Abstract
R388 conjugative relaxase TrwC acts as a site-specific recombinase, promoting recombination between two cognate oriTs on double-stranded DNA substrates. The relaxosome component TrwA is also required for efficient recombination. In this work we present data on the in vivo control of this reaction by host proteins that affect local DNA topology. In the absence of TrwA, binding of integration host factor (IHF) to the oriT keeps the recombination levels low, probably by keeping the relaxosome complex, formed at recombination locus 1, in a “closed” conformation. In an IHF-deficient (IHF−) background, the formation of a transcript elongation complex at this locus still hampers recombination. A mutation abating the promoter sequence at locus 1, or repression of transcription by exposure to rifampin, lifts the inhibition imposed on recombination in an IHF− background. We also observe an increase in conjugation efficiency under these conditions. Relieving the inhibition imposed by these host factors allows efficient levels of recombination between short oriT loci in the absence of TrwA. The presence of TrwA counteracts these inhibitory effects. TrwA would then activate both recombination and conjugation by switching the conformation of the relaxosome to an “open” form that exposes single-stranded DNA at the nic site, promoting the initial TrwC nicking reaction.
Bacterial conjugation is a mechanism for horizontal gene transfer between bacteria. The conjugative machinery is currently comprehended as three distinct functional modules: the relaxosome, which is the nucleoprotein complex that processes DNA for transfer; a type IV secretion system, which provides the transmembrane conduit for the DNA transfer; and the coupling protein, which links the relaxosome to the secretion system (21). Conjugative DNA processing by the relaxosome starts by a strand-specific nicking of the plasmid oriT at the nic site by the action of the relaxase protein. By a specialized DNA replication process, the plasmid DNA is subsequently transferred as a single-stranded substrate to the recipient cell, piloted by the relaxase, which presumably recircularizes the DNA (22).
In the conjugative plasmid R388, the relaxosome is formed by the oriT plus three proteins: the accessory proteins TrwA and integration host factor (IHF) and the relaxase TrwC (27). The conjugation accessory protein TrwA is a 53-kDa tetrameric DNA binding protein. It has been assigned to the ribbon-helix-helix family of proteins (25), which are associated with transcriptional repression processes (1). TrwA binds specifically to two sites at oriT, sbaA and sbaB, lying on direct repeats 1 and 2 and on inverted repeat 4 (IR4), respectively (26) (Fig. 1). TrwA binding to oriT leads to an increase in TrwC nicking activity as well as the transcriptional repression of the trwABC operon. Deletion of trwA shows a 105-fold reduction in assays of mobilization of an oriT-containing plasmid (26).
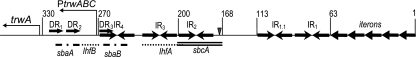
FIG. 1.
Schematic representation of R388 oriT. Coordinates are given as in reference 19. Arrows indicate the presence of iterons, direct repeats (DRs) and IRs. The nic site is shown as a vertical arrowhead. TrwA and IHF binding sites are underlined with dashed and dotted lines, respectively. The TrwC binding site is underlined with a double line.
IHF is a heterodimeric protein, encoded by genes himA and hip (10, 24). IHF bends DNA upon binding to a 30- to 35-bp region containing a 13-bp consensus sequence, 5′-WATCAAN4TTR-3′ (13). IHF binding to DNA participates in the formation of high-order DNA-protein complexes needed in various DNA metabolic processes. Footprinting assays revealed that there are two IHF binding sites at R388 oriT, an ihfA site (bp 203 to 215) and an ihfB site (bp 277 to 289) (27). These sites are placed alternating with TrwA binding sites within the promoter region of the trwABC operon (Fig. 1). No significant difference in the conjugation frequency of R388 in IHF-deficient (IHF−) strains used either as donor or recipient cells is observed (19), yet its binding to the oriT inhibits nic cleavage by TrwC (27). It has been proposed that IHF binding to oriT induces a structural change in the DNA topology, which in turns induces rigidity around the nic site, forming a “closed” relaxosome that impedes oriT melting and nicking by TrwC.
The relaxase TrwC is a bifunctional enzyme, comprising an N-terminal relaxase domain (N293), which displays supercoiled DNA nicking, single-stranded DNA cleavage, and DNA strand-transferase abilities, and a C-terminal domain, where DNA helicase and ATPase activities are localized (23). In addition, a third domain involving the N-terminal 600 residues has been associated with the ability of TrwC to mediate efficient oriT-oriT recombination on double-stranded DNA (dsDNA) substrates (4). Furthermore, TrwC is the only relaxase described to promote site-specific integration of a conjugatively transferred plasmid DNA into a dsDNA oriT copy present in the recipient cell (9).
