Abstract
Enterococci account for nearly 10% of all nosocomial infections and constitute a significant treatment challenge due to their multidrug resistance properties. One of the well-studied virulence factors of Enterococcus faecalis is a secreted bacterial protease, termed gelatinase, which has been shown to contribute to the process of biofilm formation. Gelatinase belongs to the M4 family of bacterial zinc metalloendopeptidases, typified by thermolysin. Gelatinase is synthesized as a preproenzyme consisting of a signal sequence, a putative propeptide, and then the mature enzyme. We determined that the molecular mass of the mature protein isolated from culture supernatant was 33,030 Da, which differed from the predicted molecular mass, 34,570 Da, by over 1,500 Da. Using N-terminal sequencing, we confirmed that the mature protein begins at the previously identified sequence VGSEV, thus suggesting that the 1,500-Da molecular mass difference resulted from a C-terminal processing event. By using mutants with site-directed mutations within a predicted C-terminal processing site and mutants with C-terminal deletions fused to a hexahistidine tag, we determined that the processing site is likely to be between residues D304 and I305 and that it requires the Q306 residue. The results suggest that the E. faecalis gelatinase requires C-terminal processing for full activation of protease activity, making it a unique enzyme among the members of the M4 family of proteases of gram-positive bacteria.
Enterococcus faecalis is a gram-positive coccus that usually occurs in pairs or short chains and is commonly found as part of the resident flora in the mammalian intestinal tract (37). In contrast to the beneficial role that enterococci play in intestinal homeostasis, these organisms are becoming increasingly important to human health as leading causes of nosocomial infections. These infections include urinary tract and abdominal infections, bacteremia, endocarditis, and wound infections (1, 9, 29). E. faecalis is ubiquitous in the environment and can withstand high salt concentrations and wide pH and temperature ranges. Enterococci are also resistant to desiccation and temperatures up to 60°C. One of the most important public health aspects of enterococci is their increasingly wide range of antibiotic resistance (14). Multidrug resistance is common among clinical isolates, leaving few therapeutic options for treating enterococcal infections.
Some strains of E. faecalis can form biofilms, which may increase their ability to colonize patients and persist at infection sites (22). Biofilms are bacterial communities growing as surface-attached aggregates encased in an exopolymer matrix. Biofilms are generally thought to be more resistant to antibiotics than the corresponding free-living bacteria, which compounds the problem of antimicrobial resistance. Moreover, recent estimates suggest that over 65% of hospital-acquired infections stem from the ability of the infecting organism to produce biofilms (17).
As biofilm formation is thought to be an environmentally responsive process, we recently examined the contributions of two-component signal transduction networks to biofilm formation by E. faecalis V583 (11). We found that of the 17 two-component response regulator mutants tested, only the fsrA response regulator mutant was significantly attenuated in biofilm formation. FsrA is part of a signal transduction system that is responsive to the accumulation of an 11-amino-acid peptide lactone, termed gelatinase biosynthesis-activating pheromone (24). This peptide is encoded by the fsrD gene and is thought to be proteolytically processed from the FsrD protein and exported by the fsrB gene product (25), and its presence in a culture is sensed by the FsrC histidine kinase. Gelatinase biosynthesis-activating pheromone sensing by the FsrC sensor kinase results in activation of the FsrA response regulator transcription factor by a phosphoryl group. The phosphorylated form of FsrA is believed to activate expression of two cotranscribed protease genes, gelE and sprE (28). Additionally, the FsrABC system was shown by microarray analysis to affect a variety of other genes, some of which potentially are involved in virulence and some of which are involved in metabolic pathways (2). As the GelE and SprE proteases are Fsr-regulated proteins, we explored the contribution of the E. faecalis proteases gelatinase and serine protease to biofilm formation (12). We found that the biofilm deficiency of an fsrA mutant was due to an inability to make gelatinase. Complementation of the fsrA mutant by gelatinase expressed from an fsr-independent promoter restored biofilm formation. Addition of purified gelatinase also restored biofilm formation to fsr-defective strains, suggesting a critical, but as-yet-unknown, role for gelatinase in biofilm formation by E. faecalis. The involvement of GelE in biofilm formation was also reported by Kristich et al. (16).
