Abstract
In the oligotrophic freshwater bacterium Caulobacter crescentus, d-xylose induces expression of over 50 genes, including the xyl operon, which encodes key enzymes for xylose metabolism. The promoter (PxylX) controlling expression of the xyl operon is widely used as a tool for inducible heterologous gene expression in C. crescentus. We show here that PxylX and at least one other promoter in the xylose regulon (PxylE) are controlled by the CC3065 (xylR) gene product, a LacI-type repressor. Electrophoretic gel mobility shift assays showed that operator binding by XylR is greatly reduced in the presence of d-xylose. The data support the hypothesis that there is a simple regulatory mechanism in which XylR obstructs xylose-inducible promoters in the absence of the sugar; the repressor is induced to release DNA upon binding d-xylose, thereby freeing the promoter for productive interaction with RNA polymerase. XylR also has an effect on glucose metabolism, as xylR mutants exhibit reduced expression of the Entner-Doudoroff operon and their ability to utilize glucose as a sole carbon and energy source is compromised.
d-Xylose is the main constituent of xylan polymers, the major component of hemicellulose in plant cell walls. This pentose, in monomeric or polymerized form, is often present in habitats containing plant-derived biomass. The aquatic bacterium Caulobacter crescentus avidly consumes d-xylose as a carbon and energy source. d-Xylose metabolism in C. crescentus proceeds via a novel pathway initiated by the enzyme xylose dehydrogenase (XDH) (2, 24). The enzymes of this pathway are encoded in the xyl operon, whose expression is induced by d-xylose (9, 16, 24). In the work presented here, we identified the transcription factor controlling expression of the xyl operon and characterized the mechanism of regulation.
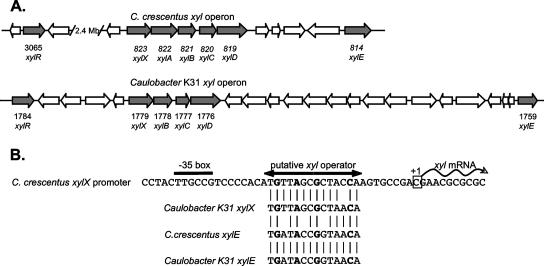
For the past decade, the xyl operon promoter (PxylX) has been the standard tool for engineering inducible gene expression in C. crescentus (1, 10, 12, 15, 20, 25, 26). This promoter is highly active when it is induced by addition of d-xylose to the medium and is virtually silent in the absence of xylose, provided that there is a single copy on the chromosome or it is in a low-copy-number plasmid vector. In its native chromosomal context, PxylX controls the expression of the five-gene xyl operon (CC0823 to CC0819) (Fig. 1) (9). A putative operator sequence in PxylX was identified by virtue of its conservation in several xylose-regulated promoters (9). This operator overlaps the −10 region of PxylX (Fig. 1), suggesting that a DNA binding protein interacting with this site would interfere with RNA polymerase holoenzyme binding and/or transcription initiation. The simplest model for control of the xyl regulon is that a repressor (“XylR”) binds to operator sites in the absence of xylose but dissociates from the operator when it is bound to xylose to allow gene expression. This model is consistent with the observation that a 4-bp mutation in the upstream half-site of the putative operator of PxylX, which did not affect the −10 region expected to interact with RNA polymerase, resulted in constitutive high-level expression of PxylX (9).
FIG. 1.
Caulobacter genomic loci involved in xylose metabolism and regulation. (A) The C. crescentus xyl operon (CC0823 to CC0819) and the surrounding region are shown on the top line. This region includes xylE (CC0814), which encodes a putative xylose transporter. The gene nomenclature is the nomenclature described previously (18, 24). The xylR gene (CC3065) is on the opposite side of the 4-Mb chromosome, and the approximate distance is indicated. In contrast, in the Caulobacter sp. strain K31 genome (shown below the C. crescentus operon and surrounding region), xylR is located very close to the xyl operon. Predicted coding regions in the K31 genome are based on the draft genome sequence and annotation publicly released by the U.S. Department of Energy's Joint Genome Institute (http://genome.jgi-psf.org/draft_microbes/cau_k/cau_k.home.html and http://genome.ornl.gov/microbial/caul/). Homologous genes were identified using BLAST through the Integrated Microbial Genomics web portal (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). (B) Sequence of the C. crescentus xyl operon promoter (PxylX) region, with the start site and putative promoter and operator motifs indicated (9, 16). The four base pairs that were altered in constitutive mutants are indicated by bold type. Predicted XylR operators found upstream of the C. crescentus xylE and Caulobacter sp. strain K31 xylX and xylE genes are aligned with the C. crescentus PxylX operator.
