structure (indexed in the RCBS Protein Data Bank under the identification number 1oi4) closely resembles that of the PhpI peptidase (1), suggesting that it might function as a peptidase. We recently cloned and purified YhbO, but we could not detect any chaperone, protease, or peptidase activities in the purified protein (1). In this study, we show that YhbO is required for the protection of bacterial cells against many environmental stresses, including oxidative, thermal, osmotic, UV, and pH stresses, and that its putative nucleophilic cysteine, C104, is required for its function in vivo.
Growth defects of the yhbO-deficient strain.
The yhbO-deficient strain was kindly provided by H. Mori (Nara Institute of Sciences and Technology, Japan) and contains yhbO disrupted by λRed in the E. coli strain BW25113 [lacIq rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1] (5), leading to strain JW3112. Disruption of the yhbO gene is not likely to exert any polar effects on the expression of vicinal genes, since yhbO is a single-gene operon (Colibri server, Pasteur Institute [http://genolist.pasteur.fr/Colibri/]). The doubling time of the yhbO-deficient strain was similar to that of its parental strain, in both Luria-Bertani (LB) medium (14) and 63 glycerol minimal medium, at 30°C, 37°C, and 43°C (not shown). The yhbO mutant, however, gave smaller colonies on LB plates (at 30, 37, and 43°C), reflecting a slight growth disadvantage compared to the control strain (not shown). The yhbO mutant also produced slightly smaller colonies on glucose (1%) LB plates incubated at 30°C under anaerobic conditions (in an anaerobic glove chamber containing less than 5 ppm O2), suggesting that it is not significantly affected by anaerobic conditions (not shown).
The yhbO mutant is sensitive to oxidative stress.
Logarithmic-phase cultures of wild-type and yhbO mutant cells grown in LB medium to an optical density at 600 nm (OD600) of 0.4 were incubated at 37°C with aeration in the presence of 15 mM or 50 mM hydrogen peroxide, and viable cell counts were periodically determined. After 90 min of exposure to 50 mM H2O2, wild-type cell counts were approximately 105 CFU per ml, while yhbO mutant counts were approximately 103 (100-fold lower) (Fig. 1A). After a similar exposure to 15 mM H2O2, the counts of the yhbO mutant were approximately 30-fold lower than those of the wild-type strain (Fig. 1A). Bacteria were not reproducibly sensitive to lower hydrogen peroxide concentrations, which is probably due to the high efficiency of the hydrogen peroxide detoxification enzymes (the KatE and KatG catalases [induced, respectively, by entry into stationary phase and by hydrogen peroxide] and the AhpC alkylhydroperoxide reductase) (9).
FIG. 1.
Increased sensitivity of the yhbO mutant to environmental stresses. Logarithmic-phase cultures of wild-type (squares) and yhbO-deficient (diamonds) cells were incubated for the indicated times, as described in the text, in the presence of 15 mM (filled symbols) or 50 mM (open symbols) H2O2 (A), at 50°C (filled symbols) or 53°C (open symbols) (B), under UV irradiation (C), at pH 2.5 (D), at pH 10.5 (E), or in the presence of 2.5 M NaCl (F), and viable cell counts were determined. Experiments were done three times, and the mean value ± standard error of the mean (SEM) was calculated.
We checked for a possible accumulation of oxidized proteins or peptides in the yhbO mutant by measuring protein and peptide carbonyls (11) as described in references 24 and 25. Logarithmic-phase cultures of wild-type and yhbO mutant cells were incubated at 37°C with aeration for 40 min in the presence of 50 mM hydrogen peroxide. Proteins from the bacterial crude extract were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and probed with the OxyBlot protein oxidation detection kit (Chemicon International, Serological Corporation). The yhbO mutant (Fig. 2A, lane 2) did not display any significant increase in protein oxidation compared with its parent (Fig. 2A, lane 1). Peptides were extracted from bacteria (bacteria were centrifuged and resuspended into 1 M acetic acid as described in reference 12) after a 50 mM hydrogen peroxide stress for 40 min and analyzed on a high-performance liquid chromatography C18 reverse-phase column equilibrated in 0.1% trifluoroacetic acid in water and eluted with a linear gradient of 0 to 100% acetonitrile containing 0.1% trifluoroacetic acid, as described elsewhere (12). We could not detect any significant difference between the peptide profile of the yhbO mutant and that of its parental strain (either before or after the hydrogen peroxide stress) (not shown). These results suggest that YhbO is not involved in the processing of oxidatively modified proteins and peptides.
