Abstract
Mycobacterium leprae, a major human pathogen, grows poorly at 37°C. The basis for its inability to survive at elevated temperatures was investigated. We determined that M. leprae lacks a protective heat shock response as a result of the lack of transcriptional induction of the alternative sigma factor genes sigE and sigB and the major heat shock operons, HSP70 and HSP60, even though heat shock promoters and regulatory circuits for these genes appear to be intact. M. leprae sigE was found to be capable of complementing the defective heat shock response of mycobacterial sigE knockout mutants only in the presence of a functional mycobacterial sigH, which orchestrates the mycobacterial heat shock response. Since the sigH of M. leprae is a pseudogene, these data support the conclusion that a key aspect of the defective heat shock response in M. leprae is the absence of a functional sigH. In addition, 68% of the genes induced during heat shock in M. tuberculosis were shown to be either absent from the M. leprae genome or were present as pseudogenes. Among these is the hsp/acr2 gene, whose product is essential for M. tuberculosis survival during heat shock. Taken together, these results suggest that the reduced ability of M. leprae to survive at elevated temperatures results from the lack of a functional transcriptional response to heat shock and the absence of a full repertoire of heat stress response genes, including sigH.
Mycobacterium leprae infection is responsible for approximately 500,000 new cases of leprosy annually (32). One-fourth of these leprosy patients will suffer from some form of nerve damage due to the predilection of this obligate intracellular pathogen for the Schwann cells of peripheral nerves (2, 21, 22). M. leprae also causes a natural, systemic infection in nine-banded armadillos (Dasypus novemcinctus), whose core body temperature is 33 to 34°C (1, 19). This obligate pathogen has been shown to maintain its metabolic activity in axenic medium (30), cultured murine and human macrophages (9, 13), and primary rat Schwann cells (8) for several weeks at 33°C but rapidly loses this activity at 37°C. A possible explanation for the inability of M. leprae to survive at elevated temperatures is that it is unable to mount a protective heat stress response.
The heat stress response in Mycobacterium tuberculosis, a close relative of M. leprae, is an adaptive pathway involved in the survival of bacteria that are exposed to increased ambient temperature. It is characterized by genome-wide transcriptional changes, resulting in the induction of over 100 genes, including several sigma factors (sigB, sigE, and sigH), all of the major, highly conserved heat shock protein genes, and in particular, those genes in the HSP70 and HSP60 operons (28). SigH together with the RNA polymerase core enzyme (RNAP) orchestrates the initiation of this response (16, 18), and the heat shock regulators HspR and HrcA negatively regulate this response (18, 28). The resulting heat shock proteins ensure appropriate folding, translocation, and assembly of polypeptide structures and the degradation of protein aggregates that form at elevated temperatures.
In contrast to M. tuberculosis, virtually nothing is known about the heat stress response mechanism in M. leprae. This bacterium has lost >50% of its protein-coding capacity due to gene deletion or other mutational events, leaving its genome with only a limited repertoire of intact sigma factor genes (sigA, sigB, sigC, and sigE) (3, 4). However, functional studies have not been conducted to determine if sigB and sigE can transcriptionally respond to heat stress without a functional SigH or whether their gene products are capable of responding to heat stress. In addition, no comprehensive search of the M. leprae genome for the presence of the highly induced heat shock response genes found in M. tuberculosis has been conducted.
To begin to understand M. leprae's reduced ability to survive at elevated temperatures, we performed quantitative reverse transcription-PCR (qRT-PCR) to examine the transcriptional response of specific sigma factor genes and heat shock genes to defined heat stress. The capability of M. leprae's sigE to respond to heat stress was analyzed using an M. tuberculosis sigE knockout mutant. Bioinformatics tools were implemented to determine the presence of M. leprae homologs for the highly induced genes and regulatory components found in M. tuberculosis during heat shock.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
M. leprae Thai-53 was propagated and maintained in serial passage in the hind footpads of athymic nude mice (Hsd: athymic nude; Harlan, Indianapolis, IN) as previously described (30). All M. tuberculosis and Mycobacterium smegmatis strains were grown using either liquid Middlebrook 7H9 or solid 7H11 media (BD, Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase and 0.05% Tween 80 and appropriate antibiotics when required (streptomycin, 30 μg/ml; kanamycin, 50 μg/ml; hygromycin, 100 μg/ml; zeocin, 50 μg/ml) as previously described (references 17 and 33, respectively). Escherichia coli XL-1 Blue supercompetent cells (Stratagene, La Jolla, CA) were grown in Luria-Bertani broth (BD, Difco) supplemented after transformation with kanamycin (50 μg/ml) at 37°C in shaking cultures.
M. leprae heat stress conditions.
The appropriate conditions to study heat stress in M. leprae were determined by adding 1 × 108 M. leprae Thai-53 cells to prewarmed 7H12 medium (BACTEC 12B medium; BD Diagnostic Systems) and held at 33°C or 37°C in 5% CO2 for up to 24 h or at 42°C for 2 h in 5% CO2. At 2, 4, 8, 16, and 24 h post-onset of heat stress, bacterial viability was determined using Buddemeyer radiorespirometry (6). The appropriate heat stress level was then determined as that which induces 10 to 15% loss of viability compared to that of bacteria held at 33°C for the same amount of time. M. leprae cells were then exposed to these heat stress conditions, immediately put on ice, pelleted at 10,000 × g for 10 min at 4°C, resuspended in 70% ethanol, and stored at −20°C to stop transcription and preserve bacterial RNA (31).
