Abstract
The bacterial transposon Tn7 has a pathway of transposition that preferentially targets conjugal plasmids. We propose that this same transposition pathway recognizes a structure or complex found during filamentous bacteriophage replication, likely by targeting negative-strand synthesis. The ability to insert into both plasmid and bacteriophage DNAs that are capable of cell-to-cell transfer would help explain the wide distribution of Tn7 relatives.
Tn7 is a 14-kb bacterial transposon that only activates its transposition machinery when specific targets are found in the cell (reviewed in references 5 and 19). Tn7 can use two transposition pathways that recognize different types of target sites with distinct but overlapping sets of transposon-encoded proteins, TnsA, TnsB, TnsC, TnsD, and TnsE (TnsABCDE). The TnsABC proteins constitute the core transposition machinery that normally interacts with one of the two target site-selecting proteins, TnsD or TnsE, to carry out transposition. Transposition with the TnsABC+D proteins catalyzes Tn7 transposition into a single site found in Escherichia coli and a wide variety of other bacteria, called the attachment site (attTn7). TnsD is a sequence-specific DNA binding protein that recognizes a sequence in the C-terminal-encoding portion of glmS but directs transposition events into the attTn7 site in the terminator of glmS transcription. Transposition into the attTn7 site occurs at a single position and in a single orientation with no detectable negative effect on the host (8, 10, 11). Transposition via the TnsABC+E proteins does not recognize any particular DNA sequence but instead preferentially directs transposition into mobile plasmids, called conjugal plasmids, by recognizing an aspect of transfer-associated DNA replication (17, 18, 27). Presumably, the TnsE-mediated pathway would facilitate the dissemination of Tn7 to new hosts, while the TnsD-mediated pathway would provide a “safe haven” once in a new bacterial host.
The TnsABC+E transposition machinery recognizes a structure or complex found during active conjugal DNA replication. TnsE-mediated transposition events preferentially occur into the conjugal plasmid (>90%) in the cell, even though in these experiments the plasmid only comprises ∼1% of the DNA in the cell (17, 27). Nonconjugal plasmids are not preferred targets, and mobilizable plasmids are only targets when actively mobilized (27). Genetic results show that TnsE-mediated transposition occurs into conjugal plasmids in recipient cells, likely by recognizing a component of lagging-strand DNA synthesis during rolling-circle DNA replication. In the donor cell, leading-strand DNA synthesis displaces the strand that is then transferred to the recipient bacterium (7, 26). Because single-stranded DNA is introduced into the recipient cell in the 5′-to-3′ direction, and because it is replicated as it enters the cell, DNA replication in the recipient cell must continually be reprimed in a discontinuous process. Tn7 displays an orientation bias in which the right end of the transposon is juxtaposed with the 3′ end of the nascent lagging strand of conjugal plasmid replication. The same orientation bias with regard to the lagging strand occurs when TnsE directs transposition to targets found in chromosomal replication (17). We presume that something is special about lagging-strand DNA synthesis found during conjugation that preferentially attracts TnsE-mediated transposition.
Tn7 is remarkably prevalent in a variety of environments (16). While conjugal plasmids are abundant in environmental settings, additional targets beyond actively conjugating plasmids may exist that could facilitate the dissemination of Tn7 and its relatives. Bacteriophage P1 replication was previously shown to not be a TnsE-mediated transposition target (27); however, it is unknown whether the filamentous bacteriophage are a productive target.
TnsD-mediated transposition can occur into the genome of an M13 derivative carrying attTn7.
One potential complication in determining if M13 is a transposition target is that the majority of the M13 genome encodes functions essential for bacteriophage growth. Therefore, we inserted 2.3 kb of exogenous DNA that would not be required for bacteriophage growth. We chose to clone the attTn7 region of the E. coli chromosome into M13mp18 using PCR because it included the specific site normally utilized by TnsD-mediated transposition that could serve as a positive control in our experiments (subsequently called M13-attTn7). When the M13-attTn7 bacteriophage was grown on a Tn7 donor strain containing a mini-Tn7 element with the TnsABC+D proteins provided in trans (JF55 pOX-Gen pCW4miniMu#76), bacteriophage conferring resistance to kanamycin (the transposon marker) were readily isolated (Tables 1 and 2). In this assay, bacteriophage were grown on the Tn7 donor strain for 2 hours to allow a 100-fold increase in bacteriophage. To determine the number of mini-Tn7-containing M13-attTn7 bacteriophage, 1.6 × 109 bacteriophages were used to infect 5 × 108 actively growing E. coli XL1-Blue cells as a reporter strain. M13 bacteriophage with mini-Tn7 was detected by the ability to confer kanamycin resistance. Transposition frequency was determined by dividing the number of CFU found on LB medium supplemented with kanamycin (50 μg/ml), tetracycline (20 μg/ml), and nalidixic acid (5 μg/ml) by the total number of infectious bacteriophage particles used to infect the host (tetracycline and nalidixic acid were used to counterselect against residual Tn7 donor cells in the bacteriophage lysate).