GelE was originally characterized by Mäkinen et al. (18) as an extracellular Zn metalloprotease with activity against a number of substrates, including the insulin B chain, azo dye-impregnated collagen (azocoll), and the pheromones and inhibitor peptides involved in conjugative plasmid transfer in E. faecalis. GelE was also shown to function in clearing the bacterial cell surface of misfold proteins and in activating an autolysin (41). Gelatinase is synthesized as a 509-amino-acid prepropolypeptide which is subject to cleavage of the 192 amino acids at the amino-terminal end comprising the presequence or signal sequence and the prosequence.
In this study, an apparent difference between the predicted and actual molecular masses of the purified gelatinase revealed that gelatinase undergoes C-terminal proteolytic processing for full maturation. The role of this processing in gelatinase activity was examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. faecalis strains and plasmids used in this study are listed in Tables 1 and 2. Oligonucleotide primers used for plasmid construction are shown in Table S1 in the supplemental material. Plasmid pML28 was constructed from the vector pAT28 (39) by inserting a 369-bp EcoRI-BamHI fragment carrying the promoter of the aphA-3 kanamycin resistance gene (40). The gelE coding sequence and its ribosome binding site were then cloned in pML28 as a BamHI fragment to obtain pML29 (12). For construction of pML33, the mature gelE coding sequence was first cloned as an NdeI-XhoI fragment in the pET21 expression vector (Novagen) using oligonucleotide primers MGelNT-NdeI and MGelCT-XhoI, yielding plasmid pML52, which encoded a GelE protein with a six-His tag extension. A fragment of this plasmid was generated by PCR amplification using oligonucleotide primers MGelNT-Nde and pET20downPst (the latter primer hybridizes to the pET21 vector in a region downstream of the six histidine codons). This fragment was digested with NcoI, which naturally occurs in the gelE sequence, and cloned in the pML29 vector digested with NcoI and SalI (the latter enzyme blunted with Klenow polymerase) to replace the wild-type 3′ end of the gene with the histidine tag-modified version. Plasmid pML34 was constructed by cloning the PCR amplification product obtained with oligonucleotide primers GelE5′ and GelECT-14BamHI, digested with BamHI, in the BamHI site of plasmid pML28. Plasmid pML37 was derived from pML29 in which the 3′ end of the wild-type gene was replaced by the 3′ end of a gelE gene missing the last 14 amino acids but fused to six histidine codons. This construct was first generated in the pET21 expression vector (Novagen) via BamHI-XhoI cloning of a PCR fragment amplified with oligonucleotides MGelENT-NdeI and GelECT-14XhoI, yielding plasmid pML56. Plasmid pML56 was used as a template for PCR amplification with oligonucleotide primers NGelENT-NdeI and pET20downPst. The resulting fragment was digested with NcoI and ligated to pML29 digested with XbaI (treated with Klenow polymerase to obtain a blunt end) and NcoI. Plasmids pML42 and pML43 were constructed by cloning PCR-amplified fragments obtained with oligonucleotide primers MGelNT-NdeI and GelEIHis2 and with oligonucleotide primers MGelNT-NdeI and GelEQHis2, respectively, digested with NcoI and XbaI in similarly digested pML29. PCR products were obtained by amplification of E. faecalis V583 genomic DNA.
TABLE 1.
E. faecalis strains used in this study
| Strain | Relevant genotype | Relevant phenotype | Origin |
|---|---|---|---|
| V583 | Parental | GelE+ | Clinical isolate |
| FA2-2 | fsrC | GelE− | Lab strain |
| EF 129 | fsrC aphA-3 promoter | GelE− Spcr | pML28 → FA2-2 |
| EF 130 | fsrC aphA-3 promoter-gelE | GelE++ Spcr | pML29 → FA2-2 |
| EF 131 | fsrC aphA-3 promoter-gelE (His tag) | GelE++ Spcr | pML33 → FA2-2 |
| EF 132 | fsrC aphA-3 promoter-gelE (C-terminal 14 amino acids) | GelE++ Spcr | pML34 → FA2-2 |
| EF 133 | fsrC aphA-3 promoter-gelE (C-terminal 14 amino acids, His tag) | GelE++ Spcr | pML37 → FA2-2 |
| EF 134 | fsrC aphA-3 promoter-gelE (C-terminal 13 amino acids, His tag) | GelE++ Spcr | pML42 → FA2-2 |
| EF 135 | fsrC aphA-3 promoter-gelE (C-terminal 13 amino acids, His tag) | GelE++ Spcr | pML43 → FA2-2 |
| EF 136 | fsrC aphA-3 promoter-gelE(E303P) | GelE++ Spcr | pML38 → FA2-2 |
| EF 137 | fsrC aphA-3 promoter-gelE(D304P) | GelE++ Spcr | pML39 → FA2-2 |
| EF 138 | fsrC aphA-3 promoter-gelE(I305P) | GelE++ Spcr | pML40 → FA2-2 |
| EF 139 | fsrC aphA-3 promoter-gelE(Q306P) | GelE+ Spcr | pML41 → FA2-2 |
| EF 140 | fsrC aphA-3 promoter-gelE(E137Q) | GelE− Spcr | pML44 → FA2-2 |
| EF 141 | fsrC aphA-3 promoter-gelE(E137Q) (His tag) | GelE− Spcr | pML45 → FA2-2 |
TABLE 2.