FIG. 2.
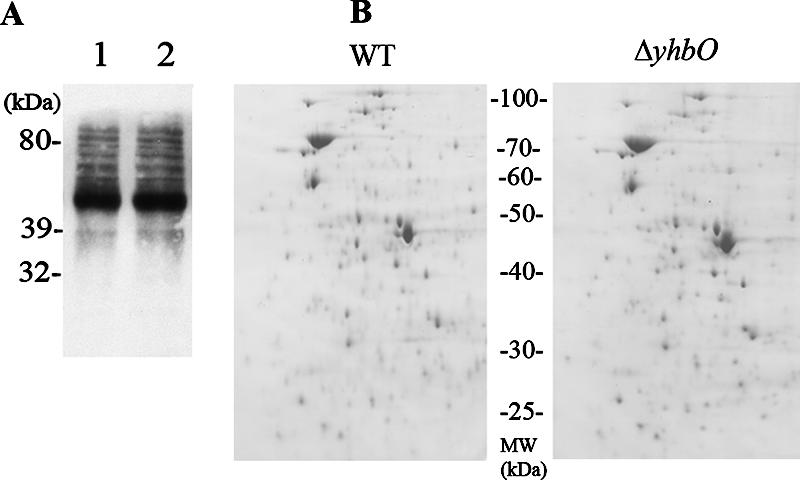
Unchanged protein metabolism in the yhbO mutant. (A) Detection of oxidatively modified proteins from E. coli cells after a 50 mM H2O2 challenge. Logarithmic-phase cells from the parental strain (lane 1) and from the yhbO mutant (lane 2) were incubated for 40 min at 37°C in the presence of 50 mM H2O2, and crude extracts were prepared, electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel (10 μg protein in each lane), transferred to a polyvinylidene difluoride membrane, and immunoassayed for protein carbonyls using the OxyBlot kit. (B) 2D gel electrophoresis of insoluble protein fractions after heat shock. The yhbO mutant and its parent were grown at 30°C in Luria-Bertani medium to logarithmic phase and shifted to 53°C for 90 min. Insoluble proteins (15,000 × g pellet) were analyzed by 2D gel electrophoresis, as described in reference 4, and stained with Coomassie blue (the pH 4 to 7 gradient is from left to right). The amount of protein loaded onto each gel corresponds to identical amounts of bacteria (pellet fractions contain membrane proteins and aggregated protein material [4]).
Hence, although YhbO is required for oxidative stress resistance, and despite its strong homology with several peptidases, it does not appear to affect the metabolism of proteins and peptides in oxidatively stressed cells.
The yhbO mutant is sensitive to thermal stress.
Bacteria (wild-type strain BW 25113 and yhbO mutant JW 3122) were cultured in LB broth (Difco) at 37°C with aeration until the OD600 reached 0.4. They were then shifted to a shaking water bath at 50°C or 53°C, and viable cell counts were periodically determined by plating bacteria overnight on LB agar plates. The yhbO mutant displayed an increased sensitivity to temperature stress (Fig. 1B). After 30 min of exposure to a temperature of 53°C, wild-type cell counts were approximately 108 CFU per ml, while yhbO mutant counts were approximately 106 CFU per ml (100-fold lower), suggesting that YhbO is important for heat stress resistance. When bacteria were incubated for 30 min at 50°C, yhbO mutant counts were approximately 10-fold lower than wild-type cell counts (Fig. 1B). The effects of heat stresses of up to 48°C were not greater in the mutant than they were in the wild-type strain (not shown). Thus, like Hsp31 and ClpB, YhbO helps E. coli to survive to severe heat stresses (in contrast with the DnaK chaperone, which is required for bacterial growth from 42°C onwards [4]).
