Abstract
Expression of minigenes encoding tetra- or pentapeptides MXLX or MXLXV (E peptides), where X is a nonpolar amino acid, renders cells erythromycin resistant whereas expression of minigenes encoding tripeptide MXL does not. By using a 3A′ reporter gene system beginning with an E-peptide-encoding sequence, we asked whether the codons UGG and GGG, which are known to promote peptidyl-tRNA drop-off at early positions in mRNA, would result in a phenotype of erythromycin resistance if located after this sequence. We find that UGG or GGG, at either position +4 or +5, without a following stop codon, is associated with an erythromycin resistance phenotype upon gene induction. Our results suggest that, while a stop codon at +4 gives a tripeptide product (MIL) and erythromycin sensitivity, UGG or GGG codons at the same position give a tetrapeptide product (MILW or MILG) and phenotype of erythromycin resistance. Thus, the drop-off event on GGG or UGG codons occurs after incorporation of the corresponding amino acid into the growing peptide chain. Drop-off gives rise to a peptidyl-tRNA where the peptide moiety functionally mimics a minigene peptide product of the type previously associated with erythromycin resistance. Several genes in Escherichia coli fulfill the requirements of high mRNA expression and an E-peptide sequence followed by UGG or GGG at position +4 or +5 and should potentially be able to give an erythromycin resistance phenotype.
Macrolides are antibacterial agents whose primary mode of action is inhibition of protein synthesis (4, 20, 35). The best-studied macrolide, erythromycin, binds to a site in 23S rRNA on the large ribosomal subunit close to the peptidyl transferase center, near the entrance to the nascent peptide exit tunnel (34). Erythromycin blocks the elongation step of translation during early rounds of protein synthesis. The drug has a high affinity for ribosomes with nascent peptides shorter than five amino acids, but it does not bind to ribosomes that carry long nascent peptide chains (6). It has been suggested that erythromycin sterically blocks elongation of the nascent peptide chain (35) and promotes dissociation of peptidyl-tRNA (16, 17), thereby increasing the rate of peptidyl-tRNA drop- off (14).
Expression of a short open reading frame in 23S rRNA encoding five amino acids followed by a stop codon confers resistance to erythromycin by a mechanism that depends strongly on both the sequence and the length of the peptide (31). Studies addressing structural features of erythromycin resistance peptides (E peptides) revealed that only short peptides, ranging in size from four to six amino acids, with certain amino acid sequences can confer erythromycin resistance. Among E-peptide sequences there is a consensus sequence, MXLXV (where X is an unknown residue), that confers high levels of erythromycin resistance (33). Sequence analysis revealed that the third amino acid, a leucine in a majority of E peptides, was the most conserved (33). A model for cis-acting E peptides was proposed where the newly synthesized E peptide remains tightly bound to the ribosome and prevents binding of erythromycin to its binding site (33). However, physiological conditions under which the minigene encoded in 23S rRNA is expressed have not yet been identified.
Recently, it was shown that NGG codons (CGG, AGG, UGG, or GGG) at positions +2 to +5, with AUG at +1, are associated with a strongly decreased expression of a reporter gene. These NGG codons give normal gene expression if located at position +7 or later, thus defining an early NGG-specific codon window (11). Later work suggested that the reason for the low gene expression associated with early NGG codons could be ascribed to peptidyl-tRNA drop-off (10, 11). The drop-off phenomenon is associated with active translation of the NGG codons since drop-off disappears if such a codon is shifted out of its correct reading frame. Also codons like GGN or GNG (where N is Pr, C, or U) fail to promote significant peptidyl-tRNA drop-off. The reason for this NGG-dependent peptidyl-tRNA drop-off is not clear, and it is not known at what translational step this event is triggered.
In this study we show that the NGG codons UGG and GGG at +4 or +5 in an induced reporter gene encoding a potential E peptide as its N-terminal part give improved growth in the presence of erythromycin. We used this erythromycin resistance phenotype to analyze the formation of functional E peptides generated by a drop-off event on NGG early codons. This assay enabled us to further characterize the event of peptidyl-tRNA drop-off on early NGG codons. Our results suggest that the amino acid encoded by an early UGG (tryptophan) or GGG (glycine) is incorporated into the growing peptide chain before drop-off occurs. Thus, the drop-off reaction should take place after peptide bond formation and in connection with translocation or after peptidyl-tRNA is transferred to the ribosomal P site. An important finding is that E-peptide-mediated erythromycin resistance might be more prevalent than previously thought. It may occur not only as a result of translation of a minigene buried within the 23S rRNA sequence but also as a consequence of an abortive translation event on UGG or GGG codons in the early coding region of mRNA.
MATERIALS AND METHODS
Chemicals and kits.
Chemicals used in this work were of the highest available quality grade. Ampicillin and erythromycin were purchased from Sigma. Restriction enzymes and T4 DNA ligase were from Fermentas and New England BioLabs. Plasmid DNA was prepared using a GFX Micro Plasmid prep kit. The GFX PCR and Gel Band Purification kits were used for gel band extractions (Amersham Bioscience, Buckinghamshire, United Kingdom). DNA sequencing was performed by MWG Biotech AH. Immunoglobulin G (IgG)-Sepharose was from Amersham Pharmacia Biotech. Isopropyl-β-d-thiogalactopyranoside (IPTG) was purchased from Sigma.
Plasmid constructions.