Plasmids used in this study
| Vector | Description | Predicted gelatinase C-terminal sequence |
|---|---|---|
| pML28a | Shuttle vector with aphA-3 promoter | |
| pML29b | GelE | GAKEDIQVNQPSESVLVNE |
| pML33 | GelE plus His6 tag | GAKEDIQVNQPSESVLVNELEHHHHHH |
| pML34 | GelE without C-terminal 14 amino acids | GAKED |
| pML37 | GelE without C-terminal 14 amino acids plus His tag | GAKEDLEHHHHHH |
| pML42 | GelE without C-terminal 13 amino acids plus His tag | GAKEDILEHHHHHH |
| pML43 | GelE without C-terminal 12 amino acids plus His tag | GAKEDIQLEHHHHHH |
| pML38 | GelE with glutamic acid-to-proline substitution at position 303 | GAKPDIQVNQPSESVLVNE |
| pML39 | GelE with aspartic acid-to-proline substitution at position 304 | GAKEPIQVNQPSESVLVNE |
| pML40 | GelE with isoleucine-to-proline substitution at position 305 | GAKEDPQVNQPSESVLVNE |
| pML41 | GelE with glutamine-to-proline substitution at position 306 | GAKEDIPVNQPSESVLVNE |
| pML44 | GelE with glutamate at position 137 mutated to Gln | GAKEDIQVNQPSESVLVNE |
| pML45 | GelE with glutamate at position 137 mutated to Gln plus His6 tag | GAKEDIQVNQPSESVLVNELEHHHHHH |
Strains were cultured in Todd-Hewitt broth (THB) or M17 medium (Difco Laboratories). Escherichia coli XL10 Gold (Stratagene) was used for plasmid construction and propagation. Strains were cultivated in Luria-Bertani broth. The antibiotics used for selection in E. coli and E. faecalis were spectinomycin (150 and 750 μg/ml, respectively) and tetracycline (15 μg/ml). Screening for gelatinase production was carried out on THB agar plates containing 1.5% skim milk. Electroporation was carried out as described previously (5).
Site-directed mutagenesis.
Plasmid pML29 was subjected to site-directed mutagenesis using a QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and mutagenic primers to replace residues surrounding the putative C-terminal processing site with proline or the active site E137 with glutamine (see Table S1 in the supplemental material). Plasmid pML33 was used as template to generate pML45.
Gelatinase purification.
For all GelE proteins, most of the purification steps were performed at 4°C; the only exception was hydrophobic interaction chromatography, which was performed at room temperature. The procedure used was the procedure described by Hancock and Perego (12).
Protein determination, electrophoretic techniques, and Western blotting.
The protein concentration was determined with a bicinchoninic acid reagent kit (Pierce), using bovine serum albumin as a standard. Enzyme purification was monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a 12% polyacrylamide separating gel. Concentrated (20×) culture supernatants from overnight cultures were run on SDS-PAGE gels and stained with Coomassie blue or transferred to a polyvinylidene difluoride membrane. GelE was detected with anti-His (C-terminal) monoclonal antibody (Novagen) using an ECL Western blotting detection kit (Amersham Biosciences).
Protease activity assay.
Azocoll (18) was suspended at a final concentration of 5 mg/ml in the assay buffer (50 mM Tris [pH 7.8], 1 mM CaCl2) and incubated for 2 h at 37°C with vigorous shaking. The solution was filtered (Whatman 1MM paper), and the precipitate was resuspended immediately in the same volume of fresh buffer. Stirring was rapid enough to obtain what appeared to be a uniform suspension. Tubes (diameter, 13 mm) were prewarmed at 37°C for 15 min before the reaction was initiated by addition of enzyme. Assays were stopped by immersing tubes in an ice-water bath. Chilled tubes were then centrifuged, and the optical densities at 550 nm of supernatant fractions that were diluted 1:2 were measured. Data were obtained for every time point in triplicate.
