Abstract
The hyperthermophilic archaeon Archaeoglobus fulgidus strain 7324 has been shown to grow on starch and sulfate and thus represents the first sulfate reducer able to degrade polymeric sugars. The enzymes involved in starch degradation to glucose 6-phosphate were studied. In extracts of starch-grown cells the activities of the classical starch degradation enzymes, α-amylase and amylopullulanase, could not be detected. Instead, evidence is presented here that A. fulgidus utilizes an unusual pathway of starch degradation involving cyclodextrins as intermediates. The pathway comprises the combined action of an extracellular cyclodextrin glucanotransferase (CGTase) converting starch to cyclodextrins and the intracellular conversion of cyclodextrins to glucose 6-phosphate via cyclodextrinase (CDase), maltodextrin phosphorylase (Mal-P), and phosphoglucomutase (PGM). These enzymes, which are all induced after growth on starch, were characterized. CGTase catalyzed the conversion of starch to mainly β-cyclodextrin. The gene encoding CGTase was cloned and sequenced and showed highest similarity to a glucanotransferase from Thermococcus litoralis. After transport of the cyclodextrins into the cell by a transport system to be defined, these molecules are linearized via a CDase, catalyzing exclusively the ring opening of the cyclodextrins to the respective maltooligodextrins. These are degraded by a Mal-P to glucose 1-phosphate. Finally, PGM catalyzes the conversion of glucose 1-phosphate to glucose 6-phosphate, which is further degraded to pyruvate via the modified Embden-Meyerhof pathway.
Archaeoglobus fulgidus, which represents the first isolated sulfate reducer in the domain of archaea, was isolated by Stetter et al. (62, 64) from hydrothermal areas near Volcano (Italy). With a temperature optimum of 83°C, it belongs to the group of hyperthermophiles (63). Later, A. fulgidus strain Z (76) and strain 7324 (4) were isolated from Vulcano and from hot oil field waters of the North Sea (Norway), respectively. A. fulgidus belongs to the euryarchaeotal branch of the archaea, being closely related to methanogens and more distantly related to the Thermococcales (64).
Most metabolic studies have been performed with A. fulgidus strain VC16 (66); the complete genome of this organism has been sequenced (28). A. fulgidus VC16 grows preferentially chemoorganoheterotrophically, e.g., with lactate and sulfate. Lactate was found to be completely oxidized to 3 CO2 via a modified acetyl coenzyme A (acetyl-CoA)/carbon monoxide dehydrogenase pathway (43). The A. fulgidus type strain VC16 does not contain genes of sugar degradation and, accordingly, was not able to grow on sugars. In contrast, it was found that A. fulgidus strain 7327 can grow on a polymeric sugar, i.e., starch, and sulfate as a carbon and energy source. This is the first report of the growth of a sulfate reducer on a polymeric sugar (30). Thus far, there are only a few reports on sugar-utilizing sulfate reducers, i.e., on glucose or fructose (10, 47, 75). This inability of sugar degradation in most sulfate reducers might reflect by their specific ecological role in the anaerobic carbon cycle, where they utilize the products of the primary fermenters, e.g., organic acids, alcohols, H2/CO2, etc. (18).
In A. fulgidus strain 7324, starch was incompletely oxidized to acetate and CO2, while sulfate is reduced to H2S. Enzyme studies revealed that starch degradation to acetate proceeds via glucose and pyruvate. Glucose is degraded via a modified Embden-Meyerhof pathway similar to that described for the hyperthermophilic archaea Pyrococcus and Thermococcus (30, 61); the pathway includes unusual enzymes, such as ADP-dependent glucokinase and phosphofructokinase and glycerinaldehyde 3-phosphate:ferredoxin oxidoreductase (30, 61). Formation of acetate from pyruvate involves pyruvate:ferredoxin oxidoreductase and ADP-forming acetyl-CoA synthetase (30). The latter enzyme couples the conversion of acetyl-CoA to acetate to ATP generation and constitutes the mechanism of acetate formation typical for the archaeal domain (61). Several enzymes of this pathway have been purified and characterized (17, 25, 31, 44). Thus far, the enzymes of starch degradation to intermediates of the Embden-Meyerhof pathway, i.e., glucose or glucose 6-phosphate, have not been identified in A. fulgidus.
Starch degradation in most organisms, including the hyperthermophilic archaeon Pyrococcus furiosus, proceeds via the combined action of amylases and amylopullulanases. These enzymes catalyze the extracellular conversion of starch to maltooligosaccharides, maltose and glucose, which are transported into the cell via specific transport systems (2, 5, 12). Here, we present evidence that A. fulgidus strain 7324 utilizes an unusual pathway of starch degradation via cyclodextrins as intermediates. The pathway involves the extracellular action of cyclodextrin glucanotransferase (CGTase) and the intracellular cyclodextrin degradation to glucose 6-phosphate via cyclodextrinase (CDase), maltodextrin phosphorylase (Mal-P), and phosphoglucomutase (PGM). Starch degradation via cyclodextrins as intermediates has been described thus far only for two organisms, the bacterium Klebsiella oxytoca and the archaeon Thermococcus sp. strain B1001 (15, 20). Here, we report the purification and characterization of enzymes catalyzing the starch degradation to glucose 6-phosphate via cyclodextrins from A. fulgidus strain 7324. Both the pathway and the enzymes differ in various aspects from those of K. oxytoca and Thermococcus sp. strain B1001.
MATERIALS AND METHODS
Growth of A. fulgidus and P. furiosus.
A. fulgidus strain 7324 (DSM 8774) grown under anaerobic conditions at 76°C in a 100-liter Biostat fermentor on a medium described by Möller-Zinkhan et al. (42) containing starch (1 g liter−1) and yeast extract (0.5 g liter−1) as the carbon and energy sources. In addition, A. fulgidus strain 7324 and strain VC16 (DSMZ 4304) were grown on 2 g of α- and β-cyclodextrin liter−1, respectively, in standard medium containing yeast extract (0.5 g liter−1). P. furiosus (DSMZ 3638) was grown strictly anaerobically at 90°C in a 100-liter Biostat fermentor on a medium described by Fiala and Stetter containing starch (1 g liter−1) as the carbon and energy source (14). Cells were harvested at the beginning of the stationary growth phase and stored at −20°C.
Preparation of the cell extracts, pellet fraction, and concentrated growth supernatant.
The cells were resuspended in 100 mM Tris-HCl (pH 8.0) or 50 mM potassium phosphate buffer (pH 7.0). For preparation of cell extracts the suspension was passed three times through a French pressure cell at 1.5 × 108 Pa and centrifuged (15 min at 14,000 rpm at 4°C). The pellet was resuspended in 50 mM sodium acetate (pH 5.0) for determination of cell associated proteins (9). For determination of the extracellular enzymes α-amylase, pullulanase, and CGTase, the growth supernatant was concentrated by filtration (0.4-μm pore size), ultrafiltration (exclusion size, 30 kDa), centrifugation in Centricon tubes (exclusion size, 30 kDa), and vacuum centrifugation at 50°C (9, 29). In addition, growth supernatant and resuspended pellet were applied to a starch column (19); proteins were eluted with 2 M NaCl.
Determination of enzyme activities in crude extracts.
Enzyme assays were performed at 50 and 80°C, respectively. Coupled enzyme assays containing auxiliary enzymes were performed between 50 and 60°C. The auxiliary enzymes were added shortly before start of the reaction, and it was ensured that these enzymes were not rate limiting. One unit of enzyme activity is defined as 1 μmol of substrate consumed or product formed per min. For assays using iodine for determination of starch conversion 1 U is defined as 1% starch conversion per min.
(i) α-Amylase and pullulanase activities.
α-Amylase and pullulanase activities were determined as the liberation of reducing sugars from starch and pullulan, respectively, by using a 3,5-dinitrosalicylic acid (DNS) assay as described by Brown et al. (9). Crude extracts, the pellet fraction, and the growth supernatant were tested. The respective samples of starch-grown P. furiosus (DSMZ 3638), prepared by the same procedure as that used for A. fulgidus, were used as a positive control.
(ii) CGTase activity.
CGTase activity was determined by monitoring the decrease of starch concentration with iodine. The assay contained 0.5% soluble starch in 50 mM sodium acetate (pH 5.5) with 1 mM CaCl2 and protein (crude extract, pellet fraction, or growth supernatant). After incubation for 5 to 60 min, a 20-μl aliquot of the assay mixture was added to 1 ml of iodine solution. Another aliquot was used to exclude the formation of reducing sugars by using the DNS assay. Activity was found only in the pellet fraction and not in crude extracts and growth supernatant, indicating extracellular or membrane-bound CGTase.
(iii) CDase activity.
CDase activity was measured by using the DNS assay as the liberation of reducing sugars from α-cyclodextrin. The assay contained 0.5% α-cyclodextrin in 50 mM sodium acetate (pH 4.5) and 5 to 150 μg of protein and was incubated for 1 to 15 min.
(iv) Mal-P activity.
Mal-P activity was determined as phosphate-dependent degradation of maltoheptaose to glucose-1-phosphate, which was converted to glucose-6-phosphate via PGM. Glucose-6-phosphate dehydrogenase was used to couple this reaction with the reduction of NADP+ at 80°C in a discontinuous assay. The incubation solution (250 μl each) contained 50 mM potassium phosphate buffer (pH 6.8), 2.5 mM MgCl2, 1 mM maltoheptaose, and 5 to 200 μg of protein. After incubation for 5 to 30 min, 750 μl of detection solution (50 mM potassium phosphate buffer [pH 6.8], 2.5 mM MgCl2, 1 mM NADP+, 2 U of PGM, and 1 U of glucose-6-phosphate dehydrogenase) were added, followed by incubation at an ambient temperature.
(v) PGM activity.
PGM activity was measured as glucose-1,6-bisphosphate-dependent conversion of glucose-1-phosphate to glucose-6-phosphate by coupling this reaction to the formation of NADP+ with glucose-6-phosphate dehydrogenase using a discontinuous assay. The incubation solution (250 μl each) contained 50 mM HEPES (pH 8.2), 5 mM MgCl2, 10 mM glucose-1-phosphate, 0.15 mM glucose-1,6-bisphosphate, and 5 to 200 μg of protein. After incubation for 5 to 30 min at 80°C, 750 μl of detection solution (50 mM Tris-HCl [pH 8], 1 U of glucose-6-phosphate dehydrogenase, 1 mM NADP+) was added, followed by incubation at an ambient temperature.
Purification of enzymes.
Cell extract was purified from frozen cells suspended in 50 mM Tris-HCl (pH 8.0) containing 30 mM NaCl, 5 mM EDTA, and 1 mM dithioerythritol. Upon passing through a French pressure cell at 1.3 × 108 Pa, cells were disrupted and centrifuged for 90 min at 100,000 × g at 4°C. All chromatographic steps were performed at 10°C.