The oriT-specific recombination activity has been previously characterized (4). In addition to the catalytic activity of TrwC, TrwA was described to be necessary for efficient recombination. Two distinct recombination loci, i.e., minimal oriT sites needed for efficient recombination, were described, oriT1 and oriT2. The two recombination loci showed different DNA requirements. Intriguingly, both could be deleted of TrwA binding sites without affecting the reaction efficiency. These substrates without the sba sites behave as better substrates for TrwC-mediated recombination than full-length oriTs in the absence of TrwA. Deletion of the sba sites also removes the IHF binding sites, ihfA and ihfB. Based on the previously described inhibition of TrwC nicking activity by IHF (27) and the positive effect of deletion of the IHF binding sites on recombination, it has been proposed that IHF binding to oriT inhibits TrwC-mediated recombination (4).
The ability to promote site-specific recombination between two cognate oriTs in the absence of conjugation is rare in relaxases, although it is not unique. As for TrwC (4, 9, 20), a similar oriT-oriT recombination ability has been described for three additional relaxases, NikB of the IncI1 plasmid R64 (14) and the relaxases of Enterococcus faecalis plasmids pAD1 (12) and pAMα1 (11). In the case of the oriT recombination catalyzed by NikB, the reaction on dsDNA substrates was strictly dependent on the presence of protein NikA, which is homologous to TrwA (14). Conversely, on single-stranded DNA (ssDNA) substrates NikB was sufficient to promote recombination, although NikA does accelerate the reaction. A hypothetical role for NikA in the generation of ssDNA for efficient recombination was proposed by those authors (14). Similarly, in TrwC-mediated recombination, we have observed a dependence on situations such as replication or transcription that favors ssDNA exposure at recombination locus oriT1 (4).
In this work we characterize the roles of host factors in TrwC-mediated recombination on dsDNA substrates. We describe a regulatory module, in which TrwA and IHF proteins act as enhancer and inhibitor of the reaction, respectively. Additionally, we show how the formation of an active transcription elongation complex at oriT1 is inhibitory for site-specific recombination. We obtained elevated recombination efficiencies on substrates containing short oriT copies in the absence of TrwA in an IHF− background. The conjugation frequency was also enhanced under these conditions. Overall, the data presented suggest that TrwA counteracts the effect of host factors that maintain the relaxosome in a “closed” topological conformation for TrwC nicking at oriT, which is presumably a limiting step for the initiation of both conjugation and recombination processes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli strain DH5α (16) was used as a host for the recombination assays on a wild-type IHF (IHF+) background. For the IHF− background, we constructed strain CIG1 (see below). Bacterial plasmids used in this work are listed in Table 1. Luria-Bertani broth was used for bacterial growth and was supplemented with agar for solid culture. Selective media included antibiotics at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 25 μg/ml; kanamycin (Km), 50 μg/ml; nalidixic acid (Nx), 20 μg/ml; streptomycin (Sm), 300 μg/ml; spectinomycin (Sp), 100 μg/ml; and trimetroprim (Tp), 20 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was supplied at a concentration of 60 μg/ml.
TABLE 1.
Plasmids used in this work
| Plasmid | Description | Construction,a reference, or source |
|---|---|---|
| pCIG1028 | oriT1(1-402)-oriT2(1-330); Cmr Kmr | 4 |
| pCIG1043 | oriT1(1-402)-oriT2(1-200); Cmr Kmr | 4 |
| pCIG1044 | oriT1(1-402)-oriT2(1-270); Cmr Kmr | pCIG1028 (Kpn1+BamH270) |
| pCIG1046 | oriT1(1-200)-oriT2(1-200); Cmr Kmr | 4 |
| pCIG1047 | oriT1(1-270)-oriT2(1-330); Cmr Kmr | pCIG1028 (Xba1+Hind270) |
| pCIG1049 | oriT1(1-200)-oriT2(1-330); Cmr Kmr | 4 |
| pCIG1050 | oriT1(1-270)-oriT2(1-270); Cmr Kmr | pCIG1044 (Xba1+Hind270) |
| pCIG1077 | pKK223-3::PABC-trwA-trwL; Apr | pSU1529 (TrwA-pmob-Eco+TrwA-stop-Eco)b |
| pCIG1083 | oriT1(168-200)-oriT2(1-191); Cmr Kmr | 4 |
| pCIG1106 | oriT1(−10 box mut)-oriT2(1-330); Cmr Kmr | pCIG1028 (Xba1+−10boxXho) |
| pCIG1108 | oriT1(1-330)-oriT2(1-330); Cmr Kmr | pCIG1028 (Xba1+Hind330) |
| pET29c | Cloning vector; Kmr | Novagen |
| pET::trwA | pET3a::trwA; Apr | 4 |
| pET::trwAC | pET3a::PABC-trwAC; Apr | 9 |
| pKD20 | λRed plasmid; Apr | 5 |
| pSU1376 | pSU19::oriT(1-330); Cmr | 19 |
| pSU1529 | pKK223-3::trwB-trwL; Apr | 18 |
| pSU1621 | pET3a::trwC; Apr | 17 |
| R388 | IncW natural conjugative plasmid; Tpr | 6 |
Plasmids were made by insertion of a PCR-amplified fragment into the indicated plasmid. Oligonucleotides used for the PCR are indicated in parentheses and are described in Table 2.