DNA manipulations and sequencing.
All recombinant DNA techniques were performed according to standard procedures using E. coli as an initial host. DNA restriction and modifying enzymes were obtained from New England Biolabs (Ipswich, MA) and used according to the manufacturer's suggested protocols. PCR amplicons were generated from either M. leprae or M. tuberculosis genomic DNA or cDNA. To confirm the sequences of amplicons, the DNA sequence of each PCR product was determined using automated DNA sequencing (LSU Genelab, Baton Rouge, LA). These sequences were then aligned to those of the M. leprae TN strain (http://genolist.pasteur.fr/Leproma/) or M. tuberculosis H37Rv (http://www.sanger.ac.uk/Projects/M_tuberculosis/) using ClustalW (http://www.ebi.ac.uk/clustalw/).
Cloning M. leprae sigE into mycobacterial sigma factor knockout mutants.
A 2.9-kb fragment, including sigE (ML1076) and its putative anti-sigma factor rseA (ML1077), was amplified from M. leprae Thai-53 DNA using the GeneAmp XL-PCR kit (Applied Biosystems, Foster City, CA) and primers (see Table S3 in the supplemental material) by PCR and subcloned into the StuI-digested pSM316 vector. The resultant recombinant plasmid (pML7677) was transformed into E. coli XL-1 Blue cells. A clone containing the M. leprae sigE fragment was identified by PCR analysis and direct DNA sequencing, and plasmid DNA was amplified, purified, and electroporated into electrocompetent M. tuberculosis ST28 (MtbΔsigE) and M. smegmatis RH244 (MsmegΔsigE) single-knockout mutants and an M. smegmatis RH315 (MsmegΔsigE/ΔsigH) double-knockout mutant (Table 1) using previously described procedures (17, 33). After appropriate antibiotic selection, crude cell lysates were made of several positive clones, and these preparations were analyzed for the presence of M. leprae sigE using the M. leprae sigE-specific primer set and PCR. Positive M. tuberculosis clones were then analyzed for the presence of an interrupted (hph-containing) M. tuberculosis sigE gene by using an M. tuberculosis sigE-specific primer set and PCR (see Table S3 in the supplemental material), which was designed to span the hph gene in the MtbΔsigE mutant and MtbΔsigE/MlepsigE-complemented mutant. The entire M. leprae sigE insert was PCR amplified and sequenced from positive clones of the MtbΔsigE/MlepsigE-complemented mutant, MsmegΔsigE/MlepsigE-complemented mutant, and MsmegΔsigE/ΔsigH/MlepsigE-complemented mutant.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| M. leprae Thai-53 | M. leprae isolate obtained from skin biopsy of leprosy patient in Thailand and continuously serially passaged in nude mice | NHDP |
| M. tuberculosis strains | ||
| H37Rv | Virulent M. tuberculosis isolate derived from human lung isolate H37Rv by W. Steenken, 1934, NY | ATCC 25618 |
| ST28 (MtbΔsigE) | H37Rv sigE::hph (Hgmr) (SigE−); result of homologous recombination of pSM270 construct into sigE of M. tuberculosis in chromosome of H37Rv | 17 |
| ST29 (MtbΔsigE/MtbsigE) | ST28 sigE::hph (Hgmr) (SigE−) + H37Rv (SigE); result of insertion of pSM316 (Kmr Strr) construct into chromosome of ST28 | 17 |
| ST28/pML7677 (MtbΔsigE/MlepsigE) | ST28 sigE::hph (Hgmr Kmr) (SigE−) + M. leprae (SigE); result of insertion of pML7677 (Kmr StrrM. leprae SigE) construct into ST28 chromosome | This work |
| M. smegmatis strains | ||
| mc2155 | High-frequency transformation strain of M. smegmatis derived from wild-type strain mc26 | 24 |
| RH244 (MsmegΔsigE) | mc2-155 sigE::zeo (Zeor) (SigE−); result of recombination of a pRH1264Z construct into sigE of chromosome of mc2155 | 18 |
| RH315 (MsmegΔsigE/ΔsigH) | RH244 sigH::aph (Zeor Kmr) (SigE− SigH−); result of recombination of pRH1317 into sigH of chromosome of RH244 | 18 |
| RH244/pML7677 (MsmegΔsigE/MlepsigE) | RH244 sigE::zeo (Zeor Kmr) attB::sigE; result of insertion of pML7677(Kmr) into chromosome of RH244 | This work |
| RH315/pML7677 (MsmegΔsigE/ΔsigH/MlepsigE) | RH315 sigE::zeo sigH::aph (Zeor Kmr) attB::sigE; result of insertion of pML7677 (Kmr) into chromosome of RH315 | This work |
| E. coli XL-1 Blue | recA1 endA1 supE44 thi-1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pSM316 | E. coli mycobacterial suicide vector which inserts into the attB site of mycobacteria; sacB Strr | 17 |
| pML7677 | pSM316 with M. leprae sigE and rseA genes and promoter region inserted into the Stu1 site | This work |
Tissue culture and M. tuberculosis infection.