We developed a genetic screen that identified the C. crescentus xylR gene and confirmed the location of the operator site for the xyl operon. Electrophoretic gel mobility shift assays demonstrated experimentally that DNA binding by XylR, a member of the LacI family of transcription factors, is sensitive to the presence of d-xylose. We also observed an unexpected regulatory connection between xylose and glucose metabolism that is distinctly different from any regulatory connection observed previously in bacteria.
MATERIALS AND METHODS
Bacterial strains and media.
C. crescentus strains were routinely grown in PYE broth or on PYE agar (6) supplemented with the following antibiotics as necessary: kanamycin (20 μg/ml in agar and 5 μg/ml in broth), oxytetracycline (2 μg/ml in agar and 1 μg/ml in broth), and nalidixic acid (20 μg/ml in agar). When indicated, cultures were grown on M2 minimal salts medium (6) containing glucose or xylose (10 mM). Escherichia coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar (21) supplemented with the following antibiotics as necessary: kanamycin (50 μg/ml) and oxytetracycline (10 μg/ml).
Construction of a conditional clpX mutant strain.
Two strains that were used for genetic analysis of xylose regulation were constructed by inserting the suicide plasmid pUJ174 (tetracycline resistant) or pUJ167 (kanamycin resistant) into the xylX (CC0823) locus. These plasmids carry a translational fusion of the complete, functional coding sequence of the clpX gene to the first 18 bp of the xylX coding sequence. The chromosomal clpX locus was then eliminated, as described by Jenal and Fuchs (12), such that clpX expression was effectively dependent on xylose induction. Growth of the resulting strains, UJ200 (tetracycline-resistant integrant of pUJ174) and UJ270 (kanamycin-resistant integrant of pUJ167), was dependent on the presence of d-xylose in the medium to induce expression of the essential clpX gene.
Transposon mutagenesis to identify the xylose repressor.
The mini-Tn5 transposon (5) was delivered on a suicide plasmid (pUT-Km1) into UJ200 by conjugation. Transposon insertion mutants were selected on PYE agar plates supplemented with nalidixic acid (20 μg/ml) and kanamycin (20 μg/ml). Potential xylR::Tn5 insertions were verified by transducing the Tn5 marker into strain LS1280, which harbors a xylX::gusA insertion (16), and selecting for kanamycin resistance. The site of Tn5 insertion was determined for transductants that showed constitutive (xylose-independent) expression of gusA. Tn5 insertion sites were mapped by direct sequencing of chromosomal DNA using the following oligonucleotide primers complementary to the ends of the kanamycin resistance cassette of Tn903: primer 698 (TCT AGA GTC GAC CTG CAG GC) and primer 699 (TAC CGA GCT CGA ATT CGG CC). Sequencing reaction mixtures (total volume, 20 μl) contained 500 ng of genomic DNA as the template and 10 pmol of the sequencing primer, and the manufacturer's protocol (Big Dye; Perkin-Elmer) was used with the following modifications: the annealing temperature was raised to 58°C, and the number of cycles was increased to 99. The sequencing reactions were performed with an ABI Prism 310 DNA sequencer or an ABI Prism 3100 Avant genetic analyzer. To avoid complications from the Pxyl::clpX fusion, subsequent experiments were performed with strains in which the disrupted xylR locus had been transduced with bacteriophage φCr30 into the wild-type NA1000 or CB15 strain background.
Enzyme assays.
The regulatory effects of xylR mutations were tested by assaying chromosomally expressed XDH activity, glucuronidase activity from a xylX::gusA insertion, and β-galactosidase activity from plasmid reporter constructs. Preparation of cell extracts and a spectrophotometric assay of NAD+-dependent XDH activity were carried out as described previously (24). Glucuronidase activity was assayed (16) in the LS1280 strain background, into which xylR::Tn5 loci had been transduced, using cultures grown in PYE medium in the presence or absence of 10 mM xylose. To examine the activity of PxylX and PxylE using lacZ transcriptional fusions, the promoter regions of these genes (including at least 200 bp upstream of the start codon) were cloned into the pRKlac290 vector, a low-copy-number (three to five copies per cell) tetracycline-resistant plasmid. (Plasmid pCS225, the PxylX reporter, has been described previously [16].) β-Galactosidase activity was determined by the standard Miller assay (16) using cultures grown in PYE medium in the presence or absence of 10 mM d-xylose or d-glucose. For comparison, cultures of wild-type C. crescentus strain NA1000 containing pCS225 or the pRKlac290 vector alone were grown under identical conditions and assayed.
Electrophoretic mobility shift assays.