For pCIG1077, a PCR-amplified fragment containing trwA under the control of its own promoter was inserted into pSU1529, and the orientation was selected so that trwA was transcribed in the same direction as the adjacent trwB and trwC genes.
Plasmids and strain constructions.
E. coli strain CIG1 was constructed by chromosomal inactivation of the hip gene on a DH5α strain by the method of Datsenko and Wanner (5). Oligonucleotides Sp_hip_5 and Sp_hip_3 (Table 2) were used to amplify an Sp resistance cassette from a mini-Tn5 Sm/Sp cassette DNA template (8) containing at the ends 40 bp homologous to the 5′ and 3′ regions of the target gene sequence. One hundred nanograms of the PCR product was transformed into arabinose-induced DH5α cells harboring a plasmid, pKD20, coding for an l-arabinose-inducible λRed recombinase. Transformed cells were grown at 30°C and plated on LB agar plus Sp. Colonies were tested for positive inactivation of hip by PCR analysis with oligonucleotides Hip5 and Hip3 (Table 2).
TABLE 2.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| −10boxXho | AACAAGCTTTCCCGTAGTGTTACTGTAGTGGTTCACTCGAGGCATTTACAAGGGGT |
| BamH270 | AACGGATCCATTGTAGTGGCATAA |
| Hind270 | CCAAAGCTTATTGTAGTGGCATAACAC |
| Hind330 | AACAAGCTTCCTCTCCCGTAGTGTTAC |
| Hip3 | AACGAAAGGGTGAAAACTG |
| Hip5 | AAGCTTTCAAAGCAGCTAA |
| Kpn1 | CCAGAATTCATGTAACTAGGTACCCTCATTTTCTGC |
| Sp_hip_3 | CAAGTTTGAGTAAAAAACTTAACCGTAAATATTGGCGCGAAGACATTATTTGCCGACTAC |
| Sp_hip_5 | ATGACCAAGTCAGAATTGATAGAAAGACTTGCCACCCAGCGTAACGGCGCAGTGGCGGTT |
| Xba1 | CCATCTAGACTCATTTTCTGCATCAATC |
| TrwA_pmob_Eco | CCAGAATTCCTACAATATTGCCGCAAC |
| TrwA_stop_Eco | TCAGAATTCAATCCTCCTTCCCCTC |
The added restriction sites that were used for cloning into the same sites of the vector plasmids are underlined.
Plasmids were constructed using standard methods (29). Their construction is outlined in Table 1. Inserts were obtained by PCR with primers (Table 2) incorporating restriction sites adequate for ligation into the same sites of the vector. Restriction enzymes, shrimp alkaline phosphatase, and T4 DNA ligase were purchased from Fermentas. Vent polymerase was purchased from New England Biolabs. DNA sequences of all cloned PCR segments were determined.
Recombination assays.
All substrate plasmids to test recombination are derivatives of plasmid pCIG1028 (4), which carries two directly repeated copies of R388 oriT separated by a Kmr gene and a lacIq gene. Intramolecular recombination was tested as described previously (4). Briefly, a lacZΔM15 strain harboring the substrate plasmid (pCIG1028 or its derivatives) and a helper plasmid coding for TrwC (plus TrwA when indicated) was grown as described previously (4) and plated on selective media with X-Gal. Recombination between the two oriT copies of the substrate plasmid induces expression of the downstream lacZα gene. Recombination frequency is estimated by the number and aspect of blue colonies. The strict correlation between recombination and the appearance of blue color was previously confirmed by DNA restriction analysis (4).
Rif treatment.