(i) CGTase.
CGTase was purified from the membrane fraction from 60 g of starch-grown cells. The pellets obtained from centrifugation at 100,000 × g were resuspended in 50 mM sodium acetate (pH 5.0) containing 0.1% sodium dodecyl sulfate (SDS), incubated at 4°C for 30 min, and centrifuged for 90 min at 100,000 × g at 4°C. The supernatant was then adjusted to 2 M ammonium sulfate in 50 mM Tris-HCl (pH 8.0) containing 1 mM DTE (buffer A) and applied to a phenyl-Sepharose column (60 ml) equilibrated in 2 M ammonium sulfate in buffer A. Protein was eluted with a decreasing ammonium sulfate gradient in buffer A. Fractions containing the highest specific CGTase activity were concentrated by ultrafiltration on a 10-kDa filter. The concentrated protein suspension was loaded to a Superdex 200 column (16/60, 120 ml) equilibrated in 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 1 mM DTE. Protein was eluted at a flow rate of 1 ml min−1. The fractions containing the highest CGTase activity were pooled, adjusted to 2 M ammonium sulfate in buffer A, and applied to a phenyl-Sepharose column (15 ml). Protein was eluted with a decreasing ammonium sulfate gradient in buffer A for 300 ml. Fractions containing the highest specific activity were diluted in 50 mM Tris-HCl (pH 9.0) containing 1 mM DTE and applied to a UNO Q1 column. Proteins were eluted with a NaCl gradient from 0 to 2 M NaCl in 50 mM sodium acetate (pH 5.8) with 1 mM DTE. Fractions containing the highest activities were adjusted to 2 M ammonium sulfate in buffer A and applied to a Resource Phe column (Pharmacia, Freiburg, Germany). Elution was performed by a stepwise gradient (decrease of 5% ammonium sulfate for 1 ml each). CGTase-containing fractions were concentrated by ultrafiltration on a 10-kDa filter to 0.5 ml applied to a Bioprep SE 1000/17 column, equilibrated in 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 1 mM DTE. Protein was eluted at a flow rate of 0.5 ml min−1. Fractions containing CGTase activity were adjusted to 2 M ammonium sulfate in 50 mM BisTris (pH 7.0) with 1 mM DTE and applied to a Resource Phe column; elution was performed by a step gradient (decrease of 5% ammonium sulfate for 1 ml each). Fractions containing CGTase activity exhibited two protein bands in an SDS gel in approximately the same ratio. This partially purified CGTase was used for measurements and determination of the N-terminal amino acid sequence. The activity of the protein could be stabilized by the addition of 1 mM DTE but not by the addition of NaCl (0.1 to 1 M). Calcium chloride at 1 mM increased the activity fivefold.
(ii) CDase.
CDase was purified from cells grown on 66 g of starch. The supernatant obtained at 100,000 × g was applied to a Q-Sepharose column (430 ml) equilibrated to 50 mM Tris-HCl (pH 9.0) with 1 mM DTE (buffer B). Proteins were eluted with a NaCl gradient (0 to 2 M) in buffer B. Fractions containing the highest CDase activity were diluted in 50 mM Tris-HCl (pH 8.0) (buffer C) and applied to a DEAE-Sepharose column (100 ml). Elution was performed by using an NaCl gradient from 0 to 1 M in buffer C. Fractions containing the highest specific CGTase activity were concentrated by ultrafiltration on a 10-kDa filter. The concentrated protein suspension was loaded onto a Superdex 200 column (16/60, 120 ml) equilibrated in 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 1 mM DTE. Protein was eluted with a flow rate of 1 ml min−1. Fractions containing CDase activity were diluted with 50 mM sodium acetate (pH 5.0) (buffer D) and applied to a UNO S1 column. Protein was eluted with NaCl (0 to 1 M) in buffer D. Fractions with the highest CDase activity were adjusted to pH 8.0 with buffer C and applied to a UNO Q1 column. Protein was eluted with NaCl (0 to 1 M) in buffer C. The fraction containing the highest CDase activity was applied to a Bioprep SE 1000/17 column, equilibrated to 50 mM Tris-HCl (pH 7.4) with 150 mM NaCl. Elution was performed with a flow of 0.5 ml min−1. At this stage the enzyme was essentially pure. The purification procedure is summarized in Table 1.
TABLE 1.
Purification of CDase from A. fulgidus strain 7324
| Purification step | Protein (mg) | Activitya (U) | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1,820 | 121 | 0.066 | 100 | 1 |
| Q-Sepharose | 684 | 118 | 0.17 | 97.5 | 2.6 |
| DEAE-Sepharose | 199 | 116 | 0.58 | 95.9 | 8.8 |
| Superdex 200 16/60 | 3.7 | 9 | 2.4 | 7.4 | 36.4 |
| UNO S1 | 1 | 8.3 | 8.3 | 6.8 | 125 |
| UNO Q1 | 0.12 | 1.21 | 10 | 1 | 151 |
| Bioprep SE 1000/17 | 0.03 | 1.08 | 36 | 0.9 | 545 |
Activity was measured at 80°C as the formation of reducing sugars from 0.5 % α-cyclodextrin in 50 mM sodium acetate (pH 4.5).
(iii) Mal-P.
Mal-P was purified from 40 g of starch-grown cells. The supernatant obtained at 100,000 × g was adjusted to pH 5.0 and applied to an SP-Sepharose column (75 ml) equilibrated in buffer D. Proteins were eluted with a NaCl gradient from 0 to 2 M in 50 mM Tris-HCl (pH 9.0) (buffer E). Fractions containing the highest Mal-P activity were diluted in 50 mM potassium phosphate (pH 6.8) (buffer F) and applied to a Q-Sepharose column (20 ml) equilibrated in buffer E. Proteins were eluted with a NaCl gradient from 0 to 2 M in buffer F. Fractions containing the highest Mal-P activity were adjusted to 2 M ammonium sulfate in 50 mM Tris-HCl (pH 7.0) (buffer G) and applied to a phenyl-Sepharose column (20 ml). Proteins were eluted with an ammonium sulfate gradient from 2 to 0 M in buffer G. Fractions containing Mal-P activity were adjusted to 2 M ammonium sulfate in buffer G again and applied to a Resource Eth column. Proteins were eluted with an ammonium sulfate gradient from 2 to 0 M in buffer G. The fraction containing the highest activity was applied to a Bioprep SE 1000/17 column, equilibrated to 50 mM Tris-HCl (pH 7.4) with 150 mM NaCl. Elution was performed with a flow of 0.5 ml min−1. At this stage the enzyme was essentially pure. The purification procedure is summarized in Table 2.
TABLE 2.
Purification of Mal-P from A. fulgidus strain 7324
| Purification step | Protein (mg) | Activitya (U) | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 2,002 | 30 | 0.015 | 100 | 1 |
| SP-Sepharose | 40.8 | 1.8 | 0.045 | 6.1 | 3 |
| Q-Sepharose | 11.4 | 2.3 | 0.2 | 7.6 | 13 |
| Phenyl-Sepharose | 2.3 | 1.1 | 0.49 | 3.7 | 33 |
| Resource Eth | 0.5 | 1 | 2 | 3.3 | 133 |
| Bioprep SE 1000/17 | 0.03 | 0.25 | 8.3 | 1.6 | 573 |
Activity was determined at 60°C in the discontinuous assay as the formation of glucose-1-phosphate from 0.75 mM maltoheptaose in 50 mM potassium phosphate buffer (pH 6.8) with 2.5 mM MgCl2.
(iv) PGM.
PGM was purified from 66 g of starch-grown cells. The supernatant obtained at 100,000 × g was applied to a Q-Sepharose column (430 ml) equilibrated in 50 mM Tris-HCl (pH 9.0) with 1 mM DTE (buffer H) to separate PGM activity from CDase activity. Proteins were eluted with a NaCl gradient (0 to 1 M in buffer H). Fractions containing highest PGM activity were adjusted to 2 M ammonium sulfate in 50 mM Tris-HCl (pH 8.0) with 1 mM DTE (buffer I) and applied to a phenyl-Sepharose column (15 ml). Proteins were eluted with an ammonium sulfate gradient from 2 to 0 M in buffer I. Fractions with the highest PGM activity were adjusted to 2 M ammonium sulfate in buffer I again and applied to a Resource Eth column. Elution was performed with an ammonium sulfate gradient (2 to 0 M in buffer I). The fraction containing the highest activity was applied to a Bioprep SE 1000/17 column, equilibrated to 50 mM Tris-HCl (pH 7.4) with 150 mM NaCl and 1 mM DTE. Elution was performed with a flow of 0.5 ml min−1. Fractions containing PGM activity were adjusted to pH 8.0 with buffer I and applied to a UNO Q1 column. Protein was eluted with a NaCl gradient (0 to 1 M in buffer I). At this stage the enzyme was essentially pure. The purification procedure is summarized in Table 3.
TABLE 3.
Purification of PGM from A. fulgidus strain 7324
| Purification step | Protein (mg) | Activitya (U) | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1,820 | 65.9 | 0.036 | 100 | 1 |
| Q Sepharose | 240 | 16.8 | 0.07 | 25.5 | 1.9 |
| Phenyl Sepharose | 6.9 | 0.81 | 0.117 | 1.2 | 3.25 |
| Resource Eth | 0.95 | 0.69 | 0.866 | 1 | 24 |
| Bioprep SE 1000/17 | 0.15 | 0.52 | 3.4 | 0.79 | 94.4 |
| UNO Q1 | 0.015 | 0.066 | 4.4 | 0.1 | 122 |
Activity was determined at 50°C in the continuous assay as the formation of glucose 6-phosphate in 50 mM HEPES (pH 8.2) with 5 mM MgCl2, 1 mM glucose-1-phosphate, 0.125 mM glucose-1,6-bisphosphate, 1 mM NADP+, and 1 U of glucose-6-phosphate dehydrogenase.
Isolation of the CGTase gene.
The CGTase gene was isolated and amplified by PCR from genomic DNA from A. fulgidus strain 7324 by using primers homologous to the nucleotide sequence derived from the N-terminal amino acid sequence of the CGTase from A. fulgidus strain 7324 and from the C-terminal sequence of the α-glucanotransferase (GTase) from Thermococcus litoralis (75): the forward primer had the sequence (5′→3′) ATG GAA AA(AG) ATA AAC TTC ATA TTT GGC ATC, and the reverse primer had the sequence TCA AAG CTC CCT GAA CCT TAC CGT G. The PCR contained Proof start polymerase (QIAGEN, Hilden, Germany), purified DNA, primer, buffer, deoxynucleoside triphosphate, and MgSO4 according to the specifications of the supplier. The resulting PCR product was cloned via U-A cloning into vector pdrive (QIAGEN) according to the supplier's instructions and introduced into Escherichia coli JM109 for plasmid amplification and conservation. The plasmid AFGTpdrive was sequenced by using a BigDye terminator sequencing kit (ABI Prism; Applied Biosystems, Warrington, England). The whole procedure was done three times in independent experiments. The CGT gene was cleaved from pdrive by use of the restriction enzymes BamHI and XhoI and ligated into the vector pET17b (Novagen). The resulting plasmid, AFGTpET17b, was transformed in E. coli BL21 Codon Plus (Stratagene).