C. crescentus strains were harvested from 500-ml PYE medium cultures during log-phase growth (optical density at 600 nm, 0.3 to 0.5). Cells were harvested by centrifugation using a cooled Beckman JA-10 rotor at 5,000 rpm at 4°C for 15 min. Cell pellets were washed once in 40 ml of lysis buffer (50 mM sodium phosphate [pH 7.0], 50 mM NaCl, 1 mM EDTA, 10% glycerol), centrifuged again, and resuspended in 5 ml of lysis buffer containing 10 μg/ml lysozyme. Cells were disrupted by sonication, and cell debris was removed by centrifugation at 13,000 rpm for 15 min. Supernatant (cell extract) was transferred to a sterile tube and stored on ice for immediate use or frozen at −70°C. The protein concentration was determined using a Bio-Rad protein assay kit. In gel shift experiments 20 μg of protein per lane was used. The target duplex DNA molecule consisted of two complementary 35-base oligonucleotides based on the xylX promoter region (5′-CGT CCC CAC ATG TTA GCG CTA CCA AGT GCC GAC GA and 5′-TCG TCG GCA CTT GGT AGC GCT AAC ATG TGG GGA CG; putative XylR operator underlined). The oligonucleotides were synthesized and labeled with Cy5 fluorophores at the 5′ end by Operon Technologies (Alameda, CA). The oligonucleotides were annealed by mixing equimolar amounts, denaturing the preparations in a thermocycler for 5 min at 95°C, and then cooling them slowly to room temperature for 1 h. Binding reaction mixtures (15 μl) contained 2.5 ng of annealed duplex operator DNA, 3 μl of 5× binding buffer (final concentrations, 150 mM KCl, 0.1 mM dithiothreitol, 0.1 mM EDTA, and 10 mM Tris [pH 7.4]), 1 μg of poly(dI-dC), and 20 μg of protein. Sugars were added to the concentrations described below. The binding reaction mixtures were incubated at room temperature for 10 min. Gel loading dye was added to each reaction mixture, and complex formation was analyzed on 6% nondenaturing polyacrylamide gels made and run with 1× Tris-borate-EDTA buffer. Gels were run at 100 V for 50 min and then visualized wet with a Typhoon imaging system (Molecular Dynamics). For quantitative analysis the ImageQuant software was used. Background counts were determined in lanes to which no protein had been added and were subtracted from the counts for lanes that included cell extracts.
Isolation of suppressor mutations in the operator site of PxylX.
Spontaneous mutations in which UJ200 reacquired the ability to grow in the absence of xylose were selected by plating on PYE agar. cis-acting point mutations in the xyl operator that disabled binding of XylR to PxylX were expected to be rare compared to spontaneous loss-of-function mutations in xylR. To distinguish the cis- and trans-acting mutations, the PxylX region was transduced into strain UJ270 by using bacteriophage φCR30 (27) and selecting for tetracycline resistance from the integrated pUJ174. For strains in which the xylose-independent growth phenotype was linked to tetracycline resistance, the location of the mutation was identified by sequencing the xylX promoter region in the transduced strain.
Molecular cloning of xylR.
The C. crescentus CC3065 locus and the homologous gene from Caulobacter sp. strain K31 (Caulo_DRAFT_1784) were amplified using PCR. Genomic DNA was purified from PYE medium cultures of each species using a Qiagen DNeasy kit by following the manufacturer's instructions for gram-negative bacteria. PCR primers were designed to anneal at least 100 bp outside the annotated stop and start codons of the coding regions and to incorporate an XhoI restriction site at one end of the amplified product and a HindIII site at the other end to facilitate cloning. The primer sequences were as follows: CC_3065_HindIIIfor, GTC AGC TAA GCT TCG CCG CCG CAC AGG ATG ATC GC; CC_3065_XhoIRev, GTC AGC TCG AGC GAC ACA AGC CGC GCC CCC TCA G; K31_1784_HindIIIFor, GTC AGC TAA GCT TCG GCG TGG TCG GGA CAG GCT TGA; and K31_1784_XhoIRev, GTC AGC TCG AGG CCG ACC GCG CCG CAT GCT CTA. GoTaq thermostable DNA polymerase (Promega Corp., Madison, WI) was used for PCR. PCR products were analyzed on 1% agarose gels, extracted using a Qiagen gel extraction kit, digested with the XhoI and HindIII restriction enzymes, ligated into XhoI/HindIII-digested pMR20 plasmid DNA, and electroporated into E. coli strain S17.1 with selection for oxytetracycline resistance. After identification of clones with the desired inserts, plasmids carrying the cloned xylR gene were transferred into C. crescentus wild-type and mutant strains by conjugation (6), with selection for oxytetracycline and nalidixic acid resistance.
RESULTS
Genetic identification of xylR in C. crescentus.
Previously published data (9, 16) suggested that the xyl operon was regulated by a repressor. In order to identify the gene encoding the “xylose repressor” (xylR), C. crescentus strains in which xylR null mutations could be positively selected were engineered. In strain UJ200, the clpX gene is expressed ectopically from the xyl operon promoter (PxylX), and the native clpX locus is eliminated. In this strain, clpX expression is normally dependent on xylose induction. ClpX is an essential ATPase subunit of the ClpXP protease, which in C. crescentus is required for cell cycle progression (12). As expected, the engineered strain grows only in the presence of xylose.
