Abstract
The survival of a bacterium with a depleted oxygen or nutrient supply is important for its long-term persistence inside the host under stressful conditions. We studied a gene, dps, from Mycobacterium smegmatis, encoding a protein, Dps (for DNA binding protein from starved cells), which is overexpressed under oxidative and nutritional stresses and provides bimodal protection to the bacterial DNA. Characterization of the dps promoter in vivo is therefore important. We cloned a 1-kb putative promoter region of the dps gene of M. smegmatis in an Escherichia coli-Mycobacterium shuttle vector, pSD5B, immediately upstream of the lacZ gene. Promoter activities were assayed in vivo both in solid medium and in liquid cultures by quantitative β-galactosidase activity measurements. To characterize the minimal promoter region, a 200-bp fragment from the whole 1-kb sequence was further cloned in the same vector, and in a similar way, β-galactosidase activity was quantitated. Primer extension analysis was performed to determine the +1 transcription start site of the gene. Point mutations were inserted in the putative promoter sequences in the −10 and −20 regions, and the promoter sequence was confirmed. The promoter was not recognized by purified M. smegmatis core RNA polymerase reconstituted with purified Mycobacterium tuberculosis σA or σB during multiple- and single-round in vitro transcription assays. Promoter-specific in vivo pull-down assays with an immobilized 1-kb DNA fragment containing the dps promoter established that extracellular function sigma factors were associated with this starvation-inducible promoter. Single-round transcription at the dps promoter further supported the idea that only core RNA polymerase reconstituted with σF or σH can generate proper transcripts.
The regulation of gene expression in mycobacteria is an important area of research in order to address the basic biology of a gram-positive organism, as well as to understand the various mechanisms of pathogenicity. Promoter identification and analysis in mycobacteria, therefore, are essential and have improved significantly with the help of various reporter technologies (16). The approach involves in vitro fusion of probable promoter and other regulatory regions just upstream of genes whose products are stable and easily assayable. Such simple systems can be used to monitor the effects of mutations in promoter elements, as well as to follow their responses to various environmental signals. Promoterless plasmids containing the Escherichia coli lacZ gene, encoding β-galactosidase (12), have been used as candidates for a reporter assay, with various lactose derivatives as substrates (3, 6, 16, 20, 21).
We attempted here to generate a lacZ construct with the dps promoter in the E. coli-Mycobacterium shuttle vector pSD5B (16). This vector is promoterless to begin with, and thus, when the intrinsic lacZ is placed in frame downstream of the dps promoter, it can be regulated as a function of the expressibility of the dps gene. Dps has been reported to be a nucleoid-like DNA binding protein capable of in vitro oligomerization under optimum conditions (25). It has been identified under glucose-limiting conditions or in stationary phase in mycobacteria (2). Its major function is to protect the genomic DNA under stress encountered during the stationary phase (2, 15). We have shown that DNA protection occurs in two different ways, and we called this bimodal protection (14, 15).
Thus, in our system, it would be interesting to assess what is the minimum promoter element necessary to regulate lacZ expression as a function of the glucose concentration in the growth medium. Eventually, it would help to utilize such a promoter element in regulated gene expression in mycobacteria.
We further attempted here to discover the nature of the sigma factor(s) that regulates the transcription of the dps gene. Thirteen sigma factors have been identified in mycobacteria (7, 11); however, clear annotation of all the sigma factors in Mycobacterium smegmatis is still not available in the The Institute for Genomic Research (TIGR) database, although the sequence of the whole M. smegmatis genome is known (33). On the other hand, all 13 sigma factors are identified in the Mycobacterium tuberculosis database (TIGR) (31). The sequence homology among various subunits of M. tuberculosis and M. smegmatis RNA polymerases (Table 1) prompted us to reconstitute a heterologous system of M. smegmatis core RNA polymerase (α2ββ′ω) and various M. tuberculosis sigma factors to generate a functionally active holo-RNA polymerase that could recognize the dps promoter element. It should be mentioned here that the dps gene is present in M. smegmatis and other mycobacteria, but not in M. tuberculosis. We believe that a heterologous transcription system like that shown here can address the basic issues concerning the structure of the promoter element and its regulation and that it is useful. The RNA polymerase used here was genetically modified with a histidine tag at the C-terminal end of the β′ subunit and purified through a single Ni column. The core RNA polymerase was isolated using a phosphocellulose column. The identification of proteins trapped in the immobilized promoter-containing template and subsequent single-round in vitro transcription helped in characterizing the sigma factors responsible for recognizing the dps promoter.
TABLE 1.
Comparison of core subunits of RNA polymerase and annotated sigma factorsa
| Subunit name | Species | Global alignment score | CLUSTAL W alignment score | % Identity
|
|||
|---|---|---|---|---|---|---|---|
|
E. coli
|
M. smegmatis
|
||||||
| SIMb | LALIGNc | SIMb | LALIGNc | ||||
| α | M. tuberculosis | 2,035 | 1,916 | 90.9 | 90.9 | ||
| β | 7,943 | 6,663 | 91.7 | 91.4 | |||
| β′ | 7,943 | 7,463 | 91.4 | 91.4 | |||
| ω | 539 | 485 | 83.0 | 79.1 | |||
| σA | 1,697 | 2,217 | 60.3 | 32.8 | 79.8 | 76.7 | |
| σB | 1,475 | 1,790 | 43.7 | 33.7 | 93.7 | 92.3 | |
| σF | 972 | 1,237 | 29.7 | 27.8 | 83.8 | 80.4 | |
| σH | 202 | 391 | 27.5 | 14.1 | 38.8 | 32.6 | |
σJ, σL, σM, σC, σD, σK, σI, σG, and σE were found to be putative in the mc2155 genome; hence, sequence homology couldnot be performed.