Molecular mass determination.
The molecular masses of the native protease and mutants of this protease were determined using matrix-assisted laser desorption ionization—time of flight mass spectrometry. The analysis was performed at the Mass Spectroscopy Core Facility at The Scripps Research Institute.
Biofilm formation.
Biofilm formation on polystyrene was quantified as previously described (38). Briefly, E. faecalis strains were grown overnight at 37°C in M17 medium supplemented with 0.5% lactose and spectinomycin (500 μg/ml) when appropriate. Each culture was diluted 1:100 in M17 medium, and 200 μl of the resulting cell suspension was used to inoculate sterile 96-well flat-bottom polystyrene microtiter plates. After incubation for 24 h at 37°C, wells were gently washed three times with 200 μl of phosphate-buffered saline, dried in the inverted position, and stained with 1% crystal violet for 15 min. The wells were rinsed again, and the crystal violet was solubilized in 200 μl ethanol-acetone (80:20, vol/vol). The optical density at 550 nm was determined using a microplate reader. Each assay was performed in triplicate and repeated three times.
Autolysis assay.
E. faecalis FA2-2 strains containing pML28, pML29, pML34, and pML41 were grown overnight at 37°C in 2.5 ml THB containing 600 μg/ml of spectinomycin. Twenty-five microliters of each overnight culture was inoculated into 2.5 ml SM17 medium containing spectinomycin and 3% glycine (the appropriate final concentration of glycine in the medium was obtained using a filter-sterilized 25% glycine stock solution) and grown overnight at 37°C. Then 1.5 ml of the overnight culture was centrifuged at 13,000 rpm for 3 min. The supernatants were discarded, and the pellets were washed three times in ice-cold sterile distilled water. After the third wash, the pellets were resuspended in 10 mM sodium phosphate buffer (pH 6.8). Two hundred microliters of the suspended cells was dispensed into a 96-well plate, and the optical density at 600 nm at 37°C was determined for 9 h at 30-min intervals. A statistical analysis (Student's t test) was carried out with the SAS program.
RESULTS
Molecular mass of mature gelatinase.
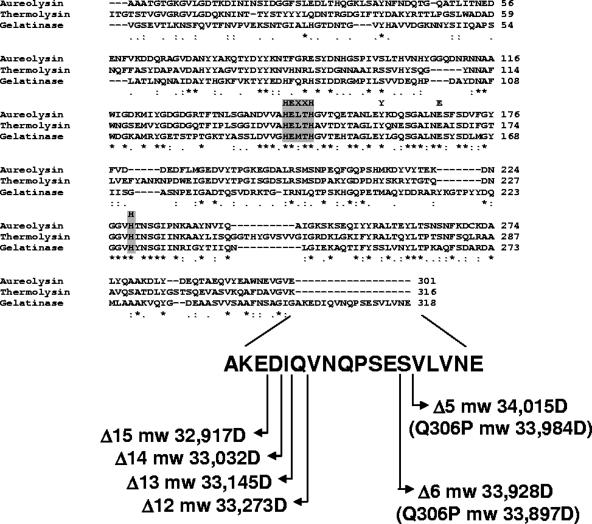
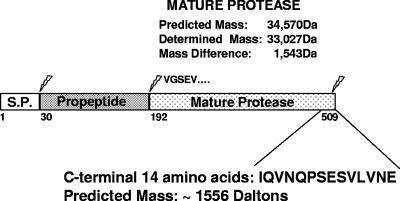
The E. faecalis protease gelatinase belongs to the M4 family of bacterial zinc metallopeptidases typified by thermolysin of Bacillus thermoproteolyticus (27) and aureolysin of Staphylococcus aureus (30). An amino acid sequence alignment of the mature portions of thermolysin, aureolysin, and gelatinase (Fig. 1) revealed that gelatinase had an 18-amino-acid extension at the carboxy-terminal end. Some M4 family members from bacteria belonging to the family Vibrionaceae require processing at the carboxy terminus, in addition to two processing events at the amino terminus, to become fully active (15, 23). For this reason we investigated the possibility that the enterococcal gelatinase undergoes a carboxy-terminal processing event by determining the molecular weight of the mature enzyme purified from the culture supernatant. As shown in Fig. S1 in the supplemental material, mass spectrometry analysis of purified gelatinase revealed a molecular mass of 33,030 Da, which was 1,540 Da less than the predicted molecular mass of the protein (34,570 Da) based on its deduced amino acid sequence. N-terminal sequencing revealed that the purified, mature protein began with the previously determined sequence “VGSEV” (36) (data not shown), suggesting that the molecular mass difference had to result from a C-terminal processing event(s) that, based on the expected molecular masses of proteins lacking the last 12 to 15 amino acids, is consistent with cleavage of the last 14 amino acids from the carboxy-terminal end (Fig. 2).