Plasmids were constructed using standard recombinant DNA techniques (25). The 3A′ translation assay system was used as a reporter system for appropriate codon variants (3). The assay system is based on expression of genes which code for three (3A′) and two (2A′) identical, engineered antibody binding B-domains of protein A from Staphylococcus aureus. The antibody binding domain was originally called Z (19) but is here referred to as A′ (3A′ is the whole gene) in order to avoid confusion with lacZ (18). A′ gene-derived proteins are stable, nonfunctional, and nontoxic for the Escherichia coli host bacterium, and they can be easily isolated quantitatively by affinity chromatography on IgG-Sepharose (3). The plasmid pHN98, a derivative of pDA348 (22), carrying the 3A′ test gene plus the 2A′ internal control gene, was used as vector for cloning of complementary deoxyoligonucleotides into the SwaI/SalI restriction site, generating the gene variants used in this work. The test gene 3A′ and the control gene 2A′ are under separate control by the Ptrc promoter (11, 30). The 3A′ gene constructs are preceded by a Shine-Dalgarno (SD+) sequence (5′-AAGGAGG-3′) eight nucleotides upstream of the AUG start codon, which is followed by the DR-A downstream region (29) or by an E-peptide-encoding sequence. The appropriate codon variants downstream of the AUG initiation codon are followed by the rest of the 3A′ gene sequence. For comparison, plasmid pEG998 (10, 11) with a non-SD (SD−) sequence (5′-AAAUAAAU-3′) upstream of the AUG initiation codon was also used. Plasmids carrying gene variants were transformed into strain XAc (25).
DNA sequencing.
The recombinant DNA sequences obtained by subcloning were sequenced by MWG Biotech AG. The sequencing primer pHG1 used is complementary to a unique sequence (5′-TACGCAAADCGCCTCTC) 389 bases upstream of the initiation codon of the 3A′ gene.
Protein A′ assay and growth conditions.
Overnight cultures of transformants with plasmids carrying the 3A′ reporter system (3) in LB medium with 100 μg/ml ampicillin were used to inoculate fresh medium. IPTG was added to a final concentration of 1 mM. The exponentially growing cultures were cooled, and the cells were harvested by centrifugation at an optical density at 590 nm (OD590) of 0.4, followed by resuspension in 1 ml of 10× TST (0.5 M Tris [pH 7.4], 2.5 M NaCl, 0.5% Tween 20) buffer (3). The cells were lysed by incubation at 95°C for 5 min, and cell debris was eliminated by centrifugation. The 3A′ and 2A′ proteins were purified from the supernatant fraction using IgG-Sepharose (Amersham) minicolumns and a vacuum manifold system (Promega). The A′ proteins were eluted from the column with 0.1 ml of 0.5 M acetic acid (pH 3.2). The eluted protein samples were dried in a Savant SpeedVac Plus SC110A (Telechem International, Inc. Sunnyvale, CA) and dissolved in sample loading buffer. After denaturation at 95°C for 5 min, A′ proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% acrylamide) (25). The bands were quantified by scanning using an Image Reader 1000, version 1.2 (FujiFilm Life Science, Japan). The protein ratios were obtained by using the Image Gauge program, version 4.0. Protein A′ values were normalized for molecular weight and presented as relative expression (3A′/2A′). The 3A′ value represents the test gene and that of 2A′ represents the internal control gene in the same plasmid (3). Each molar ratio represents the mean of at least three independent measurements.
Growth in the presence of erythromycin.
Overnight cultures of plasmid-carrying transformants with the 3A′ reporter system (XAc with the appropriate plasmid) grown in LB medium containing 100 μg/ml ampicillin were diluted to an OD590 of 0.01 with fresh medium containing 100 μg/ml ampicillin and 1 mM IPTG. Erythromycin was added to a concentration of 75 μg/ml, and the growth of cultures was monitored at regular time intervals by measuring the OD590.
For measurements of relative growth, cultures prepared as described above were grown until the OD of the corresponding control culture grown only in the presence of ampicillin and IPTG reached an OD590 of 1.00. At this time, ODs of test cultures with erythromycin were measured and normalized relative to the control culture (33).
Peptidyl-tRNA drop-off experiments.
Growth of a strain MB01 with temperature-sensitive peptidyl-tRNA hydrolase (Pth) [MG1655 zdh-925::Tn10 pth(Ts)] carrying different plasmid constructs was assayed as described previously (10). The cells with pHN98-derived plasmids were grown overnight at 30°C in LB medium with 100 μg/ml ampicillin. Fresh cultures were grown until the OD590 reached 0.4 and were then split into two subcultures in fresh medium, giving an OD590 of 0.1. IPTG (1 mM) was added to the cultures, and growth of both cultures at 37.5°C was continued for another 3.5 h. Growing cultures were monitored by measurements of the OD590 as a function of time.
Bioinformatics.
E. coli K-12 (12) RefSeq (21) data (accession number NC_000913.2) were downloaded from NCBI on 20 August 2007. The 4,267 genes annotated in EcoGene 2.0 (23) (file NC_000913.fna) were screened for sequences with Ile or Leu codons at position +3 and GGG or TGG codons at positions +4 or +5 using scripts depending on BioPerl (28). mRNA expression data relative to genomic DNA were taken from Bernstein et al. (2). Percentiles of expression values under each condition (using 1,803 data points for the LB condition and 2,845 data points for the M9 condition) were calculated from the empirical cumulative distribution of the expression data using the R ecdf function (21a). Initiation regions of genes (positions −35 to +20) were initially searched for ribosomal binding sites using a custom model derived from the refined flexible model of Shultzaberger and collaborators (27), converted into a duration hidden Markov model [using the following parameters: discarding the +4 position of the SD profile and the −1, −2, and −3 positions of the initiation region profile; no insertions allowed and no deletions allowed within profiles; genomic background composition for null and spacer emissions; loop probability of 49/50 for mute states; entry probabilities split evenly before the SD and the initiation region profiles [details to be published elsewhere]) for use with HMMER, version 2.3.2 (9) which was calibrated with fixed sequences of length 50 and run with an E-value of 100; results were then inspected manually.
RESULTS AND DISCUSSION
Early NGG codons are associated with decreased gene expression and peptidyl-tRNA drop-off also in an E-peptide-encoding sequence context.