Eight kanamycin-resistant strains were isolated after strain UJ200 was mutagenized with mini-Tn5 and mutants able to grow without xylose were selected. To verify that xylose-independent growth of the mutant strains was specifically due to a transposon insertion, the two phenotypes (xylose-independent growth and kanamycin resistance) were mapped genetically by transduction using bacteriophage φCr30 (27). For several independent mutant strains in which complete linkage was observed, the transposon insertion site was mapped by DNA sequencing to gene CC3065 (Fig. 1 and 2). CC3065, referred to here as xylR, encodes a LacI family transcriptional repressor with an N-terminal helix-turn-helix DNA binding domain and a C-terminal sugar binding domain. The C. crescentus XylR sequence is aligned with the E. coli LacI sequence in Fig. 2. The greatest conservation occurs in the N-terminal DNA binding domain and hinge, but detectable similarity is evident throughout.
FIG. 2.
Caulobacter xylose repressor, XylR. The predicted polypeptide product of C. crescentus xylR (CC3065; GenBank accession number AAK25027) is aligned with the putative polypeptide products of Caulobacter sp. strain K31 xylR (gene 1784 in the draft sequence available in September 2007) and E. coli lacI (GenBank accession number AAA24457) (21). The locations of the Tn5 insertions in the xylR1.2::Tn5 and xylR4.6::Tn5 mutants are each indicated by a line under the residue encoded by the codon in which the Tn5 inserted. HTH, helix-turn-helix.
To avoid complications from the Pxyl::clpX fusion, subsequent experiments were performed with two strains in which disrupted xylR alleles (xylR4.6::Tn5 and xylR1.2::Tn5, whose insertion locations are shown in Fig. 2) had been transduced into a clean wild-type strain background, generating strains CS816 (xylR1.2::Tn5) and CS817 (xylR4.6::Tn5). Although the insertion sites are separated by nearly 600 bases, with the xylR4.6 insertion nearer the N-terminal DNA binding domain and xylR1.2 nearer the C terminus, the strains had similar phenotypes, and subsequent results suggested that both disrupted alleles are effectively null alleles.
The assumption of our selection scheme was that disruption of xylR would allow sufficient expression of the PxylX-clpX fusion for the engineered strain to grow in the absence of xylose. In this mutant, other genes of the xyl operon, including the xylB (CC0821) (Fig. 1) gene encoding XDH, are also predicted to be expressed constitutively. To verify this, XDH activity was examined in wild-type and mutant strains (Table 1). Significant XDH activity was seen in CB15 only when it was grown in the presence of xylose, as described by other workers (19, 25). In contrast, CS816 showed high XDH activity in the absence of xylose. When the intact xylR gene was expressed from a low-copy-number plasmid in a xylR mutant strain, repression of XDH activity in the absence of xylose was restored. Complementation by an episomal copy of xylR showed that the regulatory defect was not due to polar effects on other genes caused by the Tn5 insertion. Further evidence that XylR is responsible for transcriptional regulation of PxylX was obtained from an analysis of a lacZ reporter fused to this promoter (Table 2). β-Galactosidase activity was very low in the wild-type strain background in the absence of xylose but was constitutively high in the mutant strain.
TABLE 1.
XDH is constitutively expressed in a xylR mutant
| Strain background | Complementing plasmid | d-Xylose induction | XDH activity (nmol NADH produced min−1 mg protein−1) |
|---|---|---|---|
| CB15 | − | 0.7 ± 0.6 | |
| + | 17.9 ± 5.4 | ||
| CS816 (xylR1.2::Tn5) | − | 39.7 ± 6.7 | |
| + | 29.4 ± 7.4 | ||
| CS816 (xylR1.2::Tn5) | pDT2 (containing xylR from CB15) | − | 0.5 ± 0.4 |
| + | 23.0 ± 9.7 | ||
| CS816 (xylR1.2::Tn5) | pDT4 (containing xylR from K31) | − | 0.6 ± 0.5 |
| + | 19.0 ± 7.4 |
TABLE 2.