FIG. 1.
Amino acid sequence alignment of members of the M4 family of bacterial metalloproteases. The amino acid sequences of aureolysin of S. aureus, thermolysin of B. thermoproteolyticus, and gelatinase of E. faecalis were aligned using the ClustalW program. The active site residues are indicated by the gray boxes. Symbols: asterisks, identical residues in all sequences; colons, conserved substitutions; periods, semiconserved substitutions. The sequence of the C-terminal 18 amino acids is shown at the bottom along with the calculated molecular masses of GelE mutant proteins lacking the last 5 (Δ5), 6 (Δ6), 12 (Δ12), 13 (Δ13), 14 (Δ14), and 15 (Δ15) amino acids. The predicted molecular masses (mw) were obtained with the ExPASy pI-Mw tool. The predicted molecular masses of the GelE Q306P mutant proteins lacking the last five or six amino acids are indicated in parentheses. D, daltons.
FIG. 2.
Schematic representation of gelatinase proteolytic processing. The gelE gene encodes a 509-amino-acid protein with a probable signal peptide (S.P.) that is 29 amino acids long. The propeptide sequence extends from residue 30 to residue 191, and the mature gelatinase starts at residue V192. The predicted masses of the mature protein and C-terminal peptide were determined with the ExPASy pI-Mw tool. The molecular weight of the mature protein purified from culture supernatants was determined by mass spectrometry.
Gelatinase is processed at the C-terminal end.
In order to determine the site of processing of the gelatinase enzyme, we expressed the wild-type protein and C-terminal deletion mutants from an FsrA-independent promoter (aphA-3 [39]) on a replicative vector in the FA2-2 strain, which is gelatinase deficient due to the absence of a functional fsr system (12) (Table 2).
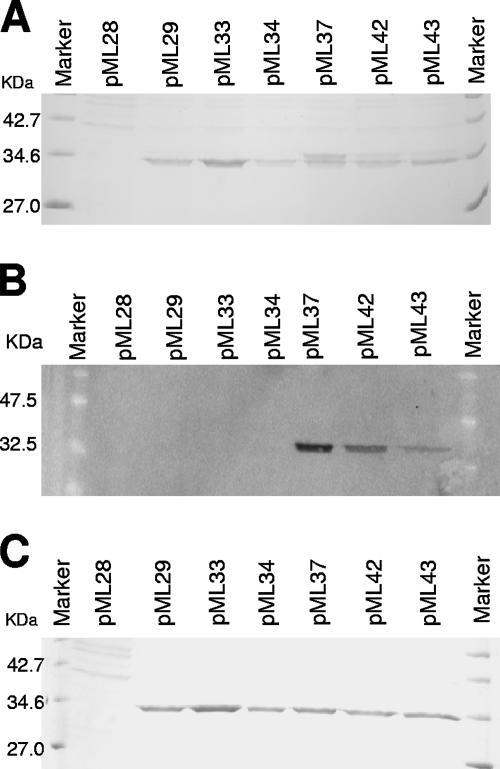
Complementation of the gelatinase-deficient phenotype was first tested on THB-milk agar plates (data not shown) in order to ensure that an active gelatinase was expressed. Then 20×-concentrated culture supernatants were analyzed for the presence of gelatinase protein by SDS-PAGE and Western blotting. As shown in Fig. 3A, while strain FA2-2 carrying the pML28 vector, which contained only the aphA-3 promoter, did not produce gelatinase, the strain carrying plasmid pML29, which expressed the wild-type gelE gene, secreted a protein with a molecular mass of approximately 34 kDa. A protein of similar size was produced by strains carrying plasmids pML33 and pML34, which expressed a wild-type GelE protein with a six-histidine extension at the C-terminal end and a GelE protein lacking the C-terminal 14 amino acids, respectively. The culture supernatant of the strain carrying plasmid pML37 (expressing a GelE protein lacking the C-terminal 14 amino acids but with a six-His tag fused to the D304 residue via a Leu-Glu linker) produced a protein band that was the same size as the band observed in the samples described above and an additional band at a slightly higher molecular weight. A similar band pattern was obtained with the culture supernatants of the strains carrying plasmids pML42 and pML43, which expressed GelE fused to the six-His tag at the I305 and Q306 residues, respectively. Notably, the concentration of the higher-molecular-weight band relative to the lower-molecular-weight band decreased in the order pML37 > pML42 > pML43. Analysis of the same supernatants by Western blotting using an anti-C-terminal His tag antibody revealed that only the truncated GelE proteins expressed by plasmids