MATERIALS AND METHODS
Bacterial strains, medium, and growth conditions.
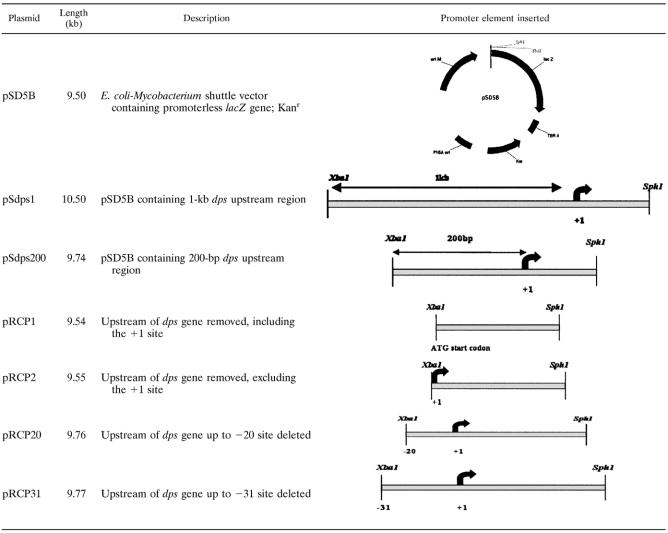
All the plasmids used in the present study are listed (see Table 3). The M. smegmatis wild-type strain mc2155 (33) was used in all experiments. The bacteria were grown in MB7H9 medium supplemented with 2% glucose, 25 μg/ml kanamycin, and 0.05% Tween 80. Plate cultures were grown in liquid medium with 1.5% agar. For quantitative β-galactosidase assays in liquid cultures, 2 mM o-nitrophenyl-β-d-galactopyranoside (ONPG) was used as a substrate. However, for plate assay of lacZ, 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside-containing MB7H9 plates were used.
TABLE 3.
Plasmid constructs containing several deletion constructs of the dps promoter
A transcriptional fusion construct of a putative 1-kb promoter of the dps gene and lacZ and promoter induction assay.
A set of two primers, Prisat1 and Prisat2 (Table 2), was used to amplify the 1-kb fragment containing upstream, as well as downstream, sequence (up to +46 bp) from the +1 start site of mc2155 genomic DNA. The PCR product was digested with XbaI and SphI and was then cloned in a promoterless E. coli-Mycobacterium shuttle vector, pSD5B, with a lacZ reporter system (16) previously digested with the same set of restriction enzymes. The resulting recombinant plasmid, pSdps1 (Table 3), therefore had the lacZ reporter gene just downstream of the dps promoter.
TABLE 2.
Primers used in the present study
| Primer name | Sequence (5′-3′) |
|---|---|
| Prisat1 | GGCATCGATCCCCTGCTACCG |
| Prisat2 | GCCACATCGCTCGCCTTCTTG |
| Prisat200 | GGGATGGTCGAGTTGCTCGAAC |
| dpsrev | CAGCGTCGCGATGCGCTCGGCCACC |
| Map2 | GGAAGTGATTCCTCCGGATATCG |
This plasmid was then electroporated to mc2155 competent cells, cultured to mid-log phase (optical density at 600 nm, 0.7) in MB7H9 medium supplemented with 2% glucose, harvested, washed once with phosphate-buffered saline, and then transferred to MB7H9 medium supplemented with either 2% or 0.02% glucose, and cells were harvested at different time points as required. Promoter induction was assayed by β-galactosidase activity (lacZ expression) in liquid culture using ONPG as described previously (27). The activity was represented in Miller units and was calculated as follows: activity (Miller units) = 1,000 × A420/(time [min] × volume [culture] × optical density at 600 nm).
All the readings were taken in triplicate and averaged. In each case, mc2155 transformed with the empty vector pSD5B was taken as the negative control (data not shown).
Subcloning and characterization of a minimal 200-bp promoter region from the 1-kb promoter.
A set of two primers, Prisat200 and Prisat2 (Table 2), was used to amplify the minimal 200-bp promoter fragment, along with the same downstream stretch of sequence from the pSdps1 plasmid. The PCR product was digested with XbaI and SphI and was then cloned in a promoterless E. coli-Mycobacterium shuttle vector, pSD5B, with a lacZ reporter system (16) previously digested with the same set of restriction enzymes. The resulting recombinant plasmid, pSdps200 (Table 3), therefore had the lacZ reporter gene just downstream of the dps promoter. Promoter induction under starvation (0.02% glucose) was assayed using liquid cultures of mc2155 cells transformed with pSdps200, similarly to mc2155-pSdps1.
Mapping of the transcription start site by the primer extension method.