We asked whether early NGG codons in the context of an E-peptide-encoding sequence (AUG AUA CUU GUA NGG…), following the initiation codon, influence gene expression. For comparison, we also analyzed the influence of early NGG codons on protein 3A′ expression for the related DR-A sequence context (AUG AAA GCA AUU NGG GUA…) downstream of the initiation codon (10) (Fig. 1). The DR-A sequence context was previously shown to be associated with a high level of expression of the 3A′ reporter gene, both with or without an upstream SD sequence (29). Both gene variants in an SD− DR-A context or SD+ MILV context are followed by the proximal 3A′ gene sequence (UCG ACU GGC CAA AGU CUA…). The sequence that follows early NGG codons was previously shown not to influence NGG codon-induced peptidyl-tRNA drop-off (11).
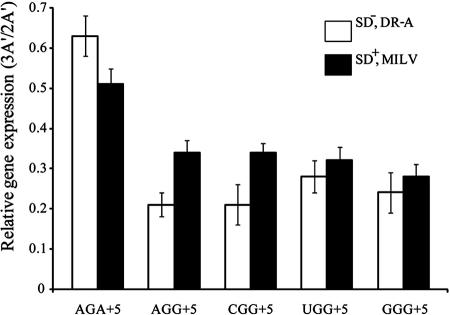
FIG. 1.
Influence on reporter gene expression by NGG codons in the early coding region in a DR-A or an E-peptide-encoding sequence context. Expression of the reporter gene 3A′ variants with an SD−, DR-A context (11) (empty columns) or with an SD+, MILV context (see text for sequences). The DR-A-related constructs have a SD− sequence upstream of the reporter gene together with different +5 codons in the downstream region denoted DR-A, as indicated. The MILV-related constructs are preceded by a canonical SD sequence positioned eight nucleotides upstream of the translation initiation site. The MILV peptide-encoding sequence is followed by indicated +5 codons. Both the DR-A- and MILV-related variants constitute the proximal part of the 3A′ gene. More complete sequence information on E-peptide variants is given in Table 1. 3A′ represents the test gene, and 2A′ represents the internal control gene in the same plasmid. Results are expressed as molar ratios (3A′/2A′) with mean values based on at least three independent measurements ± standard error.
Using the indicated test systems, as expected, almost no protein 3A′ expression was observed for minigene variants based in the DR-A codon context or encoding MIL and MILV peptides (E peptides) followed by a UAA stop codon at the +4 or the +5 position (data not shown) (10, 11). 3A′ expression was observed to be the highest in cells expressing a peptide with an AGA codon at +5 in both analyzed sequence contexts. The gene variants with NGG codons at the +5 position all showed decreased protein 3A′ expression levels, in agreement with a previous report that early NGG codons are associated with low gene expression as the result of peptidyl-tRNA drop-off (10, 11). Thus, decreased gene expression by NGG codons previously observed for the DR-A sequence context was also observed in the case of an E-peptide-encoding sequence.
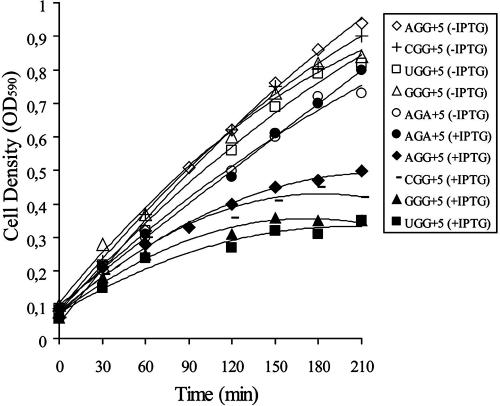
Early NGG codons in a DR-A sequence context give peptidyl-tRNA drop-off and cause growth inhibition at an elevated temperature of a pth(Ts) mutant strain as a result of insufficient Pth activity. The thermosensitive Pth in this strain is inactivated even at 37°C (10). We analyzed whether NGG codons placed after an E-peptide-encoding sequence in the proximal part of the 3A′ reporter gene are also associated with growth inhibition of this pth(Ts) strain. Cultures were pregrown at 30°C, followed by dilution into fresh medium without or with 1 mM IPTG for induction of the 3A′ gene with its proximal E-peptide-encoding sequence, and incubation was continued at 37.5°C (10). As shown in Fig. 2, induction by IPTG of test genes with a GGG, CGG, UGG, or AGG codon at position +5 inhibits growth of the pth(Ts) strain. In the absence of induction, an inhibitory effect by early NGG on growth of the pth(Ts) strain is not observed. It was noted that the arginine codon AGA at position +5 did not inhibit growth of the strain in a normal broth medium even in the presence of IPTG induction (Fig. 2). This is in agreement with previous results that AGA in the early coding region is not associated with peptidyl-tRNA drop-off (10). A comparison of strains carrying NGG codons in either an MILV sequence-encoding context with SD+ or a DR-A context with an SD− sequence (10) shows a similar extent of growth arrest of the strain with its pth-encoded temperature-sensitive Pth enzyme (not shown). Taken together, the results suggest that the decreased gene expression associated with early NGG codons at position +5 is the result of peptidyl-tRNA drop-off also in genes encoding an E-peptide sequence and not only in the case of the earlier described DR-A sequence context (10).
FIG. 2.
Growth of pth(Ts) strains with plasmids having NGG codons at position +5 in the 3A′ reporter gene. Growth was determined for pth(Ts) strain carrying plasmids with the reporter gene 3A′, with the SD+ sequence and the E-peptide-encoding context in the beginning of the gene, as described in the legend of Fig. 1, and the indicated codons at position +5. Growth was at 37.5°C in LB medium either in the absence of IPTG induction (open symbols)or following induction with 1 mM IPTG at time zero (filled symbols).
Influence on growth in the presence of erythromycin by potential E-peptide-encoding sequences.
The effect on E-peptide activity by NGG codons in the coding context of a potential E-peptide-encoding sequence in the beginning of a 3A′ reporter gene was analyzed further. E peptides should be of a very narrow size range of four to six amino acid residues (33), and both tripeptides and heptapeptides have little or no activity in protection against erythromycin binding (33). The efficiency of an E peptide to prevent ribosomal binding of erythromycin depends on the amino acid sequence, in particular, the identity of the C-terminal residue. E peptides with Leu or Ile at the third position and a nonpolar, hydrophobic amino acid at the C terminus give the highest activity in preventing ribosomal binding of erythromycin (33).