Bioinformatic analysis.
The genome of A. fulgidus strain VC16 was examined for the conserved regions of the glycosyl hydrolase (GH) 13 family and for the enzymes of the pathway described here by using BLAST (3) and online genome search tools (http://www.tigr.org).
Expression in E. coli and purification of recombinant CGTase.
For expression, cells were grown in Luria-Bertani medium at 37°C. Expression was initiated by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 0.4 mM). After 18 h of further growth, the cells were harvested by centrifugation. Cells were disrupted by passage through a French pressure cell. After centrifugation, the supernatant was heat precipitated at 70°C for 60 min and centrifuged again (20,000 × g at 4°C for 30 min). The resulting solution was applied to a Superdex 200 equilibrated in 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl, 1 mM DTE, and 1 mM CaCl2. Elution was performed at a flow rate of 1 ml min−1. Eluted protein was essentially pure at this stage. The enzyme was stabilized by the addition of 1 mM DTE and 1 mM CaCl2 during the purification procedure.
Determination of the N-terminal amino acid sequence.
The purified protein was run on a 12% polyacrylamide gel and blotted onto a polyvinylidene difluoride membrane. N-terminal microsequencing on a model 473A sequencer (Applied Biosystems) was carried out as described by Meyer et al. (39).
Determination of native molecular mass.
Gel filtration chromatography was carried out at ambient temperature on a Superdex 200 (50 mM Tris-HCl containing 150 mM NaCl; 1 ml min−1). A calibration for the column was performed with dextran blue (2,000 kDa), ovalbumin (43 kDa), and chymotrypsin (25 kDa) from Amersham Pharmacia Biotech UK, Ltd. (Little Chalfont, United Kingdom), and amylase (200 kDa) and bovine serum albumin (fraction V, 66 kDa) from Sigma.
Determination of enzyme activities and kinetic parameters of purified enzymes.
Characterization of enzymes included the determination of kinetic parameters, the optimum pH and temperature, and temperature stability. The influence of cations and product formation were analyzed. Kinetic parameters were calculated from Lineweaver-Burk plots. The long-term stability against the temperature of the purified enzymes, as well as potential stabilizing additives (1 M NaCl, 1 M ammonium sulfate, 1 to 5 mM CaCl2, or 1 M MgCl2) were tested in sealed vials that were incubated at temperatures between 80 and 100°C for up to 120 min; the optimal pH was used. The vials were then cooled on ice for 10 min, and the remaining enzyme activity was tested at the optimal temperature.
(i) CGTase.
CGTase activity (starch → cyclodextrins) was determined at 80°C as the decrease in starch as determined by an iodine assay (as indicated below). The assay contained 200 μl of 0.5% soluble starch in 50 mM sodium acetate (pH 5.8) containing 1 mM CaCl2 and 1 mM DTE and protein. The pH dependence of the enzyme was measured between pH 4.0 and 8.0 using either 100 mM sodium acetate (pH 4.0 to 6.0) or 100 mM potassium phosphate buffer (pH 6.0 to 8.0), each containing 1 mM DTE and 1 mM CaCl2. The influence of CaCl2 (0 to 5 mM) was tested at 80°C in 50 mM sodium acetate (pH 5.8) with 1 mM DTE. The formation of α- and β-cyclodextrin was determined with methyl red and phenolphthaleine assays, respectively. Therefore, the standard assay was incubated for 18 h at 60°C; aliquots were withdrawn and used for analysis.
Transglycosylation experiments were performed using 10 mM glucose, maltose, and maltooligosaccharides as substrates.
(ii) CDase.
CDase activity (cyclodextrin → maltooligodextrin) was determined as the liberation of reducing sugars from α-, β-, or γ-cyclodextrin at 80°C using the DNS assay. The standard assay (200 μl) contained 0.5% cyclodextrin in 50 mM sodium acetate (pH 4.5) and protein. The pH dependence of the enzyme was measured between pH 4.0 and 7.0 using 100 mM sodium acetate. The temperature dependence of the enzyme activity was measured between 20 and 90°C in 100 mM sodium acetate (pH 4.5). The cation specificity (0.1 to 10 mM each) was examined by using a standard test system at 80°C upon incubation in 20 mM EDTA for 30 min. Product formation from cyclodextrins was studied by thin-layer chromatography (TLC). Each assay contained 1.25% cyclodextrin and 50 mM sodium acetate (pH 4.5) and protein and was incubated for 30 min to 24 h at 80°C. A portion (5 μl) of each was applied to the matrix. Residual activity measured to ensure the availability of active enzyme.
(iii) Mal-P.
Mal-P activity [(glucose)n + Pi → (glucose)n − 1 + glucose-1-phosphate] was measured at 80°C as phosphate-dependent conversion of maltoheptaose to glucose-1-phosphate, which was converted to glucose-6-phosphate. This reaction was coupled to the reduction of NADP+ using glucose-6-phosphate dehydrogenase. The discontinuous assay contained 50 mM potassium phosphate buffer (pH 6.8), 2.5 mM MgCl2, 1 mM maltoheptaose, and protein; the detection solution contained 50 mM potassium phosphate (pH 6.8), 2.5 mM MgCl2, 1 mM NADP+, 2 U of PGM, and 1 U of glucose-6-phosphate dehydrogenase. After incubation at 80°C, the vials were cooled on ice, and detection solution was added; incubation was then performed at an ambient temperature. For determination of substrate specificity, maltoheptaose was exchanged for different maltooligodextrins and polysaccharides (1 to 50 mM) by using a standard discontinuous test system. Phosphate was replaced by pyrophosphate, AMP, ADP, ATP, and pyridoxal phosphate (0.1 to 10 mM). The pH dependence of the enzyme was measured between pH 4.0 and 9.0 using either 100 mM sodium acetate (pH 4.0 to 5.0), piperazine (pH 5 to 7.5), triethanolamine (pH 7.5 to 8.5), and HEPES (pH 8.5 to 9.0). The temperature dependence of the enzyme activity was measured between 25 and 90°C in 100 mM potassium phosphate (pH 5.8). The following effectors of α-glucan phosphorylases were tested at 80°C (10 μM to 5 mM each): AMP, ADP, ATP, glucose, α-cyclodextrin, β-cyclodextrin, ammonium sulfate, sodium sulfate, sodium vanadate, and sodium molybdate. The coenzyme pyridoxal phosphate was detected by measuring a protein spectrum in 50 mM potassium phosphate (pH 5.8) containing 200 mM NaCl and quantified with phenylhydrazine hydrochloride (71). Pyridoxal phosphate from Roche Diagnostics was used as a calibration standard.
(iv) PGM.
PGM activity (glucose-1-phosphate ⇆ glucose-6-phosphate) was measured at 50°C as the formation of glucose-6-phosphate in the presence of glucose-1,6-bisphosphate. This reaction was coupled to the reduction of NADP+ using glucose-6-phosphate dehydrogenase. The assay contained 50 mM HEPES (pH 8.2), 1 mM DTE, 10 mM MgCl2, 1 mM NADP+, 1 mM glucose-1-phosphate, 0.125 mM glucose-1,6-bisphosphate, 1 U of glucose-6-phosphate dehydrogenase, and protein. For temperatures above 50°C, a discontinuous assay was used. The incubation solution contained 50 mM HEPES (pH 8.2), 1 mM DTE, 5 mM MgCl2, 10 mM glucose-1-phosphate, 0.125 mM glucose-1,6-bisphosphate, and protein. After incubation the vials were cooled on ice, and detection solution (50 mM Tris-HCl [pH 8.0], 1 U of glucose-6-phosphate dehydrogenase, 1 mM NADP+) was added, followed by incubation at an ambient temperature. The conversion of glucose-6-phosphate to glucose-1-phosphate was examined by using TLC. The assay contained 50 mM HEPES (pH 8.2), 5 mM MgCl2, 0.125 mM glucose-1,6-bisphosphate, and 5 or 10 mM glucose-6-phosphate. After incubation for 15 to 90 min at 80°C, aliquots were withdrawn, and 2- to 5-μl portions were applied to a TLC plate. The activation of PGM activity by the addition of 5 to 20 mM cysteine, imidazole, or histidine was examined in a standard assay at 50°C. Thus, protein was preincubated with the respective additive and MgCl2 for 20 min at 50°C. The pH dependence of the enzyme was measured between pH 4.0 and 9.0 by using either 100 mM sodium acetate (pH 4.0 to 6.0), triethanolamine (pH 6.0 to 8.0), or Tris-HCl (pH 8.0 to 9.0). The temperature dependence of the enzyme activity was measured between 35 and 90°C in 100 mM HEPES (pH 8.2) in the discontinuous assay. The stability versus temperature was measured at 80°C in the presence or absence of MgCl2. Inhibition by vanadate was tested at 50°C by the addition of 0 to 100 μM sodium vanadate. Compensation by 1 to 50 mM EDTA was tested.
Analytical methods. (i) DNS assay for the determination of reducing sugars.
Samples (200 μl; up to 2% substrate) were mixed with 200 μl of DNS reagent (2 N NaOH, 30 g of sodium potassium tartrate, 1 g of DNS, and 100 ml of double-distilled water [filtrated]) and boiled for 5 min. The extinction at 546 nm was measured after the addition of 600 μl of water (40). Glucose (7 mM) was used as a standard.
(ii) Iodine assay for the determination of starch.
Starch was determined as starch-iodine complexes. A total of 1 ml of iodine solution (0.01% iodine and 0.1% potassium iodide in 3.8 mM HCl) was mixed with a 20-μl sample (up to 0.5% starch). Extinction was measured at 660 nm.
(iii) Determination of cyclodextrins.
α-Cyclodextrin was measured as the decrease of extinction at 505 nm as indicated by the formation of methyl orange-α-cyclodextrin inclusion complexes. The assay contained sample (up to 5 mM α-cyclodextrin), 10 μl of 0.6 N HCl, 15 μl of 1 mM methyl orange in 50 mM sodium phosphate buffer (pH 6.0), and double-distilled water at 1,000 μl. β-Cyclodextrin was measured as the decrease of extinction at 550 nm as indicated by the formation of phenolphthaleine-β-cyclodextrin inclusion complexes. The assay contained 50 μl of sample, 0.5 ml of phenolphthaleine solution (1.5 parts 3.8 mM phenolphthaleine in 98% ethanol and 50 parts carbonate buffer [pH 10.5]), and 0.7 ml of double-distilled water.