The +1 transcription start site of the dps gene was identified by the primer extension method (15). M. smegmatis transformed with pSdps1 was grown for 60 h in MB7H9 medium supplemented with 0.02% glucose and harvested. Total RNA was then isolated using an RNA minikit (Auprep). A total of 10 to 12 μg of RNA was used to prepare cDNA using a primer end labeled with [γ-32P]ATP (Board of Radiation and Isotope Technology, Bombay, India), Map2 (Table 2), and RevertAid Moloney murine leukemia virus reverse transcriptase (Fermentas). Sequencing-grade Taq DNA polymerase and dideoxynucleoside triphosphates were obtained from Promega. The primer was designed approximately 71 bp downstream of the +1 site. A sequencing ladder was prepared by the dideoxy-mediated chain termination method and was run using the Fmol DNA cycle-sequencing system (Promega) with an annealing temperature of 50°C. A 10% polyacrylamide gel containing 6 M urea was run to resolve the sequencing product. The gel was dried, and a picture was taken using a phosphorimager (FLA2000; Fujifilm).
Identification of the promoter region by site-directed mutations of bases in the −10 and −20 regions.
Several upstream deletion constructs were made from mc2155 genomic DNA by specific PCR primers in the upstream region proximal to the +1 start site (Table 3). The aim was to discover the minimal promoter sequence for the dps gene. The PCR conditions were 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min for 30 cycles. mc2155 cells transformed with pRCP1, pRCP2, pRCP20, and pRCP31 were grown in liquid MB7H9 medium (0.02% glucose) for 72 h, and promoter activity was assayed similarly by lacZ expression. mc2155-pSdps200 was taken as a positive control.
Site-specific point base mutations were further carried out by the Quickchange protocol (Stratagene) in the −20 (TCGAAC) and −10 (GAAACG) regions of the promoter. The PCR conditions were 98°C for 10 s, 65°C for 30 s, and 72°C for 3 min (25 to 30 cycles), using pSdps200 as a template. All mutations were confirmed by DNA sequencing. β-Galactosidase activity was assayed in liquid cultures to compare the promoter induction under starvation.
Growth phase-dependent variation of Dps protein in vivo.
mc2155 cell pellets were collected at different time points during growth and lysed. Crude cell lysates were resolved on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels and then transferred electrophoretically to polyvinylidene difluoride membranes (Amersham Pharmacia). The membranes were probed with affinity-purified Dps antibody diluted 2,500-fold. The reactive proteins were identified using a goat anti-rabbit antibody conjugated to horseradish peroxidase. The protein bands were developed with amino ethyl carbazole as the substrate.
Promoter-specific pull-down assay and identification of associated sigma factors.
A 1.1-kb linear PCR-amplified dps promoter was incubated at 37°C for 45 min with 10 to 20 μM biotin-11-dUTP (Fermentas), 100 μM deoxynucleoside triphosphate mix (Sigma), and 0.6 unit of Klenow fragment (Fermentas) in buffer A (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.2 mM EDTA), and unincorporated biotin-dUTP and deoxynucleoside triphosphates were removed by ethanol precipitation. Biotinylated dps promoter fragments were pooled in 25 μl of double-distilled water. Twenty-five microliters of biotin-dps DNA fragments was then mixed with 250 μl of streptavidin-agarose beads (Sigma) for 1 hour at room temperature with constant stirring. Biotinylation of DNA was checked by characteristic UV and fluorescence measurements. The ratio between biotin and DNA was found to be nearly 1:1. Crude cell lysates were taken from wild-type mc2155 cells and grown for 72 h in 0.02% glucose-supplemented MB7H9 medium, and then DNA binding proteins were precipitated with polyethyleneimine. Protein extraction steps from the pellet were performed by 0.4 M ammonium sulfate precipitation, and the pellet obtained in this step was dissolved in buffer B (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM potassium glutamate, 10% glycerol, 0.5 mM EDTA, and 0.1% Triton X-100) with 10 μl of dps-biotin-streptavidin-agarose beads and kept for binding overnight at 4°C with constant mixing. The beads were allowed to settle on ice for 10 min, the supernatant was removed, and the beads were washed three times with buffer B. Bound proteins were then eluted with buffer C (20 mM Tris-HCl, pH 7.9 at 4°C, 100 mM NaCl, 0.2 mM EDTA, and 0.1 mM dithiothreitol [DTT] [TGED] plus 0.4 M NaCl) and were subjected to Western blotting with sigma antibodies (obtained from Astrazeneca) and mass spectrometric identification.
Reconstitution of RNA polymerase holoenzymes from M. tuberculosis SigF and SigH with core RNA polymerase from M. smegmatis.