E. coli strains CIG1and DH5α harboring plasmid pSU1621 (coding for TrwC) and pCIG1028, a substrate plasmid with full-length oriTs, were grown at 37°C overnight to saturation on a selective LB broth containing Km and Ap. Since the Km resistance cassette is encoded within the recombination-excised DNA, selection with Km keeps the recombined plasmids below detection levels. After the overnight incubation, 100 μl of a 10−6 culture dilution was plated on LB agar plus Ap, Cm, X-Gal, and different sub-MIC levels of rifampin (Rif) (0, 0.5, 1, and 2 μg/ml). Recombination frequency is measured as a simple percentage of recombinant (blue) with respect to nonrecombinant (white) colonies. All colonies showing blue sectors are counted as positive recombinants.
Conjugation assays.
Standard mating assays were performed as described previously (15). Strains DH5α and CIG1 (IHF+ and IHF−, respectively) were used as donor and/or recipient strains. Recipient cells contained plasmid pET29c in order to confer Km resistance for positive selection of transconjugants. Plasmid pSU1376, coding for a 330-bp R388 oriT, was mobilized with plasmid pCIG1077 (containing all of the transfer region of R388 except oriT) or pSU1529 (containing all of the transfer region of R388 except oriT and trwA).
RESULTS
Inhibitory role of IHF in TrwC-mediated recombination.
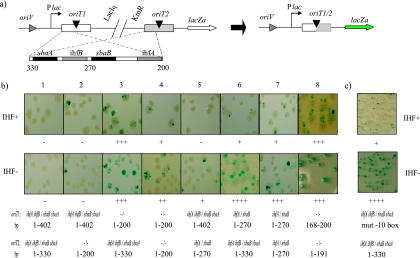
TrwC-mediated recombination in substrates containing only base pairs 1 through 200 of oriT was reported to be more efficient than that in substrates containing full-length oriTs in the absence of TrwA, suggesting an inhibitory effect associated with the oriT region from bp 200 to 330 (4). Deletion of bp 200 to 300 involves IHF binding sites, ihfA and ihfB (Fig. 1). It has been suggested that in the presence of IHF, the relaxosome is in a “closed” conformation and nicking by TrwC is inhibited and that the presence of TrwA would counteract IHF relaxosome inhibition (27). To investigate the possible role of IHF in TrwC-mediated recombination, we constructed an IHF− DH5α derivative by introducing a Sp resistance cassette disrupting the hip gene in a DH5α strain. We transformed substrates lacking sba and ihf sites [oriT(1-200)] at position oriT1 and/or oriT2 (Fig. 2a) into isogenic IHF+ and IHF− strains and tested them for their ability to host TrwC recombination in both the presence and absence of TrwA in the helper plasmid. In addition, we tested substrates harboring deletions involving only ihfB and sbaA [oriT(1-270)]. In the presence of TrwA, all substrates behaved very efficiently in IHF− and IHF+ backgrounds, giving rise to a 100% recombination frequency (data not shown) as measured by the color phenotype. However, a negative effect of IHF on recombination is evident in the absence of TrwA (Fig. 2b, compare top and bottom panels). While plasmid pCIG1028, harboring the full-length oriTs, was not affected by the IHF background (Fig. 2b, panel 1, top and bottom), substrate plasmids with oriT1(1-270) were far better substrates for recombination in IHF− strains (panels 6 and 7). To a lesser extent, this enhanced recombination phenotype was also observed when oriT2 harbored the deletion of ihfB and sbaA [oriT2(1-270)] (panel 5). A very mild effect (if any) was observed in substrate plasmids containing oriT1(1-200) (panels 3 and 4), and virtually no effect was observed with oriT2(1-200) (panel 2). Since in substrate plasmid pCIG1028 oriT1 is 402 bp long and oriT2 is 330 long, we constructed plasmid pCIG1108, with oriT1(1-330), and confirmed that it did not behave significantly differently from pCIG1028 (data not shown). Thus, binding of IHF to the region from bp 200 to 270 of oriT inhibits recombination.
FIG. 2.
(a) Schematic view of a substrate construction. oriT1 is the first oriT encountered by the replication fork, represented by a gray triangle. nic sites are pictured as black triangles, lying on the DNA strand to be nicked by the relaxase. sba and ihf sites are indicated with black and gray rectangles, respectively. Numbers represent oriT coordinates. The expected product of recombination is depicted to the right. Deletion of the intervening DNA segment between the two nic sites eliminates a lacIq repressor gene and places the lacZα gene close to the lactose promoter. As a result, the lacZα gene is expressed from the recombined substrates. (b) Recombination on substrates harboring different oriT deletions involving sba and ihf sites. The reactions are all in the absence of TrwA and in strains DH5α (top row) or CIG1 (bottom row), which provide the IHF+ or IHF− background, respectively. The extent of DNA present at each oriT copy is indicated below each column, together with the corresponding genotype. + and − symbols represent the estimated relative recombination. (c) Recombination on a substrate containing oriT1 harboring a mutation in the −10 box, under the same conditions as for panel b.