(iv) Determination of sugars by TLC.
Mono- and oligosaccharides were separated on silica 60 plates (Merck) with butanol-ethanol-water (5:3:2) as the mobile phase. For visualization, the plates were dipped in 5% H2SO4 and backed for 15 min at 120°C. Sugars appear as dark spots on a light surface. In addition, plates were dipped in Mandelin's reagent (0.6 g of ammonium metavanadate in 22.5 ml of A. bidest, 2.5 ml of concentrated H2SO4, and 25 ml of acetone) and backed for 5 min at 120°C. Sugars appear as light spots on a yellow surface. A mixture of saccharides (1 to 13 glucose units, 0.1%) was used as a standard. To distinguish panose and isopanose, a hydrolysis by glucoamylase was performed, and aliquots were applied to a TLC plate (23).
(v) Determination of sugar phosphates by TLC.
Sugar phosphates were separated on cellulose plates (Merck) with isobutyric acid-ammonia-water (66:1:33) as a mobile phase. Plates were dipped in ammonium thiocyanate (1% in acetone), dried for 5 min in hot air, and dipped in iron(III) chloride solution (50 mg in 100 ml of acetone). Sugar phosphates appear as light spots on a dark red surface.
(vi) Determination of protein.
Protein was determined by the Bradford method (8), using bovine serum albumin as a standard.
Source of material.
Yeast extract was from Difco (Stuttgart, Germany). Tris was from Biomol (Hamburg, Germany). Enzymes and coenzymes were from Boehringer (Mannheim, Germany). If not mentioned otherwise, all other chemicals were reagent grade and were obtained from E. Merck (Darmstadt, Germany) or Serva (Heidelberg, Germany). Gases (N2, 5.0; CO; N2/CO2, 80%/20%) were from Linde (Hamburg, Germany). A. fulgidus strains 7324 (DSM 8774) and strain VC16 (DSMZ 4304) and P. furiosus (DSMZ 3638) were from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). All of the fast protein liquid chromatography materials and columns used here were obtained from Pharmacia (Freiburg, Germany) and Bio-Rad (Munich, Germany).
Nucleotide sequence accession number.
The sequence of the CGTase gene was deposited in GenBank under the accession number EU138451.
RESULTS
Enzymes of starch degradation via cyclodextrins in cell extracts of A. fulgidus strain 7324.
In extracts of starch-grown A. fulgidus strain 7324, activities of typical enzymes of the classical starch degradation pathways, i.e., α-amylase or amylopullulanase, were not found. For comparison, we analyzed starch-grown P. furiosus cells, using the same conditions as for A. fulgidus, and found both α-amylase (0.8 U mg−1) and amylopullulanase (0.15 U mg−1) at activities similar to those previously described (59). Instead, in A. fulgidus strain 7324 the following enzyme activities of an unusual starch degradation pathway could be detected: extracellular CGTase (0.09 U mg−1 at 80°C) and intracellular CDase (0.02 U mg−1 at 80°C), Mal-P (0.015 U mg−1 at 55°C), and PGM (0.04 U mg−1 at 55°C). All of these enzymes were induced three- to fivefold after growth on starch compared to lactate, indicating a functional a role in starch degradation. CGTase activity was not detected in lactate-grown cells, indicating an even higher degree of induction. Furthermore, CGTase activity was not found in crude extracts, suggesting the absence of intracellular CGTase. The presence of these inducible enzymes indicates that starch is degraded in A. fulgidus strain 7324 to glucose-6-phosphate via the extracellular formation of cyclodextrins.
Growth on cyclodextrins.
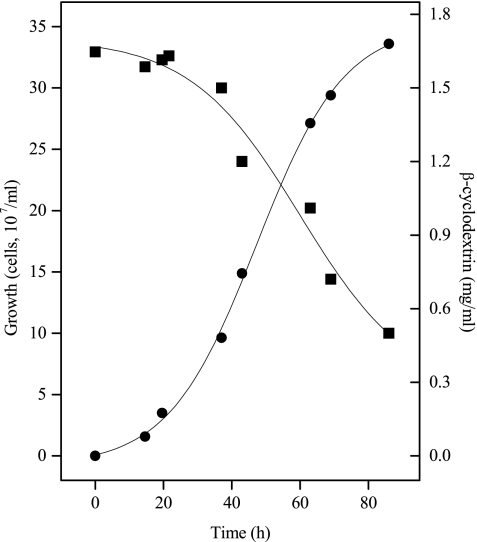
In accordance with the starch degradation via cyclodextrins, we found that A. fulgidus strain 7324 grew well on cyclodextrins and sulfate as a carbon and energy source. A typical growth curve is shown in Fig. 1. The organism grew exponentially with a doubling time of about 10 h up to maximal cell densities of 3.5 × 108 to 4 × 108 cells ml−1. Lower densities were obtained with α-cyclodextrin (2 × 108 cells ml−1). For comparison, the organism grew on starch with a doubling time of about 4 to 5 h up to 1 × 108 to 1.5 × 108 cells ml−1 (30). Neither growth on cyclodextrins nor growth on any sugar could be demonstrated for A. fulgidus strain VC16.
FIG. 1.
Growth of A. fulgidus strain 7324 on β-cyclodextrins at 76°C on medium containing 2 g of cyclodextrin liter−1, 0.5 g of yeast extract liter−1, and 30 mM sulfate. •, Cell number per ml; ▪, cyclodextrin concentration.
Purification and characterization of enzymes of starch degradation.
The purification and characterization of the enzymes of starch degradation to glucose-6-phosphate from starch-grown cells of A. fulgidus strain 7324 are described below.
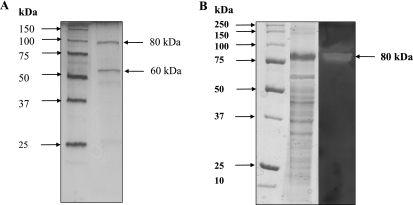
CGTase. (i) Partial purification.
CGTase was partially purified from starch-grown cells after elution by 0.1% SDS from membrane fractions by six purification steps. In this way the enzyme was purified ∼333-fold to a specific activity of 30 U mg−1 at 80°C with a yield of 1.4%. After the last chromatographic step, two bands were apparent in SDS-polyacrylamide gel electrophoresis (PAGE) gels (Fig. 2A). By using a zymogram the band at 80 kDa was identified to contain CGTase activity (Fig. 2B). The specific activity with 0.5% starch as a substrate and in the presence of calcium (1 mM) was 30 U mg−1, forming α- and β-cyclodextrin as products. CGTase showed a temperature optimum of 83°C and a pH optimum of 7. In the absence of calcium, the activity was fivefold lower. The N-terminal amino acid sequence of the subunit was (M)EKINFIFGIHNHQPLGNF (19 amino acids). This sequence was, with the exception of one amino acid, identical to that of an GTase from T. litoralis (72). Significantly less similarity was found for the N-terminal amino acid sequences of other archaeal GTases and CGTases.
FIG. 2.
Purification and activity of CGTase from A. fulgidus strain 7324. (A) SDS-PAGE, with silver staining of partially purified CGTase. Lanes: 1, standard; 2, 1 μg of protein after Resource Phe. (B) Zymogram of CGTase from A. fulgidus strain 7324. Partially purified protein was used to identify the band containing CGTase activity. Lanes: 1, standard; 2, 10 μg of protein after the addition of Superdex 200 stained with Coomassie brilliant blue; 3, 10 μg of protein after the addition of Superdex 200 with activity staining.
(ii) Identification and sequencing of the CGTase gene.
Based on the high identity to the N-terminal amino acid sequence of a GTase from T. litoralis, primers were constructed to amplify the respective genes from A. fulgidus strain 7324. The resulting PCR product was 1,980 bp, according to 659 amino acids and a calculated molecular mass of 77.88 kDa of the gene product. The gene sequence showed highest similarity to the sequence of GTase from T. litoralis (93.6%). Comparison of the deduced amino acid sequence showed a 1% difference, comprising only conserved substitutions, and thus indicating functional identity. In the genome of A. fulgidus strain VC16, no homologous ORF could be identified.
(iii) Recombinant CGTase.
A CGTase gene from A. fulgidus strain 7324 was cloned and overexpressed in E. coli BL21 Codon Plus. The resulting protein had a molecular mass of 80 kDa, which corresponds to the calculated molecular mass. The apparent native molecular mass of 170 kDa, as determined by gel filtration, indicates a homodimeric structure of CGTase. The protein catalyzed the conversion of starch with a Vmax of 6 U mg−1 in the presence of calcium and DTE. The resulting products were α- and β-cyclodextrin, with β-cyclodextrin as main product. CGTase also exhibited a low transglycosylation activity, e.g., using a mixture of maltose and cyclodextrins as substrates, CGTase generated glucose and maltooligodextrins (data not shown). The optimum pH and temperature of the recombinant protein were similar to those determined for the enzyme purified from A. fulgidus cells. CGTase was stable against thermal denaturation, not losing activity upon incubation at 80°C for 120 min. At 83°C the enzyme showed a half-life of 60 min. CGTase was dependent on divalent cations. The highest activity was observed with calcium (100%), which could be replaced by magnesium (90%), manganese (85%), iron (63%) and, less efficiently, by nickel, cobalt, and zinc (<10%).
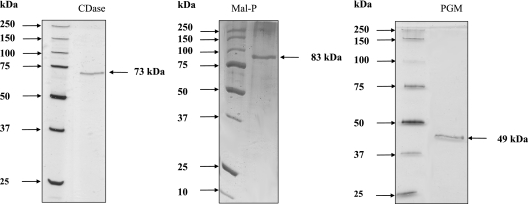
Characterization of CDase.
CDase was purified 550-fold using six chromatographic steps with a yield of 0.9% and a specific activity of 36 U mg−1 (80°C) (Table 1). SDS-PAGE revealed only one subunit, with an apparent molecular mass of 73 kDa (Fig. 3). The apparent molecular mass of native CDase was approximately 70 kDa, indicating monomeric structure. The N-terminal amino acid sequence of the subunit could not be determined due to a blocked N terminus. CDase from A. fulgidus strain 7324 was highly specific for cyclodextrins as substrates: γ-cyclodextrin was the preferred substrate of the enzyme, as indicated by the 10- to 15-fold-higher catalytic efficiency versus α- and β-cyclodextrin (Table 4). No activity was measured with starch, pullulan, amylose, amylopectin, or oligosaccharides (G2 to G17). Rate dependence on the concentration of all cyclodextrins showed Michaelis-Menten kinetics with the apparent Vmax and Km values shown in Table 4.
FIG. 3.
Purification of CDase, Mal-P, and PGM from A. fulgidus strain 7324 by SDS-PAGE, with silver staining. Lanes: 1, standard; CDase, 0.3 μg of protein after Bioprep SE 1000/17; Mal-P, 0.2 μg of protein after Bioprep SE 1000/17; PGM, 0.3 μg of protein after UNO Q1.