In vitro purification of His6-tagged holo-RNA polymerase (obtained from strain SM07, made by Raju Mukherjee) from M. smegmatis (24 h in 2% glucose plus MB7H9 medium) and N-terminal His6-tagged SigH from M. tuberculosis (in pRSET-a vector [Astrazeneca]) was performed using the Qiagen Ni-nitrilotriacetic acid affinity matrix according to the manufacturer's instructions. For this purpose, RNA polymerase was first purified from 500 ml of actively growing culture of M. smegmatis, following the same protocol as reported earlier (26) up to polyethyleneimine precipitation, and then by ammonium sulfate fractionation. Subsequently, the partially purified enzyme fractions were passed through a Qiagen Ni-nitrilotriacetic acid column for affinity purification (details of this preparation will be published elsewhere). N-terminal glutathione S-transferase-tagged SigF from M. tuberculosis (in the pGEX-3X vector [Astrazeneca]) was also purified using a glutathione-Sepharose affinity matrix. All of the proteins were dialyzed in TGED plus 50% glycerol. Purification of core RNA polymerase from the His6-tagged holoenzyme was carried out on a phosphocellulose ion-exchange column (P11; Whatman) equilibrated with TGED and 50 mM KCl. The flowthrough and wash fractions contained σA, and the core subunits were bound to the matrix. Elutions were done in TGED plus 500 mM KCl when the core polymerase (α2ββ′ω) was eluted out. The eluted fractions were checked in 10% SDS-polyacrylamide gel electrophoresis (PAGE) and by Western analysis with σA antibody. A reconstitution experiment was performed by incubating the M. smegmatis core RNA polymerase and M. tuberculosis σA, σF, and σH at 1:2 molar ratio in buffer R (50 mM Tris-HCl, pH 7.9 at 4°C, 0.1 M KCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mM EDTA) at 37°C for 30 min and were resolved in an 8% native PAGE and silver stained. The reconstituted holoenzyme bands were then cut out and subjected to SDS-PAGE analysis and also silver stained.
In vitro transcription assay.
A multiple-round in vitro transcription was performed to measure the RNA polymerase activity as described previously (23). The assay mixture contained 40 mM Tris-HCl, pH 8.0; 200 mM NaCl; 10 mM MgCl2; 0.1 mM EDTA; 14 mM 2-mercaptoethanol; 200 μM (each) of ATP, CTP, and GTP; 50 μM UTP; 2 μCi of [3H]UTP (Perkin Elmer); and 1.5 μg calf thymus DNA per 100 μl. The calf thymus DNA assay mixture was used as a control for nonspecific transcription. An assay mixture containing σA-specific acetamidase gene promoter DNA (2 kb) was used as a positive control for specific transcription. DNA (1 kb) containing the dps promoter was then subjected to specific transcription with RNA polymerase of M. smegmatis containing sigma A, F, and H from M. tuberculosis (at a DNA/holopolymerase molar ratio of 1:10). The enzymes were all incubated with the assay mixture for the optimum time at 37°C and then spotted on DE-81 papers (Whatman) presoaked in 5 mM EDTA to stop the reaction. The dried filters were washed twice in 5% Na2HPO4 buffer for 15 min, twice in deionized water for 5 min, and finally once with ethanol and dried. The filters were then placed into scintillation vials filled with Ultima Gold scintillation fluid (Packard) and counted.
An in vitro single-round runoff transcription assay was performed using conditions modified from those described earlier (18). Purified σF, σH, and σA proteins from M. tuberculosis were incubated with M. smegmatis core RNA polymerase in a 2:1 molar ratio at 37°C for 30 min in transcription buffer [500 mM Tris-HCl, pH 7.8 at 37°C, 30 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT, 250 μg/ml bovine serum albumin (nuclease free), 500 mM NaCl]. To the reconstituted holoenzymes, DNA template (at a protein/DNA molar ratio of 10:1) and a mixture of heparin, [α-32P]UTP, and four NTPs were added, and the mixture was incubated for 10 min at 37°C. The concentrations of nucleotides and heparin in the final reaction volume were 0.15 mM ATP, 0.15 mM GTP, 0.15 mM CTP, 0.15 mM UTP, 2 μCi [α-32P]UTP, and 200 μg heparin/ml. The DNA template for in vitro transcription was prepared by PCR using primers Prisat1 and dpsrev (Table 2), with mc2155 genomic DNA as a template. A linear fragment of 317 bp, which generated a transcript of 234 bp, was subsequently used for the assay. Known σF- and σH-dependent promoters, usfXp1 (in pYZ99 vector; Bose Institute, Kolkata, India) and sigBP (in pARC8176 vector [Astrazeneca]), respectively, were used as positive controls for in vitro transcription. All samples were electrophoresed in a 10% denaturing polyacrylamide gel containing 6 M urea and analyzed by phosphorimager (FLA2000; Fujifilm).
RESULTS
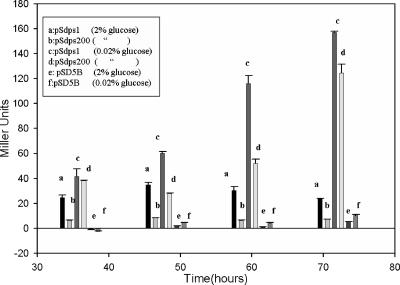
The full-length 1-kb dps promoter and minimal 200-bp dps promoter fragment show comparable promoter activities under glucose starvation.