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Construction or reference(s) |
|---|---|---|
| NLC28 | araD139 Δ(argF-lac)169 rpsL150 relA flhD5301 deoC1 ptsF25 rbsR22 e14− Δ(fimB-fimE)632::IS1 Δ(fruK-yeiU)725 Valr | 3, 12, 20 |
| JF39 | NLC28 attTn7::mini-Tn7 Kanr | P1(JF400) × JF31a |
| JF55 | NLC28 recA56 attTn7::mini-Tn7 Kanr | This workb |
| JF31 | NLC28 ilvD-500::Tn10 | P1 (CAG18431) X NLC28 |
| JF400 | NLC28 pCW4 mini-Mu#76 pEMΔ | This work |
| CAG18431 | MG1655 ilvD-500::Tn10 | 15, 23 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 glnV44 relA1 lac [F′ proAB lacIq (lacZ)ΔM15 Tn10] | Stratagene |
P1 transduction was via linkage between ilvD and the chromosomal attTn7 site. Donor phage was grown on strain JF400, which allows transposition into the chromosomal attTn7 site from plasmid pEMΔ (this table and Table 2) (1). The mini-Tn7 Kanr transposition events in the donor cell attTn7 site could be cotransduced to the recipient strain using the linked wild-type ilvD gene.
TABLE 2.
Plasmids and bacteriophages used in this study
| Plasmid or bacteriophage | Relevant informationa (reference) |
|---|---|
| pCW4 miniMu#76 | Tetracycline-resistant (Tetr) pACYC184 derivative expressing TnsABC+D (25) |
| pCW15 | Chloramphenicol-resistant (Camr) pACYC184 derivative encoding TnsABC (25) |
| pJP103 | pTA106 (pSC101) derivative encoding TnsE; low-level TnsE expression from the native tnsE promoter; ampicillin resistant (Ampr) (17) |
| pJP104 | pTA106 (pSC101) derivative encoding TnsE; high TnsE expression from a lac promoter; Ampr (17) |
| pGEM-attTn7 | 3,372-bp attTn7-containing fragment amplified using PCR primers JEP17 (5′-ACA TGG GAT GAG GAG ATA ACA TAA TCT CCC-3′) and JEP18 (5′-TTG TAA TGC CGG ATG CGG CGT AAA ACA CCG-3′) and cloned using the Promega pGEM-T Easy Vector system I kit |
| M13-attTn7 | 2,258-bp NsiI-EcoRV fragment subcloned from pGEM-attTn7 into M13mp18, replacing the HincII-PstI fragmentb |
| pEMΔ | Contains a mini-Tn7 element, a Kanr cassette flanked by left and right Tn7 ends (1) |
| pACYC184 | p15A replicon; Tetr Camr (4) |
| pTA106 | pSC101 replicon; Ampr (17) |
| M13mp18 | M13 bacteriophage cloning derivative (28) |
| pOX-Gen | F plasmid derivative; Genr (9) |
Sequencing confirmed that transposition events were occurring at the single predicted position and orientation within the fragment and made the expected 5-bp target site duplication (5). The frequency of kanamycin-resistant bacteriophages was 4.8 × 10−7 (kanamycin resistance-conferring bacteriophages per total bacteriophages) after 2 hours of growth. The frequency of kanamycin-resistant bacteriophages modestly increased at 4 and 6 hours, to 6.2 × 10−7 and 7.1 × 10−7, respectively. No kanamycin-resistant bacteriophage was found with a vector-only control (i.e., lacking transposition functions). This experiment indicated that M13 could tolerate the insertion of a mini-Tn7 element and that the attTn7 site was functional. This was consistent with a previous result with full-length Tn7; however, in the experiment presented here we can rule out any contribution of TnsE or other known transposon-encoded proteins in targeting an attTn7 site in M13 (21).