TABLE 4.
Kinetic constants of CDase from A. fulgidus strain 7324 at 80°C and pH 4.5
| Parameter | α-Cyclodextrin | β-Cyclodextrin | γ-Cyclodextrin |
|---|---|---|---|
| Vmax (U mg−1) | 22 | 42 | 21 |
| Km (mM) | 0.26 | 0.63 | 0.02 |
| Vmax/Km | 84.6 | 66.6 | 1,050 |
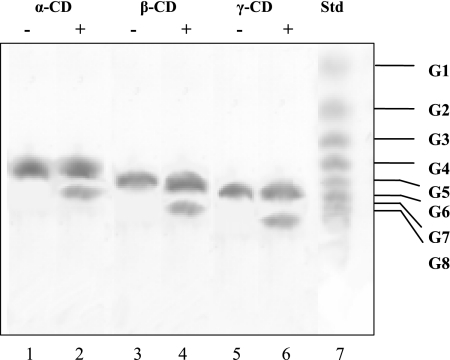
TLC experiments revealed that CDase from A. fulgidus strain 7324 linearized α-, β-, and γ-cyclodextrin to the respective oligosaccharides, i.e., maltohexaose, maltoheptaose, and maltooctaose (Fig. 4). Subsequent hydrolytic degradation to smaller maltooligodextrins was not observed, even after incubation for up to 24 h. This catalytic property is of interest, since other CDases degrade cyclodextrins to the level of maltose and glucose. CDases activity was dependent on divalent cations. EDTA treatment (20 mM) resulted in a complete loss of activity, which could be restored by the addition of Ca2+, Co2+, and Mg2+ (each 10 mM), respectively. However, Ni2+, Cu2+, and Zn2+ inhibited CDase activity at concentrations of 1 mM, as described as a typical feature of CDases (50). The temperature optimum for CDase was 80°C with an Arrhenius activation energy of 150 kJ mol−1 (linear part, 40 to 75°C). CDase showed high thermostability: the enzyme did not lose activity at 80°C upon incubation for 2 h. At 90°C, the enzyme showed a half-life of 50 min. The addition of (NH4)2SO4, NaCl, CaCl2, or MgCl2 (1 M each) could not stabilize the enzyme against heat inactivation. The pH optimum for CDase was 4.5; 50% of the activity was demonstrated at pH 3.3 and 6.0.
FIG. 4.
Determination of reaction products of CDase from the substrates α-, β-, and γ-cyclodextrin as analyzed by TLC on silica gel 60. The assay contained 0.5% of the respective cyclodextrin and 2 μg of protein (+) in 50 mM sodium acetate (pH 4.5) incubated at 80°C for 1.5 h. −, Control assays without protein. The sugars were stained with 5% H2SO4 after 15 min of baking at 120°C. Lanes 1 and 2, α-cyclodextrin; lanes 3 and 4, β-cyclodextrin; lanes 5 and 6, γ-cyclodextrin; lane 7, standard for glucose units. Note that cyclodextrins showed smaller migration distances than the respective maltooligosaccharides, which corresponded to the glucose unit standard.
Characterization of Mal-P.
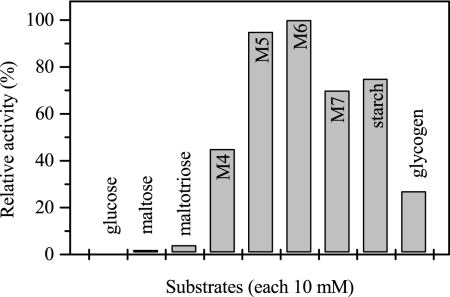
Mal-P was purified 575-fold via five chromatographic steps with a yield of 1.6% and a specific activity of 8.3 U mg−1 (at 60°C) (Table 2). SDS-PAGE revealed only one subunit with an apparent molecular mass of 83 kDa (Fig. 3); the apparent molecular mass of native Mal-P was approximately 350 kDa, indicating a homotetrameric structure. The N-terminal amino acid sequence of the subunit was determined to be METVVNXIKSKLPENLEGLLDLAY (24 amino acids). An alignment of this sequence with the sequences of all known maltodextrin and glycogen phosphorylases from archaea showed highest similarity (23 amino acids identical) to the Mal-P from T. litoralis (72). Lower similarities were found for the enzymes from Thermococcus zilligii and Sulfolobus solfataricus (<50% [data not shown]). No homologous open reading frame was found in A. fulgidus strain VC16. Mal-P contained pyridoxal phosphate (0.8 mol of pyridoxal phosphate per mol of native protein), a typical cofactor of phosphorylases. Mal-P catalyzed the phosphate-dependent cleavage of maltooligodextrins forming glucose-1-phosphate. Mal-P accepted maltooligodextrins with 3 to 17 glucose units and starch and glycogen as substrates (Fig. 5). The rate dependency of Mal-P from these substrates and from phosphate followed Michaelis-Menten kinetics. The highest catalytic efficiency was shown for maltoheptaose (Fig. 6A) and maltohexaose. Starch and glycogen were converted with significant lower catalytic efficiencies due to the high apparent Km values (Table 5).
FIG. 5.
Substrate spectrum of Mal-P from A. fulgidus strain 7324. The relative activity of 100% corresponds to 34 U mg−1. M4, maltotetraose; M5, maltopentaose; M6, maltohexaose; M7, maltoheptaose. The standard deviation was <5%.
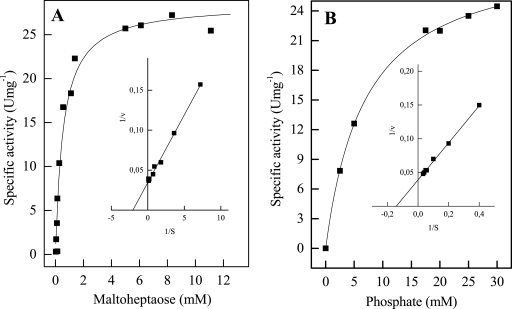
FIG. 6.
Rate dependence of Mal-P from A. fulgidus strain 7324 on the concentration of maltoheptaose (A) and phosphate (B). The inset graphs show double reciprocal plots of the rates versus the corresponding substrate concentrations.
TABLE 5.
Kinetic constants of Mal-P from A. fulgidus strain 7324 at 80°C and pH 5.8
| Parameter | Maltohexaose | Maltoheptaose | Glycogen | Starch |
|---|---|---|---|---|
| Vmax (U mg−1) | 34 | 25 | 14 | 30 |
| Km (mM) | 1 | 0.47 | 42.9 | 4.9 |
| Vmax/Km | 34 | 53.2 | 0.33 | 6.12 |
The apparent Km value for phosphate was 7.2 mM with maltoheptaose as a substrate. The Vmax value was 25 U mg−1 (Fig. 6B). Other than phosphate, the enzyme does not accept other phosphorylated compounds, such as AMP, ADP, ATP, pyrophosphate, and pyridoxal phosphate. Typical eukaryotic phosphorylase inhibitors (AMP, glucose, ammonium sulfate, sodium sulfate, cyclodextrins, vanadate, and molybdate) and activators (ADP and ATP) did not significantly affect the rate of Mal-P from A. fulgidus. The temperature optimum of Mal-P was 80°C, with an Arrhenius activation energy of 53.6 kJ mol−1 (linear part 30 to 75°C). Mal-P was highly thermostable. At 80°C the enzyme did not lose activity upon incubation for 120 min. At 95°C the half-life of the enzyme was 350 min. The pH optimum of Mal-P was 6.8; at pH 5 and 9, 50% activity was demonstrated.
Characterization of PGM.
PGM was purified 122-fold using five chromatographic steps, with a yield of 0.1% and a specific activity of 4.4 U mg−1 (at 50°C) (Table 3). SDS-PAGE revealed one subunit of 49 kDa (Fig. 3). The apparent molecular mass of native PGM was approximately 200 kDa, indicating a homotetrameric structure. The N-terminal amino acid sequence of the subunit was determined to be M(X)KIFGTFGVRGIANE (15 amino acids). This N-terminal amino acid sequence showed high similarity to the N-terminal amino acid sequence of PGM from Thermococcales and less similarity to the open reading frame AF0458 in A. fulgidus type strain VC 16 (9 of 15 amino acids). Preincubation of PGM with cysteine, histidine, or imidazole resulted in an increase in enzyme activity. The greatest increase was obtained with 20 mM histidine (fivefold activation), which was therefore added to the purified enzyme prior to characterization. PGM catalyzed the reversible conversion of glucose-1-phosphate to glucose-6-phosphate. Activity was dependent on the cofactor glucose-1,6-bisphosphate. The rate dependence on glucose-1-phosphate (Km = 0.49 mM) and the cofactor (Km = 34 μM) showed Michaelis-Menten kinetics with Vmax values of 32 U mg−1. PGM required divalent cations for activity. With Mg2+, which was most effective, an apparent Km value of 52 μM was measured. The enzyme also catalyzed the reverse reaction, the conversion of glucose-6-phosphate to glucose-1-phosphate as shown by thin-layer chromatography. The phosphate analog vanadate inhibited PGM. The addition of 80 μM vanadate resulted in a complete loss of activity; inhibition of 50% of that obtained with 30 μM vanadate. The temperature optimum of PGM was 81°C, with an Arrhenius activation energy of 78.9 kJ mol−1 (25 to 80°C). PGM was highly thermostable in the presence of Mg2+ (5 mM), and a significant loss of activation could not be determined after 120 min of incubation. In the absence of magnesium, the enzyme exhibited a significantly lower half-life of 30 min upon incubation at 80°C. The pH optimum of the enzyme was 8.2; 50% activity was observed at pH 6.0 and 9.3.
DISCUSSION
The enzymes of starch degradation to glucose-6-phosphate and glucose were studied in A. fulgidus strain 7324. The enzymes of the classical starch degradation pathways, e.g., α-amylase and amylopullulanase, as found in most starch-utilizing organisms (5), could not be detected in A. fulgidus. Instead, we present evidence here that this organism utilizes an unusual novel starch degradation pathway via cyclodextrins as intermediates. This pathway comprises a CGTase that catalyzed the extracellular formation of cyclodextrins from starch and the subsequent intracellular degradation of cyclodextrins to glucose-6-phosphate, involving CDase, Mal-P, and PGM. All of these enzymes were induced after growth on starch, indicating the functional involvement of starch degradation.