pML37, pML42, and pML43 maintained the histidine tag fused to the D304, I305, and Q306 residues, respectively (Fig. 3B). The intensities of the bands detected corresponded to the intensities of the higher-molecular-weight bands observed in the Coomassie blue-stained gel shown in Fig. 3A. (i.e., pML37 > pML42 > pML43). Since no His tag was detected in the supernatant of the strain expressing the GelE wild-type protein with the histidine tag fused to the last residue (E318; pML33), these results indicated that maturation of GelE occurred via cleavage at the C-terminal end and that this process was progressively delayed by deletion of the last 12 to 14 amino acids. Nevertheless, incubation of the same proteins for 12 h at 37°C resulted in the presence of only one form of GelE, corresponding to the mature form, in all preparations (Fig. 3C). This indicated that maturation could eventually occur even when the C-terminal 14 amino acids were replaced by the L-E-H6 amino acid sequence.
FIG. 3.
Analysis of the protein profiles for the culture supernatants of GelE derivatives. The supernatants were concentrated 20-fold and loaded on a 12% polyacrylamide gel for Coomassie blue staining (A) or for Western blot analysis using anti-C-terminal His tag antibody (B). (C) Protein profile of the same supernatants after overnight incubation al 37°C.
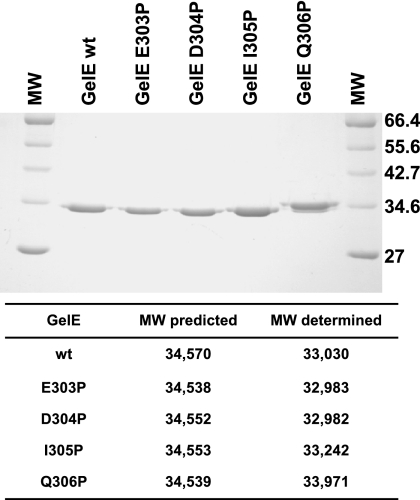
Proline substitution at Q306 affects gelatinase activity.
Mass spectrometry determination of the molecular mass of mature wild-type gelatinase from culture supernatant revealed a discrepancy of approximately 1,500 Da with the predicted value (Fig. 2). Because the predicted molecular mass of the last 14 amino acids at the carboxy-terminal end of gelatinase is 1,556 Da (Fig. 2), we investigated whether any of the residues at positions 16 (E303), 15 (D304), 14 (I305), and 13 (Q306) from the C-terminal end had a critical role in the processing of the protease. These four residues were individually changed to proline by site-directed mutagenesis, and the resulting proteins were purified from culture supernatants of strain FA2-2 together with the wild-type form of GelE. SDS-PAGE analysis of 1 μg of each protein revealed that while the GelE E303P, D304P, and I305P mutant proteins had an electrophoretic mobility equal to that of the wild-type protease, the mobility of the Q306P mutant protein was slightly retarded (Fig. 4). A mass spectrometry analysis carried out with the five proteins indicated that the E303P and D304P proteins had molecular masses in the range of the molecular mass obtained for the wild-type protein, suggesting that these substitutions did not affect the processing of the C-terminal end. The I305P mutant protein seemed to be approximately one amino acid larger than the wild-type GelE, while the molecular weight of the Q306P protein indicated that five or six additional amino acids were present compared to the wild-type protein (Fig. 4).
FIG. 4.
Analysis of the electrophoretic mobility of GelE proline substitution mutant proteins. Gelatinases purified from culture supernatants of strain FA2-2 expressing the wild-type protein (GelE wt) and the E303P, D304P, I305P, and Q306P mutant proteins were run on a 12% SDS-PAGE gel and stained with Coomassie blue. The broad range protein marker (New England Biolabs) (lanes MW) was included in the analysis. The molecular weights of the five proteins were determined by matrix-assisted laser desorption ionization—time of flight mass spectrometry. MW, molecular weight.
