Abstract
The response of exponentially growing Desulfovibrio vulgaris Hildenborough to pH 10 stress was studied using oligonucleotide microarrays and a study set of mutants with genes suggested by microarray data to be involved in the alkaline stress response deleted. The data showed that the response of D. vulgaris to increased pH is generally similar to that of Escherichia coli but is apparently controlled by unique regulatory circuits since the alternative sigma factors (sigma S and sigma E) contributing to this stress response in E. coli appear to be absent in D. vulgaris. Genes previously reported to be up-regulated in E. coli were up-regulated in D. vulgaris; these genes included three ATPase genes and a tryptophan synthase gene. Transcription of chaperone and protease genes (encoding ATP-dependent Clp and La proteases and DnaK) was also elevated in D. vulgaris. As in E. coli, genes involved in flagellum synthesis were down-regulated. The transcriptional data also identified regulators, distinct from sigma S and sigma E, that are likely part of a D. vulgaris Hildenborough-specific stress response system. Characterization of a study set of mutants with genes implicated in alkaline stress response deleted confirmed that there was protective involvement of the sodium/proton antiporter NhaC-2, tryptophanase A, and two putative regulators/histidine kinases (DVU0331 and DVU2580).
Sulfate-reducing bacteria (SRB) are ubiquitous in nature and play an important role in global carbon and sulfur cycling. Their habitat range includes freshwater, marine, and hypersaline aquatic systems, cold oceanic sediments, the deep subsurface, hydrothermal vents, and hot springs (11, 26, 35). Although long thought to have a relatively restricted catabolic range, this functionally defined assemblage is now recognized to be remarkably versatile. SRB mediate the degradation of aromatic compounds once thought to be refractory to anaerobic degradation, including benzene (2, 3, 8, 22, 23), and reduce a variety of metals, including radionuclides (19, 21, 24, 35). For these reasons they have also been studied for possible use in the bioremediation of environments contaminated with organic and metal pollutants.
Desulfovibrio vulgaris Hildenborough is one of the better-characterized SRB. This gram-negative deltaproteobacterium, isolated 60 years ago from clay soil in Hildenborough, Kent (United Kingdom), has served as one of the principal models for resolving the physiological and genetic basis of sulfate respiration. The recent completion of its genome sequence (14) has enabled genome-wide expression studies (6, 13, 27, 36, 37) that are now beginning to resolve its adaptive response to changing environmental parameters. Although this information is essential for predicting its behavior in possible applications for bioremediation, information about the range of conditions that support D. vulgaris growth or survival remains scarce.
Alkaline environments are common in nature (e.g., alkaline groundwater, lakes, and intestinal segments of some higher organisms) and in sites contaminated by human activity (29). There are some data documenting the presence of SRB in alkaline environments (1), but little is known about specific adaptive mechanisms of these bacteria or even whether mechanisms common to better-characterized organisms, such as Escherichia coli and Bacillus subtilis, are used by SRB. The adaptive strategies used by other microbes include (i) increased proton pumping by ATP synthase, (ii) increased metabolic acid production through amino acid deaminases and sugar fermentation, (iii) changes in cell surface properties, and (iv) increased expression and activity of monovalent cation/proton antiporters (9, 29, 33). Among these strategies, monovalent cation/proton antiporters are thought to play a central role in alkaline pH homeostasis in many bacteria (29).
In order to better resolve similarities and differences in the adaptation of Desulfovibrio species, we used genome-wide transcription profiling to characterize the response of D. vulgaris to an upshift in the pH of its growth medium. Adaptive mechanisms suggested by transcriptional analysis were then examined by characterizing a study set of mutants with genes implicated in the alkaline stress response deleted. Together, these analyses revealed a response system mechanistically similar to the systems of better-characterized species.
MATERIALS AND METHODS
Cell growth and pH upshift conditions.
Two-liter bottles containing 1.8 liters of defined piperizine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-buffered medium LS4D (13, 27) amended with 60 mM lactate and 50 mM sulfate were inoculated with exponentially grown cultures (1:10) to generate three biological replicate pairs consisting of a control and a test culture. The headspace of the vessels was 5% H2, 5% CO2, and 90% N2. An alkaline stress (pH 10) was applied to each test culture at mid-exponential growth (optical density at 600 nm, ca. 0.4) by addition of 35 ml of 2 M KOH with vigorous stirring. The control of each pair was treated by addition of 35 ml of sterile anoxic distilled water. Samples (300 ml) were taken for the gene expression analysis from each bottle at 30, 60, 120, and 240 min after addition of the stressor. The zero time samples were taken prior to stressor addition. The sample biomass was immediately chilled to 4°C during collection by pumping the sample out of the vessel with a peristaltic pump through a 7-m-long stainless steel tube immersed in an ice bath. Following centrifugation at 11,000 × g for 10 min at 4°C and decanting of the medium, the cell pellet was resuspended in 50 ml of anoxic 10 mM phosphate-buffered saline and cells were collected again by centrifugation. The cell pellets were immediately frozen in liquid N2 and stored at −80°C until RNA isolation.
To determine the growth rates of mutants at different pH values, cells were grown in B3 medium (containing [per liter] 0.1 g of NaCl, 0.1 g of MgCl2·6H2O, 0.1 g of CaCl2·2H2O, 0.5 g of NH4Cl, 0.1 g of KCl, 1.4 g of Na2SO4, 1 g of Na2S, 0.001 g of resazurin, 1 ml of 1 M K2HPO4, 1 ml of trace minerals, 1 ml of Thauer's vitamins, 1 ml of 1 M cysteine, and 1 ml of 1 M Na2S). The following buffering agents were used: 25 mM sodium bicarbonate for pH 7; 50 mM Tris for pH 7.5; and 50 mM glycine for pH 8.0 and 9.0. In all media 50 mM lactate and 40 mM sodium sulfate were used.
Analytical methods.
The concentrations of organic acids (lactate, pyruvate, acetate, formate, and fumarate) and inorganic ions (sulfate and phosphate) in culture media were determined using a Dionex 500 system equipped with an AS11HC column. In some cases the concentrations of organic acids were also measured using a high-performance liquid chromatograph equipped with an HPX 78 (Bio-Rad) column. Hydrogen concentrations were determined with an RGD2 reduction gas detector (Trace Analytical) with a 60/80 Mole Sieve 5A column (6 ft by 0.125 in.) with N2 as the carrier gas.
Mutant construction.