TnsE-mediated transposition occurs into the M13 genome.
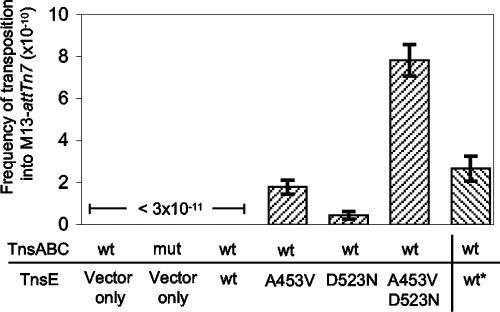
When M13-attTn7 was grown for 2 hours on a host that expressed the TnsABC+E proteins, we were also able to identify kanamycin resistance-conferring bacteriophage (Fig. 1). The wild-type TnsE protein allowed transposition at a frequency of about 3 × 10−10 when expressed from a lac promoter (Fig. 1). We also examined the ability of a series of high-activity mutants that were isolated previously in a genetic screen (17). The mutant proteins are toxic when expressed at anything but very low levels and are therefore introduced on a vector with a very low copy number and expressed from the native tnsE promoter (17). Despite the lower expression levels, the increased activity mutants directed transposition into M13 at levels higher than the wild-type protein. In fact, we found that transposition could not be detected when wild-type TnsE was expressed at this level; wild-type TnsE needed to be expressed at higher levels to give any kanamycin resistance-conferring bacteriophage in this assay (Fig. 1 and data not shown).
FIG. 1.
Frequency of Tn7 transposition into the M13-attTn7 genome in E. coli in various genetic backgrounds. Wild-type TnsABC was expressed from pCW15 (wt) or a pCW15 derivative with the TnsCA225V mutation (mut) (24). pTA106 was included as a vector control (vector only). Wild-type or mutant derivatives of TnsE, TnsEA453V (A453V), TnsED523N (D523N), or the double mutant TnsEA453V+D523N (A453V D523N) were expressed from pJP103. Wild-type TnsE was expressed with pJP104 (wt*). M13-attTn7 bacteriophage containing mini-Tn7 in strains expressing wild-type TnsABC without TnsE, in the mutant TnsABCA225V, or with TnsABC with wild-type TnsE expressed from pJP103 was never identified (<3 × 10−11). A very low background of spontaneous nalidixic acid-resistant cells (∼10−11) was found in the assay where the F′ Tn10 had mated into residual host strains from the XL1-Blue strain. These were easily identifiable because they contained all of the JF55 chromosomal and plasmid makers (i.e., resistance to rifampin, chloramphenicol, and ampicillin but sensitivity to gentamicin via loss of the pOX-Gen by replicon exclusion). Restriction analysis and DNA sequencing absolutely confirmed that the mini-Tn7 element was not contained on the M13-attTn7 plasmid. Error bars show the standard errors of the means (n = 9).
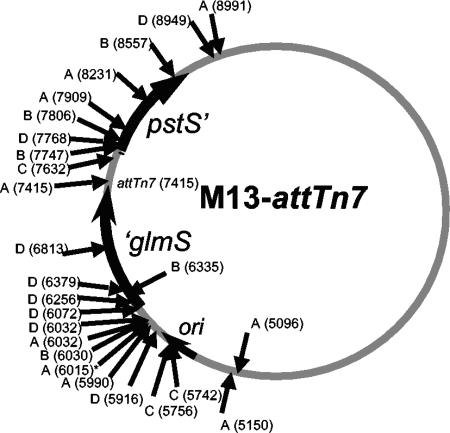
DNA sequencing revealed that TnsE-mediated transposition could occur throughout a large portion of the M13-attTn7 bacteriophage genome. This included 15 independent insertion events in the M13mp18 backbone and 10 independent insertions into the cloned fragment from the E. coli chromosome (Fig. 2). We assume that transposition events in the remainder of the M13 genome could not be detected because they would result in bacteriophage that could not be further propagated, a requirement for this assay. As expected, we were never able to isolate kanamycin resistance-conferring bacteriophage when the M13-attTn7 bacteriophage was grown with only the wild-type TnsABC; the core TnsABC machinery will not normally allow transposition without the TnsD or TnsE protein. We also tested whether random Tn7 transposition events could target the bacteriophage by using a mutant TnsABCA225V core machinery. The TnsABCA225V machine does not require TnsD or TnsE for activation in vivo or in vitro and appears to allow random transposition (2, 18). We were unable to recover any kanamycin-resistant M13-attTn7 bacteriophage using TnsABCA225V.