These results suggested that a proline at position 305 may have a role in positioning the cleavage site but that Q306 is the most critical residue for processing of the 14 C-terminal amino acids. Replacement of Q306 with proline in fact resulted in an alternative cleavage event that most likely deleted the C-terminal five amino acids.
Activity of GelE mutants with synthetic substrates and in biofilm formation.
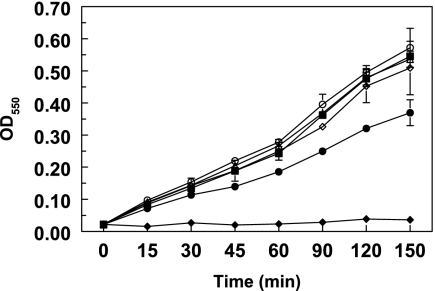
In order to compare the activities of the four site-specific mutants of GelE (E303P, D304P, I305P, and Q306P), the purified proteins were assayed in three independent experiments using azocoll as a substrate. All proteinases were still able to degrade azocoll, but the Q306P mutant protein was consistently less active than the other proteins (Fig. 5), showing a 30% reduction in protease activity. The results suggested that correct processing of the last 14 amino acids at the C-terminal end of GelE is necessary for full activity of the protein.
FIG. 5.
Hydrolysis of azocoll by GelE and site-specific mutants as a function of time measured by the absorbance of released azocoll dye. The assay was carried out with FA2-2 strains carrying plasmids pML28 (vector alone) (♦), pML29 (GelE wild type) (▪), pML38 (GelE E303P) (▵), pML39 (GelE D304P) (○), pML40 (GelE I305P) (⋄), and pML41 (GelE Q306P) (•). The data are the averages of two independent experiments; the error bars indicate the standard deviations. Each data point is the average of triplicate measurements. OD550, optical density at 550 nm.
The previously described role of gelatinase in biofilm formation prompted us to test whether the proline substitutions had any effect on biofilm formation. As shown in Fig. S2 in the supplemental material, none of the mutant proteins affected the ability of strain FA2-2 to form biofilms, but the sensitivity of the assay may not have been high enough to detect the small enzymatic differences between these proteins.
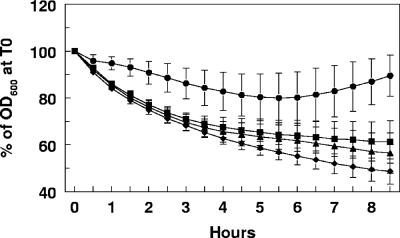
GelE Q306P mutation affects cell autolysis.
GelE has been shown to be involved in the maturation of the E. faecalis muramidase-1 autolysin in vitro (33), and GelE-producing strains have been shown to have increased autolysis (41). For these reasons we investigated whether processing of the C-terminal end of gelatinase had any effect on cell autolysis. Strains derived from FA2-2 carrying plasmid pML28 (vector), pML29 (GelE wild type), pML34 (GelE-CT14), or pML41 (GelE Q306P) were subjected to an autolysis assay as described in Materials and Methods. The results (Fig. 6) consistently and reproducibly indicated that the strain expressing the GelE Q306P mutant protein had a slightly higher rate of autolysis than the strain expressing the GelE wild-type protein (P < 0.05). Notably, the strain expressing the GelE protein lacking the last 14 amino acids showed a slightly lower, although not statistically significant, level of autolysis than the wild-type protein-expressing strain (P < 0.36). Consistent with the results reported by Waters et al. (41), the strains expressing GelE had a higher autocatalytic rate than the control strain not expressing the enzyme.
FIG. 6.
Autolysis assay with FA2-2 strains expressing gelatinase. The autolysis assay was carried out with FA2-2 strains carrying plasmids pML28 (vector alone) (•), pML29 (GelE wild type) (▴), pML34 (GelE-CT14aa) (▪), and pML41 (GelE Q306P) (♦). Samples were analyzed at 30-min intervals as described in Materials and Methods. OD600, optical density at 550 nm; T0, time zero.
These results suggest that although the Q306P mutation reduced the protease activity of GelE, the inability to process the C-terminal end increased the ability of the enzyme to access or activate the target autolysin.
Active site mutation affects the GelE N- and C-terminal processes.