Bacterial mutants and strains are listed in Table 1. Deletion mutant JW381 (ΔnhaC-2) was constructed by marker exchange with a mutagenic plasmid as described elsewhere (4). The primers used for mutagenic plasmid construction were 5′-TATGGCAGATGTCAATGCCGAAGT-3′ and 5′-AAGACTGTAGCCGTACCTCGAATCTAATGTAGGCTCCAGTGGCCGA-3′ for the upstream region and 5′-ACGGCTTCCACGTCAACTATCTCA-3′ and 5′-AATCCGCTCACTAAGTTCATAGACCGTAGGGAAGGGCTACCTGAGGC-3′ for the DNA region downstream of the gene. The kanamycin resistance marker replacing nhaC-2 is also flanked by sequence bar codes unique to this deletion, with 5′-GCCGACAGAGCTTGAGATA-3′ at the promoter-proximal end of the kanamycin marker and 5′-AGCCTGGAACAGCTATACAC-3′ at the distal end.
TABLE 1.
Strains of D. vulgaris Hildenborough used in this study
| Strain | Putative gene | Annotation | Strain description | Source |
|---|---|---|---|---|
| ATCC 29579 | Not applicable | Wild type | Parent of deletion and plasmid insertion mutants | ATCC |
| JW381 | DVU3108 | nhaC | ΔnhaC::ntp MP+ | This study |
| JW391 | DVU2204 | tnaA | tnaA::pMO391 MP+b | This study |
| JW3024 | DVU2580 | Regulator | Tn5-RL27 insertionc | This study |
| JW3011 | DVU0331 | Histidine kinase | Tn5-RL27 insertion | This study |
| JW801 | Megaplasmid deleted (MP−)a | Wild type | Parent strain of transposon mutants | G. Voordow |
MP, 202-kb megaplasmid endogenous to D. vulgaris Hildenborough (= ATCC 29579).
pMO391 is pCR8/GW/TOPO (Invitrogen) with an internal tnaA fragment from D. vulgaris used to create an insertional mutation through homologous recombination (Spr).
Tn5-RL27 is a modified high-frequency insertion transposon (19).
Insertion mutant JW391 was constructed using plasmid pMO391 bearing a spectinomycin resistance gene (Spr). pMO391 is pCR8/GW/TOPO (Invitrogen) with an internal tnaA (DVU2204) fragment from D. vulgaris that was used to create an insertional mutation of tnaA through homologous recombination. The primers used for strain construction were tnaA350Fd (5′-ACAAGCCCGTCTTCATCTCCAACT-3′) (forward) and tnaA1273Rv (5′-TGTAGTCCATGTGGTCGTTGGTGT-3′) (reverse). The transposon mutants were generated by conjugation between D. vulgaris and E. coli BW20767(pRL27) (19). The conjugation procedure was a modification of the method of Fu and Voordouw (12). Cultures of D. vulgaris were grown to mid-exponential phase and combined at a 3:1 or 6:1 ratio with the E. coli donor grown to early exponential phase in LC medium (1.0%[wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). Mating mixtures were concentrated by centrifugation. The concentrated cells were placed onto filter disks (pore size, 0.22 μm; GSWP; Millipore Billerica, MA), and the disks were placed on the surface of solidified LS4 (LS4D with 1% [wt/vol] yeast extract added) and incubated for 16 h at 34°C. Then the cells were washed from the membrane with 2 ml LS4 medium. After 6 h of incubation, antibiotic G418 (400 μg/ml) was added to select for the transposon mutants and nalidixic acid (200 μg/ml) was added to select against the E. coli donor. Then cells were spread onto LS4 agar (100 to 500 μl/plate) with both antibiotics. The plates were incubated in the anaerobic chamber at 34°C for at least 4 days to allow the colonies to grow. The chromosomal localization of the transposon insertions was identified by sequencing DNA after semirandom PCR amplification. For semirandom PCR, a variation of a protocol described by Chun et al. (7) was used. One microliter of a 50-μl boiled single-colony suspension in distilled H2O was used as the template DNA in a 20-μl PCR mixture containing primer tpnRL17-1 (5′-AACAAGCCAGGGATGTAACG-3′) (19) and either primer CEKG 2A (5′-GGCCACGCGTCGACTAGTACN10AGAG-3′), primer CEKG 2B (5′-GGCCACGCGTCGACTAGTACN10ACGCC-3′), or primer CEKG 2C (5′-GGCCACGCGTCGACTAGTACN10GATAT-3′). One microliter of a 1:5 dilution of this reaction mixture was used as the template DNA for a second PCR performed with primers tpnRL17-2 (5′-AGCCCTTAGAGCCTCTCAAAGCAA-3′) and CEKG 4 (5′-GGCCACGCGTCGACTAGTAC-3′). For the first reaction, the thermocycler conditions were 94°C for 2 min, followed by six cycles of 94°C for 30 s, 42°C for 30 s (with the temperature reduced 1°C per cycle), and 72°C for 3 min and then 25 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min; for the second reaction, the thermocycler conditions were 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min. Samples that produced distinct bands on an agarose gel after the second reaction were cleaned with a PCR purification kit (Qiagen, Valencia, CA) and sequenced by using primer tpnRL17-1. The chromosomal locations of the insertions were identified by BLAST analysis of the sequences adjacent to the transposon compared with the complete genome.
Microarray construction, hybridization, and image analysis.
DNA microarrays covering 3,482 of the 3,551 protein-encoding sequences of the D. vulgaris genome annotated by TIGR (14) were designed, constructed, and validated with 70-mer oligonucleotide probes as previously described (6, 13, 27). Briefly, all oligonucleotides were commercially synthesized without modification by MWG Biotech Inc. (High Point, NC), prepared in 50% (vol/vol) dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO), and spotted onto UltraGAPS glass slides (Corning Life Sciences, Corning, NY) using a BioRobotics Microgrid II microarrayer (Genomic Solutions, Ann Arbor, MI). For each oligonucleotide probe there were two replicates on a single slide. After printing, the oligonucleotide probes were fixed onto the slides by UV cross-linking (600 mJ of energy) according to the protocol of the manufacturer of the UltraGAPS glass slides (Corning Life Science).
Total RNA extraction, purification, and labeling were performed independently for each cell sample using previously described protocols (6, 13). Briefly, total cellular RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) by following the manufacturer's protocol. RNA extracts were purified according to the RNeasy mini kit (Qiagen, Valencia, CA) instructions, and on-column DNase digestion was performed with the RNase-free DNase Set (Qiagen) to remove genomic DNA contamination according to the manufacturer's procedure. Labeling of cDNA targets from purified total RNA was carried out using the reverse transcriptase reaction with random hexamer priming and the fluorophore Cy5-dUTP (Amersham Biosciences, Piscataway, NJ). Genomic DNA was extracted from D. vulgaris cultures at stationary phase and labeled with the fluorophore Cy3-dUTP (Amersham Biosciences). The efficiency of labeling was routinely monitored by measuring the absorbance at 260 nm (for DNA concentration), 550 nm (for Cy3), or 650 nm (for Cy5).