THP-1 macrophage-like cells (ATTC TIB-202) were obtained, expanded, and maintained as specified elsewhere (http://www.atcc.org). THP-1 cells were stimulated to differentiate into adherent, well-spread macrophages by the addition of 100 nM phorbol myristate acetate and maintained for 3 days at 37°C, 5% CO2. These cells were washed in medium and used to determine the ability of M. tuberculosis H37Rv and its derivatives to survive in these cells at appropriate time points postinfection by developing M. tuberculosis killing curves as previously described (17).
Killing curves of mycobacterial host strains and their derivatives.
Mid-log-phase cultures of M. tuberculosis H37Rv and its derivatives were subjected to heat shock conditions (45°C) or sodium dodecyl sulfate (SDS) treatment (0.05%) as previously described (17). Mid-log-phase cultures of M. smegmatis were subjected to heat stress conditions (50°C) or SDS treatment (0.01%) as previously described (33). Dilutions of bacteria were plated on 7H11 agar plates and incubated at 37°C to determine the number of CFU prior to stress (T0), and 50-μl samples were diluted in 7H9 and plated to determine the number of CFU poststress. Results are expressed as the percentage of survivors with respect to T0.
Purification of mycobacterial RNA and reverse transcription of cDNA.
Mycobacterial RNA was purified using a previously described protocol (31). DNA was removed from these preparations using the Turbo DNA-free kit (Ambion, Austin, TX). DNA-free RNA aliquots were then stored at −80°C until use. Total mycobacterial RNA (1 μg) was converted to cDNA using the Advantage RT-for-PCR kit (BD BioSciences, Clontech, Mountain View, CA), using random hexamers or gene-specific primers where appropriate (see Table S3 in the supplemental material). Controls for DNA contamination consisted of 1 μg RNA from each strain incubated with the reverse transcription reagents, excluding the reverse transcriptase. Template cDNA was also made from BALB/c mouse spleen total RNA (BD Biosciences, Clontech).
PCR and RT-PCR.
PCR amplification of mycobacterial DNA or cDNA was performed by obtaining gene sequences from either Leproma (http://genolist.pasteur.fr/Leproma) or TubercuList (http://www.sanger.ac.uk/Projects/M_tuberculosis/), designing primers using Omiga 2.0 DNA and Protein Sequence Analysis (Primer Design) software (Oxford Molecular Ltd., Cambell, CA) (see Table S3 in the supplemental material) and using recommended primer annealing temperatures described by this software and 35 cycles of standard PCR. Mycobacterial DNAs were used as positive controls, and RT-negative reactions, buffer, and mouse cDNA were used as negative controls for each assay. Gene transcripts were observed in ethidium bromide-stained agarose gels using a GelDoc2000 gel analyzer (Bio-Rad).
Quantitative real-time RT-PCR assays for quantitative assessment of gene expression.
Transcript levels of mycobacterial genes were determined using M. leprae cDNA and real-time qRT-PCR using a 7300 sequence detection system (Applied BioSystems) with specific primer sets and probes designed from gene sequences as defined by the Primer Express software (Applied BioSystems) (see Table S3 in the supplemental material). Quantitative analysis of data was accomplished using the standard curve comparative method. Data were normalized for potential template concentration variation by dividing each gene transcript value by that of the 16S rRNA value from the same template. Due to the limits of the TaqMan assay, only values that were at least twofold induced were considered different.
Bioinformatics and comparative genomics.
Nucleic acid and deduced amino acid sequences for comparative analysis were obtained from Leproma (http://genolist.pasteur.fr/Leproma) or TubercuList (http://genolist.pasteur.fr/TubercuList/). Alignments were performed using ClustalW (http://www.ebi.ac.uk/clustalw/). NCBI Blastn (http://www.ncbi.nlm.nih.gov/BLAST/) was used to search for specific promoter and other regulatory sequences. The ExPASy Translate tool (http://au.expasy.org/tools/dna.html/) was used to translate M. leprae pseudogenes. Protein domains were identified using ExPASy Procite Database of Protein Families and Domains website (http://us.expasy.org/prosite/).
Statistical analysis of data.
Statistical comparisons of the data for this study were made using three replicates from at least two separate experiments. Analysis was performed using InStat V.3 (GraphPad Software) using nonparametric analysis and the unpaired t test with Welch correction.
RESULTS
Sensitivity of M. leprae to heat stress.
To establish appropriate conditions for heat stress of M. leprae, purified nude mouse-derived M. leprae Thai-53 cells (Table 1) were exposed to different temperatures for various time periods, and the effects of these conditions on bacterial viability were determined using radiorespirometry. Exposure of M. leprae to 37°C for 2 h resulted in a 5% loss in bacterial viability, and exposure of M. leprae to 37°C for longer periods resulted in a greater loss of viability (Fig. 1a). Exposure of M. leprae to 45°C for 2 h resulted in a significant loss of viability (>55%) compared to bacteria maintained at 33°C (Fig. 1a). Based on these data, exposure of M. leprae to 37°C for 4 h was chosen for subsequent heat shock experiments to allow the majority of the bacterial population to remain viable and be able to transcriptionally respond to heat stress conditions. This temperature is ∼5°C above the optimal temperature for short-term survival of this organism in vitro (30). However, some experiments were also conducted at 42°C for 2 h to ensure that the proper conditions for heat stress on protein folding would occur (10).
FIG. 1.