The 1-kb upstream region of the dps gene, when fused to the upstream region of the lacZ reporter in a promoterless E. coli-Mycobacterium shuttle vector, pSD5B (16), showed significant promoter activity in mc2155 cells. The promoter activity was observed only under glucose starvation, i.e., in 0.02% glucose-supplemented MB7H9 medium, and was maximal around 60 to 72 h of growth (Fig. 1, bars c). It was fivefold more than that in 2% glucose (Fig. 1, bars a). The quantitation of the promoter activity was carried out in liquid cultures through ONPG activity assays. It was then calculated in terms of Miller units (see Materials and Methods) (27). All the experiments were performed under identical conditions in triplicate. Next, a promoter-reporter construct, pSdps200 (Table 3), was made by PCR amplification of the 200-bp minimal upstream promoter fragment using a set of two primers, Prisat200 and Prisat2 (Table 2), in pSD5B. Promoter activity assays were followed in liquid cultures similarly to that of mc2155-pSdps1. From Fig. 1, it is evident that this region was sufficient to show significant promoter induction, comparable to that observed with the 1-kb promoter region, under starvation conditions (Fig. 1, bars d). No significant promoter induction was found in 2% glucose-supplemented MB7H9 medium (Fig. 1, bars b).
FIG. 1.
Comparison of promoter induction of pSdps1 (bars a) and pSdps200 (bars b) in MB7H9 medium supplemented with 2% glucose and 0.02% glucose, pSdps1 (bars c), and pSdps200 (bars d). The error bars indicate standard deviations.
Identification of the transcription start site (+1) by primer extension analysis.
Primer extension analysis was performed to map the +1 transcription start site of the M. smegmatis dps gene (Fig. 2). Here, the transcription initiation site (+1) was found to be a “G” base that was 8 bases upstream from the dps translation start codon, ATG. The sequence had been read in a reverse complementary way, as a reverse primer specific to the vector backbone was used, and the cDNA (Fig. 2, lane P) was around 93 nucleotides in length.
FIG. 2.
Identification of the +1 transcription start site by primer extension analysis. Total RNA was isolated from M. smegmatis cells transformed with pSdps1 and grown for 72 h in 0.02% glucose-supplemented MB7H9 medium. The cDNA was synthesized and run in lane P, along with a sequencing reaction of the top strand with a reverse primer represented by C, T, A, and G. The sequence is shown on the right, and the +1 site is circled.
Characterization of the promoter sequence.
In order to identify the upstream promoter region of the dps gene, we made several upstream deletion constructs (Table 3). pRCP20 and pRCP31 were generated by removing all the bases upstream up from the putative −20 bases and −31 bases, respectively. This was achieved by keeping the position the same for the reverse primer at the 3′ end while shifting the forward primer to different locations. In all cases, mc2155 genomic DNA was used as the template during PCR. Two negative controls were also constructed, one of which contained no bases upstream from the +1 start site (pRCP1) and hence was equivalent to the empty vector, pSD5B, while the other one had only the +1 transcription site (pRCP2) (Table 3). An ONPG activity assay was performed in 0.02% glucose-supplemented MB7H9 medium. The results (see Table S1 in the supplemental material) indicated significant promoter induction with pRCP20. The promoter containing upstream −20 base pairs was therefore sufficient to give activity comparable to that of pSdps200 and pRCP31. Cells transformed with pSD5B, pRCP1, and pRCP2 did not show any activity, as expected. Point base mutations in these upstream regions were then carried out at every first, third, and sixth base of the −20 (TCGAAC) and −10 (GAAACG) sequences (Fig. 3A) by site-directed mutagenesis (see Materials and Methods). All of the single-base mutation constructs are listed in Fig. 3A. As is evident from Fig. 3B, mutations in the hexameric stretch TCGAAC (−20) severely affected lacZ expression, as observed in ONPG assays in liquid cultures of the bacteria grown for 72 h in 0.02% glucose (lanes b, c, and d). This indicated that the dps gene is controlled by a promoter element located 20 bases upstream from the +1 start site.
FIG. 3.
(A) Promoter sequence as identified by site-directed mutagenesis of pSdps200; point base mutations in the −10 and −20 regions are shown in boldface and underlined. (B) Effects of specific mutations (first, third, and sixth bases) in −10 and −20 upstream sequences from the +1 site of the dps gene on promoter activity. Differential lacZ expression in liquid cultures of M. smegmatis transformed with variants of pSdps200 is shown. The error bars indicate standard deviations. The cells were grown in MB7H9 medium supplemented with 0.02% glucose and harvested at 72 h.
In a separate set of experiments, we looked at the Dps protein expression profile at different time points of bacterial growth under glucose-fed, as well as glucose-starved, conditions. Figure 4 shows the protein expression levels at different stages of growth probed with anti-Dps antibody. It was clear that Dps, being a starvation-induced protein, was expressed in late stationary phase, which more or less mimicked starvation conditions, as most of the carbon source was used up; however, the expression was basal under carbon-fed conditions. Therefore, we concluded that Dps was induced under glucose starvation at both the transcriptional and translational levels.
FIG. 4.
Growth phase-dependent intracellular expression profile of Dps protein in wild-type mc2155 cultures under glucose-starved (0.02%) and -fed (2%) conditions in MB7H9 medium. P, in the first lane (indicated by the arrow), represents pure His6-tagged Dps protein as a positive control.
The dps promoter is not recognized by either σA or σB: a new set of ECF sigma factors is required.