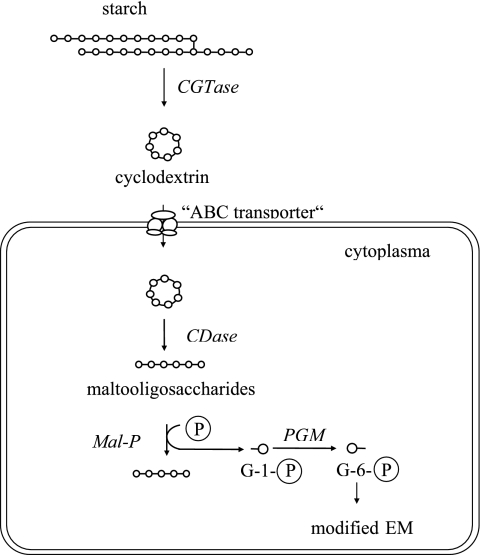
The pathway of starch degradation in A. fulgidus as depicted in Fig. 7, is similar to that proposed for two other organisms, the bacterium K. oxytoca (15) and the archaeon Thermococcus sp. strain B1001 (20). As in A. fulgidus, both organisms form extracellular cyclodextrins from starch by means of a CGTase. In K. oxytoca and Thermococcus sp. strain B1001, the cyclodextrins formed are transported via ABC transporter into the cell. In analogy, we assume that cyclodextrin uptake in A. fulgidus proceeds also via an ABC-like transport system. The pathway of intracellular conversion of cyclodextrins in A. fulgidus showed significant differences from the pathways in K. oxytoca and Thermococcus sp. strain B1001. Klebsiella hydrolyzes cyclodextrins via maltodextrins to glucose-1-phosphate and glucose by an intracellular GTase, a Mal-P, and a PGM (15). In the hyperthermophilic archaeon Thermococcus sp. strain B1001, cyclodextrins were completely hydrolyzed to glucose and maltose by the action of CDase without the functional involvement of Mal-P and PGM (20). It should be mentioned that in K. oxytoca, rather than in Thermococcus sp. strain B1001, the classical pathway of starch degradation via α-amylase and amylopullulanase is operative in parallel (15, 48). Other than A. fulgidus and Thermococcus sp. strain B1001, the only hyperthermophilic archaeon studied in detail with respect to starch degradation is P. furiosus. For this organism it has been shown by microarray analyses that a single amylopullulanase is the only starch-acting enzyme. Other GHs, such as CGTase, that were annotated in the genome turned out not to be functionally involved in starch degradation (33).
FIG. 7.
Proposed pathway of starch degradation to glucose-6-phosphate in the hyperthermophilic sulfate-reducing archaeon A. fulgidus strain 7324. The ABC transporter was postulated in analogy to K. oxytoca and Thermococcus sp. strain B1001 (15, 20). G-1-P, glucose-1-phosphate; G-6-P, glucose-6-phosphate. The formation of glucose by CGTase is not shown. Glucose and glucose-6-phosphate are degraded via a modified Embden-Meyerhof pathway to pyruvate.
In accordance with the extracellular formation of cyclodextrins, A. fulgidus strain 7324 grew well on cyclodextrins and sulfate as a carbon and energy source. Growth was most effective on β-cyclodextrin, which is the cyclodextrin species formed at the highest rates by the CGTase of A. fulgidus. Utilization of cyclodextrin as a carbon source for growth has been reported only for a few bacteria and archaea (5).
The starch degradation pathway described here for A. fulgidus is the first report on polymeric sugar degradation in a sulfate-reducing organism. Other sulfate reducers do not appear to have this ability. In accordance, the available genomes of sulfate reducers do not contain genes encoding enzymes of polymeric sugar degradation, e.g., GHs (35). This inability to grow on polymeric sugars is in accordance with the ecological function of sulfate reducers as typical organisms of the terminal part of the anaerobic carbon cycle. As such, they usually utilize the products, e.g., organic acids, alcohols, and H2/CO2, formed by primary fermenters (18). However, the unusual starch degradation pathway via cyclodextrins may enable A. fulgidus to compete effectively against other starch-degrading organisms, which usually do not utilize cyclodextrins as substrates. Cyclodextrins are not easily hydrolyzed by the common starch-degrading enzymes and, moreover, cyclodextrins are described to be even effective inhibitors of these enzymes (37).
The enzymes of starch degradation to glucose-6-phosphate from A. fulgidus strain 7324 are discussed below with respect to their catalytic, molecular, and thermophilic properties.
CGTase.
CGTases catalyze the formation of cyclic, nonreducing maltooligosaccharides (cyclodextrins) from starch. Since cyclodextrins are of biotechnological interest, CGTases are well-characterized enzymes, in particular from Bacillus species (69). However, the defined physiological role of CGTases has not been identified in most cases (7, 69). A distinct function of CGTase as part of a starch degradation has been shown here for A. fulgidus strain 7324 and, previously, for CGTase from K. oxytoca and Thermococcus sp. strain B1001 (15, 65). Recently, recombinant CGTases from T. kodakaraensis (51) and from P. furiosus (76) were characterized after heterologous expression in E. coli. However, the CGTase gene from P. furiosus was shown not to be functionally involved in starch degradation on basis of microarray experiments (33).
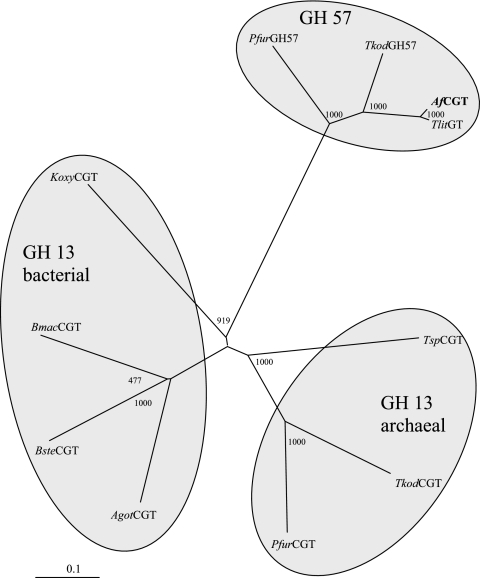
The N-terminal amino acid sequence of CGTase from A. fulgidus strain 7324 showed greatest similarity to that of a GTase from T. litoralis (24, 72). Using this N-terminal amino acid sequence and the C-terminal sequence of the GTase from T. litoralis, we succeeded in the amplification of the CGTase gene from A. fulgidus strain 7324. The complete gene sequence showed 93.6% identity to GTase from T. litoralis. An alignment of the deduced amino acid sequence of the CGTase from A. fulgidus strain 7324 and the respective sequences of the known archaeal CGTases, as well as GTase from T. litoralis, is shown in Fig. 8. Usually, CGTases belong to the large, very conserved GH 13 family (“α-amylase family”) (21, 37). Surprisingly, only parts of the conserved catalytic residues of this family are found in the A. fulgidus sequence. Moreover, the sequence comparison revealed that CGTase from A. fulgidus should be classified into GH 57 family (21) since it contains all conserved specific catalytic residues of GH 57 family, as well as the typical substrate binding sites, based on the crystal structure of GTase from T. litoralis (22). The attribution of A. fulgidus CGTase as a member of the GH 57 family was also supported by the phylogenetic analysis, as shown in Fig. 9. CGTases of the GH 13 family and GTases from the GH 57 family form separate clusters. CGTase from A. fulgidus clearly clustered into the GH 57 family. The GH 57 family comprises mainly GTases but only a few enzymes with CGTase activity.
FIG. 8.
Alignment of deduced amino acid sequence of CGTase from A. fulgidus strain 7324 (Af) and archaeal (cyclodextrin)GTases. The dots represent the substrate binding sites of the GH 57 family, the squares show the proposed calcium binding sites, and the arrowheads indicate the conserved catalytic residues of the GH 57 family. The four conserved regions typical for enzymes of the GH 13 family are indicated by boxes and designated I, II, III, and IV. The invariant amino acids of the GH 13 family within the conserved regions are marked by asterisks. The alignment was constructed by using CLUSTAL X (68). Identical residues are indicated by dark shading; conserved substitutions are indicated by light shadings. NCBI accession numbers: Tl, T. litoralis D88253; Tk, T. kodakaraensis (AB072372); Pf, P. furiosus ABA33720.1; Tsp, Thermococcus sp. strain B1001 BAA88217.1.
FIG. 9.
Phylogenetic relationship of CGTase from A. fulgidus strain 7324 (AfCGT) with members of the GH 13 and GH 57 families. The tree was constructed by the neighbor-joining method of CLUSTAL X (68). The given numbers represent bootstrapping values according to the neighbor-joining method. NCBI accession numbers: T. litoralis (TlitGT), D88253; Thermococcus sp. strain B1001 (TspCGT), BAA88217.1; T. kodakaraensis (TkodCGT), BAB78538.1; P. furiosus CGTase (PfuCGT), ABA33720.1; A. gottschalkii (AgotCGT), CAH61550.1; K. oxytoca (KoxyCGT), P08704; Geobacillus stearothermophilus (BsteCGT), CAA41770.1; Bacillus macerans (BmacCGT), P04830; P. furiosus alpha-amylase (PfuGH57), AAA72035.1; T. kodakaraensis 4-α-glucanotransferase (TkodGH57), BAA22062.1.
This was unexpected, since GTases usually catalyze the intracellular interconversion of linear sugars rather than the formation of cyclic sugars. In contrast to this sequence-based classification, comparison of activity patterns clearly defines the A. fulgidus enzyme as a functional CGTase. The catalytic function of GTase of T. litoralis is still not finally clarified: the enzyme has been reported to be involved only in the interconversion of linear sugars (72), whereas Jeon et al. (24) described an additional CGTase activity forming cyclodextrins besides the formation of linear saccharides. However, the A. fulgidus strain 7324 CGTase catalyzed the formation of α- and β-cyclodextrins from starch. β-Cyclodextrin is the main product, thus classifying the enzyme as β-CGTase. In accordance with this, A. fulgidus strain 7324 grows preferentially on β-cyclodextrin. β-CGTases are also reported for T. kodakaraensis and several Bacillus strains (27, 51). Using starch as the sole substrate of the CGTase, the A. fulgidus enzyme catalyzed the formation of cyclodextrins only. However, as described for other CGTases and GTases (51, 69), the A. fulgidus enzyme also shows transglycosylation activity when cyclodextrins and maltooligodextrins are used as substrates. For example, CGTase catalyzes the conversion of α-cyclodextrin and maltose to glucose and maltoheptaose. The formed glucose probably enters the modified Embden-Meyerhof pathway and might explain the glucokinase induction during growth on starch (31).
Like most CGTases (6, 69), with exception of the monomeric CGTase from Anaerobranca gottschalkii (67), the A. fulgidus enzyme shows homodimeric structure with subunits of ∼80 kDa. As described for bacterial CGTases (69), the Archaeoglobus enzyme was stimulated fivefold by calcium ions. Accordingly, conserved calcium binding sites were found in the sequence of CGTase (Fig. 8). The enzyme showed a temperature optimum of 83°C in accordance with the growth optimum of A. fulgidus. The pronounced thermophilic properties might be advantageous considering the application of CGTase in the biotechnological production of cyclodextrins.