We investigated the extent of heat-induced protein aggregation in both the wild-type and yhbO-deficient strains. Cells were grown at 30°C and then subjected to heat shock treatment for 90 min at 53°C. After ultrasonic disruption of cells, the insoluble cell fraction was isolated by centrifugation at 15,000 × g, and the pellets were analyzed by 2D gel electrophoresis, as described in reference 4. The heat shock treatment at 53°C did not lead to a significant increase in protein aggregation in the yhbO mutant strain compared to the wild-type strain (Fig. 2B), as opposed to what has been observed in a dnaK strain (4). This result is consistent with the hypothesis that YhbO is devoid of any chaperone activity (1).
Since YhbO displays a strong structural homology with archaeal peptidases and with the E. coli chaperone/peptidase Hsp31 (6, 20), whose mutant accumulates peptides (12), we checked whether the yhbO mutant accumulates peptides upon heat shock. Peptides were extracted from bacteria after a heat shock treatment for various times (5 to 60 min) at 53°C and analyzed on a high-performance liquid chromatography C18 reverse-phase column as described above (12, 31). We could not detect any difference between the peptide profile of the yhbO mutant and that of its parental strain (either after bacterial growth at 30°C or after a heat shock at 53°C) (not shown).
The yhbO mutant is sensitive to UV irradiation.
Logarithmic-phase cultures of wild-type and yhbO-deficient cells were exposed to UV light. Five-milliliter cultures were transferred to a 10-cm-diameter petri dish, placed under a germicidal lamp (254 nm, 1 J/m2), and sampled periodically for up to 10 min. After exposure, samples were kept on ice until viable cell counts were determined. After 10 min, wild-type cell counts were approximately 107 CFU per ml, while mutant counts were less than 106 CFU per ml (20-fold less than those of the wild-type strain) (Fig. 1C).
The yhbO mutant is sensitive to acid and alkaline pHs.
Logarithmic-phase cultures of wild-type and yhbO mutant cells were incubated at 37°C in Luria broth medium (pH 7.2) with aeration until the OD600 reached 0.4 and then shifted to LB medium at pH 2.5 or 10.5, and viable cell counts were periodically determined on LB plates at pH 7. After 60 min of exposure at pH 2.5, wild-type cell counts were approximately 4 × 107 CFU per ml, while yhbO mutant counts were less than 2 × 106 CFU per ml (around 20-fold less) (Fig. 1D). After 23 min of exposure at pH 10.5, wild-type cell counts were approximately 2 × 105 CFU per ml, while yhbO mutant counts were approximately 6 × 103 CFU per ml (around 30-fold less) (Fig. 1E). We conclude, therefore, that YhbO is also required for bacterial resistance to extreme pHs. The involvement of YhbO in acid stress resistance is consistent with its overexpression during acid stress (28).
The yhbO mutant is slightly sensitive to salt stress and insensitive to cold stress.
Logarithmic-phase cultures of wild-type and yhbO mutant cells were incubated at 37°C in Luria broth medium with aeration until the OD600 reached 0.4. Salt stress was achieved by adding NaCl to reach a concentration of 2.5 M, and viable cell counts were done at various times during approximately 3 h. After 2 h of exposure to 2.5 M NaCl, wild-type cell counts were approximately 107 CFU per ml, while yhbO mutant counts were reproducibly three- to fivefold lower (Fig. 1F). The involvement of YhbO in resistance to osmotic stress is consistent with its overexpression during salt stress (27). To test the cold stress sensitivity of the yhbO mutant, we incubated logarithmic-phase cultures of wild-type and yhbO-deficient cells at 37°C in Luria broth medium with aeration until the OD600 reached 0.4. The cultures were diluted with growth medium, and equal quantities of cells were plated on petri dishes. The plates were then sealed in plastic bags to prevent drying and stored at 4°C. At different times (up to several days), the number of colonies that survived was measured after an overnight incubation of the plates at 37°C. We found no significant difference between the sensitivities of the yhbO-deficient strain and its parent to cold (not shown). Similarly, when the yhbO mutant and its parental strain, grown in rich medium to an OD600 of 0.4, were transferred from 37°C to 10°C, we observed for both strains a 4-h lag that preceded the resumption of exponential growth at a generation time of 12 h (not shown).