To simplify TrwC-mediated recombination to its maximum, we assayed recombination substrates harboring most permissive oriT deletions in the presence or absence of TrwA and IHF. Previous deletion analysis (4) showed that substrates harboring bp 168 to 200 at oriT1 (i.e., containing the nic site and TrwC binding site sbcA) and bp 1 to 191 at oriT2 (containing only the proximal arm of IR2 and the oriT region 3′ to the nic site) were sufficient for efficient recombination in the presence of TrwA. We tested plasmid pCIG1083, containing these minimal sites, in different IHF and TrwA backgrounds. This substrate recombined efficiently in the absence of TrwA, independently of the IHF− background (Fig. 2b, panel 8).
IHF effect in conjugation.
An in vivo role of IHF in R388 conjugation was previously discarded, since no effect on conjugation frequencies was observed by using IHF− strains as either donors or recipients (19). These experiments were performed in all cases in the presence of TrwA. We have suggested a role of IHF as a repressor of TrwC nicking activity in recombination, while TrwA lifts IHF inhibition. This would imply that if a similar regulatory mechanism was imposed on relaxosome processing during R388 conjugative transfer, it would not be detected in the presence of TrwA. We set up mating assays in the presence or absence of TrwA in different IHF backgrounds in donor and/or recipient cells. Transfer frequencies in the absence of TrwA were about 104-fold lower than those obtained in the presence of TrwA, regardless of the IHF background, as previously reported. Interestingly, in the absence of TrwA, transfer frequencies from IHF− donors were higher. In spite of the variability between different experiments, this increase was observed in every experiment. Table 3 shows the ratio of frequencies obtained between IHF− and IHF+ donor strains: in the absence of TrwA, the ratios indicate an average increase of 5- to 10-fold with IHF− cells as donors. In the presence of TrwA, there was no significant difference when donor cells were either IHF+ or IHF−.
TABLE 3.
Ratios of conjugation frequencies using IHF− and IHF+ donor strains
| Recipient strain | Conjugation frequency ratio (IHF−/IHF+)a:
|
|
|---|---|---|
| With TrwA | Without TrwA | |
| IHF+ | 0.97 (0.25-1.79) | 11.68 (3.58-19.77) |
| IHF− | 1.70 (0.93-2.47) | 5.31 (3.23-7.39) |
Mobilization assays were performed as described in Materials and Methods. Results represent the ratio between the frequencies of transconjugants per donor cell using an IHF− strain (CIG1) versus an IHF+ strain (DH5α) as donor cells. Data are the means from four or five independent experiments; 95 % confidence intervals are given in parentheses.
Effect of transcription on recombination.
Substrates with deletions involving bp 270 to 330, particularly when present in oriT1, showed positive recombination in the absence of IHF (Fig. 2b, panels 6 and 7). Surprisingly, no recombination was observed under these conditions in the wild-type substrate (Fig. 2b, panel 1). Hence, we can assume that the region from bp 270 to 330 in oriT1 exerts an inhibitory effect on the reaction which is relieved only when TrwA is present, independently of the presence of IHF. Interestingly, this oriT region also contains the putative promoter region for the trwABC operon (Fig. 1) (18, 19, 26). It is known that TrwA binding to the oriT results in transcriptional repression of the trwABC operon (26). TrwA would likely alter the normal binding of the RNA polymerase to the promoter region and hence repress transcription. We hypothesized that binding of RNA polymerase or the formation of an active transcription complex and subsequent elongation could be affecting the DNA topology at oriT1 and therefore impeding TrwC-mediated recombination. This effect would be eliminated in the presence of TrwA.
The proposed promoter for the trwABC operon is situated at bp 273 and 296 of oriT (−35 and −10 boxes, respectively). To test a possible effect of transcription from this promoter on the recombination reaction, we introduced a mutation converting this putative −10 box (TAGGAT) to an XhoI restriction site (CTCGAG) in oriT1, which would impede RNA polymerase binding. We tested this plasmid (pCIG1106) as a substrate for recombination. Figure 2c shows how recombination is strongly enhanced in IHF− backgrounds in the absence of TrwA. No enhancing effect was obtained by mutating the promoter in the presence of IHF.