To hybridize a single glass slide, the Cy5-dUTP-labeled cDNA probes obtained from stressed or unstressed cultures were mixed with equal amounts of the Cy3-dUTP-labeled genomic DNA (6, 27). Labeled genomic DNA (Cy3) was used as a control and as the common reference to cohybridize with labeled RNA (Cy5) samples for each slide. After washing and drying, the microarray slides were scanned using the ScanArray Express microarray analysis system (Perkin Elmer). The fluorescence intensity of both the Cy5 and Cy3 fluorophores was analyzed with the software ImaGene, version 6.0 (Biodiscovery, Marina Del Rey, CA).
Microarray analysis.
Arrays were scanned using the scanning laser confocal fluorescence microscope of the ScanArray microarray analysis system (GSI Lumonics), and hybridization signal intensities were quantitated using the software ImaGene (Biodiscovery). Statistical analysis of the microarray data was performed as described elsewhere (6), and cluster analysis was performed using TIGR Multi Experiment Viewer (MeV) (32). To assess the significance of the normalized log ratio, a Z score was calculated by using the following equation:
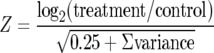 |
where 0.25 is a pseudovariance term. Log ratios and Z values for all genes detected in this study are listed in Table S1 in the supplemental material. For the microarray experiments, each log2 value represents the average of the results of three hybridization experiments using cDNA derived from two different cultures of D. vulgaris.
Protein sequence analysis.
For protein sequence analysis, the programs Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html) (15), LipoP 1.0 Server (http://www.cbs.dtu.dk/services/LipoP) (17), and SignalP 3.0 (10) from the Expasy database were used. The genomes and open reading frames of D. vulgaris and the gene contents of different sequenced genomes were analyzed using Microbesonline (http://www.microbesonline.org/) and NCBI databases.
Microarray data accession number.
All microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE8197.
RESULTS AND DISCUSSION
General features of the physiological and transcriptional response to elevated pH.
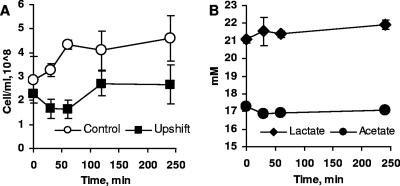
The initial studies examined the general physiological response of D. vulgaris to an abrupt increase in pH (Fig. 1). Lactate consumption stopped immediately following the pH shift, as did production of acetate (Fig. 1B). Hydrogen accumulation was not detected during the stress. However, viable cell numbers remained constant during the 2 h following the shift (Fig. 1A), as demonstrated by enumerating cells on agar plates and immediate resumption of growth measured by optical density following transfer to fresh neutral medium. During this period of exposure to elevated pH, altered expression of approximately 400 genes was observed. At 30, 60, 120, and 240 min following the shift, transcription of 78, 122, 178, and 184 genes, respectively, was significantly up-regulated (log2 signal intensity change greater than 1 and Z score greater than 2). The transcription of comparable numbers of genes was down-regulated at least twofold at the same time points (175, 267, 210, and 183 genes, respectively). The majority of the up-regulated genes fell into the following COG functional categories: amino acid biosynthesis, energy metabolism, and signal transduction systems. A large fraction of the down-regulated genes were assigned to signal transduction, transcription, and phage-related categories.
FIG. 1.
Growth and pH changes during pH upshift. (A) Cell numbers. (B) Lactate and acetate concentrations in the growth medium of the stressed culture. The data are means of triplicate cultures, and the observed variability was within 10%. The error bars indicate standard deviations of the means (n = 3).
Using K-means clustering analysis with the Euclidian distance, all genes were assigned to 30 groups based on their patterns of expression. Graphs representing all groups are shown in Fig. S1 in the supplemental material. Genes comprising each group are listed in Table S2 in the supplemental material. In our initial analysis we focused on cluster 22 (44 genes), which was comprised of genes that were the most highly up-regulated at all time points; cluster 19, which consisted of genes that were only moderately up-regulated (122 genes); cluster 4, which was comprised of the most highly down-regulated genes (57 genes); and cluster 21, which consisted of genes which were moderately down-regulated (100 genes) at all time points.
Genes up-regulated during exposure to high pH.
Elevated pH is generally thought to stress the cell through alkalinization of the cytoplasm, reduction of the membrane potential, and damage to proteins and the cell envelope (9, 33; for reviews, see references 29 and 31). The cell responds by pumping protons inside, importing or synthesizing compounds to acidify the cytoplasm, and activating systems of protein repair or degradation. These processes are all reflected in the response of E. coli to elevated pH. In alkali-stressed D. vulgaris, genes encoding ATPase synthase, Na+/H+ antiporter NhaC-2 (DVU3108), chaperones and proteases such as DnaK (DVU0811; log2 R = 0.8 to 1.5), ATP-dependent Clp protease subunit B (DVU1874; log2 R = 1.2 to 1.5), and ATP-dependent protease La (DVU3303) were found among other highly up-regulated genes (Table 2).
TABLE 2.