We also performed the stress sensitivity experiments described in the above sections with stationary-phase cells, but the yhbO mutant was no more significantly affected by the different stresses than it was during the logarithmic phase (not shown).
Complementation of the yhbO mutant by its wild-type allele, but not by the C104A mutant.
In order to ascertain that the stress-sensitive phenotypes of the yhbO mutant are indeed due to the loss of YhbO, we complemented the mutant with its wild-type allele. The wild-type yhbO gene was transferred from the pET-21-a-yhbO high-yield expression vector (1) to pBAD33, a tightly controlled expression vector under the control of the arabinose PBAD promoter, yielding pBAD33-yhbO (7). Then, since YhbO possesses a putative catalytic triad centered around cysteine 104 (22), we constructed the C104A mutant in order to establish whether this residue is relevant to the activity in vivo of YhbO (cysteine in position 104 was substituted for alanine by site-directed mutagenesis in vitro [Stratagene QuikChange kit]) of the appropriate codon in the pBAD33-yhbO plasmid. The forward primer used to create the mutation was 5′-CCGGTGTTTGCCATCGCCCACGGCCCGCAGTTGCTG-3′, and the reverse primer was of the same length and complementary to the forward primer. Strain BW25113 was used for transformation of the new expression vector constructs, and clones were checked by DNA sequencing. The wild-type strain and the yhbO mutant either uncomplemented (i.e., transformed with the pBAD33 control plasmid) or complemented with the wild-type or with the C104A yhbO allele were challenged with a hydrogen peroxide stress. Bacteria were grown in 63 minimal medium supplemented with 0.4% glycerol as a carbon source and 3 μM arabinose for induction of YhbO (this arabinose concentration, which yields a moderate expression of YhbO, was found to be optimal for complementation studies, and the 63 minimal medium gave a more controllable expression of the pBAD operon than the LB medium). As shown in Fig. 3A, the wild-type yhbO allele efficiently complemented the yhbO mutant, displaying a sensitivity to H2O2 stress close to that of the wild-type strain. In contrast, the C104A mutant was unable to complement efficiently the yhbO mutant. When bacteria were challenged by a thermal stress for 30 min at 53°C, the viability of the yhbO mutant was 600-fold lower than that of the parental strain; the wild-type allele efficiently complemented the yhbO mutant, and the C104A allele was unable to do so (Fig. 3B). These results suggest that Cys104 is required for the activity in vivo of YhbO.
FIG. 3.
Complementation of the yhbO mutant by its wild-type allele, but not by the C104A mutant. Logarithmic-phase cultures of wild-type cells (filled circles) and yhbO-deficient cells either uncomplemented (empty circles) or complemented with the wild-type yhbO allele (filled squares) or with the C104A yhbO allele (filled triangles) were incubated for 90 min in the presence of the indicated hydrogen peroxide concentration (A) or for the indicated times at 53°C (B), and viable cell counts were determined. Experiments were done three times, and the mean value ± standard error of the mean (SEM) was calculated.
YhbO is a general stress protein.