Expression of promoter-lacZ fusions in wild-type and xylR mutant strains
| Strain background | Promoter driving lacZ | d-Xylose induction | β-Galactosidase activity (Miller units) |
|---|---|---|---|
| CB15 | xylX | − | 5.8 ± 9.4 |
| + | 765 ± 47 | ||
| xylE | − | 294 ± 54 | |
| + | 713 ± 114 | ||
| edo | − | 1,730 ± 326 | |
| + | 1,930 ± 501 | ||
| CS816 (xylR1.2::Tn5) | xylX | − | 1,170 ± 249 |
| + | 1,000 ± 87 | ||
| xylE | − | 986 ± 370 | |
| + | 931 ± 347 | ||
| edo | − | 1,180 ± 390 | |
| + | 922 ± 344 |
The C. crescentus xylose regulon contains over 50 genes whose expression is stimulated by d-xylose, many of which share a conserved putative operator motif with the xyl operon (9). As a test of whether xylR may control the expression of other members of this regulon, we examined expression of CC0814, which is located near the xyl operon but is not part of the same transcription unit (Fig. 1). CC0814 encodes a predicted membrane-spanning protein related to the XylE xylose:H+ symporter of E. coli (GenBank accession number AAA79016; 36% identity over 490 amino acids with the CC0814 protein; E = 3 × 10−64) (4). Preliminary experimental evidence supported the hypothesis that the CC0814/xylE gene is involved in xylose uptake, as a knockout mutant was defective in growth on and transport of d-xylose (data not shown). Published microarray data showed that there was a 3.5-fold increase in expression of this gene in M2 xylose medium compared with M2 glucose medium (9). Expression of C. crescentus xylE was examined using a plasmid-borne promoter fusion to lacZ. Expression of the xylE promoter also became xylose independent in a xylR mutant strain (Table 2). The fact that PxylE expression is not as tightly regulated as PxylX expression in wild-type C. crescentus could result from differences in the operator sequence (Fig. 1B) or operator positioning within the promoter. The PxylE transcription start site has not been determined, and Caulobacter promoter elements are often difficult to predict purely from the sequence, so it is difficult to say at this point how XylR might affect PxylE.
Gel shift analysis of XylR.
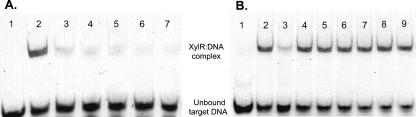
An electrophoretic gel mobility shift assay was used to investigate the interaction of XylR with target DNA. The target DNA, a fluorescently labeled duplex oligonucleotide representing the xylX promoter region, was designed so that it was centered on the putative operator element, a 14-bp palindromic sequence identified based on its conservation upstream of 15 genes whose expression is induced by xylose (9). Previous work showed that a multiple-base change in the left side of this sequence renders Pxyl constitutively active (9). In the xyl operon promoter, this sequence overlaps the “−10” region upstream of the transcription initiation site (16). Cell extracts from C. crescentus CB15 formed a discrete complex (Fig. 3A, lane 2) with the synthetic target DNA. When extracts from two distinct xylR mutant strains were used in the gel shift assay under identical conditions, over 80% of the signal was eliminated from the complex (Fig. 3A, lanes 4 and 6). The residual DNA-protein complex observed with extracts from the xylR mutant strains may reflect in vitro interactions of the target DNA with other proteins, whose significance in vivo is unknown. The intensity of the PxylX-protein complex with extracts from wild-type CB15 was similarly reduced in the presence of 1 mM d-xylose (Fig. 3A, lane 3), consistent with a model in which interaction of XylR with d-xylose dramatically reduces the affinity of XylR for the operator site. Titration of the PxylX-XylR complex with various concentrations of d-xylose indicated that under these conditions the dissociation constant for d-xylose is in the range from 20 to 50 μM (data not shown), consistent with published observation that 20 μM d-xylose activates PxylX expression in vivo, but not to its maximal extent (16).
FIG. 3.
Gel mobility shift analysis of XylR binding to the PxylX operator. Binding reactions and electrophoresis conditions are described in Materials and Methods. (A) Lane 1, no cell extract added (negative control); lane 2, CB15 extract; lane 3, CB15 extract with 1 mM d-xylose; lane 4, CS816 (xylR1.2::Tn5) extract; lane 5, CS816 (xylR1.2::Tn5) extract with 1 mM d-xylose; lane 6, CS817 (xylR4.6::Tn5) extract; lane 7, CS817 (xylR4.6::Tn5) extract with 1 mM d-xylose. (B) Lanes 2 to 9 all contained cell extract from CB15. Lane 1 contained the negative control with no cell extract added, and lane 2 contained the positive control with CB15 cell extract and no added sugar. For the binding reactions shown in lanes 3 to 9, various sugars or amino acids were present at a concentration of 1 mM (lane 3, d-xylose; lane 4, l-xylose; lane 5, d-arabinose; lane 6, l-arabinose; lane 7, d-ribose; lane 8, d-glucose; lane 9, l-glutamate).
To investigate the specificity of the XylR interaction with the inducer, several related pentoses (l-xylose, l-arabinose, d-arabinose, and d-ribose) were tested to determine their effects on the protein-DNA complex. At a concentration of 1 mM, only d-xylose induced nearly complete dissociation of XylR from target DNA (Fig. 3B). Minor reductions in the XylR-DNA complex level were seen in the presence of some other pentoses, although there was considerable variation between experiments. d-Ribose and l-arabinose, which resulted in 9 and 15% reductions in the XylR-DNA complex level (Fig. 3B), respectively, were also found to induce moderate expression of the PxylX-lacZ fusion and XDH activity in CB15 in vivo (unpublished data). The extent to which these sugars bind XylR directly will be determined in future experiments with purified XylR. The results of the experiments presented here do not rule out the possibility that the inducing ligand for XylR is a d-xylose (or related pentose)-derived metabolite, analogous to the situation with allolactose and E. coli LacI.