We observed initially that there was no transcription signal during in vitro reaction at the dps promoter with purified M. smegmatis holo-RNA polymerase containing the principal sigma factor (1, 4). This raised the question of whether any other sigma factors are necessary to recognize the dps promoter. However all extracellular function (ECF) sigma factor genes are not yet annotated for the M. smegmatis genome in the TIGR database. Thus, we first developed a pull-down assay over an immobilized DNA template in order to identify the nature of the sigma factors associated with the transcription complex. Subsequently, in vitro transcription at the dps promoter was carried out with M. smegmatis core RNA polymerase reconstituted with M. tuberculosis sigma factors.
We first attempted to immobilize the DNA template by incorporating a biotinylated nucleotide at its terminal. The biotin-avidin interaction is known to be one of the strongest noncovalent interactions, with a Ka (association constant) value of 1013 to 1015 M−1 (13), and has wide application in protein-DNA interactions. Biotinylation of a PCR-amplified 1-kb promoter fragment was carried out with Klenow polymerase, and the labeled promoter was further immobilized over streptavidin-agarose beads. Subsequently, mc2155 cells were grown in MB7H9 medium supplemented with 0.02% glucose and harvested at 72 h. DNA binding proteins were precipitated with 10% polyethyleneimine and further fractionated by ammonium sulfate precipitation. The total protein mixture was then incubated with the dps-biotin-streptavidin beads, associated proteins were eluted out, and the fractions were subjected to immunoblotting against M. smegmatis α and M. tuberculosis β′ and sigma antibodies. As is evident from Fig. 5A, the core subunits were present and two ECF sigma factors, σF and σH, were detectable in the sample (Fig. 5B, E1 and E2). These sigma factors are known to be involved in persistence and heat shock and oxidative stress responses in M. tuberculosis (5, 24, 30). We also noticed growth phase-dependent expression of σF and σH in M. smegmatis culture under 0.02% glucose conditions, but not when the culture was grown in 2% glucose (not shown), further supporting our previous results. To further confirm that dps was not recognized by σA, pull-down assays were performed with two other promoters, those for rel (17) and the acetamidase gene (28), and probed with σA antibody. It is clear from Fig. 5C that σA was not present with the dps promoter, whereas both rel and the acetamidase gene were σA specific, which further supported our contention that dps was recognized directly by stationary-phase-induced ECF sigma factors. It should be mentioned here that we probed the pull-down assay eluate in the same way with other sigma factor antibodies from M. tuberculosis, and none of them showed any positive result.
FIG. 5.
(A) Identification of the core RNA polymerase subunits in the eluted fractions of biotin-streptavidin pull-down assays with the dps promoter at 48 (lanes a) and 72 (lanes b) hours under glucose-depleted conditions. The eluates were probed with M. tuberculosis β′ antibody and M. smegmatis α antibodies. In both cases, lanes S contain the purified holo-RNA polymerase as a standard. (B) Identification of new ECF family sigma factors σH and σF associated with the dps promoter by promoter-specific biotin-streptavidin pull-down assays. The eluted fractions, E1 and E2, were treated with antibodies against M. tuberculosis σH and σF, respectively. (C) Western blot with σA antibody done to check the sensitivity of the dps promoter with holo-RNA polymerase containing σA. Lane 1, pure σA; lanes 2 to 4, pull-down fractions with the 1.6-kb rel promoter (lane 2), the 2-kb acetamidase gene promoter (lane 3), and the 1-kb dps promoter (lane 4).
Multiple- and single-round in vitro runoff transcription assay with σF-, σH-, and σA-reconstituted RNA polymerases.
To determine whether the dps promoter is recognized by σF- or σH-containing RNA polymerases, the following multiple- and single-round transcription assays were carried out. Reconstitution of M. smegmatis core RNA polymerase with the M. tuberculosis sigma factors A, F, and H resulted in a protein band upon 8% native PAGE analysis. The band, when subjected to 12% SDS-PAGE analysis, showed the presence of all the core subunits, along with the sigma factors with which they were reconstituted (see Fig. S1 in the supplemental material). We carried out multiple-round transcription assays at the dps promoter with reconstituted RNA polymerases, as described in Materials and Methods. The amounts of [3H]UTP incorporated in the transcripts were monitored to quantify the transcription activities. No significant transcription was observed at the dps promoter when it was incubated with σA-reconstituted polymerase, whereas the acetamidase gene resulted in appreciable radioactive signal. Transcription from the dps promoter, on the other hand, was initiated by both σF- and σH-containing polymerases (see Fig. S2 in the supplemental material).
When the dps promoter containing linear DNA was subjected to single-round runoff transcription in the presence of heparin (18), an expected length of 234 bp was noticed with σF- and σH-containing core RNA polymerase (Fig. 6B). It is also clear from the figure that σA-containing RNA polymerase is incapable of initiating transcription at this promoter. In addition, in vitro transcriptions with known M. tuberculosis σA-, σF-, and σH-dependent promoters, rel (17), usfXp1 (9), and sigB (30), respectively, were performed as positive controls, and as is evident from Fig. 6C, D, and E, they indeed showed runoff transcripts of the expected lengths. Also, in the presence of rifampin, the transcription reactions were all found to be inhibited (Fig. 6), thereby confirming the formation of true RNA transcripts in all cases. These experiments conclusively established that the dps promoter can be transcribed only by ECF sigma factors, σF and σH.
FIG. 6.