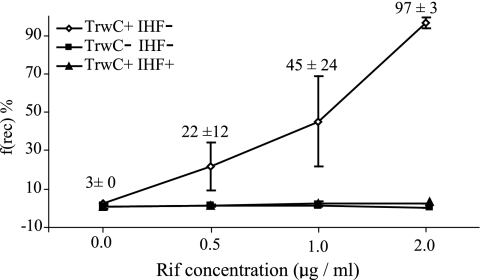
The observed inhibitory effect of the mutation in the −10 box sequence could be caused by RNA polymerase binding to oriT or by the formation of an active RNA elongation complex. The antibiotic Rif blocks the elongation of the RNA transcript at the 5′ end without affecting its binding to the promoter (2). Thus, the effect of Rif could allow us to distinguish between the two possibilities. We tested the effect on recombination of subinhibitory concentrations of Rif on substrates containing full-length oriTs in the absence of TrwA. Cultures were grown to saturation in liquid media selective for the substrate (Km) and the helper plasmid (Ap). Selecting the substrate plasmid with Km rather than with Cm would force the selection of cells harboring unrecombined substrates. After the overnight incubation, the cells were plated on increasing subinhibitory concentrations of Rif plus X-Gal, and blue colonies were counted. While no fully blue colonies were obtained, very significantly, cell cultures plated on Rif-containing media started to show incipient blue colonies, characterized by the presence of blue sectors and dots. These were counted as recombinants, to give us a measure of the frequency of recombination. Figure 3 shows the effect of increasing levels of Rif (0, 0.5, 1, and 2 μg/ml) on TrwC-mediated recombination in both IHF+ and IHF− backgrounds. In the absence of IHF, increasing concentrations of Rif lead to an increasing number of these partially blue colonies. A Rif concentration of 0.5 μg/ml was sufficient to increase the frequency of recombinants from 3 to 22%. When 1 μg/ml of Rif was supplied to the medium, 45% of colonies were monitored as recombinants, and when the concentration reached 2 μg/ml, 97% recombination was obtained. No significant difference was observed when Rif was added in the absence of TrwC or with an IHF+ strain (Fig. 3).
FIG. 3.
Effect of sub-MIC levels of Rif in TrwC-mediated recombination. CIG1 or DH5α strains harboring a full-length oriT substrate and a helper plasmid coding for TrwC, or TrwA only as a control, were plated in LB agar plus 0, 0,5, 1, or 2 μg/ml Rif and X-Gal. The frequency of recombination [f(rec)%] was calculated as described in Materials and Methods. Values are means from two to five independent experiments ± standard deviations.
DISCUSSION
The site-specific recombination reaction mediated by the R388 relaxase TrwC has been previously characterized (4), in terms of both the catalytic activity and the DNA sequence requirements. In this work, we provide evidence that this reaction is positively regulated by TrwA and negatively regulated by IHF and RNA polymerase. We propose that binding of these factors to oriT1 produces IHF-mediated DNA bending and active transcription, which exert topological constraints that inhibit TrwC nicking at oriT1, while TrwA binding would counteract these effects, favoring the initiation of the reaction at oriT1.
In the absence of TrwA, the enhanced recombination of substrates lacking the region from bp 200 to 330 of oriT suggested the presence of some inhibitory factor within this oriT region (4). The region comprises TrwA and IHF binding sites (Fig. 1), and IHF is known to inhibit TrwC cleavage in the relaxosome (27). In fact, in the absence of TrwA, we observe an inhibitory role of IHF in recombination. When recombination is tested in an IHF− background, recombination is strongly enhanced in substrates harboring oriT1(1-270) (Fig. 2b, panels 6 and 7), which still conserves an IHF binding site. This suggests that the inhibitory role of IHF is exerted prominently by binding to this recombination locus and thus probably acting in the initiation of the reaction. Substrates lacking both ihfA and ihfB sites [oriT(1-200) show little difference according to the IHF background, which would support the idea that the inhibitory effect exerted by IHF occurs mainly through binding to its putative binding sites within the oriT. The fact that the role of IHF is not observed in the presence of TrwA suggests a role of TrwA opposed to that of IHF, in a way similar to that by which nic cleavage of supercoiled DNA by TrwC is enhanced by TrwA and inhibited by IHF (27).