Selected up-regulated genes during pH 10 stress
| Gene | Description | Expression change (log2 ratio) ata:
|
|||
|---|---|---|---|---|---|
| 30 min | 60 min | 120 min | 240 min | ||
| DVU0258 | Sensory transduction histidine kinase related | 0.96 | 0.95 | 1.24 | 2.28 |
| DVU0260 | Response regulator | 0.96 | 1.30 | 1.23 | 1.25 |
| DVU0261 | Universal stress protein family | 0.59 | 0.73 | 0.48 | 0.31 |
| DVU0262 | Hypothetical protein | 0.53 | 1.25 | 0.95 | 1.29 |
| DVU0263 | Tetraheme cytochrome c3 | 0.22 | 0.41 | 0.20 | 0.69 |
| DVU0264 | Ferredoxin, 2[4Fe-4S] | 0.79 | 0.77 | 0.94 | 0.88 |
| DVU0265 | 25.3-kDa protein in hmc operon | 1.61 | 2.08 | 1.54 | 1.56 |
| DVU0331 | Putative histidine protein kinase | 1.36 | 1.18 | 1.83 | 2.23 |
| DVU0667 | HD domain protein | 2.28 | 1.52 | 1.74 | 1.69 |
| DVU0692 | Molybdopterin oxidoreductase | 1.09 | 1.21 | 1.52 | 2.01 |
| DVU0693 | Respiratory nitrate reductase, beta subunit (narH) | 1.09 | 1.52 | 1.48 | 2.01 |
| DVU0694 | Molybdopterin oxidoreductase, molybdopterin binding subunit | 2.17 | 1.91 | 1.01 | 1.97 |
| DVU0774 | ATP synthase F1, epsilon subunit (atpC) | 1.77 | 2.57 | 2.20 | 3.21 |
| DVU0775 | ATP synthase F1, beta subunit (atpD) | 1.11 | 1.55 | 1.63 | 2.27 |
| DVU0776 | ATP synthase F1, gamma subunit (atpG) | 0.87 | 1.60 | 1.13 | 2.26 |
| DVU0777 | ATP synthase F1, alpha subunit | 0.38 | 0.67 | 0.97 | 1.49 |
| DVU0778 | ATP synthase F1, delta subunit | 0.77 | 1.03 | 1.31 | 1.86 |
| DVU0779 | ATP synthase F0, B subunit | 0.20 | 0.27 | 0.49 | 1.48 |
| DVU0780 | ATP synthase F0, B subunit | 0.41 | 0.23 | 0.28 | 1.02 |
| DVU1030 | Universal stress protein family | 1.19 | 0.97 | 1.14 | 1.64 |
| DVU1035 | Glucokinase (glk) | 1.09 | 2.03 | 2.11 | 1.82 |
| DVU1198 | Riboflavin synthase, beta subunit (ribH) | 1.94 | 2.05 | 1.59 | 2.03 |
| DVU1446 | Heptosyltransferase family | 2.98 | 2.30 | 2.56 | 1.63 |
| DVU2367 | Acyl-(acyl carrier protein)-UDP-N-acetylglucosamine O-acyl transferase | 0.74 | 0.73 | 0.97 | 0.26 |
| DVU2368 | (3-R)-Hydroxymyristoyl-(acyl carrier protein) dehydratase (fabZ) | 1.64 | 1.90 | 1.87 | 2.42 |
| DVU2369 | UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase | 0.78 | 0.79 | 1.07 | 1.07 |
| DVU2370 | Outer membrane protein OmpH, putative | 1.58 | 1.80 | 1.62 | 1.64 |
| DVU2526 | Periplasmic (NiFe) hydrogenase, large subunit | 1.68 | 1.24 | 1.42 | 1.98 |
| DVU2571 | Ferrous iron transport protein B | 1.15 | 1.42 | 1.53 | 1.20 |
| DVU2572 | Ferrous iron transport protein A, putative | 1.25 | 1.36 | 1.80 | 1.76 |
| DVU2816 | Efflux system protein | 1.04 | 1.80 | 1.87 | 1.95 |
| DVU3025 | Pyruvate-ferredoxin oxidoreductase | 0.64 | 0.54 | 0.70 | 1.19 |
| DVU3026 | l-Lactate permease family protein | 0.01 | 0.10 | 0.18 | 0.59 |
| DVU3027 | Glycolate/lactate oxidase, subunit GlcD | 0.88 | 0.73 | 0.93 | 1.25 |
| DVU3028 | Glycolate oxidase, iron-sulfur subunit | 1.57 | 2.07 | 1.92 | 2.30 |
| DVU3029 | Phosphate acetyltransferase | 0.13 | 0.13 | 0.13 | 0.46 |
| DVU3030 | Acetate kinase | 0.45 | 0.40 | 0.51 | 0.62 |
| DVU3035 | Methyl-accepting chemotaxis protein, putative | 1.41 | 1.74 | 1.31 | 1.56 |
| DVU3081 | Integral membrane protein, putative | 0.27 | 2.38 | 1.35 | 2.64 |
| DVU3108 | Na+/H+ antiporter NhaC (nhaC) | 1.36 | 1.60 | 1.41 | 2.10 |
| DVU3110 | l-Aspartate oxidase, putative | 0.91 | 1.19 | 1.35 | 2.45 |
| DVU3298 | Hypothetical protein | 1.38 | 1.86 | 1.09 | 2.15 |
| DVU3299 | Membrane protein | 0.67 | 0.74 | 1.73 | 1.76 |
| DVU3300 | Hypothetical protein | 0.68 | 2.27 | 0.84 | 3.57 |
| DVU3301 | Hypothetical protein | 0.88 | 3.36 | 1.53 | 3.70 |
| DVU3303 | ATP-dependent protease La, putative | 0.87 | 0.21 | 1.03 | 1.97 |
| DVU3304 | Sensor histidine kinase | 0.62 | 0.41 | 0.66 | 1.11 |
Log2 gene expression ratios at different times after the pH was changed compared with the control at the same times.
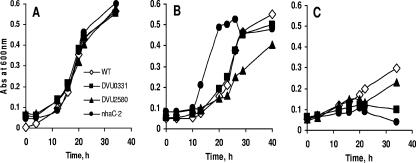
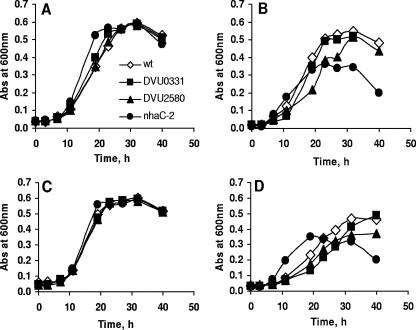
The D. vulgaris gene coding for the putative Na+/H+ antiporter NhaC-2 (DVU3108) is of particular interest. The role of Na+/H+ antiporters in regulation of intracellular pH homeostasis and survival at high pH has been demonstrated for a number of bacteria (5, 16, 28, 29). Although several genes in D. vulgaris have been identified as putative Na+/H+ antiporters, only the expression of nhaC-2 changed substantially in response to high pH. Our analysis of an nhaC-2 deletion mutant demonstrated that there was increased sensitivity to pH 8.9 relative to the wild type, which was accentuated at higher concentrations of sodium chloride (Fig. 2 and 3). Increased cell lysis in stationary phase was observed for this mutant but not for the wild type (Fig. 3B and D). This phenotype is similar to that of an E. coli nhaA-deficient mutant (29).
FIG. 2.
Growth characteristics of selected D. vulgaris mutants at elevated pH with 50 mM glycine buffer. (A) pH 7.0. (B) pH 7.9. (C) pH 8.9. The data are the means of triplicate cultures, and the observed variability was within 10%. WT, wild type.
FIG. 3.