FIG. 2.
Representation of the 9,505-bp M13-attTn7 bacteriophage genome and the position of the TnsE-meditated mini-Tn7 insertion events. Numbering follows the previously established convention for M13mp18 (28). The M13mp18 sequence extends from bp 8529 to 9505/1 to 6266. The M13 origin (ori; bp 5487 to 5867), ′glmS (bp 6267 to 7371), pstS′ (bp 7708 to 8529), and the position of TnsD-mediated insertion events (attTn7) are indicated. Independent transposition events are indicated by arrows either inside (left-to-right insertion events) or outside (right-to-left insertion events) the circle. The positions of mini-Tn7 insertion events are indicated in parentheses along with the strain background according to the following letter code: A, pCW15 (TnsABC) pJP104 (TnsEwt); B, pCW15 pJP103 (TnsEA453V); C, pCW15 pJP103 (TnsED523N); D, pCW15 pJP103 (TnsEA453V+D523N). The position and orientation of individual transposition events were determined by isolating the replicative (double-stranded) form of the bacteriophage and sequencing from the left end of the element using a primer complementary to this end. To confirm that the process of transposition was responsible for relocating the element into the bacteriophage, we sequenced a subset of the insertions from both ends and in all cases identified a 5-bp duplication that is indicative of Tn7 transposition. The asterisk indicates where identical insertions were found in this experiment that are likely siblings.
The TnsE-mediated transposition events occurred at many different positions in M13, consistent with previous results with the TnsE pathway of transposition; TnsE-mediated transposition is not specific for any particular DNA sequence (Fig. 2). However, the TnsE-mediated insertions occurred with a striking right-to-left orientation bias that was reminiscent of a bias with DNA replication identified in conjugal plasmids and in the chromosome (17). By comparing the Tn7 orientation bias found in the chromosome and conjugal plasmids with these new results with M13, we can surmise which process of the M13 life cycle is targeted.
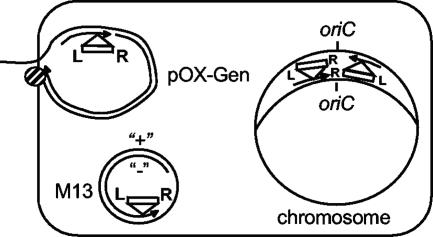
Previous results suggest that recessed 3′ ends are recognized by TnsE, which targets Tn7 into the adjacent duplex DNA in a specific right-to-left orientation. The observations presented here are consistent with this hypothesis and indicate that negative-strand DNA synthesis attracts TnsE-mediated transposition events into the M13 genome (Fig. 3) (17, 19). Filamentous bacteriophages are packaged as single-stranded DNA circles that must be made double-stranded when introduced into a new host (14). Negative-strand DNA synthesis is required to produce the complementary DNA strand after bacteriophage infection. Negative-strand synthesis is further required for a short window of time (∼20 min) to make additional copies of the replicative form of the bacteriophage genome. After a sufficient number of the replicative forms of the M13 genome have accumulated in the cell, positive-strand replication generates single-stranded, circular copies of the genome, which are coated with a bacteriophage-encoded single-strand DNA binding protein, gp5, before extrusion through the cell envelope.
FIG. 3.
Orientation of TnsABC+E transposition events in a replicating chromosome, F plasmid (pOX-Gen), and M13. TnsE-mediated transposition events occur in a single orientation with lagging-strand DNA replication in the chromosome and the F plasmid derivative pOX-Gen (19). The orientation bias found in this work suggests that negative-strand DNA synthesis attracts Tn7 insertion events in the TnsABC+E pathway (see text for details). Rectangles represent Tn7, with the right (R) and left (L) ends of the element indicated. Arrowheads show the 3′ ends of the DNA strands. The negative and positive strands of M13 are indicated. The F plasmid TraI protein is shown with a hatched circle bound to the 5′ end of the conjugating plasmid. On the chromosome, the origins of chromosomal DNA replication (oriC) and replication by lagging-strand DNA replication are indicated.
Frequency and targeting of transposition.