Viability analysis of M. leprae and real-time RT-PCR analysis of M. leprae genes using RNA from nude mouse-derived M. leprae held in prewarmed medium at 33°C or 37°C for 4 h or 42°C for 2 h. (a) Percent relative viability values were obtained from 7-day Buddemeyer radiorespirometry readings (counts per minute) from M. leprae held at 37°C or at 45°C divided by that of M. leprae held at 33°C for each time period and each temperature. Values represent the mean and standard deviation of at least six observations for each time. A single asterisk denotes that the percent relative viability values between the 2-h 37°C incubation and other indicated times are considered significantly different at P < 0.001; a double asterisk denotes that the percent relative viability values between the 2-h 37°C incubation and other indicated times are considered significantly different at P < 0.05. (b and c) The increases of mRNA levels for sigma factor gene transcripts (b) and heat shock gene transcripts (c) due to heat stress were obtained by normalizing each template for potential template concentration differences by dividing each of the mRNA values by that of the 16S rRNA value from a 1:10−5 dilution of the same cDNA template and dividing these values by those of those of M. leprae held at 33°C.
M. leprae gene transcription during heat stress.
To determine if heat stress induced the expression of heat shock-responsive sigma factors and heat shock genes in M. leprae, transcript levels for sigB, sigE, groES (HSP60 operon), dnaK (HSP70 operon), htpG, and hsp18 genes were compared at 33°C, 37°C, and 42°C using qRT-PCR. There was no significant induction (P > 0.05) of sigma factor genes sigB and sigE (Fig. 1b) or heat shock genes (Fig. 1c) in heat-stressed bacteria at 37°C or 42°C compared to bacteria held at 33°C.
Complementation of the MtbΔsigE mutant.
The lack of induction of the M. leprae sigE during heat shock suggests a defect in its regulation under these stress conditions. However, for the overall heat shock response to be initiated, SigE and SigB must be capable of recognizing and binding to specific heat shock promoters and inducing heat shock genes in response to elevated temperatures. To determine if the sigE gene product is functionally capable of responding to heat shock stress conditions, M. leprae's sigE and its putative anti-sigma factor (rseA) (Fig. 2a) were cloned into pSM316, a mycobacterial vector that integrates into the L5 attB site of the mycobacterial chromosome (12). This construct was electroporated into MtbΔsigE, which lacks a functional sigE but contains all the other functional sigma factor genes, including sigH (Table 1). The resultant strain (MtbΔsigE/MlepsigE-complemented mutant) had the expected disruption of the M. tuberculosis sigE gene (indicated by its hygromycin-resistant phenotype), possessed a chromosomal copy of the pML7677 plasmid (indicated by its kanamycin-streptomycin-resistant phenotype), and displayed similar growth characteristics as those of H37Rv (data not shown). PCR analysis of the M. tuberculosis and M. leprae sigE in both the MtbΔsigE mutant and the MtbΔsigE/MlepsigE-complemented mutant demonstrated the presence or absence of the appropriate sigE alleles (Fig. 2b).
FIG. 2.
M. leprae sigE fragment for complementation of MtbΔsigE and MsmegΔsigE and MsmegΔsigEΔsigH mutants. (a) Map of the PCR amplicon containing the sigE and rseA genes of M. leprae. Large arrows indicate the direction of transcription of each gene; the 410-bp PCR amplicon for detection of M. leprae sigE is shown as a dotted line; small arrows indicate positions of primers. (b) Genotype analyses of M. tuberculosis H37Rv and derivatives were accomplished using M. tuberculosis sigE and M. leprae sigE PCR assays. The predicted sizes of PCR amplicons for M. tuberculosis sigE under these conditions were 605 bp for the intact M. tuberculosis sigE gene and 2.1 kb for the hygromycin-interrupted M. tuberculosis sigE gene. Lanes 1 to 4, results of PCR amplification of the M. tuberculosis sigE gene from M. tuberculosis H37Rv, MtbΔsigE, MtbΔsigE/MtbsigE-complemented mutant, and MtbΔsigE/MlepsigE-complemented mutant, respectively; lane 5, negative buffer control; lane 6, 100-bp DNA ladder (Promega); lanes 7 to 10, results of PCR of the M. leprae sigE from M. tuberculosis H37Rv, MtbΔsigE, MtbΔsigE/MtbsigE-complemented mutant, and MtbΔsigE/MlepsigE-complemented mutant, respectively; lane 11, negative buffer control.
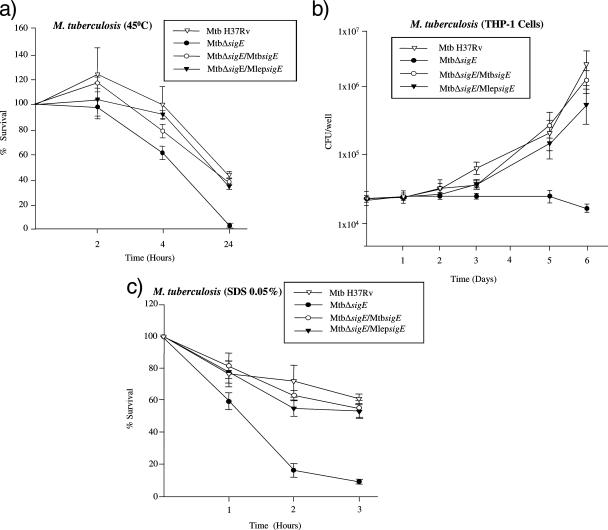
The sensitivity of M. tuberculosis H37Rv and its derivatives to heat shock (45°C) over time was determined. Results confirmed that the MtbΔsigE mutant was the most sensitive to heat stress at each time interval (Fig. 3a). The sensitivity of the MtbΔsigE/MlepsigE-complemented mutant to heat was comparable to that of the H37Rv and MtbΔsigE/MtbsigE-complemented mutant. M. leprae sigE was also able to complement the MtbΔsigE mutant's growth defect in THP-1 macrophage-like cells and susceptibility to detergent stress (Fig. 3b and c, respectively). The data suggested that M. leprae's sigE could function as efficiently as that of M. tuberculosis in the MtbΔsigE mutant.