Growth characteristics of selected D. vulgaris mutants at elevated pH and sodium chloride concentrations with 50 mM Tris buffer. (A) pH 7.0 and 40 mM NaCl. (B) pH 7.9 and 40 mM NaCl. (C) pH 7.0 and 80 mM NaCl. (D) pH 7.9 and 80 mM NaCl. The data are the means of triplicate cultures, and the observed variability was within 10%. wt, wild type.
The DVU3110 gene located in close proximity to nhaC-2 and transcribed in the same direction was also highly up-regulated at high pH. This gene was predicted to encode a putative flavin adenine dinucleotide-binding oxidoreductase/l-aspartate oxidase with a bona fide homolog present in only one other Desulfovibrio strain (D. vulgaris DP4; >99% identity; GenBank accession number CP000527.1). The next most closely related amino acid sequence (42.8% identity) was found in the genome of Magnetospirillum magneticum AMB-1(GenBank accession number AP007255.1), annotated as a member of the succinate dehydrogenase/fumarate reductase protein family (COG1053). The absence of genes typically associated with characterized succinate dehydrogenases and fumarate reductases, such as genes for b-type cytochromes and small membrane anchor proteins in the vicinity of DVU3110, does not allow us to suggest that DVU3110 codes for a protein with similar activities. In addition, neither fumarate nor succinate was detected in cultural medium of D. vulgaris exposed to pH 10 (data not shown). Nonetheless, the presence of a gene encoding 4Fe-4S ferredoxin (DVU3109) immediately downstream of DVU3110 suggests that the latter might encode for an oxidoreductase. An identical gene context surrounds the DVU3108 homolog in D. vulgaris DP4. Since an ortholog of DVU3110 has not been identified in the sequenced genome of another Desulfovibrio species (D. desulfuricans G20; GenBank accession number CP000112.1), a response to elevated pH via the combined activities of the oxidoreductase/l-aspartate oxidase and NhaC may be a species-specific strategy. Of additional note, genes identified as l-aspartate oxidase genes in the genomes of many Gammaproteobacteria (for instance, E. coli, Nitrosococcus oceanii, and Methylococcus capsulatus) are immediately upstream of a gene for a putative sigma E, suggesting that there is more general involvement of this type of reductase/l-aspartate oxidase in the stress response.
In addition to increased transcription of the gene encoding l-aspartate oxidase, a number of genes involved in amino acid synthesis and metabolism were also consistently up-regulated. Genes encoding tryptophan synthase subunits α and β (DVU0471 and DVU0470) and other members of the tryptophan operon (DVU0460 to DVU0469) were slightly or moderately up-regulated (Table 3). In addition, transcription of a gene annotated as a tryptophanase gene (DVU2204) was elevated after 240 min of pH stress (log2 R = 1.3). The orthologous gene in E. coli (tnaA) was previously shown to be up-regulated at pH 9 (25, 34), and growth of a D. vulgaris mutant with this gene deleted was diminished at pH 8 (data not shown). The concerted increase in transcription of genes for biosynthesis and transport of amino acids other than tryptophan (genes for cysteine synthase A, dihydrodipicolinate reductase involved in lysine biosynthesis, isopropylmalate dehydratase involved in leucine biosynthesis, and homoserine dehydrogenase involved in aspartate biosynthesis) suggests that D. vulgaris employs multiple components of amino acid metabolism for survival at high pH (Table 3).
TABLE 3.
Gene expression profile of selected genes involved in amino acid metabolism
| Gene | Description | Expression change (log2 ratio) ata:
|
|||
|---|---|---|---|---|---|
| 30 min | 60 min | 120 min | 240 min | ||
| DVU0085 | Tryptophan synthase, beta subunit | 1.36 | 1.75 | 1.90 | 2.45 |
| DVU0086 | Hypothetical protein | 1.18 | 1.88 | 0.84 | 3.00 |
| DVU0285 | Imidazole glycerol phosphate synthase | 0.88 | 1.03 | 1.10 | 0.15 |
| DVU0286 | Imidazole glycerol phosphate synthase, cyclase subunit | 1.79 | 1.39 | 1.53 | 0.66 |
| DVU0339 | d-Isomer-specific 2-hydroxyacid dehydrogenase family | 1.01 | 0.85 | 1.20 | 1.24 |
| DVU0460 | Predicted phospho-2-dehydro-3-deoxyheptonate aldolase | −0.08 | 0.04 | 0.15 | 0.57 |
| DVU0461 | Predicted 3-dehydroquinate synthase | 0.36 | 0.11 | 0.54 | 0.42 |
| DVU0462 | Chorismate mutase/prephenate dehydratase | −0.04 | 0.26 | 0.73 | 0.47 |
| DVU0463 | 3-Phosphoshikimate 1-carboxyvinyltransferase | 0.52 | 0.34 | 0.93 | 0.46 |
| DVU0464 | Prephenate dehydrogenase | −0.04 | 0.12 | 0.56 | 1.02 |
| DVU0465 | Anthranilate synthase, component I | 0.87 | 0.20 | 0.51 | 0.48 |
| DVU0466 | Anthranilate synthase, glutamine amidotransferase component | 0.18 | 0.37 | 0.08 | 0.07 |
| DVU0467 | Anthranilate phosphoribosyltransferase | 1.16 | 0.70 | 1.18 | 0.97 |
| DVU0468 | Indole-3-glycerol phosphate synthase | 1.01 | 0.85 | 1.41 | 1.32 |
| DVU0469 | N-(5-Phosphoribosyl)anthranilate isomerase | 0.72 | 0.93 | 1.27 | 0.99 |
| DVU0470 | Tryptophan synthase, beta subunit | 0.61 | 0.66 | 0.71 | 0.74 |
| DVU0471 | Tryptophan synthase, alpha subunit | 0.99 | 1.16 | 1.15 | 1.31 |
| DVU0663 | Cysteine synthase A | 0.83 | 1.31 | 1.65 | 0.82 |
| DVU0890 | Homoserine dehydrogenase | 1.33 | 1.55 | 1.69 | 1.19 |
| DVU1466 | Acetylglutamate kinase | 1.43 | 1.01 | 1.36 | 1.04 |
| DVU1585 | Vitamin B12-dependent methionine synthase family protein | 1.53 | 1.66 | 1.27 | 1.00 |
| DVU1609 | Dihydrodipicolinate reductase | 1.31 | 1.27 | 1.13 | 0.73 |
| DVU1610 | Glutamine-dependent NAD+ synthetase | 1.51 | 1.09 | 1.52 | 1.37 |
| DVU2981 | 2-Isopropylmalate synthase | 0.49 | 0.65 | 0.48 | 0.34 |
| DVU2982 | 3-Isopropylmalate dehydratase, large subunit, putative | 0.89 | 0.87 | 1.11 | 0.70 |
| DVU2983 | 3-Isopropylmalate dehydratase, small subunit | 0.60 | 0.76 | 1.03 | 0.30 |
| DVU2984 | Conserved hypothetical protein | 0.96 | 0.97 | 1.30 | 1.00 |
| DVU3048 | Aspartate semialdehyde dehydrogenase | 1.09 | 1.30 | 1.73 | 1.50 |
| DVU3371 | 5-Methyl-homocysteine S-methyltransferase | 0.54 | 1.55 | 2.18 | 1.87 |
Log2 gene expression ratios at different times after the pH was changed compared with the control at the same times.