We wanted to find out if peptidyl-tRNA drop-off at early NGG codons could give rise to functional E peptides. To do this, variants of the reporter gene 3A′ encoding a potential E-peptide sequence as the N-proximal part, with NGG codons at positions +4 or +5, were constructed and studied for their influences on bacterial growth in the presence of erythromycin (Table 1). All sequences analyzed have a Leu at position +3 since this amino acid is required for resistance to erythromycin, provided the peptide is of appropriate length (33). For comparison, we also analyzed gene variants with a UAA stop codon following the E-peptide-encoding sequence. Since expression of such minigenes is known to be associated with erythromycin resistance (33), IPTG was added for induction of the reporter gene with different early codons, and bacterial doubling times were determined. With IPTG and in the absence of erythromycin, all analyzed strains had similar doubling times of 40 min (data not shown). In the presence of IPTG and 75 μg/ml of erythromycin, expression of the reporter gene with UGG or GGG at +4 was associated with doubling times of 130 and 140 min, respectively (Table 1). Other analyzed strains had doubling times of more than 160 min in the presence of erythromycin. For some of the analyzed strains, it was not possible to obtain a fair estimate of doubling times in the presence of antibiotic because growth was almost totally inhibited. A doubling time of 90 min was measured for cells carrying constructs with a UAA stop codon at positions +5 or +6. Interestingly, a similar doubling time of 100 min was found for cells having a construct with a UGG codon instead of the UAA at +5. Thus, an early UGG or GGG in the induced reporter gene, even in the absence of a stop codon, gives significantly improved growth in the presence of erythromycin.
TABLE 1.
Gene variants investigated in this work
| Plasmida | Nucleotide sequence of gene variant | Amino acid sequence | Doubling time (min)b |
|---|---|---|---|
| pGM412 | AUG AUA CUU UAA | MIL | No growth |
| pGM483 | AUG AUA CUU GGA | MILG | ND |
| pGM484 | AUG AUA CUU GGC | MILG | ND |
| pGM485 | AUG AUA CUU GGU | MILG | ND |
| pGM413 | AUG AUA CUU GGG | MILG | 140 |
| pGM493 | AUG AUA CUU GGG UAA | MILG | ND |
| pGM494 | AUG AUA CUU AGA | MILR | 200 |
| pGM486 | AUG AUA CUU CGG | MILR | 180 |
| pGM503 | AUG AUA CUU CGG UAA | MILR | ND |
| pGM487 | AUG AUA CUU UGG | MILW | 130 |
| pGM505 | AUG AUA CUU UGG UAA | MILW | ND |
| pGM414 | AUG AUA CUU GUA UAA | MILV | 90 |
| pGM480 | AUG AUA CUU GUA GGA | MILVG | ND |
| pGM481 | AUG AUA CUU GUA GGC | MILVG | ND |
| pGM482 | AUG AUA CUU GTA GGU | MILVG | ND |
| pGM417 | AUG AUA CUU GUA GGG | MILVG | 150 |
| pGM490 | AUG AUA CUU GUA GGG UAA | MILVG | ND |
| pGM428 | AUG AUA CUU GUA AGA | MILVR | No growth |
| pGM476 | AUG AUA CUU GUA AGG | MILVR | ND |
| pGM415 | AUG AUA CUU GUA CGG | MILVR | 160 |
| pGM492 | AUG AUA CUU GUA CGG UAA | MILVR | ND |
| pGM416 | AUG AUA CUU GUA UGG | MILVW | 100 |
| pGM491 | AUG AUA CUU GUA UGG UAA | MILVW | ND |
| pGM574 | AUG AGU CUU AAA GUA UAA | MSLKV | 90 |
| pGM575 | AUG AGU CUU AAA GUA | MSLKV | No growth |
| pGM585 | AUG AGU CUU AAA GUA CUA UAA | MSLKVL | ND |
| pGM577 | AUG AGU CUU AAA GUA CUA CGG | MSLKVLR | No growth |
| pGM578 | AUG AGU CUU AAA GUA CUA CGG UAA | MSLKVLR | No growth |
| pGM581 | AUG AGU CUU AAA GUA CUA GGG | MSLKVLG | No growth |
| pGM582 | AUG AGU CUU AAA GUA CUA GGG UAA | MSLKVLG | No growth |
| pGM576 | AUG AGU CUU AAA GUA CUA UGG | MSLKVLW | No growth |
| pGM583 | AUG AGU CUU AAA GUA CUA UGG UAA | MSLKVLW | No growth |
The plasmid pHN98 with the 3A′/2A′ reporter system, which is a derivative of pDA3480 (22) was used for subcloning of AUG downstream sequences at the restriction site SalI/SwaI, thus generating this plasmid series. The gene variants were preceded by an SD+ sequence (5′-AAGGAGG-3′) eight nucleotides (5′-AUUUAAAU-3′) upstream of the AUG translation initiation site. All gene variants listed in the table are followed by the rest of the 3A′ gene sequence (UCG ACU GGC CAA AGU CUA … ). Plasmids were transformed into the standard E. coli strain XAc [Δ(lacproB) argE ara gyrA rpoB thi] (5).
Doubling time in LB medium in the presence of 100 μg/ml ampicillin, 1 mM IPTG, and 75 μg/ml of erythromycin is measured for the set of representative strains. For peptidyl-tRNA drop-off experiments, selected plasmids were transformed into the MB01 strain [MG1655 zdh-925::Tn10 pth(Ts)] (10). No growth, absence of growth after an overnight incubation; ND, not determined.