FIG. 3.
Survival curves of M. tuberculosis H37Rv and its derivatives after exposure to various environmental and intracellular stresses. Each experiment was performed using triplicate samples, and data for at least two experiments are shown. The values represent the mean and the standard deviation obtained for each point. (a) Bacteria were exposed to 45°C for up to 24 h, and results are expressed as the percentage of viable bacteria (CFU) with respect to T0 (prior to stress exposure). (b) Growth of M. tuberculosis H37Rv and its derivatives in human THP-1 macrophages expressed as CFU/well. (c) Growth of M. tuberculosis H37Rv and its derivatives exposed to 0.05% SDS for up to 3 h expressed as the percentage of CFU with respect to T0 (prior to stress exposure).
Complementation of a mycobacterial ΔsigE/ΔsigH mutant.
To determine whether the M. leprae sigE gene product was capable of responding to heat stress in the absence of a functional SigH, pML7677 was electroporated into and expressed in MsmegΔsigE and MsmegΔsigE/ΔsigH double mutants (Table 1). The resultant strains possessed a chromosomal copy of the pML7677 plasmid, as determined by PCR/direct DNA sequencing of the M. leprae sigE gene fragment, and displayed similar growth characteristics to that of the wild-type strain, M. smegmatis mc2155 (data not shown). The sensitivity of M. smegmatis mc2155 and its derivatives to heat shock (50°C) over time was then evaluated. Results indicated that the M. leprae sigE was unable to complement the heat stress defect of the MsmegΔsigE/ΔsigH double knockout mutant but was able to complement the heat stress defect of MsmegΔsigE containing a functional sigH (Fig. 4a). In addition, the M. leprae sigE was able to complement the lack of detergent stress response in this mutant (Fig. 4b).
FIG. 4.
Survival curves of M. smegmatis mc2155 and its derivatives after exposure to stress conditions. The results are expressed as the percentage of CFU with respect to T0 (prior to stress exposure). Each experiment was performed using triplicate samples, and reported results are from at least two experiments. The values represent the mean and the standard deviation obtained for each point. (a) Exposure to 50°C heat stress over time; (b) exposure to 0.01% SDS detergent stress for 2 h.
sigB transcript levels in M. tuberculosis H37Rv and its derivatives.
The sigB transcript levels in M. tuberculosis H37Rv and its derivatives were analyzed by quantitative real-time RT-PCR after exposure of bacteria to heat stress. At 37°C, sigB levels in M. tuberculosis H37Rv and the MtbΔsigE/MlepsigE-complemented mutant were comparable; however, levels in MtbΔsigE were significantly lower (P < 0.001) (Fig. 5a). In contrast, sigB levels in all strains were elevated to comparable levels following 45°C heat stress (Fig. 5b).
FIG. 5.
Comparison of sigB mRNA levels in M. tuberculosis H37Rv and its derivatives, MtbΔsigE, and MtbΔsigE/MtbsigE- and MtbΔsigE/MlepsigE-complemented mutants, respectively, before and after heat stress. Data are expressed as normalized mRNA values detected by real time RT-PCR analysis using cDNA samples obtained from exponentially growing cultures at 37°C (a) and exposed to 45°C (b) for 2 h. Normalized mRNA values for gene transcripts were obtained by dividing the mRNA values by that of the 16S rRNA value (1:10−5 dilution) for the same template.
M. leprae heat shock genes.
Using a bioinformatics approach, genome-wide screening of M. leprae for the presence of homologs of the most highly induced genes found in M. tuberculosis during heat stress (28) was performed. Results showed that M. leprae lacked intact homologs for 68 of 100 of these heat-inducible genes (see Table S1 in the supplemental material). Furthermore, no homologs were found for 5 of the 10 most highly induced heat shock response genes in M. tuberculosis. This included the most highly induced of these genes, hsp/acr2, which is required for M. tuberculosis survival during heat stress.
M. leprae heat shock operons, promoters, and regulatory sequences.
Even though the majority of the heat-induced genes were absent in M. leprae, the major heat shock operons HSP70 (dnaK-grpE-dnaJ1) and HSP60 (groES-groEL1) were conserved in M. leprae (see Table S2 in the supplemental material). The organization of these operons in M. leprae was identical to that of M. tuberculosis (http://www.sanger.ac.uk/Projects/M_tuberculosis/) (Fig. 6a and b). Using RT-PCR we determined that these two sets of genes are expressed as operons in M. leprae. The HSP70 operon appeared to be transcribed as a single mRNA containing a single dnaK-grpE-dnaJ1 transcript and the HSP60 operon as a single groES-groEL1 transcript (Fig. 6c and d).
FIG. 6.