Characterization of the XylR operator sequence.
To confirm the location of the XylR operator, spontaneous point mutations in PxylX were selected that allowed xylose-independent growth of strain UJ200. Strains containing potential operator mutations were screened for by transductional mapping, which yielded roughly 50 strains in which the xylose-independent phenotype showed close linkage to the xyl locus. The xyl promoter region was amplified by PCR and sequenced for 13 strains randomly chosen from this collection. Each strain contained only a single base change, and four different mutations were identified (Fig. 1B): C to T at position −10 (three isolates), G to A at position −15 (three isolates), A to G at position −18 (three isolates), and G to A at position −21 (four isolates). The affected bases are all conserved in the 14-bp motif previously proposed to be the XylR operator (9).
XylR is conserved in Caulobacter sp. strain K31.
Four of the five genes of the C. crescentus xyl operon and the critical NAD-dependent d-xylose dehydrogenase activity are conserved in Caulobacter sp. strain K31 (14, 24) (Fig. 1), a chlorophenol-resistant groundwater isolate whose genome was recently sequenced. A gene encoding a putative xylE transporter is also highly conserved in the K31 genome (gene 1759; 83% identity with the C. crescentus xylE product over 480 amino acids). The xyl operon and the xylE-like gene are not present in the genomes of other α-Proteobacteria, including relatively closely related stalked species (Maricaulis maris, Hyphomonas neptunium, and Oceanicaulis alexandrii) and nonstalked species (e.g., Sinorhizobium meliloti, Bradyrhizobium japonicum, and Agrobacterium tumefaciens).
Given the conservation of key components of the xylose regulon between C. crescentus and Caulobacter sp. strain K31, we sought to determine whether xylose-dependent regulation is similarly conserved. XDH activity was assayed in cell extracts from cultures grown in PYE medium in the presence or absence of d-xylose. XDH activity was undetectable in extracts from K31 grown in the absence of xylose but was induced to a level comparable to the C. crescentus CB15 level in PYE medium supplemented with 1 mM d-xylose (24.3 nmol NADH produced min−1 mg−1 protein in K31, compared to 17.9 nmol NADH produced min−1 mg−1 protein for CB15). A XylR-like gene (Caulo_DRAFT_1784) is located in close proximity to the xyl operon in K31 (Fig. 1A). These genes are reciprocal best matches in the two genomes, and in their products there is 81% amino acid identity over 349 residues (Fig. 2). In light of the nearly complete conservation of the N-terminal helix-turn-helix DNA binding domain, the XylR operator is expected to be highly conserved in K31 as well. The region upstream of K31 xylX contains a sequence matching the C. crescentus xylX operator at 13 of 14 bases, including the four critical bases identified genetically (Fig. 1B). The regions upstream of xylE in the two organisms contain identical putative operator sequences that match the C. crescentus xylX operator at 10 of 14 bp, including the four critical bases.
To determine whether the putative K31 XylR is functionally interchangeable with C. crescentus XylR, the gene was amplified by PCR and cloned into the same low-copy-number plasmid vector (pMR20) used in the previous experiment. When this construct was introduced into CS816, xylose-dependent regulation of XDH expression was fully restored (Table 2), demonstrating that the K31 gene has the same regulatory function, repressing expression of the xyl operon in the absence of d-xylose.
XylR is necessary for normal glucose metabolism in C. crescentus.
The CC3065 gene was previously identified as a locus in which Tn5 insertions resulted in an inability to form colonies on M2G minimal salts agar plates containing 10 mM glucose as the sole carbon source, while the ability to grow on M2X minimal salts agar plates with d-xylose was retained (24). The xylR mutant strains isolated in the current work exhibited similar behavior, forming colonies on M2X agar but not on M2G agar. Colonies readily formed on M2GX agar (M2 agar with both glucose and xylose), demonstrating that glucose is not overtly toxic to these strains. In liquid culture, xylR mutant strains showed very little growth in M2G medium after 1 to 3 days of incubation at 30°C. Surprisingly, when growth in PYE or M2X medium was examined, supplementation of these media with glucose enhanced the growth yields similarly for both wild-type and xylR mutant strains (data not shown). This observation suggests that although xylR mutant strains do not effectively utilize glucose as a sole carbon and energy source in M2 medium, they have not completely lost the capacity to transport and metabolize glucose.