However, the model based on opposite effects of TrwA and IHF to control initiation of recombination did not explain all the results obtained. Remarkably, we observed no increase in the recombination proficiency of substrates containing full-length oriTs in IHF− backgrounds in the absence of TrwA (Fig. 2b, panel 1), while a release of IHF inhibition was observed on oriT(1-270). The oriT region from bp 300 to 330 contains the putative promoter sequence for the trwABC operon, for which TrwA is a transcriptional repressor (26). Hence, we tested whether RNA polymerase binding and the formation of an active transcription complex could affect the efficiency of recombination. For this purpose, we replaced the −10 box at oriT1, so that the RNA polymerase would not recognize and bind the promoter. This substrate behaves similarly to the substrate containing an oriT1(1-270) in IHF− strains; i.e., the mutation of the −10 box abolishes the inhibitory effect of the region from bp 270 to 330 (Fig. 2c). We also treated the cells with sub-MIC levels of Rif, which is known to alter transcription elongation but not RNA polymerase binding (2), and we observed a marked increase in recombination on substrates containing the full-length oriTs in the absence of TrwA and IHF (Fig. 3). Thus, it is the presence of an active transcription elongation complex that inhibits recombination, which could be explained by the effect on local supercoiling, since DNA superhelicity generated by transcription diffuses from its site of origin (32). It is well known that the level of supercoiling and DNA topology are involved in the assembly of nucleoprotein complexes engaged in DNA-processing reactions, in particular those implicated in site-specific recombination (28, 30). DNA transposition has also been reported to be repressed by transcription across its target sites (3, 7).
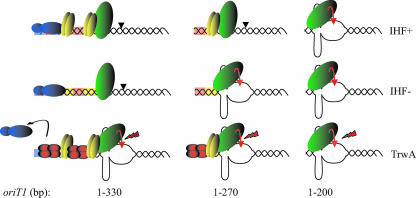
Figure 4 outlines a model by which site-specific recombination is tightly modulated by the concerted action of at least three different factors. Binding of host proteins IHF and RNA polymerase at oriT1 would impose a topological constraint, by IHF-mediated DNA bending and transcription-coupled DNA supercoiling. This constraint would maintain the relaxosome in a “closed” conformation, preventing ssDNA exposure and hence nicking by TrwC. In turn, TrwA would act as a positive regulator. Binding to the sba sites would lead to the ejection of RNA polymerase from oriT and to a switch from a “closed” to an “open” relaxosome formation, facilitating the stabilization of the cruciform at IR2 so that TrwC can firmly bind to catalyze cleavage at the nic site. This model would also be consistent with the apparent need of the complete IR2 at oriT1 but not at oriT2 (4).
FIG. 4.
Model for the role of accessory proteins in the modulation of TrwC-mediated recombination. IHF dimers are pictured in yellow, RNA polymerase is pictured in blue, TrwC is represented as a monomer in green, TrwA is represented as a tetrameric protein in red. Corresponding colored shadows indicate protein binding sites. Lightning bolts represent the enhancing effect of TrwA on TrwC activity exerted independently from its putative binding sites, a black curved arrow shows the ejection movement of RNA polymerase from the oriT caused by the action of TrwA, and red curved arrows represent TrwC catalytic nicking of the oriT at the nic site. The lengths of the oriTs at recombination locus 1 are indicated below the columns. Binding of IHF at the oriT represses nicking by TrwC. In the absence of IHF, formation of a transcription elongation complex exerts a similar inhibition. TrwA relieves the inhibition imposed both by IHF and RNA polymerase. Short sites (column 3) are constitutively in an “open” conformation, facilitating TrwC nicking.
Tight modulation of TrwC nicking at oriT (and hence of the recombination process) by these host factors could be extrapolated to the control of bacterial conjugation, since TrwC nicking at oriT is probably a key step for its initiation. This view is supported by the observation that the effects of TrwA and IHF on conjugation frequencies parallel those observed in recombination (Table 3). If this view is correct, both processes (TrwC-mediated recombination and plasmid conjugation) could be dependent on a particularly favorable host physiological condition, perhaps adjusted to the growth cycle (for instance, IHF levels are increased at stationary phase [31]) or to some environmental signal. A TrwA-free environment would be expected upon early establishment in the recipient cell. In such a situation, inhibition of further initiation of the conjugative process would make full biological sense.
Deletion of the DNA sequences involved in the control of TrwC-mediated recombination permits the attainment of elevated recombination levels in the absence of TrwA between the minimal oriT loci previously described (4). It therefore seems plausible to attain good recombination/integration levels on very simple substrates and mediated solely by TrwC in the absence of any accessory proteins, which gives TrwC the ability to be used as a biotechnological tool for targeted integration of foreign DNA introduced by conjugation into any type of susceptible recipient cell.
Acknowledgments
We are grateful to Josep Casadesús and Gabriel Moncalián for critical reading of the manuscript.
This work was supported by grant BIO2005-00689 from the Spanish Ministry of Education to M.L. C.E.C. was a recipient of a FPI predoctoral fellowship from the Spanish Ministry of Education and a postdoctoral fellowship from the Public Foundation “Marqués de Valdecilla.”