HSP70 and HSP60 operons of M. leprae. (a) Map of the HSP70 operon of M. leprae and the predicted 953-bp PCR amplicon (dashed line), representing a partial fragment of this operon for detection of its polycistronic transcript by RT-PCR analysis. Small arrows indicate locations of primers (see Table S3 in the supplemental material). Large arrows indicate the direction of transcription. (b) Map of the HSP60 operon of M. leprae and the predicted 889-bp PCR amplicon (dashed line), representing a partial fragment of this operon for detection of its polycistronic transcript by RT-PCR analysis. Small arrows indicate locations of primers (MLHsp60op-F and MLHsp60op-R) (see Table S3). Large arrows indicate directions of transcription. (c and d) Photographs of PCR amplicons from M. leprae cDNA of HSP70 and HSP60 operons, respectively. Lane 1, 1-kb DNA ladder (Promega); lane 2, M. leprae cDNA; lane 3, RT-negative control M. leprae RNA; lane 4, mouse cDNA (Clontech); lane 5, positive M. leprae DNA control; lane 6, negative buffer control.
Several heat stress response genes present in the genome of M. leprae also possessed candidate SigH/SigE promoter-like sequences in the region upstream of their coding sequences which were homologous to their counterparts in M. tuberculosis (Fig. 7a). In contrast, when the upstream region of the sigE of M. leprae was analyzed for the presence of a SigH RNAP promoter sequence, none was found. Further analysis showed that a SigH-dependent promoter sequence was located 42 bp downstream of the originally described start codon (ATG) within the coding sequence of sigE (Fig. 7b). A SigH-dependent promoter sequence of M. tuberculosis sigE has been demonstrated in a comparable location (18). The M. leprae genome was also shown to contain genes for the negative regulators of heat shock gene expression (HspR and HrcA). HAIR and CIRCE, regulatory DNA sequences to which HspR and HrcA bind, respectively, were also identified upstream of the M. leprae coding sequences of dnaK and groES and other M. leprae heat shock response genes (data not shown). These sequences were similar to those of M. tuberculosis.
FIG. 7.
(a) Comparison of putative promoter sequences upstream of selected M. leprae and M. tuberculosis sigma factor and heat shock response genes. Nucleotides representing −10 and −35 element sequences are shown in bold capitalized letters. Numbers indicate the number of nucleotides that the promoter sequence is upstream of the translational start codon of the respective gene. (b) Nucleic acid sequence for the M. leprae sigE gene, representing the original 5′ ATG translational start codon of the gene (18) and the newly identified putative −10 and −35 SigH binding regions as well as the newly identified 5′ ATG start codon and gene encoding the truncated SigE protein sequence.
DISCUSSION
At the level of promoter recognition and transcription initiation, survival of M. tuberculosis at elevated temperatures is controlled by the extracytoplasmic sigma factors SigH and SigE, acting directly to activate effectors of the heat stress response, as well as the stress response sigma factor SigB (15, 17, 18, 26, 27, 28). The result of this transcriptional response is characterized by global upregulation of at least 100 M. tuberculosis genes (28). These genes include those primarily involved with adaptation/detoxification (HSPR and HRCA heat shock regulons), heat shock regulators (HspR and HrcA), other previously identified heat shock-inducible genes (7, 15, 28), and a large number of genes which encode hypothetical proteins (28). Some of these genes (e.g., groES, dnaK, and clpB) encode heat shock response proteins that function as molecular chaperones, which ensure the appropriate folding, translocation, and assembly of polypeptides that are critical for the function of these proteins at elevated temperatures, or degradation of misfolded protein aggregates (5, 11, 14). Many of these genes are preceded by either a SigE-dependent RNAP promoter sequence (17, 23) or a SigH-dependent RNAP promoter sequence (18) and are negatively regulated by HspR and HrcA heat shock regulators (23, 28).
In the present study, we attempted to identify the molecular basis of the reduced survival of M. leprae at elevated temperatures. Under heat stress conditions (37°C or 42°C), there was no significant induction of the sigB and sigE stress response sigma factor genes or their heat shock regulons in this organism. It was reasoned that this defective heat shock response may result from a lack of functional genes that play key roles in this response. Even though sigE was not significantly induced under heat shock conditions in M. leprae, the present study suggested that this gene appears to possess a functional SigH-dependent heat shock promoter and other regulatory components and was capable of complementing the heat stress defect when expressed in mycobacterial sigE knockout mutants containing a functional SigB, SigH, and a full repertoire of heat shock proteins. The M. leprae sigE gene product was also able to restore the basal levels of sigB gene transcripts in the M. tuberculosis sigE knockout strain to that of the wild-type strain. These data strongly suggested that the SigE of M. leprae should be functionally capable of activating a heat stress response. In addition, bioinformatics data from this study suggested that the annotated sigE initiation codon from both mycobacterial species may be wrong and that the sigE translation likely starts from a second methionine codon.
No significant increases in sigB gene transcripts were observed during heat shock in M. leprae even though this alternative sigma factor was found to be virtually identical (96%) to that of M. tuberculosis. It possesses all of the functional motifs of SigB found in M. tuberculosis and an identical SigE/SigH-dependent promoter upstream of its coding sequence. These data strongly suggested that M. leprae sigB should be inducible and activate a stress response regulon similar to that of the SigB of M. tuberculosis. The absence of such activation in M. leprae suggests that M. leprae lacks the signaling mechanism to induce sigB transcription in response to heat stress.