Strains carrying constructs with UGG or GGG at positions +4 or +5 after an E-peptide-encoding sequence in the proximal part of the 3A′ reporter gene show growth in the presence of erythromycin. This finding motivated us to test further whether drop-off on early NGG codons is associated with formation of functional E peptides. To do this, we used the procedure described by Tenson and collaborators (33) and measured relative growth in the presence of IPTG and 75 μg/ml erythromycin (Fig. 3) for the set of strains carrying the plasmid sequences listed in Table 1. As controls, we included potential E-peptide-encoding sequences (MIL, MILV, MSLKV, MSLKVL, and MSLKVLV) followed by a UAA termination codon. In line with an earlier report (32), the data shown in Fig. 3 suggest that there is essentially no E-peptide activity of the short tripeptide MIL or the long heptapeptide MSLKVLV. Also in agreement with this report all three of the peptides four to six amino acid residues long show E-peptide activity.
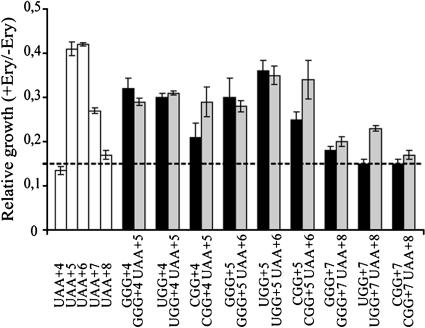
FIG. 3.
Relative growth of strains with plasmids having UAA or NGG codons GGG, UGG, and CGG at early positions. Growth was measured in the presence of 75 μg/ml of erythromycin of the XAc strain carrying plasmids with the reporter gene 3A′ with the SD+ sequence and the E-peptide-encoding context in the beginning of the test gene, as described in the notes to Table 1. Growth of cultures in the presence of IPTG and 75 μg/ml of erythromycin is normalized relative to the growth in the presence of only IPTG. Black columns represent strains with plasmids with NGG codons in the early coding region, as indicated. Gray columns represent strains with plasmids bearing corresponding constructs with the NGG codon followed by a UAA stop codon. Open columns represent strains with plasmids that have UAA at positions +4, +5, +6, +7, or +8. The peptide sequences for these constructs are MIL, MILV, MSLKV, MSLKVL, or MSLKVLV, respectively. The peptide sequence encoded by NGG codons at position +3, +4, or +7, with or without a following UAA codon, is MIL, MILV or MSLKVL, respectively. The dotted line gives the background value for total inhibition of growth by erythromycin. Five independent experiments were performed, and values are expressed as the mean ± standard error.
We investigated whether the NGG-dependent drop-off event takes place with or without incorporation of the NGG-encoded amino acid into the growing peptide, i.e., before or after the trans-peptidation reaction. If the amino acid encoded by the NGG codon at +4 is not incorporated into the growing peptide chain, the resulting tripeptide would be expected to lack E-peptide activity, as shown above (Fig. 3). In contrast, if the NGG-encoded amino acid is incorporated, the resulting peptide would be four residues long, and in the cases of GGG and UGG, E-peptide activity should be seen. We find that arginine codons at either +4 or +5 do not give any E-peptide activity (data not shown) and that there is little or no influence by MILR or MILVR on growth in the presence of erythromycin. The polar amino acid arginine at the C termini of E peptides is known not to promote E-peptide activity (33), and for that reason the arginine codons AGG and CGG were not analyzed further. Thus, even though all four NGG codons give drop-off, presumably by a similar mechanism, only GGG and UGG can for technical reasons be analyzed for amino acid addition to the nascent peptide, based on the E-peptide assay as used here.
As can be seen in Fig. 3, both GGG and UGG at +4 are associated with growth in the presence of erythromycin, whereas UAA at the same position is not. This suggests that the respective encoded amino acid, a glycine or tryptophan, respectively, is indeed incorporated into the formed nascent peptide to form a tetrapeptide. The results therefore suggest that the drop-off event associated with these NGG codons takes place after formation of the peptide bond. The abortive drop-off event associated with UGG or GGG should therefore occur from the ribosomal P site (13) or in connection with translocation to this site after the peptidyl transferase reaction has taken place. As a comparison, gene variants were constructed where GGG or UGG codons at +4 were followed by a UAA stop codon at +5. In this case release of the tetrapeptide ending by the GGG- or UGG-encoded amino acid should be complete since the UAA stop codon is very efficient. From the data shown in Fig. 3 it can be seen that the relative growth levels in the presence of erythromycin in the cases of GGG and UGG at +4 are very similar with or without a following stop codon. This suggests that the drop-off at GGG and UGG is quite high since it is comparable to the termination factor-induced peptide release at the UAA codon.
The pentapeptide MSLKV is also associated with resistance to erythromycin (Fig. 3) (33). We measured relative growth in the presence of erythromycin for a set of strains with the related E-peptide sequence MILVX, where X at +5 was encoded by GGG or UGG (Fig. 3). Strains expressing the same peptide where the encoding sequence was followed by a UAA stop codon at +6 were included for comparison (Table 1). As can be seen in Table 1, cells carrying a plasmid with a gene encoding MSLKV without a following UAA at +6 did not grow in the presence of IPTG and 75 μg/ml of erythromycin; i.e., they do not show any E-peptide activity. In contrast, cells expressing the MSLKV pentapeptide encoded by a gene with a UAA stop codon at +6 showed a relative growth value of 0.425 (Fig. 3), suggesting an E-peptide activity. Thus, the E-peptide sequence does not show activity if it is part of a longer protein sequence (14, 32, 33). It must be in the form of a short peptide since a stop codon or a drop-off-prone NGG codon after this coding sequence is required to promote growth in the presence of erythromycin (Table 1) (14, 32). The strains carrying constructs with UGG or GGG at +5 gave a similar relative growth value in the presence of erythromycin, also suggesting E-peptide activity. Interestingly, UGG or GGG at +5 with or without a UAA stop codon at +6 shows similar relative growth values (Fig. 3). Taken together, the abortive translation event induced by GGG and UGG at +4 or +5 results in generation of E peptides which functionally mimic minipeptides generated by termination at a UAA stop codon.