In C. crescentus, the Entner-Doudoroff pathway is responsible for the initial steps of glucose catabolism (9). The enzymes of this pathway are largely encoded in the edo operon, comprising genes CC2054 to CC2057. The promoter upstream of CC2057 is constitutively active in PYE medium, but expression increases roughly twofold when glucose is added or when its activity in M2G medium is compared with its activity in M2X medium (9). To examine possible effects of XylR on edo operon expression, the activities of a lacZ fusion to the edo promoter were compared in CB15 and CS816 cultures growing exponentially in PYE broth in the presence or absence of d-xylose (Table 1). Expression of this promoter was not sensitive to the presence of xylose but was reduced nearly twofold in the xylR mutant strain. If levels of the Entner-Doudoroff pathway enzymes are concomitantly reduced in a xylR mutant, a reduction in the growth rate on glucose as the sole carbon and energy source might be expected, although this may not account for the mutant's complete inability to grow on glucose in M2 medium.
DISCUSSION
d-Xylose induces expression of over 50 genes in C. crescentus (9), including an operon encoding the enzymes responsible for xylose metabolism (24). We describe here identification of a gene encoding a transcriptional repressor, XylR, which is responsible for regulation of the xylose operon. Our data are consistent with a simple regulatory mechanism: in the absence of d-xylose, XylR binds to operator sites overlapping promoters of xylose-inducible genes, preventing initiation of transcription by RNA polymerase. When d-xylose is present, it binds to XylR, stabilizing a conformation with greatly reduced affinity for DNA and freeing xylose-inducible promoters for expression. Given the evolutionary relationship of XylR to E. coli LacI, it is fitting that this mechanism mirrors the seminal model of Jacob and Monod (11) for lac operon regulation by the Lac repressor. Subsequent work on the Lac repressor and lac operon revealed somewhat greater complexity, in that the repressor actually forms a tetramer and interacts with multiple operators, looping out the intervening DNA (7, 17). Whether this is the case with XylR awaits more in-depth biochemical characterization, but there is no evidence at present that more than one operator site is involved in PxylX regulation.
The xylose binding sites of the Caulobacter XylR and XDH proteins presumably evolved independently and converged on structures specific for d-xylose, as there is no obvious sequence similarity between these proteins. It will be of interest to determine whether XylR can interact productively with pentoses such as l-arabinose and d-ribose, which gave hints of in vivo and in vitro effects on XylR. l-Arabinose is a common component of hemicellulose, although typically it is much less abundant than d-xylose. C. crescentus grows slowly on l-arabinose, and this sugar is used (albeit poorly) by XDH (24). In contrast, neither d-arabinose, d-ribose, nor l-xylose is utilized by XDH (24), nor are these compounds effective growth substrates. Interestingly, both d-ribose and l-xylose may have some effect on XylR, as they induce some expression of PxylX and XDH in vivo (data not shown). Such sugars could be useful as “gratuitous inducers” of XylR-regulated gene expression, akin to using isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer of Plac in E. coli.
The LacI family of transcription factors is abundantly represented in the C. crescentus and K31 genomes, with 11 and 15 predicted members, respectively. The closest relatives of these organisms whose genome sequences are known, M. maris and H. neptunium (both of which are stalked bacteria from marine habitats), have only five and one LacI family members, respectively. Neither species contains a xyl operon or an obvious ortholog of XylR. In C. crescentus and K31, XylR is most closely related to the product of CC2053 (C. crescentus gene nomenclature), which lies next to the operon encoding the Entner-Doudoroff pathway for glucose catabolism (9). It is reasonable to hypothesize that CC2053 controls expression of the Entner-Doudoroff operon and that XylR functionally diverged after duplication of the CC2053 progenitor in the freshwater Caulobacter lineage, after it diverged from the marine branch and acquired the pathway for xylose metabolism.
The mechanisms of both xylose metabolism and xylose-dependent transcriptional regulation in Caulobacter are distinct from other well-characterized bacterial systems. Most bacteria known to consume d-xylose utilize xylose isomerase and xylulokinase and feed the resulting xylulose-5-phosphate into the pentose phosphate pathway. In contrast, Caulobacter processes d-xylose to produce α-ketoglutarate (24). With respect to regulation, Caulobacter XylR is the first member of the LacI family shown to control xylose metabolism. The xylose repressor identified in several gram-positive bacteria (e.g., Bacillus and Lactobacillus species) behaves like Caulobacter XylR, releasing operator DNA in the presence of xylose to allow transcription (8), but it is evolutionarily derived from the ROK/NagC superfamily (3). In contrast, XylR of E. coli and related gram-negative γ-Proteobacteria is a member of the AraC family of transcriptional activators (23) and is stimulated to bind operator DNA by xylose.