We also found that when M. leprae sigE was expressed in a mycobacterial sigE/sigH knockout mutant, there was no significant increase in M. leprae's sigE transcription (data not shown), and the defective heat stress response of this mutant was not complemented by sigE. In contrast, M. leprae sigE was able to complement both M. smegmatis and M. tuberculosis sigE mutants. These data suggested that the lack of a functional sigH plays a role in the unresponsiveness of M. leprae's sigE during heat stress.
Inducible expression of sigE and sigB in mycobacteria has been shown to be regulated by SigH, and basal and inducible expression of sigB was shown to be dependent on both SigE and SigH (18). M. tuberculosis and M. smegmatis mutants lacking a functional sigH gene were found to be more susceptible to heat shock as well as other stress conditions (18, 28). These data indicated that SigH plays a central role in a network that regulates the heat stress response, allowing this organism to survive fluctuations in temperature. However, sigH is a pseudogene in M. leprae containing (i) no detectible translational start site, (ii) multiple deletions, including a 168-bp deletion of the 5′ region of the gene, (iii) no sequence similar to the consensus SigH promoter within 500 bp upstream of this pseudogene, and (iv) an in-frame stop codon, resulting from a frameshift mutation so that the encoded protein would lack both sigma 70 regions 2 and 4, which are required for promoter recognition (http://genolist.pasteur.fr/Leproma/). Our data and these sequence analyses thus strongly suggest that the absence of a functional SigH in M. leprae plays a role in the failure of sigE to be upregulated during certain stress responses and results in the unresponsiveness of M. leprae to heat stress. Unfortunately, this hypothesis could not be tested directly by introducing an intact mycobacterial sigH into M. leprae because of the lack of genetic tools and the inability to cultivate this bacterium axenically. Therefore, surrogate genetics were used to study the dependence of M. leprae SigE and SigB on a functional SigH to respond to heat stress conditions.
This study also demonstrated that the M. leprae genome lacks the majority of the heat-inducible genes found in M. tuberculosis. While the significance of this could not be directly studied, the large proportion of absent or nonfunctional heat shock-responsive genes suggested that the inability of M. leprae to survive at elevated temperatures may also be the result of inadequate effector mechanisms to respond to heat stress damage. For example, M. leprae lacks a functional hsp/acr2, the most highly induced heat stress response gene in M. tuberculosis and one that is essential for the protective heat stress response of this mycobacterial pathogen (25). In addition to hsp/acr2, the M. leprae genome lacks functional homologs for 4 others of the 10 most highly induced M. tuberculosis genes during heat stress. These data strongly suggested that M. leprae lacks a full repertoire of heat shock response genes.
M. leprae does possess HSP70 (dnaK) and HSP60 (groEL2) operons, which are among the most highly induced genes during heat shock in M. tuberculosis (28). In contrast, genes within these operons were not significantly induced in M. leprae during heat stress, even though they appeared to be intact. Both operons have candidate binding sequences for negative regulators (HAIR and CIRCE binding sequences for HspR and HrcA, respectively). In addition, HspR and HrcA appear to be expressed in M. leprae, suggesting that they are functional in this organism. Together, these data suggested that the HSP70 and HSP60 operons are intact and potentially regulated in response to stress. The lack of induction in M. leprae, however, indicates that signaling mechanisms for activating these regulons in response to heat stress are not functional in this organism. For the dnaK operon, this may result from the lack of sigH, as discussed above. For the groES-groEL operon (20), the differences in regulation between M. leprae and M. tuberculosis remain to be determined. Though our data indicate that the absence of SigH-dependent transcriptional activation of a heat stress response as well as the lack of heat shock effector genes are important in the inability of M. leprae to grow at high temperatures, other mechanisms may also play a role in temperature-restricted growth of this and other mycobacterial species.
In conclusion, the current study has provided insight into the molecular defects that likely contribute to M. leprae's reduced ability to survive at temperatures above 33°C. This defective heat stress response furthers our understanding of M. leprae's predilection for cooler peripheral regions of the human body, such as the dermal macrophages, macrophages infiltrating the aqueous humor and the iris of the eye, and Schwann cells of the peripheral nerves, where the temperature is more conducive to its survival and growth. The presence of M. leprae in these selected sites and the immunologic responses that this bacterium induces ultimately result in the characteristic pathology observed in leprosy.
Acknowledgments
We thank J. P. Pasqua and Michael Kearney for their excellent technical assistance and give special thanks to Thomas P. Gillis for his insightful discussions and excellent comments pertaining to the manuscript.