CDase.
CDases, which all belong to the GH 13 family (37), catalyze the hydrolysis of cyclic oligosaccharides. Many CDases are described for the bacteria Bacillaceae and Enterobacteriaceae (50), whereas from the domain of Archaea, only two CDases from Thermococcus sp. strain B1001 and from P. furiosus have been characterized as recombinant enzymes (20, 73). For A. fulgidus, the in vivo function of the enzyme in starch degradation was concluded from its induction after growth on starch. A. fulgidus CDase catalyzes exclusively the ring opening of cyclodextrins to the respective maltooligodextrins the rather than their further degradation to maltose and glucose, which is a typical feature of all other CDases (11, 20, 50, 73). The enzyme showed the highest catalytic efficiency for γ-cyclodextrin as a substrate, as described for CDase from K. oxytoca (13). In contrast to both archaeal and most bacterial CDases, which are homodimeric proteins (50, 70), the A. fulgidus enzyme has a monomeric structure. In accordance with the optimal growth temperature of A. fulgidus, CDase showed both a high temperature optimum and a high thermostability up to 90°C. A. fulgidus CDases required calcium ions, which seem to be tightly bound, since only harsh EDTA treatment caused a loss of activity. Tightly bound calcium ions were also reported for other members of GH 13 family (26). Copper and zinc ions, known as typical inhibitors of GH 13 family members, also inhibited the enzyme from A. fulgidus and the other known archaeal CDases and (29, 32, 34).
Mal-P (α-glucan phosphorylase).
Mal-P is a member of the α-glucan phosphorylase family, catalyzing the phosphate-dependent stepwise degradation of polymeric and oligomeric saccharides to glucose-1-phosphate. Mal-P is ubiquitous in all domains of life (60). The only characterized archaeal member of the α-glucan phosphorylase family, Mal-P from T. litoralis (72), showed high sequence similarity to the N-terminal amino acid sequence of Mal-P from A. fulgidus described here. Both archaeal Mal-P show similar catalytical properties, e.g., a preference for maltodextrins with more than four glucose units. The highest activity was obtained for maltoheptaose. The catalytic efficiencies for starch and glycogen were significantly lower compared to the maltodextrins, indicating that the archaeal enzymes represent true Mal-Ps rather than glycogen or starch phosphorylases.
In contrast to the T. litoralis enzyme, a homodimer, Mal-P from A. fulgidus was characterized as a homotetramer. Both dimeric and tetrameric forms have been described in the α-glucan phosphorylase family (49). Mal-P from A. fulgidus contained pyridoxal phosphate, the typical cofactor of glucan phosphorylases, suggesting a common catalytic mechanism (49). All bacterial α-glucan phosphorylases and both archaeal Mal-P are not subjects of allosterical regulation and thus differ from their eukaryal counterparts (60). Mal-P from A. fulgidus has been shown to be induced by the growth substrate, which has also been reported for T. litoralis and the bacterial homologues, indicating transcriptional regulation for bacterial and archaeal glucan phosphorylases (60, 72).
PGM.
PGM catalyzes the reversible conversion of glucose-1-phosphate to glucose-6-phosphate. Most PGMs belong to the bifunctional PGM/phosphomannose family (16, 36, 46, 53). Within the domain of Archaea, PGM activities were found in crude extracts of T. litoralis (72), T. kodakaraensis (52), and Methanococcus maripaludis (74). Respective genes from Thermococcus kodakaraensis and from Pyrococcus horokoshii were heterologously expressed in E. coli and characterized (1, 52). As part of the starch degradation pathway, PGM from A. fulgidus catalyzed the conversion of glucose-1-phosphate to glucose-6-phosphate, providing the substrate for the modified EM pathway (30). In accordance with this, PGM was induced during growth on starch. PGM from A. fulgidus strain 7324 and T. kodakaraensis constitutes a homotetramer of 49-kDa subunits (52). Thus, both archaeal enzymes differ from their bacterial and eukaryotic counterparts, which appear to be monomeric and dimeric proteins (16, 38, 58). PGM activity from A. fulgidus strain 7324 was dependent on the typical cofactor glucose-1,6-bisphosphate and required Mg2+, as described for most bacterial PGM (45, 57). The A. fulgidus enzyme was activated by treatment with histidine, imidazole, or cysteine. This stimulating effect, which is typical for other PGMs, was attributed to a removal of inhibiting heavy metals by these compounds (16, 41, 58). The PGM activity was inhibited by vanadate, a structure analog of phosphate, as has been reported for other PGMs (54, 55, 56).
In summary, all of the enzymes of the unusual starch degradation pathway to glucose-6-phosphate in A. fulgidus, i.e., CGTase, CDase, Mal-P, and PGM, showed highly similar catalytic and molecular properties compared to their homologues from Thermococcus species. However, these enzymes were not present in the A. fulgidus type strain VC16. Furthermore, no encoding genes of CGTase, CDase, and Mal-P and, as previously shown, of all enzymes of a modified Embden-Meyerhof pathway, as well as of amylase and amylopullulanase, have been detected in the genome of A. fulgidus strain VC16 (11, 28, 30). This is in accordance with the inability of strain VC16 to grow on starch or any sugar. The absence of starch-degrading and glycolytic genes in strain VC16 might be explained by the loss of these genes. Alternatively, A. fulgidus strain 7324 might have taken these genes by lateral gene transfer, e.g., from Thermococcales species (30, 31).
Acknowledgments
We thank H. Preidel for mass culturing A. fulgidus strain 7324 and R. Schmid (Osnabrück) for the determination of N-terminal amino acid sequences.
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (SCHO 316/8-1).
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Akutsu, J., Z. Zhang, M. Tsujimura, M. Sasaki, M. Yohda, and Y. Kawarabayasi. 2005. Characterization of a thermostable enzyme with phosphomannomutase/phosphoglucomutase activities from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. J. Biochem. 138:159-166. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., J. L. Van de Vossenberg, A. J. Driessen, and W. N. Konings. 2001. Bioenergetics and solute uptake under extreme conditions. Extremophiles 5:285-294. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beeder, J., R. K. Nilsen, J. T. Rosnes, T. Torsvik, and T. Lien. 1994. Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl. Environ. Microbiol. 60:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, H. 1977. Cyclodextrin glucanotransferase from Klebsiella pneumoniae. 2. Significance of the enzyme for the metabolism of cyclodextrins by Klebsiella pneumoniae M 5. Arch. Microbiol. 113:49-56. [DOI] [PubMed] [Google Scholar]
- 6.Bertoldo, C., and G. Antranikian. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 6:151-160. [DOI] [PubMed] [Google Scholar]
- 7.Biwer, A., G. Antranikian, and E. Heinzle. 2002. Enzymatic production of cyclodextrins. Appl. Microbiol. Biotechnol. 59:609-617. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brown, S. H., H. R. Costantino, and R. M. Kelly. 1990. Characterization of amylolytic enzyme activities associated with the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl. Environ. Microbiol. 56:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cord-Ruwisch, R., B. Ollivier, and J. Garcia. 1986. Fructose degradation by Desulfovibrio sp. in pure culture and in coculture with Methanospirillum hungatei. Curr. Microbiol. 13:285-289. [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. Life with no sugars? J. Mol. Microbiol. Biotechnol. 1:307-308. [PubMed] [Google Scholar]
- 12.Driessen, A. J., J. L. C. M. van de Vossenberg, and W. N. Konings. 1996. Membrane composition and ion-permeability in extremophiles. FEMS Microbiol. Rev. 18:139-148. [Google Scholar]
- 13.Feederle, R., M. Pajatsch, E. Kremmer, and A. Bock. 1996. Metabolism of cyclodextrins by Klebsiella oxytoca m5a1: purification and characterization of a cytoplasmically located cyclodextrinase. Arch. Microbiol. 165:206-212. [DOI] [PubMed] [Google Scholar]
- 14.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 15.Fiedler, G., M. Pajatsch, and A. Bock. 1996. Genetics of a novel starch utilization pathway present in Klebsiella oxytoca. J. Mol. Biol. 256:279-291. [DOI] [PubMed] [Google Scholar]
- 16.Galloway, C. M., and W. M. Dugger. 1994. Purification and characterization of phosphoglucomutase from peas. Physiologia Plantarium 92:479-486. [Google Scholar]
- 17.Hansen, T., B. Schlichting, J. Grötzinger, M. K. Swan, C. Davies, and P. Schönheit. 2005. Mutagenesis of catalytically important residues of cupin type phosphoglucose isomerase from Archaeoglobus fulgidus. FEBS J. 272:6266-6275. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, T. A. 1994. Metabolism of sulfate-reducing prokaryotes. Antonie Leeuwenhoek 66:165-185. [DOI] [PubMed] [Google Scholar]
- 19.Hanus, J., and J. Kucera. 1974. Starch gel for gel filtration. J. Chromatogr. 97:270-272. [Google Scholar]
- 20.Hashimoto, Y., T. Yamamoto, S. Fujiwara, M. Takagi, and T. Imanaka. 2001. Extracellular synthesis, specific recognition, and intracellular degradation of cyclomaltodextrins by the hyperthermophilic archaeon Thermococcus sp. strain B1001. J. Bacteriol. 183:5050-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280(Pt. 2):309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura, H., S. Fushinobu, B. S. Jeon, T. Wakagi, and H. Matsuzawa. 2001. Identification of the catalytic residue of Thermococcus litoralis 4-α-glucanotransferase through mechanism-based labeling. Biochemistry 40:12400-12406. [DOI] [PubMed] [Google Scholar]
- 23.Imanaka, T., and T. Kuriki. 1989. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J. Bacteriol. 171:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon, B. S., H. Taguchi, H. Sakai, T. Ohshima, T. Wakagi, and H. Matsuzawa. 1997. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis: enzyme purification and characterization and gene cloning, sequencing, and expression in Escherichia coli. Eur. J. Biochem. 248:171-178. [DOI] [PubMed] [Google Scholar]