Caulobacter seems to be unusual among bacteria in not allowing glucose to block xylose-dependent gene induction (16), as it does in both gram-positive and gram-negative enteric bacteria, although by different means (CcpA and cAMP receptor protein, respectively) (13, 22, 23). The apparent defect in glucose metabolism seen in the C. crescentus xylR mutant strains suggests there may be a regulatory connection between glucose and xylose metabolism. Reduced expression of the edo operon could contribute to this growth defect. It is also possible that elevated expression of the xylose regulon in the absence of xylose results in metabolic patterns that are harmful during glucose-dependent growth. For example, elevated expression of fructose bisphosphate aldolase, a gluconeogenic enzyme whose expression has been shown to increase threefold during growth on d-xylose (9), might create a futile cycle during growth of the xylR mutant on glucose. Further physiological characterization of the xylR mutant is needed to elucidate the role of this transcription factor in coordinating carbon metabolism. As an oligotrophic microbe adapted to scavenging multiple carbon sources simultaneously, C. crescentus is an appropriate model for exploring integrative regulation of metabolism.
Acknowledgments
This work was supported by U.S. National Science Foundation grant MCB-0317037 to C.S., as well as by Swiss National Science Foundation fellowship 3100A0-108186 to U.J. Financial support was also provided by grants to C.S. from Santa Clara University.
We gratefully acknowledge the help of the students of the 2003 and 2004 “Advanced Bacterial Genetics” summer courses at Cold Spring Harbor Laboratory, who isolated xyl and xylR mutant strains using screens developed in the Jenal lab. We also gratefully acknowledge Debbie Thurtle, Alana Okamoto, Vidyodhaya Sundaram, Andrew Spencley, Aloe Driscoll, Lindsey Herzog, Amanda Lieu, Aylene Bao, and other undergraduate students in the Bio 176 Recombinant DNA Technology course at Santa Clara University who contributed to this project.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Biondi, E. G., J. M. Skerker, M. Arif, M. S. Prasol, B. S. Perchuk, and M. T. Laub. 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 59:386-401. [DOI] [PubMed] [Google Scholar]
- 2.Brouns, S. J., J. Walther, A. P. Snijders, H. J. van de Werken, H. L. Willemen, P. Worm, M. G. de Vos, A. Andersson, M. Lundgren, H. F. Mazon, R. H. van den Heuvel, P. Nilsson, L. Salmon, W. M. de Vos, P. C. Wright, R. Bernander, and J. van der Oost. 2006. Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J. Biol. Chem. 281:27378-27388. [DOI] [PubMed] [Google Scholar]
- 3.Dahl, M. K., D. Schmiedel, and W. Hillen. 1995. Glucose and glucose-6-phosphate interaction with Xyl repressor proteins from Bacillus spp. may contribute to regulation of xylose utilization. J. Bacteriol. 177:5467-5472, 1995 Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, E. O., and P. J. Henderson. 1987. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 262:13928-13932. [PubMed] [Google Scholar]
- 5.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, A. M., T. O. Fischmann, and T. A. Steitz. 1995. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science 268:1721-1727. [DOI] [PubMed] [Google Scholar]
- 8.Gärtner, D., J. Degenkolb, J. A. Ripperger, R. Allmansberger, and W. Hillen. 1992. Regulation of the Bacillus subtilis W23 xylose utilization operon: interaction of the Xyl repressor with the xyl operator and the inducer xylose. Mol. Gen. Genet. 232:415-422. [DOI] [PubMed] [Google Scholar]
- 9.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iniesta, A. A., P. T. McGrath, A. Reisenauer, H. H. McAdams, and L. Shapiro. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA 103:10935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318-356. [DOI] [PubMed] [Google Scholar]
- 12.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laikova, O. N., A. A. Mironov, and M. S. Gelfand. 2001. Computational analysis of the transcriptional regulation of pentose utilization systems in the gamma subdivision of Proteobacteria. FEMS Microbiol. Lett. 205:315-322. [DOI] [PubMed] [Google Scholar]
- 14.Männistö, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 15.McGrath, P. T., A. A. Iniesta, K. R. Ryan, L. Shapiro, and H. H. McAdams. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124:535-547. [DOI] [PubMed] [Google Scholar]
- 16.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mossing, M. C., and M. T. Record, Jr. 1986. Upstream operators enhance repression of the lac promoter. Science 233:889-892. [DOI] [PubMed] [Google Scholar]
- 18.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poindexter, J. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Schmiedel, D., and W. Hillen. 1996. Contributions of XylR, CcpA, and Cre to diauxic growth of Bacillus megaterium and to xylose isomerase expression in the presence of glucose and xylose. Mol. Gen. Genet. 250:259-266. [DOI] [PubMed] [Google Scholar]
- 23.Song, S., and C. Park. 1997. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179:7025-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens, C., B. Christen, T. Fuchs, V. Sundaram, K. Watanabe, and U. Jenal. 2007. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 189:2181-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell-cycle regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. USA 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, J. W., and M. R. Alley. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183:5001-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West, L., D. Yang, and C. Stephens. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]