High-frequency transposition with a random transposition pathway catalyzed with the mutant TnsABCA225V machinery did not give detectable transposition into the M13-attTn7 vector (Fig. 1). Transposition with the core machinery with this TnsCA225V mutant allows transposition into the chromosome that is approximately 10-fold higher than levels found with TnsABC+E (24). The lack of observable transposition with the TnsABCA225V machinery into the M13-attTn7 vector, under conditions in which we do see transposition with TnsABC+E, suggests that the ability to target the M13 genome is specific to the TnsE pathway. This result is consistent with previous results with conjugal DNA replication (24, 27).
The low frequency of transposition into the bacteriophage genome is likely due to the limited availability of duplex DNA during the M13 life cycle; duplex DNA is required for a DNA molecule to act as a transposition target, because only the 3′ ends of the element are joined to a target DNA (6). The short amount of time that the filamentous bacteriophage normally spends as duplex DNA may partially protect this type of bacteriophage from being targets for transposition and likely explains why we were unable to detect transposition events into the M13-attTn7 vector with the random TnsABCA225V pathway of transposition (i.e., M13 may need to be actively targeted). We found that TnsABCA225V will target the double-stranded, replicative-form M13-attTn7 vector in vitro when it is the only target DNA in the reaction mixture, indicating that the vector can be a target for transposition with the mutant core machinery (A. Parks and J. E. Peters, unpublished observation). TnsABC+E transposition appears to be attracted to regions of discontinuous negative-strand synthesis, even though this type of replication only makes up a brief portion of the filamentous bacteriophage life cycle. Consistent with previous results, the high-activity TnsE mutants allowed for increased levels of transposition and occurred with the same orientation bias as transposition events with wild-type TnsE (17).
Tn7 has two pathways of transposition that facilitate the dissemination and establishment of the element within a bacterial genome while minimizing the impact on the host bacterium. Here we have described how Tn7 targets a filamentous bacteriophage genome, suggesting an expanded repertoire of vectors capable of transferring Tn7 from host to host.
Acknowledgments
We thank Nancy Craig (Johns Hopkins Medical School and Howard Hughes Medical Institute) and the E. coli Genetic Stock Center for providing bacterial strains and plasmids. We thank the members of the Peters lab for comments on the manuscript.
This work was funded by a grant from the National Science Foundation (MCB-0315316).
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Bainton, R. J., K. M. Kubo, J.-N. Feng, and N. L. Craig. 1993. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell 72:931-943. [DOI] [PubMed] [Google Scholar]
- 2.Biery, M. C., F. Steward, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig, N. L. 2002. Tn7, p. 423-456. In M. Lambowitz-Alan (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 6.Curcio, M. J., and K. M. Derbyshire. 2003. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4:865-877. [DOI] [PubMed] [Google Scholar]
- 7.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gringauz, E., K. Orle, A. Orle, C. S. Waddell, and N. L. Craig. 1988. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J. Bacteriol. 170:2832-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, R. C., and W. S. Reznikoff. 1984. Copy number control of Tn5 transposition. Genetics 107:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubo, K. M., and N. L. Craig. 1990. Bacterial transposon Tn7 utilizes two classes of target sites. J. Bacteriol. 172:2774-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKown, R. L., K. A. Orle, T. Chen, and N. L. Craig. 1988. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol. 170:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKown, R. L., C. S. Waddell, L. A. Arciszewska, and N. L. Craig. 1987. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc. Natl. Acad. Sci. USA 84:7807-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Model, P., and M. Russel. 1988. Filamentous bacteriophage, p. 375-456. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, New York, NY. [Google Scholar]
- 15.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parks, A. R., and J. E. Peters. 2007. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 189:2170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters, J. E., and N. L. Craig. 2001. Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes Dev. 15:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters, J. E., and N. L. Craig. 2000. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell 6:573-582. [DOI] [PubMed] [Google Scholar]
- 19.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2:806-814. [DOI] [PubMed] [Google Scholar]
- 20.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadri, M. I., C. C. Flores, A. J. Davis, and C. P. Lichtenstein. 1989. Genetic analysis of attTn7, the transposon Tn7 attachment site in Escherichia coli using a novel M13-based transduction assay. J. Mol. Biol. 207:85-89. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stellwagen, A., and N. L. Craig. 1997. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics 145:573-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waddell, C. S., and N. L. Craig. 1988. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 2:137-149. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins, B., and E. Lanka. 1993. DNA processing and replication during plasmid transfer between gram-negative bacteria, p. 105-136. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, NY.
- 27.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]