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Breg, J. N., J. H. van Opheusden, M. J. Burgering, R. Boelens, and R. Kaptein. 1990. Structure of Arc repressor in solution: evidence for a family of beta-sheet DNA-binding proteins. Nature 346:586-589. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 3.Casadesus, J., and J. R. Roth. 1989. Transcriptional occlusion of transposon targets. Mol. Gen. Genet. 216:204-209. [DOI] [PubMed] [Google Scholar]
- 4.César, C. E., C. Machón, F. de la Cruz, and M. Llosa. 2006. A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol. Microbiol. 62:984-996. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta, N., and R. W. Hedges. 1972. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J. Gen. Microbiol. 72:349-355. [DOI] [PubMed] [Google Scholar]
- 7.DeBoy, R. T., and N. L. Craig. 2000. Target site selection by Tn7: attTn7 transcription and target activity. J. Bacteriol. 182:3310-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper, O., C. E. César, C. Machón, F. de la Cruz, and M. Llosa. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. USA 102:16385-16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flamm, E. L., and R. A. Weisberg. 1985. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J. Mol. Biol. 183:117-128. [DOI] [PubMed] [Google Scholar]
- 11.Francia, M. V., and D. B. Clewell. 2002. Amplification of the tetracycline resistance determinant of pAMalpha1 in Enterococcus faecalis requires a site-specific recombination event involving relaxase. J. Bacteriol. 184:5187-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, D. I. 1988. Integration host factor: a protein for all reasons. Cell 55:545-554. [DOI] [PubMed] [Google Scholar]
- 14.Furuya, N., and T. Komano. 2003. NikAB- or NikB-dependent intracellular recombination between tandemly repeated oriT sequences of plasmid R64 in plasmid or single-stranded phage vectors. J. Bacteriol. 185:3871-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandoso, G., P. Avila, A. Cayón, M. A. Hernando, M. Llosa, and F. de la Cruz. 2000. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 295:1163-1172. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guasch, A., M. Lucas, G. Moncalián, M. Cabezas, R. Pérez-Luque, F. X. Gomis-Rüth, F. De La Cruz, and M. Coll. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 10:1002-1010. [DOI] [PubMed] [Google Scholar]
- 18.Llosa, M., S. Bolland, and F. de la Cruz. 1994. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J. Mol. Biol. 235:448-464. [DOI] [PubMed] [Google Scholar]
- 19.Llosa, M., S. Bolland, and F. de la Cruz. 1991. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol. Gen. Genet. 226:473-483. [DOI] [PubMed] [Google Scholar]
- 20.Llosa, M., S. Bolland, G. Grandoso, and F. de la Cruz. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J. Bacteriol. 176:3210-3217. (Erratum, 176:6414.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1-6. [DOI] [PubMed] [Google Scholar]
- 22.Llosa, M., F.-X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 24.Miller, H. I. 1984. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb. Symp. Quant. Biol. 49:691-698. [DOI] [PubMed] [Google Scholar]
- 25.Moncalián, G., and F. de la Cruz. 2004. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim. Biophys. Acta 1701:15-23. [DOI] [PubMed] [Google Scholar]
- 26.Moncalián, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270:188-200. [DOI] [PubMed] [Google Scholar]
- 27.Moncalián, G., M. Valle, J. M. Valpuesta, and F. de la Cruz. 1999. IHF protein inhibits cleavage but not assembly of plasmid R388 relaxosomes. Mol. Microbiol. 31:1643-1652. [DOI] [PubMed] [Google Scholar]
- 28.Nash, H. A. 1990. Bending and supercoiling of DNA at the attachment site of bacteriophage lambda. Trends Biochem. Sci. 15:222-227. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Stark, W. M., C. N. Parker, S. E. Halford, and M. R. Boocock. 1994. Stereoselectivity of DNA catenane fusion by resolvase. Nature 368:76-78. [DOI] [PubMed] [Google Scholar]
- 31.Valls, M., M. Buckle, and V. de Lorenzo. 2002. In vivo UV laser footprinting of the Pseudomonas putida sigma 54Pu promoter reveals that integration host factor couples transcriptional activity to growth phase. J. Biol. Chem. 277:2169-2175. [DOI] [PubMed] [Google Scholar]
- 32.Wu, H. Y., S. H. Shyy, J. C. Wang, and L. F. Liu. 1988. Transcription generates positively and negatively supercoiled domains in the template. Cell 53:433-440. [DOI] [PubMed] [Google Scholar]