The integrity of the cell envelope is also challenged at elevated pH. Changes in the expression of several genes involved in cell wall and membrane biogenesis were differentially expressed following an increase in the pH of the growth medium. Increased transcription of a gene encoding the heptosyltransferase family protein (DVU1446), involved in the synthesis of the inner core region of lipopolysaccharide, was a notable example (Table 2). One of the most highly up-regulated genes associated with cell envelope structure was fabZ (DVU2368), encoding a putative beta-hydroxyacyl-(acyl carrier protein) dehydratase. This gene, involved in fatty acid biosynthesis, is part of a predicted six-gene operon containing, in addition to fabZ, genes encoding UDP-N-acetylglucosamine O-acyltransferase (DVU2367), UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N- acyltransferase (DVU2369), outer membrane protein OmpH (DVU2370), N-acetylmuramoyl-l-alanine amidase (DVU2371), and a hypothetical protein (DVU2372). The first two enzymes are involved in lipopolysaccharide biosynthesis, while the latter two are part of the peptidoglycan biosynthetic pathway. Transcription of the other three genes in this operon (DVU2367, DVU2369, and DVU2370) was also increased at pH 10 (the log2 R values were 0.8 to 1.0, 0.78 to 1.07, and 1.6 to 1.8, respectively). Transcription of two genes (DVU2371 and DVU2372) located upstream and separated from the genes mentioned above by 151 bp was almost unchanged, suggesting that they do not belong to the same operon. The fabZ gene in E. coli is part of the sigma E regulon, which includes a sigma E consensus promoter, and is up-regulated following heat shock (30). Although the expression of this gene in E. coli was reported not to be altered by either an increase or a decrease in pH, in D. vulgaris the transcription of all genes in the operon containing the fabZ ortholog increased following the shift to higher pH. Changes in the expression of these genes in D. vulgaris have not been detected in response to other environmental stressors, including salt, nitrate, potassium chloride, and heat shock (6, 13, 27; J. D. Wall, unpublished data).
Multiple genes associated with energy generation and electron transfer reactions showed increased expression at elevated pH. For instance, DVU0692, DVU0693, and DVU0694 were significantly up-regulated after 30 min of pH stress. These genes were previously annotated as genes encoding subunits of a molybdopterin oxidoreductase with unknown specificity. Analysis of amino acid sequences predicted that the proteins encoded by DVU0694 (the first gene in the operon) and DVU0692 contain 2 and 10 transmembrane helices, respectively. Because they both also contain putative signal peptide sequences, it is likely that together they comprise a membrane-bound protein complex. The transcription of several other genes predicted to be involved in energy generation and electron transfer reactions also increased. These genes included genes encoding the large subunit of periplasmic NiFe hydrogenase isozyme 2 (DVU2526) and the Fe-S subunit of glycolate/lactate oxidase (DVU3028). In addition, genes for formate dehydrogenases (DVU0587, DVU0588, DVU2481, and DVU2482), thiosulfate reductase (DVU0179), and CO-induced hydrogenase (DVU2286 to DVU2291) were moderately up-regulated. Together with the fact that lactate consumption and acetate production were inhibited, these trends suggested that there was redirection of electron flow from sulfate reduction to unknown electron acceptors, possibly to maintain cytoplasmic redox status during the stress.
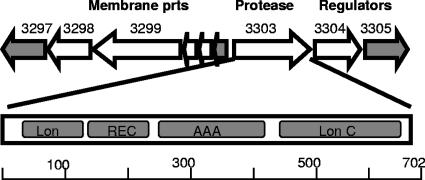
Many genes showing differential expression during alkaline pH stress code for proteins of unknown function for which BLAST analysis did not reveal any homologous proteins in publicly available databases. Thus, their possible role in surviving at high pH is obscure. Among the genes most highly up-regulated following 240 min at pH 10 were DVU3300 and DVU3301 (log2 R = 3.70 and log2 R = 3.57, respectively). These genes most likely comprise an operon containing two additional genes, DVU3298 and DVU3299, that were also up-regulated at alkaline pH (Fig. 4). Amino acid sequence analysis predicted that the proteins encoded by these genes possess at least one transmembrane segment and a signal peptide, suggesting that there is a membrane association. These genes were also previously shown to be up-regulated in D. vulgaris in response to acid, nitrate, nitrite, and sodium chloride stresses (13, 27; Wall, unpublished data).
FIG. 4.
Most highly expressed operons during pH stress, including genes encoding a putative Lon protease (DVU3303), two regulators (DVU3304 and DVU3305), and set of membrane proteins of unknown function (DVU3299 to DVU3301). Open arrows indicate genes whose expression was up-regulated; gray arrows indicate genes whose expression was unchanged. Abbreviations for domains for the Lon protease: Lon, Lon protease domain; REC, receiver domain; AAA, ATPase domain; Lon C, Lon protease C-terminal proteolytic domain. prts, proteins.
Changes in expression of signal transduction and regulatory genes.
The effect of high pH on genes categorized as signal transduction genes revealed that of the 273 genes in this category, 87 showed significant differential expression for at least one time interval, as judged by a Z score greater than 1.5. The Z score was greater than 2 for 44 of them. A peculiar feature of the D. vulgaris genome is the relatively large number of open reading frames annotated as genes encoding methyl-accepting chemotaxis proteins (MCPs). For example, E. coli has 5 MCP genes, compared with 27 MCP genes in D. vulgaris. In E. coli these proteins are involved in chemotactic responses, transducing signals to the flagellar motor. Three D. vulgaris MCP genes were up-regulated at pH 10. One of these genes (DVU3035) is located in close proximity to an operon comprised of genes presumably involved in assimilation of lactate and its conversion to acetate, a central metabolic pathway in D. vulgaris. This gene was previously shown to be up-regulated with salt stress (27). A second gene (DVU3082) was up-regulated twofold after 120 and 240 min of alkaline pH exposure. This gene was previously shown to be up-regulated with acid stress but to be unaffected by other stressors examined so far for D. vulgaris. The third gene (DVU1884), up-regulated twofold after 240 min of alkaline pH, was previously shown to be also up-regulated with heat shock and nitrate stress (6, 13). Interestingly, the latter gene is immediately downstream of a locus consisting of 14 genes (including genes encoding ClpB, DnaJ, and peptidyl-prolyl cis-trans isomerase) associated with the E. coli general stress response. Although expression of flagellar genes in D. vulgaris was decreased and the motility of the cells was suppressed, the MCPs may be involved in regulation of cellular responses other than motility. For instance, it was recently demonstrated that a chemosensory-like pathway regulates developmental gene expression in Myxococcus xanthus (18).