GGG or UGG at position +7 do not promote growth in the presence of erythromycin.
Relative growth of strains with GGG or UGG at position +7 in the E-peptide-encoding 3A′ reporter gene was measured in the presence of IPTG and 75 μg/ml of erythromycin (Fig. 3). No growth of these strains was observed even after prolonged incubation for 24 h, indicating lack of E-peptide activity. It has previously been reported that NGG codons at position +7, in contrast to earlier locations, do not decrease gene expression, and they are not associated with peptidyl-tRNA drop-off (10). Our results are thus in agreement with previous findings that heptapeptides are not effective in protecting the erythromycin-binding site on the ribosome (33).
Erythromycin resistance associated with glycine codons.
We also asked whether early glycine codons other than GGG in an E-peptide-encoding sequence context promote growth of cells in the presence of erythromycin. A construct with GGG at +4 gave a relative growth value of 0.32 (Fig. 4). A similar value was recorded in the case of cells expressing a peptide encoded by a corresponding minigene with GGG at +4, followed by UAA at +5. This suggests E-peptide activity in both cases. For constructs with GGA, GGC, or GGU at position +4, low relative growth values were found. Constructs with either GGG at +5 or GGG at +5 followed by UAA at +6 gave a higher relative growth value. These data are in line with previous results (11) that the GGG codon has a higher probability for drop-off than GGA, GGC, or GGU at positions +4 or +5. In accordance with these findings GGG, which is an NGG codon, gives higher tolerance to erythromycin than the other glycine codons GGA, GGC, and GGU, as found here.
FIG. 4.
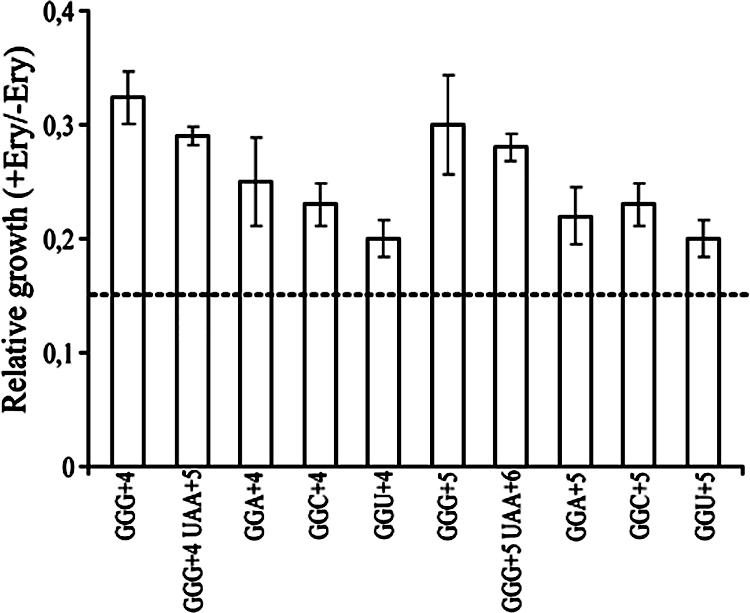
Relative growth of strains with plasmids having glycine codons at position +4 or +5 in the reporter gene. Growth in the presence of IPTG and 75 μg/ml of erythromycin (Ery) of strain XAc with appropriate plasmids carrying a 3A′ reporter gene beginning with an E-peptide-encoding sequence (as described in the legend of Fig. 3 and notes to Table 1). Values are normalized relative to the growth of cultures cultivated in the presence of only IPTG. Data for GGG constructs are taken from Fig. 3 for comparison. Five independent experiments were performed, and values are expressed as mean ± standard error.
Abortive translation by peptidyl-tRNA drop-off and erythromycin resistance.
Several peptides conferring erythromycin resistance, selected from in vivo expressed random libraries, have tryptophan or glycine at positions +4 or +5, encoded by UGG or GGG (33). This suggests that short peptides with the nonpolar amino acids glycine or tryptophan at the C terminus may form tight contacts with the ribosome, thus preventing binding of the drug and giving the phenotype of erythromycin resistance. Erythromycin inhibits protein synthesis at a step after the third amino acid has been added to the growing nascent peptide (15). At this stage, the bulky hydrophobic side chain of leucine or isoleucine at the third position may already interfere with the interaction of erythromycin with its binding site close to the peptidyl-transferase center (33), and an additional nonpolar C-terminal amino acid residue such as glycine or tryptophan could further stabilize this interaction.
Our results suggest that high expression of genes which have a codon for leucine or isoleucine at position +3 and UGG or GGG at position +4 or +5 can render cells transiently resistant to low levels of erythromycin. A bioinformatics search revealed 21 genes in E. coli K-12 with these sequence features (Table 2). Several of these genes have high mRNA expression levels during growth in either LB or M9 minimal medium (2). For instance, the lldP gene which encodes l-lactate permease and has the leucine codon CUC at position +3 and a UGG codon at +4 is upregulated in LB medium (Table 2). We found several genes with high mRNA levels that have a UGG or GGG codon at +5 and that encode inner membrane proteins (Table 2), thus providing a genetic potential for erythromycin resistance. Another example is yedI, coding for a conserved inner membrane protein, which has a UUG codon at +3 and GGG at +5 and is upregulated in M9 minimal medium. However, the highest expression within the set of genes having this specific sequence context is associated with the mdtJ gene, which encodes a multidrug efflux system transporter. It has been detected in a proteomic screen of proteins targeted to the inner membrane (7). This gene has the isoleucine codon AUU at +3 and UGG at +5 and is associated with higher expression than 84% of all analyzed mRNAs in M9 minimal medium (2) (Table 2). Many of the genes we have identified are first genes of operons annotated in RegulonDB (24) and have SD sequences, as discovered by a search with a custom model of initiation regions (see Materials and methods) and verified by eye. In particular, mtdJ has a consensus SD sequence eight bases upstream of its start codon. Thus, there are many genes in E. coli that are highly expressed at the mRNA level and carry UGG or GGG at early positions and a leucine or isoleucine codon at the +3 position. Such genes should have the potential of giving a drop-off-dependent erythromycin resistance phenotype, given appropriate physiological conditions, and it appears likely that they contribute to the basal level of erythromycin tolerance that characterizes E. coli.