Single-round heparin-resistant runoff transcription at the dps promoter carried out with M. smegmatis holo-RNA polymerase. (A) A 10% polyacrylamide gel containing 6 M urea shows an RNA ladder run separately and matched. (B) M. tuberculosis σA, σF, and σH were reconstituted with M. smegmatis core RNA polymerase to obtain the reconstituted holopolymerases. The intensity of each transcript band as obtained from phosphorimager analysis showed the expected transcripts for σF (right lane) and σH (middle lane). No transcripts were obtained with σA-reconstituted RNA polymerase (left lane). (C) In order to test single-round runoff transcription with σA-reconstituted RNA polymerase, the rel promoter was used as a positive control. The left lane shows the presence of the transcript, and the right lane shows its absence with rifampin (0.1 μg/ml). (D) Positive controls are shown using the known M. tuberculosis σH-dependent sigB promoter in the absence (left lane) and in the presence (right lane) of rifampin, as well as the σF-dependent usfXp1 promoter in the absence (middle lane) and in the presence (right lane) of rifampin. No band was obtained from the M. smegmatis dps promoter in the presence of rifampin with σH-reconstituted RNA polymerase (left lane).
DISCUSSION
The principal aim of this study was to characterize a starvation-induced mycobacterial transcription system in order to understand the mechanism regulating gene expression when bacteria enter stationary phase and eventually encounter different stresses, like oxygen and nutrient depletion and heat shock. The stationary-phase-induced expression of the dps gene and its regulation were studied extensively in E. coli (22). Various other stresses had also been shown to induce dps expression at the transcriptional or translational level (2, 25, 34). In E. coli, dps expression was found to be regulated by OxyR under oxidative stress, whereas σS is required for induction under starvation in stationary phase (2, 25). Here, we have shown that the dps promoter of M. smegmatis is induced under glucose starvation conditions and that the level of induction increases as bacteria enter the stationary phase. Under glucose-fed conditions, however, induction was negligible.
In E. coli, it is known that the ClpXP protease system degrades the mature Dps protein, thereby regulating the intracellular level of Dps at the posttranscriptional level. In mycobacteria, however, such negative-feedback control systems are not known. In all our reporter assays, we electroporated the promoter plasmid constructs into mc2155 cells, which already have a copy of the full-length dps gene in the bacterial DNA. The protein is therefore expressed from early exponential phase under starvation conditions, as well as after 60 h under glucose-fed conditions, to exert any positive control. In 2% glucose, however, significant promoter induction was not seen in any of the constructs (Table 3 and Fig. 1), so perhaps there was no positive-feedback control associated with promoter induction at the posttranscriptional level.
Systematic study of the mapping of the promoter element resulted in a putative hexameric consensus sequence, TCGAAC. The dps promoter-specific biotin-streptavidin pull-down assay (10, 19, 35) presented here identified a set of two stress-responsive sigma factors, F and H (Fig. 5B), associated with it. Later, growth-dependent expression of these two sigma factors was also monitored in mc2155 cell lysates, where concomitant expression of both proteins under stationary-phase conditions, along with Dps protein, was observed. However, sigma H expression was more than that of σF. This was indeed interesting, as it further supported the idea that bacteria adapt to various stresses encountered under dormant conditions by regulation of different subsets of genes and that ECF sigma factors play a crucial role at this stage.
Here, we have shown the purification and reconstitution of holo-RNA polymerase from M. smegmatis core enzyme and sigma factors from M. tuberculosis (32). This was necessary, as different ECF sigma genes in M. smegmatis are still not annotated. However, sequence homology among various sigma factors from mycobacteria and specific transcripts obtained using the heterologous enzymes indicated that such experiments are dependable, at least in vitro.
Recently, it has also been reported that the M. tuberculosis sigB promoter can be identified by both σF and σH; however, transcription initiates from different +1 sites (8). Reconstituted enzymes showed the specific single-round runoff transcripts at the dps promoter with σF and σH, whereas other controls showed σA-dependent transcription. We tried a sequence-based comparison of the consensus promoter sequences for σF and σH available in the literature (8, 9, 24, 29) with that obtained for the dps promoter in this study (Fig. 7), and more similarities were found with σH-dependent promoters (30). Our observation is biologically significant for two main reasons. First, we have developed an efficient single-round in vitro transcription with His-tagged reconstituted RNA polymerase, which can be regulated as a function of glucose or salt concentration in the medium. We believe that this will help a great deal in studying mycobacterial transcription, both in vivo and in vitro, and even at the single-molecule level. On the other hand, reconstitution of heterologous transcription machinery with the M. smegmatis promoter and core polymerase and M. tuberculosis sigma factors indicates the identical natures of RNA polymerase subunits, which could eventually help in understanding the structure-function relationships of this important enzyme.
FIG. 7.
Comparison of promoter consensus sequences identified by σF and σH factors in the known usfXp1 and sigB promoters from M. tuberculosis, respectively, and the dps promoter from M. smegmatis (this study).
Acknowledgments
We thank the Department of Biotechnology, Government of India, for financial support. R.P.C. is supported by a senior research fellowship from the Council for Scientific and Industrial Research, Government of India.