- 25.Johnsen, U., T. Hansen, and P. Schönheit. 2003. Comparative analysis of pyruvate kinases from the hyperthermophilic archaea Archaeoglobus fulgidus, Aeropyrum pernix, and Pyrobaculum aerophilum and the hyperthermophilic bacterium Thermotoga maritima: unusual regulatory properties in hyperthermophilic archaea. J. Biol. Chem. 278:25417-25427. [DOI] [PubMed] [Google Scholar]
- 26.Kamitori, S., A. Abe, A. Ohtaki, A. Kaji, T. Tonozuka, and Y. Sakano. 1999. Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 Å resolution and alpha-amylase 2 (TVAII) at 2.3 Å resolution. J. Mol. Biol. 318:443-453. [DOI] [PubMed] [Google Scholar]
- 27.Kaulpiboon, J., V. Rimphanitchayakit, and P. Pongsawasdi. 2004. Identification of an alkaline-tolerant cyclodextrin-metabolizing bacterium and characterization of its cyclodextrinase gene. J. Basic Microbiol. 44:374-382. [DOI] [PubMed] [Google Scholar]
- 28.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 29.Koch, R., R. Zablowski, A. Spreinat, and G. Antranikian. 1990. Extremely thermostable amylolytic enzyme from the archaebacterium Pyrococcus furiosus. FEMS Microbiol. Lett. 71:21-26. [Google Scholar]
- 30.Labes, A., and P. Schönheit. 2001. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming). Arch. Microbiol. 176:329-338. [DOI] [PubMed] [Google Scholar]
- 31.Labes, A., and P. Schönheit. 2003. ADP-dependent glucokinase from the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324. Arch. Microbiol. 180:69-75. [DOI] [PubMed] [Google Scholar]
- 32.Laderman, K. A., B. R. Davis, H. C. Krutzsch, M. S. Lewis, Y. V. Griko, P. L. Privalov, and C. B. Anfinsen. 1993. The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 268:24394-24401. [PubMed] [Google Scholar]
- 33.Lee, H. S., K. R. Shockley, G. J. Schut, S. B. Conners, C. I. Montero, M. R. Johnson, C. J. Chou, S. L. Bridger, N. Wigner, S. D. Brehm, F. E. Jenney, Jr., D. A. Comfort, R. M. Kelly, and M. W. Adams. 2006. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 188:2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leveque, E., S. Janecek, B. Haye, and A. Belarbi. 2000. Thermophilic archaeal amylolytic enzymes. Enzyme Microb. Technol. 26:3-14. [Google Scholar]
- 35.Liolios, K., N. Tavernarakis, P. Hugenholtz, and N. C. Kyrpides. 2006. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 34:D332-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lytovchenko, A., L. Sweetlove, M. Pauly, and A. R. Fernie. 2002. The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215:1013-1021. [DOI] [PubMed] [Google Scholar]
- 37.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 38.Maino, V. C., and F. E. Young. 1974. Regulation of glucosylation of teichoic acid. I. Isolation of phosphoglucomutase in Bacillus subtilis 168. J. Biol. Chem. 249:5169-5175. [PubMed] [Google Scholar]
- 39.Meyer, C., R. Schmid, P. C. Scriba, and M. Wehling. 1996. Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur. J. Biochem. 239:726-731. [DOI] [PubMed] [Google Scholar]
- 40.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426. [Google Scholar]
- 41.Milstein, C. 1961. Inhibition of phosphoglucomutase by trace metals. Biochem. J. 79:591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Möller-Zinkhan, D., G. Börner, and R. K. Thauer. 1989. Function of methanofuran, tetrahydromethanopterin, and coenzyme F420 in Archaeoglobus fulgidus. Arch. Microbiol. 152:362-368. [Google Scholar]
- 43.Möller-Zinkhan, D., and R. K. Thauer. 1990. Anaerobic lactate oxidation to 3 CO2 by Archaeoglobus fulgidus via the carbon monoxide dehydrogenase pathway: demonstration of the acetyl-CoA carbon-carbon cleavage reaction in cell extracts. Arch. Microbiol. 153:215-218. [Google Scholar]
- 44.Musfeldt, M., and P. Schönheit. 2002. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184:636-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Najjar, V. A. 1955. Phosphoglucomutase from muscle, p. 294-299. In Methods in enzymology. Academic Press, Inc., New York, NY.
- 46.Naught, L. E., and P. A. Tipton. 2001. Kinetic mechanism and pH dependence of the kinetic parameters of Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Arch. Biochem. Biophys. 396:111-118. [DOI] [PubMed] [Google Scholar]
- 47.Ollivier, B., R. Cord-Ruwisch, E. C. Hatchikian, and J. L. Garcia. 1988. Characterization of Desulfovibrio fructosovorans sp. nov. Arch. Microbiol. 149:447-450. [Google Scholar]
- 48.Pajatsch, M., A. Böck, and W. Boos. 1998. Enzymatic preparation of radiolabeled linear maltodextrins and cyclodextrins of high specific activity from [14C]maltose using amylomaltase, cyclodextrin glucosyltransferase and cyclodextrinase. Carbohydr. Res. 307:375-379. [DOI] [PubMed] [Google Scholar]
- 49.Palm, D., R. Goerl, and K. J. Burger. 1985. Evolution of catalytic and regulatory sites in phosphorylases. Nature 313:500-502. [DOI] [PubMed] [Google Scholar]
- 50.Park, K. H., T. J. Kim, T. K. Cheong, J. W. Kim, B. H. Oh, and B. Svensson. 2000. Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim. Biophys. Acta 1478:165-185. [DOI] [PubMed] [Google Scholar]
- 51.Rashid, N., J. Cornista, S. Ezaki, T. Fukui, H. Atomi, and T. Imanaka. 2002. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 184:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rashid, N., T. Kanai, H. Atomi, and T. Imanaka. 2004. Among multiple phosphomannomutase gene orthologues, only one gene encodes a protein with phosphoglucomutase and phosphomannomutase activities in Thermococcus kodakaraensis. J. Bacteriol. 186:6070-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray, W. J., Jr., J. W. Burgner, and C. B. Post. 1990. Characterization of vanadate-based transition-state-analogue complexes of phosphoglucomutase by spectral and NMR techniques. Biochemistry 29:2770-2778. [DOI] [PubMed] [Google Scholar]
- 54.Ray, W. J., Jr., and E. J. Peck, Jr. 1972. Phosphomutase, p. 407-477. In P. D. Boyer (ed.), The enzymes, vol. VI. Academic Press, Inc., New York, NY. [Google Scholar]
- 55.Ray, W. J., Jr., and C. B. Post. 1990. The oxyvanadium constellation in transition-state-analogue complexes of phosphoglucomutase and ribonuclease: structural deductions from electron-transfer spectra. Biochemistry 29:2779-2789. [DOI] [PubMed] [Google Scholar]
- 56.Ray, W. J., Jr., and J. M. Puvathingal. 1990. Characterization of a vanadate-based transition-state-analogue complex of phosphoglucomutase by kinetic and equilibrium binding studies: mechanistic implications. Biochemistry 29:2790-2801. [DOI] [PubMed] [Google Scholar]
- 57.Ray, W. J., Jr., and G. A. Roscelli. 1964. A kinetic study of the phosphoglucomutase pathway. J. Biol. Chem. 239:1228-1236. [PubMed] [Google Scholar]
- 58.Ray, W. J., Jr., and G. A. Roscelli. 1966. Activation and inhibition in the phosphoglucomutase system. J. Biol. Chem. 241:2596-2602. [PubMed] [Google Scholar]
- 59.Rüdiger, A., P. L. Jorgensen, and G. Antranikian. 1995. Isolation and characterization of a heat-stable pullulanase from the hyperthermophilic archaeon Pyrococcus woesei after cloning and expression of its gene in Escherichia coli. Appl. Environ. Microbiol. 61:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schinzel, R., and B. Nidetzky. 1999. Bacterial alpha-glucan phosphorylases. FEMS Microbiol. Lett. 171:73-79. [DOI] [PubMed] [Google Scholar]
- 61.Siebers, B., and P. Schönheit. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in archaea. Curr. Opin. Microbiol. 8:695-705. [DOI] [PubMed] [Google Scholar]
- 62.Stetter, K. O. 1988. Archaeoglobus fulgidus gen. nov., sp. nov.: a new taxon of extremely thermophilic archaebacteria. Syst. Appl. Microbiol. 10:172-173. [Google Scholar]
- 63.Stetter, K. O. 2006. Hyperthermophiles in the history of life. Philos. Trans. R. Soc. London B Biol. Sci. 361:1837-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stetter, K. O., G. Lauerer, M. Thomm, and A. Neuner. 1987. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science 236:822-824. [DOI] [PubMed] [Google Scholar]
- 65.Tachibana, Y., A. Kuramura, N. Shirasaka, Y. Suzuki, T. Yamamoto, S. Fujiwara, M. Takagi, and T. Imanaka. 1999. Purification and characterization of an extremely thermostable cyclomaltodextrin glucanotransferase from a newly isolated hyperthermophilic archaeon, a Thermococcus sp. Appl. Environ. Microbiol. 65:1991-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thauer, R. K., and J. Kunow. 1995. Sulfate-reducing archaea, p. 33-48. In N. Clark (ed.), Biotechnology handbook. Plenum Publishing Company, Ltd., London, England.
- 67.Thiemann, V., C. Donges, S. G. Prowe, R. Sterner, and G. Antranikian. 2004. Characterization of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch. Microbiol. 182:226-235. [DOI] [PubMed] [Google Scholar]
- 68.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tonkova, A. 1998. Bacterial cyclodextrin glucanotransferase. Enzyme Microb. Technol. 22:678-686. [Google Scholar]
- 70.Turner, P., A. Labes, O. H. Fridjonsson, G. O. Hreggvidson, P. Schönheit, J. K. Kristjansson, O. Holst, and E. N. Karlsson. 2005. Two novel cyclodextrin-degrading enzymes isolated from thermophilic bacteria have similar domain structures but differ in oligomeric state and activity profile. J. Biosci. Bioeng. 100:380-390. [DOI] [PubMed] [Google Scholar]
- 71.Wada, H., and E. E. Snell. 1961. The enzymatic oxidation of pyridoxine and pyridoxamine phosphates. J. Biol. Chem. 236:2089-2095. [PubMed] [Google Scholar]
- 72.Xavier, K. B., R. Peist, M. Kossmann, W. Boos, and H. Santos. 1999. Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J. Bacteriol. 181:3358-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, S. J., H. S. Lee, J. W. Kim, M. H. Lee, J. H. Auh, B. H. Lee, and K. H. Park. 2006. Enzymatic preparation of maltohexaose, maltoheptaose, and maltooctaose by the preferential cyclomaltooligosaccharide (cyclodextrin) ring-opening reaction of Pyrococcus furiosus thermostable amylase. Carbohydr. Res. 341:420-424. [DOI] [PubMed] [Google Scholar]
- 74.Yu, J. P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zellner, G., P. Messner, H. Kneifel, and J. Winter. 1989. Desulfovibrio simplex sp. nov., a new sulfate-reducing bacteria from a sour whey digester. Arch. Microbiol. 152:329-334. [Google Scholar]
- 76.Zellner, G., E. Stackebrandt, H. Kneifel, P. Messner, U. B. Sleytr, E. Conway de Macario, H.-P. Zabel, K. O. Stetter, and J. Winter. 1989. Isolation and characterization of a thermophilic, sulfate-reducing archaebacterium, Archaeoglobus fulgidus strain Z. Syst. Appl. Microbiol. 11:151-160. [Google Scholar]