TABLE 2.
E. coli genes with a Leu or Ile codon at position +3 and GGG or UGG at position +4 or +5
| Codon at position:a
|
Geneb | Blattner no. | Expression in the indicated medium (percentile)c
|
Description | |||
|---|---|---|---|---|---|---|---|
| +3 | +4 | +5 | LB | M9 | |||
| AUU | UAU | UGG | mdtJ | b1600 | 28 | 84 | Multidrug efflux system transporter |
| UUG | GCC | GGG | yedI | b1958 | 19 | 71 | Conserved inner membrane protein |
| AUC | UCC | UGG | nhaB | b1186 | 25 | 59 | Sodium:proton antiporter |
| UUG | AGA | UGG | emtA | b1193 | 42 | 58 | Lytic murein endotransglycosylase E |
| CUU | AAG | UGG | iscX | b2524 | 58 | 44 | Conserved protein |
| AUU | CCG | UGG | yheU | b3354 | 31 | 44 | Conserved protein |
| UUA | GAA | UGG | rhtB | b3824 | ND | 32 | Neutral amino acid efflux system |
| AUA | GAC | UGG | gltJ | b0654 | ND | ND | Glutamate and aspartate transporter subunit |
| CUG | AUG | UGG | yfbJ | b2258 | ND | ND | Predicted inner membrane protein |
| CUA | AUC | UGG | yoeB | b4539 | NA | NA | Toxin of the YoeB-YefM toxin-antitoxin system |
| CUG | CAA | GGG | ycjU | b1317 | NA | NA | Predicted beta-phosphoglucomutase |
| CUA | CCC | UGG | nuoM | b2277 | NA | NA | NADH:ubiquinone oxidoreductase membrane subunit M |
| CUG | UGG | cobC | b0638 | 20 | 55 | Predicted alpha-ribazole-5′-P phosphatase | |
| AUC | GGG | xylB | b3564 | 18 | 52 | Xylulokinase | |
| UUA | UGG | astD | b1746 | 35 | 50 | Succinylglutamic semialdehyde dehydrogenase | |
| AUU | UGG | yaiI | b0387 | ND | 43 | Conserved protein | |
| CUG | UGG | ygfX | b2896 | 52 | 32 | Predicted protein | |
| CUU | UGG | argH | b3960 | ND | 13 | Argininosuccinate lyase | |
| UUA | UGG | ytfN | b4221 | ND | 8 | Conserved protein | |
| CUC | UGG | lldP | b3603 | 86 | 3 | l-Lactate permease | |
| AUU | UGG | hycA | b2725 | NA | NA | Regulator of the transcriptional regulator FhlA | |
All codons ending with GG are statistically under-represented at the beginning of E. coli genes, especially GGG (26). However, the combination of a Leu or Ile codons at +3 and a UGG or GGG (in boldface) codon at +4 or +5 is neither statistically under- nor overrepresented using overall codon usage as a guide, nor does there seem to be any statistical departure from independence in their cooccurrence.
Based on gene annotations in EcoGene 2.0.
Values are the percentile rank of the relative mRNA level under a given growth condition as assayed by Bernstein et al. (2). The values are on a scale of 0 to 100 and show the percentage of all genes assayed with lower relative mRNA expression levels than the given gene. ND, no data are available for a gene under a given condition; NA, the gene was not assayed.
A recent molecular model for peptide-mediated erythromycin resistance suggests that the E peptide interacts with erythromycin and destabilizes its interaction with 23S rRNA (14). The pentapeptide is removed from the peptidyl-tRNA by a class 1 release factor at the termination step, giving erythromycin ejection from the ribosome at a high probability. Synthesis of the longer heptapeptide with the same five N-terminal amino acids leads neither to ejection of erythromycin nor to drug resistance (14). Our data suggest that even in the absence of termination, an abortive translation event on early UGG or GGG codons can give rise to peptidyl-tRNA where the peptide moiety functions as an E peptide. A very intriguing possibility which deserves further experimental evaluation is based on data suggesting an interaction between erythromycin and the conserved leucine residue (14) in the E peptide. Due to this interaction, peptidyl-tRNA could drop off from the ribosome together with the drug, which would remove the antibiotic from the ribosome and result in a transient erythromycin resistance phenotype.
Conclusions.
Our data suggest that peptidyl-tRNA drop-off on early UGG and GGG codons takes place after the encoded amino acid has been incorporated into the growing nascent peptide. Such drop-off gives rise to a peptidyl-tRNA where the peptide moiety functionally mimics a minigene peptide product known to be associated with erythromycin resistance. Even though it appears likely, for technical reasons this conclusion cannot be drawn for the other NGG codons AGG and CGG, both coding for arginine. It appears possible that E-peptide-mediated resistance to erythromycin might be more prevalent than previously thought. Although it was demonstrated that expression of a minigene buried in the 23S rRNA caused low-level resistance to erythromycin (31), the actual physiological conditions under which this particular minigene is expressed are not known.
A bioinformatic screening revealed several natural E. coli genes that fulfill the criteria: highly expressed at the mRNA level, encoding leucine or isoleucine at position +3, and having a GGG or UGG codon at positions +4 or +5. Such genes should have the potential for transient erythromycin resistance under appropriate physiological conditions.
So far, phenotypes associated with translation errors are poorly understood and have not been extensively studied because of their noninheritable nature. Nevertheless, they can represent a source of phenotypic heterogeneity, which is always present to various degrees in clonal populations (8) and which refers to epigenetic sources of population variation that does not involve changes in the genome (1). The results presented in this study therefore offer an insight into one possible mechanism of generation of heterogeneity in a population by an abortive translation event that results in transient antibiotic resistance as a response to physiological conditions.