The sigma antibodies for ECF sigma factors and the sigB promoter were provided by Astrazeneca. We also thank Manikuntala Kundu, Bose Institute, Kolkata, India, for providing the strain containing the usfXp1 promoter. His6-tagged RNA polymerase was purified from an M. smegmatis strain (SM07) obtained from Raju Mukherjee.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agarwal, N., and A. K. Tyagi. 2006. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res. 34:4245-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., R. G. Barletta, C. O. Theon and R. E. Andrews, Jr. 1997. Identification of Mycobacterium paratuberculosis gene expression signals. Microbiology 143:921-928. [DOI] [PubMed] [Google Scholar]
- 4.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcription apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-σ factors antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 6.Cirillo, J. D., R. G. Barletta, B. R. Bloom and W. R. Jacobs, Jr. 1991. A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J. Bacteriol. 173:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagles, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seegar, J. Skelton, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrel. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Dainese, E., S. Rodrigue, G. Delogu, R. Provvedi, L. Laflamme, R. Brzezinski, G. Fadda, I. Smith, L. Gaudreau, G. Palu, and R. Manganelli. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor σL and roles in virulence and in global regulation of gene expression. Infect. Immun. 74:2457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMaio, J, Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, W., S. Jin, T. Tong, H. Zhao, F. Fan, M. J. Antinore, B. Rajasekaran, M. Wu, and Q. Zhan. 2002. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 277:8061-8067. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler, A. V., and I. Zabin. 1983. Purification structure and properties of hybrid beta galactosidase proteins. J. Biol. Chem. 258:14354-14358. [PubMed] [Google Scholar]
- 13.González, M., L. A. Bagatolli, I. Echabe, J. L. R. Arrondo, C. E. Argaraña, C. R. Cantor, and G. D. Fidelio. 1997. Interaction of biotin with streptavidin. J. Biol. Chem. 272:11288-11294. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S., and D. Chatterji. 2003. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J. Biol. Chem. 278:5235-5241. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, S., and D. Chatterji. 2005. Stress responses in mycobacteria. IUBMB Life 57:149-159. [DOI] [PubMed] [Google Scholar]
- 16.Jain, S., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1997. Construction of shuttle vectors for genetic manipulation and molecular analysis of mycobacteria. Gene 190:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain, V., S. Sujatha, A. Ojha., and D. Chatterji. 2005. Identification and characterization of rel promoter element of Mycobacterium tuberculosis. Gene 351:149-157. [DOI] [PubMed] [Google Scholar]
- 18.Kajitani, M., and A. Ishihama. 1983. Determination of the promoter strength in a mixed transcription system: promoters of lactose, tryptophan and ribosomal protein L10 operons from Escherichia coli. Nucleic Acids Res. 11:671-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, M., C. Uchida, Y. Takano, M. Kitagawa, and Y. Okano. 2004. Cell cycle-dependent regulation of the human aurora B promoter. Biochem. Biophys. Res. Commun. 316:930-936. [DOI] [PubMed] [Google Scholar]
- 20.Knipfer, N., L. Nooruddin, and T. E. Shrader. 1998. Developement of an α-complementation system for mycobacterial promoter analysis. Gene 217:69-75. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, D., R. Srivastava, and B. S. Srivastava. 1998. Genetic rearrangements leading to disruption of heterologous gene expression in mycobacteria: an observation with E. coli β-galactosidase in Mycobacterium smegmatis and its implication in vaccine development. Vaccine 16:1212-1215. [DOI] [PubMed] [Google Scholar]
- 22.Lomovskaya, O. L., J. P. Kidwell, and A. Martin. 1994. Characterization of the sigma 38-dependent expression of a core Escherichia coli starvation gene, pexB. J. Bacteriol. 176:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe, P. A., D. A. Hager, and R. R. Burgess. 1979. Purification and properties of the σ subunit of Eschericia coli DNA dependent RNA polymerase. Biochemistry 18:1678-1686. [DOI] [PubMed] [Google Scholar]
- 24.Manganelli, R., R. Proveddi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. σ Factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the non-specific DNA binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathew, R., M. Ramakanth, and D. Chatterji. 2005. Deletion of the gene rpoZ encoding the omega subunit of RNA polymerase in Mycobacterium smegmatis results in fragmentation of the β′ subunit in the enzyme assembly. J. Bacteriol. 187:6565-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 28.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuber. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 29.Raman, S., R. Hazra, C. Dascher, and R. N. Husson. 2004. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186:6605-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman, S., T. Song, X. Puyang, S. Bardarov, S. W. Jacobs, Jr., and R. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigue, A., J. Brodeur, P. E. Jacques, A. L. Gervais, R. Brzezinski, and L. Gaudreau. 2007. Identification of mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigue, S., R. Provvedi, P. Jacques, L. Gaudreau, and R. Manganelli. 2006. The σ factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30:926-941. [DOI] [PubMed] [Google Scholar]
- 33.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:911-919. [DOI] [PubMed] [Google Scholar]
- 34.Stephani, K., D. Weichart, and R. Hengge. 2003. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol. Microbiol. 49:605-614. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, C., T. Basta, E. D. Jensen, and M. W. Klymkowsky. 2003. The β-catenin/Veg-T-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 130:5609-5624. [DOI] [PubMed] [Google Scholar]