This project was partially funded by NIH/NIAID RO1 AI044856 (awarded to I.S.), RO1 AI37901 (to R.N.H.), and NIH/NIAID contract Y1 AI-2646-01.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Boily, P., and F. M. Knight. 2004. Cold-induced fever and peak metabolic rate in the nine banded armadillo (Dasypus novemcinctus). Physiol. Biochem. Zool. 77:651-657. [DOI] [PubMed] [Google Scholar]
- 2.Clements, B. R., and D. M. Scollard. 1996. Leprosy, p. 9.1-9.28. In G. L. Mandell and R. Fekety (ed.), Atlas of infectious diseases, vol. 8. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 3.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive decay of the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 4.Eiglmeier, K., J. Parkhill, J. N. Honore, T. Garnier, F. Tekaia, A. Telenti, P. Klatser, K. D. James, N. R. Thomson, P. R. Wheeler, C. Churcher, D. Harris, K. Mungall, B. G. Barrell, and S. T. Cole. 2001. The decaying genome of Mycobacterium leprae. Lepr. Rev. 72:387-398. [PubMed] [Google Scholar]
- 5.Fernandes, N. D., Q.-L. Wu, D. Kong, X. Puyang, S. Garg, and R. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzblau, S. G., A. N. Biswas, P. Jenner, and M. J. Colston. 1992. Double blind evaluation of BACTEC and Buddemeyer-type radiorespirometric assays for in vitro screening of antileprosy agents. Lepr. Rev. 63:125-133. [DOI] [PubMed] [Google Scholar]
- 7.Gomez, J. E., J. M. Chen, and W. Bishai. 1997. Sigma factors of Mycobacterium tuberculosis. Tuber. Lung Dis. 78:175-183. [DOI] [PubMed] [Google Scholar]
- 8.Hagge, D. A., S. Oby-Robinson, D. Scollard, G. McCormick, and D. L. Williams. 2002. A new model for studying the effects of Mycobacterium leprae on Schwann cell and neuron interactions. J. Infect. Dis. 186:1283-1296. [DOI] [PubMed] [Google Scholar]
- 9.Hagge, D. A., N. A. Ray, J. L. Krahenbuhl, and L. B. Adams. 2004. An in vitro model for the lepromatous leprosy granuloma: fate of Mycobacterium leprae from target macrophages after interaction with normal and activated effector macrophages. J. Immunol. 172:7771-7779. [DOI] [PubMed] [Google Scholar]
- 10.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 38:571-579. [DOI] [PubMed] [Google Scholar]
- 11.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 12.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395-405. [DOI] [PubMed] [Google Scholar]
- 13.Krahenbuhl, J. L., and L. B. Adams. 2003. The role of the macrophage in resistance to the leprosy bacillus, p. 281-302. In B. S. Zwilling and T. K. Eisenstein (ed.), Macrophage-pathogen interactions. Marcel Dekker Inc., New York, NY.
- 14.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 15.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 16.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor σ H in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:366-374. [DOI] [PubMed] [Google Scholar]
- 17.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σ E: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 18.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rees, R. J. 1985. The microbiology of leprosy, p. 31-52. In R. C. Hastings (ed.), Leprosy. Churchill Livingstone, New York, NY.
- 20.Rinke de Wit, T. F., S. Bekelie, A. Osland, T. L. Miko, P. W. Hermans, D. van Soolingen, J. W. Drijfhout, R. Schoningh, A. A. Janson, and J. E. Thole. 1992. Mycobacteria contain two groEL genes: the second Mycobacterium leprae groEL gene is arranged in an operon with groES. Mol. Microbiol. 6:1995-2007. [DOI] [PubMed] [Google Scholar]
- 21.Shetty, V. P., and N. H. Antia. 1997. Pathology of nerve damage in leprosy, p. 79-137. In N. H. Antia and V. P. Shetty (ed.), The peripheral nerve in leprosy and other neuropathies. Oxford University Press, Mumbai, India.
- 22.Shetty, V. P., K. Suchitra, M. W. Uplekar, and N. H. Antia. 1997. Higher incidence of viable Mycobacterium leprae within the nerve compared to skin among multibacillary leprosy patients released from multidrug therapy. Lepr. Rev. 68:131-138. [DOI] [PubMed] [Google Scholar]
- 23.Smith, I., W. Bishai, and V. Nagaraja. 2004. Control of mycobacterial transcription, p. 219-231. In S. Coe, D. N. McMurray, K. Eisenach, B. Gicquel, and W. Jacobs, Jr. (ed.), Tuberculosis, 2nd ed. ASM Press, Washington, DC.
- 24.Snapper, S., R. Melton, S. Mustafa, T. Kieser and W. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 25.Stewart, G. R., S. M. Newton, K. A. Wilkinson, I. R. Humphreys, H. N. Murphy, B. D. Robertson, R. J. Wilkinson, and D. B. Young. 2005. The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 55:1127-1137. [DOI] [PubMed] [Google Scholar]
- 26.Stewart, G. R., B. D. Robertson, and D. B. Young. 2004. Analysis of the function of mycobacterial DnaJ proteins by over-expression and microarray profiling. Tuberculosis (Edinburgh) 84:180-187. [DOI] [PubMed] [Google Scholar]
- 27.Stewart, G. R., V. A. Snewin, G. Walzl, T. Hussell, P. Tormay, P. O'Gaora, M. Goyal, J. Betts, I.N. Brown, and D. B. Young. 2001. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7:732-737. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Truman, R. W., and J. K. Krahenbuhl. 2001. Viable M. leprae as a research reagent. Int. J. Lepr. Other Mycobact. Dis. 69:1-12. [PubMed] [Google Scholar]
- 31.Williams, D. L., S. Oby-Robinson, T. L. Pittman, and D. Scollard. 2003. Purification of Mycobacterium leprae RNA for gene expression analysis directly from biopsy specimens of leprosy patients. BioTechniques 35:534-536, 538, 540-541. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization. 2003. World Health Organization Leprosy Elimination Project status report. http://www.who.int/lep/Reports/s20042.pdf.
- 33.Wu, Q. L., K. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]