We show in this study that YhbO protects E. coli cells against many environmental stresses. Disruption of the yhbO gene results in an increased bacterial sensitivity to oxidative, thermal, acid, alkaline, osmotic, and UV stresses, leading to a 1- to 5-log decrease in the survival yield of the mutant compared to that of its parent (the increased sensitivity to environmental stresses of the yhbO mutant is of the same order of magnitude as that of the dps mutant [16]). In accordance with its role in stress management, YhbO is overexpressed severalfold in stationary phase and during hyperosmotic stress and acid stress (27, 28). Several members of the ThiJ superfamily also function in cellular protection against environmental stresses. The chaperone/peptidase Hsp31 is involved in thermal stress protection (19), Bacillus subtilis YfkM and YraA are involved in acid stress protection (19, 26), and DJ-1 is involved in oxidative stress protection (23).
We could not detect any defect (aggregation or oxidation) in protein or peptide metabolism in the yhbO mutant, during either heat or oxidative stress. Since YhbO does not display any protease or peptidase activity with classical substrates (1), we wondered whether its sequence and structural homologies with Php1 (47% sequence identities, root mean square deviation value for C-α atoms of 0.7 Å) could be translated into functional homology (see reference 25 for a review) or whether its peptidase specificity is so narrow that its physiological substrate(s) escaped detection (experiments in vitro [1] and in vivo [this study] did not reveal any protease or peptidase activity). Our complementation experiments of the yhbO mutant with its wild-type allele (giving positive results) and with the C104A mutant (giving negative results) suggest that the increased stress sensitivity of the mutant is indeed due to the loss of YhbO and that the putative nucleophilic cysteine C104 of YhbO is important for its function. Similarly, the conserved cysteine C104 of Drosophila melanogaster DJ-1b is critical for its antioxidant function in vivo (13). Several other proteins of the DJ-1/ThiJ/Pfp1Hsp31 superfamily have yet to be biochemically characterized. Hsp31 functions as both a chaperone (11, 22) and a peptidase (12) and Pfp1 functions as a peptidase, but the physiological substrates of the latter have not yet been characterized (3, 8). The biochemical characterization of DJ-1 led to contradictory results concerning its chaperone, peptidase, redox, and gene regulation activities (18, 23, 30), and B. subtilis YfkM and YraA have not yet been characterized. Similarly, the universal stress proteins UspA, -C, -D, -E, -F, and -G, discovered many years ago, still require biochemical characterization (roles in protection against superoxide-generating agents, control of iron levels, adhesion, and cell motility have been proposed [15]).
Acknowledgments
We thank Hirotada Mori, Nara Institute of Sciences and Technology, Nara 630-0101, Japan, for the gift of the yhbO-disrupted strain and A. Kropfinger for correction of the English language.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Abdallah, J., R. Kern, A. Malki, V. Eckey, and G. Richarme. 2006. Cloning, expression, and purification of the general stress protein YhbO from Escherichia coli. Protein Expr. Purif. 47:455-460. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, S., and M. R. Cookson. 2004. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 4:6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumentals, I. I., A. S. Robinson, and R. M. Kelly. 1990. Characterization of sodium dodecyl sulfate-resistant proteolytic activity in the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl. Environ. Microbiol. 56:1992-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chattopadhyay, M. K., R. Kern, M. Y. Mistou, A. M. Dandekar, S. L. Uratsu, and G. Richarme. 2004. The chemical chaperone proline relieves the thermosensitivity of a dnaK deletion mutant at 42°C. J. Bacteriol. 186:8149-8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du, X., I. G. Choi, R. Kim, W. Wang, J. Jancarik, H. Yokota, and S. H. Kim. 2000. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-Å resolution. Proc. Natl. Acad. Sci. USA 97:14079-14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halio, S. B., I. I. Blumentals, S. A. Short, B. M. Merrill, and R. M. Kelly. 1996. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang, S., and J. A. Imlay. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, S. J., S. J. Kim, I. K. Kim, J. Ko, C. S. Jeong, G. H. Kim, C. Park, S. O. Kang, P. G. Suh, H. S. Lee, and S. S. Cha. 2003. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 278:44552-44559. [DOI] [PubMed] [Google Scholar]