The DVU0667 gene coding for an HD domain protein was among the most highly up-regulated genes in cells exposed to pH 10 for 30 min. Proteins with the HD motif belong to a superfamily of metal-dependent phosphohydrolases that include a variety of uncharacterized proteins associated with nucleotidyltransferases and helicases from Bacteria, Archaea, and eukaryotes. Increases in expression of these genes suggest that D. vulgaris may respond to high pH by activating unidentified regulatory pathways that affect RNA modifications and stability.
There are only a few genes encoding alternative sigma factors in D. vulgaris (sigma N, sigma H, and a flagellar sigma factor). As reported by Chhabra et al. (6), neither a gene for sigma E nor a gene for RpoS has been found in the genome of this bacterium. This suggests that D. vulgaris may respond to stresses that cause misfolding or degradation of cell envelope proteins differently than better-studied bacteria. Indeed, in addition to the absence of sigma E, no deg and rse orthologs have been recognized in the genome. These genes have been shown to play an important role in the response of E. coli to periplasmic stress (30, 31). Thus, D. vulgaris apparently uses a system distinct from that of E. coli to control expression of genes encoding proteases and chaperone-like proteins. One of the most highly up-regulated genes (DVU3303) encodes a protease that is homologous to Lon but has a very peculiar domain structure. This protease contains a signal receiver domain common to CheY, OmpR, NtrC, and PhoB, as well as a phosphor acceptor site for histidine kinase homologs. Its carboxyl terminus contains ATPase and Lon protease (S16) proteolytic domains (Fig. 4). This domain structure suggests that this protease can be activated by phosphorylation and transfers the signal to an unknown protein which might be subsequently functionally altered by cleavage. Immediately downstream of DVU3303 are genes for a histidine kinase (DVU3304) and a putative response regulator (DVU3305), making this gene cluster a plausible candidate for a regulatory circuit involved in the response to pH stress and other stresses, since these regulatory genes were shown to be also up-regulated under pH 5.5, nitrate, chromium, or sodium chloride stress (13, 27, J. D. Wall, unpublished data).
Two additional genes annotated as genes encoding histidine kinases (DVU0331 and DVU2580) were also up-regulated in response to elevated pH. Mutants with mutations in these two genes, JW3011 and JW3024, demonstrated increased sensitivity to elevated pH (Fig. 2). Since the genes encoding these kinases are proximal to genes for cell wall biogenesis, some of which were down-regulated, they may also be part of a more general D. vulgaris response to cell envelope stress.
Genes down-regulated during exposure to high pH.
Genes for peptidyl-prolyl cis-trans isomerase (DVU2569; log2 R = −1.4 to −1.8), l-isoaspartate O-methyltransferase (DVU1849; log2 R = −1.2 to −1.4), and peptidyl-prolyl cis-trans isomerase B-2 (DVU1873; log2 R = −0.8 to −1.3) were found to be moderately down-regulated with alkaline stress. All three of these proteins are involved in cell wall biosynthesis, and observed changes in their expression may reflect a pause in cell growth and/or adaptive changes in cell wall composition to the stress. In addition, some energy production and central metabolism genes were consistently down-regulated, including genes encoding pyruvate carboxylase (DVU1834; log2 R = −1.4 to −1.8), B12 binding domain protein/radical S-adenosylmethionine domain protein (DVU3016; log2 R = −1.5 to −2.1), desulfoferredoxin (DVU3183; log2 R = −1.0 to −1.2), ferredoxin II (DVU0305; log2 R = −1.5 to −1.9), and an l-lactate permease family protein (DVU2451; log2 R = −1.1 to −1.8). The hupD gene (DVU1923; log2 R = −0.6 to −1.6) for hydrogenase expression/formation was down-regulated after 240 min of pH 10 stress. Interestingly, 28 of the 57 genes most down-regulated during high-pH stress (log2 R range, −1.5 to −3.6; average log2 R, ∼−2) were annotated as phage or transposon related, suggesting that control of their expression might be an important aspect of the D. vulgaris stress response or part of the phage survival strategy (see Table S3 in the supplemental material).
Conclusions.
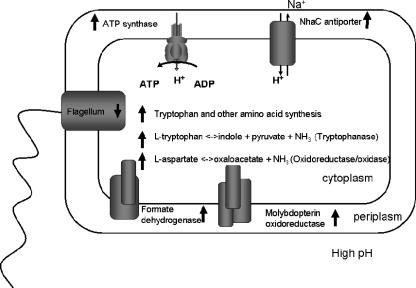
The model shown in Fig. 5 draws upon the data presented in this study and provides an overview of the response of D. vulgaris to alkaline stress. The observed increase in expression of genes for ATP synthase and the Na+/H+ antiporter NhaC-2 attenuates alkalinization of the cytoplasm via increased proton import and is similar to the responses of E. coli and other bacteria (20, 25, 29). Similarly, a decrease in expression of flagellar genes and presumptive acidification of the cytoplasm via increased expression of genes involved in amino acid metabolism are also shared responses. Increased expression of several genes for proteins potentially involved in energy generation or electron transfer reactions was also observed. These genes included genes for a formate dehydrogenase, a molybdopterin oxidoreductase, cytoplasmic Coo hydrogenase, and a periplasmic NiFe hydrogenase. The latter changes may be part of a more general strategy to retain a cellular redox state necessary to sustain cellular functions when lactate consumption and sulfate respiration are repressed.
FIG. 5.
Conceptual model of D. vulgaris response to high pH stress. The arrows next to protein, enzyme, and pathway names indicate changes in the corresponding gene expression.
Although growth was arrested at pH 10, the cells remained metabolically active, as assessed by rescue in culture and a complex transcriptional response during the period of exposure. As part of this response there appeared to be a modification of the cell envelope, indicated by modulation of the expression of genes involved in cell wall biosynthesis. Some genes in this category showed decreased expression, whereas others were up-regulated. We anticipate that ongoing studies of the cellular proteome and membrane composition will better elucidate the character and possible function of any changes in membrane structure.