Acknowledgments
This work was supported by a grant from the Swedish Science Foundation to L.A.I. and a postdoctoral fellowship from Sven and Lilly Lawski's Foundation for Natural Sciences to M.M.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Avery, S. V. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577-587. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björnsson, A., S. Mottagui-Tabar, and L. A. Isaksson. 1998. The analysis of translational activity using a reporter gene constructed from repeats of an antibody-binding domain from protein A. Methods Mol. Biol. 77:75-91. [DOI] [PubMed] [Google Scholar]
- 4.Chittum, H. S., and W. S. Champney. 1995. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr. Microbiol. 30:273-279. [DOI] [PubMed] [Google Scholar]
- 5.Coulondre, C., and J. H. Miller. 1977. Genetic studies of the lac repressor. J. Mol. Biol. 117:525-575. [DOI] [PubMed] [Google Scholar]
- 6.Cundliffe, E. 1972. The mode of action of fusidic acid. Biochem. Biophys. Res. Commun. 46:1794-1801. [DOI] [PubMed] [Google Scholar]
- 7.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 8.Dhar, N., and J. D. McKinney. 2007. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 10:30-38. [DOI] [PubMed] [Google Scholar]
- 9.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez de Valdivia, E. I., and L. A. Isaksson. 2005. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. FEBS J. 272:5306-5316. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez de Valdivia, E. I., and L. A. Isaksson. 2004. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 32:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 21 February 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol 2:2006.0007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karimi, R., M. Y. Pavlov, V. Heurgue-Hamard, R. H. Buckingham, and M. Ehrenberg. 1998. Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J. Mol. Biol. 281:241-252. [DOI] [PubMed] [Google Scholar]
- 14.Lovmar, M., K. Nilsson, V. Vimberg, T. Tenson, M. Nervall, and M. Ehrenberg. 2006. The molecular mechanism of peptide-mediated erythromycin resistance. J. Biol. Chem. 281:6742-6750. [DOI] [PubMed] [Google Scholar]
- 15.Mao, J. C., and E. E. Robishaw. 1972. Erythromycin, a peptidyltransferase effector. Biochemistry 11:4864-4872. [DOI] [PubMed] [Google Scholar]
- 16.Menninger, J. R., R. A. Coleman, and L. N. Tsai. 1994. Erythromycin, lincosamides, peptidyl-tRNA dissociation, and ribosome editing. Mol. Gen. Genet. 243:225-233. [DOI] [PubMed] [Google Scholar]
- 17.Menninger, J. R., and D. P. Otto. 1982. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob. Agents Chemother. 21:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mottagui-Tabar, S., A. Bjornsson, and L. A. Isaksson. 1994. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 13:249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson, B., T. Moks, B. Jansson, L. Abrahmsen, A. Elmblad, E. Holmgren, C. Henrichson, T. A. Jones, and M. Uhlen. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1:107-113. [DOI] [PubMed] [Google Scholar]
- 20.Oleinick, N. L. 1975. The erythromycins, p. 396-419. In J. W. Corcoran and F. E. Hahn (ed.), Antibiotics. III. Mechanism of action of antimicrobial and antitumor agents. Springer-Verlag, New York, NY.
- 21.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501—D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 22.Richter-Dahlfors, A. A., and D. I. Andersson. 1992. Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol. Microbiol. 6:743-749. [DOI] [PubMed] [Google Scholar]
- 23.Rudd, K. E. 2000. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salgado, H., S. Gama-Castro, M. Peralta-Gil, E. Diaz-Peredo, F. Sanchez-Solano, A. Santos-Zavaleta, I. Martinez-Flores, V. Jimenez-Jacinto, C. Bonavides-Martinez, J. Segura-Salazar, A. Martinez-Antonio, and J. Collado-Vides. 2006. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 34:D394—D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sato, T., M. Terabe, H. Watanabe, T. Gojobori, C. Hori-Takemoto, and K. Miura. 2001. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. (Tokyo) 129:851-860. [DOI] [PubMed] [Google Scholar]
- 27.Shultzaberger, R. K., R. E. Bucheimer, K. E. Rudd, and T. D. Schneider. 2001. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 313:215-228. [DOI] [PubMed] [Google Scholar]
- 28.Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz, C. Dagdigian, G. Fuellen, J. G. Gilbert, I. Korf, H. Lapp, H. Lehvaslaiho, C. Matsalla, C. J. Mungall, B. I. Osborne, M. R. Pocock, P. Schattner, M. Senger, L. D. Stein, E. Stupka, M. D. Wilkinson, and E. Birney. 2002. The BioPerl toolkit: Perl modules for the life sciences. Genome Res. 12:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenström, C. M., E. Holmgren, and L. A. Isaksson. 2001b. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene 273:259-265. [DOI] [PubMed] [Google Scholar]
- 30.Stenström, C. M., H. Jin, L. L. Major, W. P. Tate, and L. A. Isaksson. 2001a. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene 263:273-284. [DOI] [PubMed] [Google Scholar]
- 31.Tenson, T., A. DeBlasio, and A. Mankin. 1996. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl. Acad. Sci. USA 93:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenson, T., and A. S. Mankin. 2001. Short peptides conferring resistance to macrolide antibiotics. Peptides 22:1661-1668. [DOI] [PubMed] [Google Scholar]
- 33.Tenson, T., L. Xiong, P. Kloss, and A. S. Mankin. 1997. Erythromycin resistance peptides selected from random peptide libraries. J. Biol. Chem. 272:17425-17430. [DOI] [PubMed] [Google Scholar]
- 34.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 35.Vazquez, D. 1975. The macrolide antibiotics, p. 459-479. In J. W. Corcoran and F. E. Hahn (ed.), Antibiotics. III. Mechanism of action of antimicrobial and antitumor agents. Springer-Verlag, New York, NY.