- 11.Malki, A., R. Kern, J. Abdallah, and G. Richarme. 2003. Characterization of the Escherichia coli YedU protein as a molecular chaperone. Biochem. Biophys. Res. Commun. 301:430-436. [DOI] [PubMed] [Google Scholar]
- 12.Malki, A., T. Caldas, J. Abdallah, R. Kern, S. J. Kim, S. S. Cha, H. Mori, and G. Richarme. 2005. Peptidase activity of the Escherichia coli Hsp31 chaperone. J. Biol. Chem. 280:14420-14426. [DOI] [PubMed] [Google Scholar]
- 13.Meulener, M. C., K. Xu, L. Thomson, H. Ischiropoulos, and N. M. Bonini. 2006. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation. Proc. Natl. Acad. Sci. USA 103:12517-12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics, p. 439. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Nachin, L., U. Nannmark, and T. Nystrom. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 87:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishinaga, H., K. Takahashi-Niki, A. Andreadis, S. M. Iguchi-Ariga, and H. Ariga. 2005. Expression profiles of genes in DJ-1-knockdown and L 166 P DJ-1 mutant cells. Neurosci. Lett. 390:54-59. [DOI] [PubMed] [Google Scholar]
- 18.Olzmann, J. A., K. Brown, K. D. Wilkinson, H. D. Rees, Q. Huai, H. Ke, A. I. Levey, L. Li, and L. S. Chin. 2004. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279:8506-8515. [DOI] [PubMed] [Google Scholar]
- 19.Petersohn, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis. Microbiology 145:869-880. [DOI] [PubMed] [Google Scholar]
- 20.Quigley, P. M., K. Korotkov, F. Baneyx, and W. G. Hol. 2003. The 1.6-Å crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad. Proc. Natl. Acad. Sci. USA 100:3137-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Requena, J. R., R. L. Levine, and E. R. Stadtman. 2003. Recent advances in the analysis of oxidized proteins. Amino Acids 25:221-226. [DOI] [PubMed] [Google Scholar]
- 22.Sastry, M. S., K. Korotkov, Y. Brodsky, and F. Baneyx. 2002. Hsp31, the Escherichia coli yedU gene product, is a molecular chaperone whose activity is inhibited by ATP at high temperatures. J. Biol. Chem. 277:46026-46034. [DOI] [PubMed] [Google Scholar]
- 23.Shendelman, S., A. Jonason, C. Martinat, T. Leete, and A. Abeliovich. 2004. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLOS Biol. 2:e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamarit, J., E. Cabiscol, and J. Ros. 1998. Identification of the major oxidatively proteins in Escherichia coli exposed to oxidative stress. J. Biol. Chem. 273:3027-3032. [DOI] [PubMed] [Google Scholar]
- 25.Tian, W., and J. Skolnick. 2003. How well is enzyme function conserved as a function of pairwise identity? J. Mol. Biol. 333:863-882. [DOI] [PubMed] [Google Scholar]
- 26.Thackray, P. D., and A. Moir. 2003. SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber, A., S. A. Kögl, and K. Jung. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188:7165-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, M. A., D. Ringe, and G. A. Patsko. 2005. The atomic resolution crystal structure of the YajL (ThiJ) protein of Escherichia coli: a close prokaryotic homolog of the Parkinsonism-associated protein DJ-1. J. Mol. Biol. 353:678-691. [DOI] [PubMed] [Google Scholar]
- 30.Wood-Kaczmar, A., S. Gandhi, and N. W. Wood. 2006. Understanding the molecular causes of Parkinson's disease. Trends Mol. Med. 29:215-225. [DOI] [PubMed] [Google Scholar]
- 31.Yen, C., L. Green, and C. G. Miller. 1980. Degradation of intracellular protein in Salmonella typhimurium peptidase mutants. J. Mol. Biol. 143:21-33. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, Y., D. Liu, W. D. Kaluarachchi, H. D. Bellamy, M. A. White, and R. O. Fox. 2003. The crystal structure of Escherichia coli heat shock protein YedU reveals three potential catalytic active sites. Protein Sci. 12:2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]