Finally, our observations suggest that although the response of D. vulgaris to increased pH is similar to the response of E. coli, this response is controlled by unique regulatory circuits. The alternative sigma factors (sigma S and sigma E) contributing to this stress response in E. coli are apparently absent in D. vulgaris. Our transcriptional analysis identified several regulators that are likely part of a D. vulgaris-specific stress response system. These regulators now provide specific targets for continued biochemical and genetic characterization of the stress response system of this organism and its relationship to regulatory pathways in other bacteria, particularly within the family Desulfovibrionaceae.
Acknowledgments
We thank N. Pinel, S. Flagan, C. Walker, and K. Hillesland for careful reading of the manuscript and for valuable discussions and M. Sadilek for his invaluable assistance in performing the gas chromatography-mass spectrometry and high-performance liquid chromatography analyses. We also thank J. Jacobsen for her assistance with gene expression data analysis and handling and J. A. Ringbauer, Jr., for generating mutants. The generosity of William W. Metcalf for providing the pRL27 plasmid for mutagenesis with the Tn5-RL27 transposon is appreciated.
This work was supported by the Department of Energy's Office of Biological and Environmental Sciences under the GTL-Genomics Program via the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov).
Published ahead of print on 5 October 2007.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/
REFERENCES
- 1.Abildgaard, L., M. B. Nielsen., K. U. Kjeldesen, and K. Ingvorsen. 2006. Desulfovibrio alkalitolerans sp. nov., a novel alkalitolerant, sulphate-reducing bacterium isolated from district heating water. Int. J. Syst. Evol. Microbiol. 56:1019-1024. [DOI] [PubMed] [Google Scholar]
- 2.Aeckersberg, F., F. Bak, and F. Widdel. 1991. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156:5-14. [Google Scholar]
- 3.Beller, H. R., M. Reinhard, and D. Grbić-Galić. 1992. Metabolic by-products of anaerobic toluene degradation by sulfate-reducing enrichment cultures. Appl. Environ. Microbiol. 58:3192-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, K. S., H.-C. B. Yen, C. L. Hemme, Z. Yang, Z. He, J. Zhou, T. C. Hazen, K. H. Huang, E. J. Alm, A. P. Arkin, and J. D. Wall. 2007. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, J., A. A. Guffanti, and T. A. Krulwich. 1994. The chromosomal tetracycline resistance locus of B. subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J. Biol. Chem. 269:27365-27371. [PubMed] [Google Scholar]
- 6.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 8.DeWeerd, K. A., F. Concannon, and J. M. Suflita. 1991. Relationship between hydrogen consumption, dehalogenation, and the reduction of sulfur oxyanions by Desulfomonile tiedjei. Appl. Environ. Microbiol. 57:1929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dilworth, M. J., and A. R. Glenn. 1999. Problems of adverse pH and bacterial strategies to combat it. Novartis Found. Symp. 221:4-14. [DOI] [PubMed] [Google Scholar]
- 10.Dyrløv, J. B., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0 J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 11.Fry, N. K., J. K. Fredrickson., S. Fishbain, M. Wagner, and D. A. Stahl. 1997. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl. Environ. Microbiol. 63:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, R., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 13.He, Q., K. H. Huang, Z. He, E. J. Alm, M. W. Fields, T. C. Hazen, A. P. Arkin, J. D. Wall, and J. Zhou. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554-559. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. 374:166-168. [Google Scholar]
- 16.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juncker, A. S., H. Willenbrock, G. von Heijne, H. Nielsen, S. Brunak, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 20.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X. Wan, A. P. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickman, and J. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 188:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd, J. R., J. Ridley, T. Khiznia., N. N. Lyalikova, and L. E. Macaskie. 1999. Reduction of technetium by Desulfovibrio desulfuricans: biocatalyst characterization and use in a flowthrough bioreactor. Appl. Environ. Microbiol. 65:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londry, K. L., J. M Suflita, and R. S. Tanner. 1999. Cresol metabolism by the sulfate-reducing bacterium Desulfotomaculum sp. strain Groll. Can. J. Microbiol. 45:458-463. [PubMed] [Google Scholar]
- 23.Lovley, D. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 24.Lovley, D. R., and E. J. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer, L., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczevski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motamedi, M., and K. Pedersen. 1998. Desulfovibrio aespoeensis sp. nov., a mesophilic sulfate-reducing bacterium from deep groundwater at Aspo hard rock laboratory, Sweden. Int. J. Syst. Bacteriol. 48:311-312. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay, A., Z. He, E. J. Alm, A. P. Arkin, E. E. Baidoo, S. C. Borglin, W. Chen, T. C. Hazen, Q. He, H. Y. Holman, K. Huang, R. Huang, D. C. Joyner, N. Katz, M. Keller, P. Oeller, A. Redding, J. Sun, J. Wall, J. Wei, Z. Yang, H. C. Yen, J. Zhou, and J. D. Keasling. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padan, E., N. Maisler, D. Taglicht, R. Karpel, and S. Schuldiner. 1989. Deletion of ant in E. coli reveals its function in adaptation to high salinity and an alternative Na/H antiporter system(s). J. Biol. Chem. 264:20297-20302. [PubMed] [Google Scholar]
- 29.Padan, E., E. Bibi, M. Ito., and T. Krulwich. 2005. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717:67-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodius, V. A., W. C. Suh, G. Nanaka, and C. A. Gross. 2006. Conserved and variable functions of the sigma E stress response in related genomes. PloS Biol. 4:43-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz, N., and T. Silhavy. 2005. Sensing external stress: watchdogs of the E. coli cell envelope. Curr. Opin. Microbio. 8:122-126. [DOI] [PubMed] [Google Scholar]
- 32.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 33.Skulachev, V. P. 1999. Bacterial energetics at high pH: what happens to the H+ cycle when the extracellular H+ concentration decreases? Novartis Found. Symp. 221:200-213. [DOI] [PubMed] [Google Scholar]
- 34.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandieken, V., C. Knoblauch, and B. B. Jørgensen. 2006. Desulfovibrio frigidus sp. nov. and Desulfovibrio ferrireducens sp. nov., psychrotolerant bacteria isolated from Arctic fjord sediments (Svalbard) with the ability to reduce Fe(III). Int. J. Syst. Evol. Microbiol. 56:681-685. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, W., D. E. Culley, J. C. Scholten, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie Leeuwenhoek 89:221-237. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, W., D. E. Culley, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Oxidative stress and heat-shock responses in Desulfovibrio vulgaris by genome-wide transcriptomic analysis. Antonie Leeuwenhoek 90:41-55. [DOI] [PubMed] [Google Scholar]