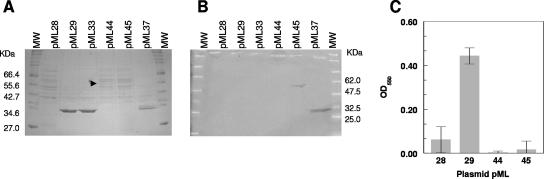
As a member of the M4 family of zinc metalloproteases, gelatinase possesses a conserved catalytic domain typified by the primary-sequence HEXXH motif responsible for coordinating zinc in the active site (Fig. 1) (13). In order to investigate the role of GelE activity in its maturation, the glutamic acid residue at position 137 in the active site was mutated to glutamine and the protein was expressed in strain FA2-2 from the aphA-3 promoter of plasmid pML28 as described above. The resulting construct, designated pML44, was also modified to express an E137Q GelE mutant protein with six His residues at the C-terminal end (plasmid pML45). The concentrated supernatants of cultures expressing these proteins were run on an SDS-PAGE gel and evaluated by Western blotting (Fig. 7). The site-specific E137Q mutation led to complete abolition of GelE activity as detected by protease activity on THB-milk plates (data not shown). Electrophoretic analysis of the culture supernatants revealed the disappearance of the mature-size protein and the concomitant accumulation of an approximately 55-kDa protein which was present at a lower level than the wild-type enzyme (expressed by plasmid pML29) (Fig. 7A), indicating that active GelE was necessary for posttranslational modification of this protein (i.e., cleavage of the propeptide). Western blot analysis carried out with an anti-C-terminal His tag antibody confirmed the presence of an approximately 55-kDa His-tagged protein in the supernatant of the cells carrying the pML45 construct, while a His-tagged protein that was the size of mature GelE was present, as expected, in the supernatant of the strain carrying plasmid pML37, as shown in Fig. 3. These results suggested that an active GelE protein was required for the multiple maturation processes at the N- and C-terminal ends involving an autocatalytic mechanism. Similar results were reported for thermolysin (19) and the Streptomyces cacaoi extracellular neutral metalloprotease Npr (4). In contrast, mutations of the zinc-binding metalloprotease motif of the LasA protease of Pseudomonas aeruginosa or the BFT toxin of Bacteroides fragilis, which belong to different protease families, were shown to affect the activity but not the propeptide processing (8, 10).
FIG. 7.
Analysis of GelE E137Q protein secretion and biofilm formation. The culture supernatants were concentrated 20-fold and loaded on a 12% polyacrylamide gel for Coomassie blue staining (A) or for Western blot analysis using anti-C-terminal His tag antibody (B). The molecular weight standards were the New England Biolabs broad range protein marker (A) and the broad range prestained protein marker (B). The arrowhead indicates the full-length GelE protein band. (C) Analysis of biofilm formation by FA2-2 strains carrying plasmids expressing wild-type GelE (pML29), GelE E137Q (pML44), and His-tagged GelE E137Q (pML45). The control strain carried only the vector pML28. The error bars indicate the standard deviations. OD550, optical density at 550 nm.
The availability of the active site mutant GelE proteins and evidence of their expression and secretion in the culture supernatant allowed us to test whether the E137Q substitution affected biofilm formation. The FA2-2 strains carrying the pML28 vector alone, the pML29 vector expressing wild-type GelE, and the pML44 and pML45 plasmids expressing the E137Q mutant protein with and without a six-His tag, respectively, were assayed for biofilm formation as described in Materials and Methods. The results (Fig. 7C) indicated that while expression of wild-type GelE induced biofilm growth, expression of the E137Q mutant proteins did not. This suggested that the protease activity, rather than the GelE protein itself, is required for biofilm formation.
DISCUSSION
The zinc metalloprotease secreted by E. faecalis, gelatinase, is one of the few characterized virulence factors of this organism. Gelatinase has been shown to contribute to disease pathogenesis in a number of model systems, including peritonitis in mice (34), endophthalmitis in rabbits (7), and nematode killing (34). In addition to gelatin, only a few substrates are known for this protease. The list of known substrates includes the enterococcal conjugative sex pheromones and a number of host-derived in vitro substrates, including the insulin B chain, endothelin, hemoglobin, fibrinogen, fibronectin, collagen, and laminin (18). In addition, gelatinase has been shown to interfere with innate immune defenses through inactivation of the antibacterial peptides LL-37 and α-defensin (31, 32). Recently, gelatinase has been shown to be relevant for in vitro translocation of E. faecalis across polarized human enterocyte-like T84 cells (42).
As a member of the M4 family of zinc metalloproteases, gelatinase possesses a conserved catalytic domain typified by the primary-sequence HEXXH motif responsible for coordinating zinc in the active site (13). In addition, these zinc metalloproteases bind three or four calcium atoms, which are thought to stabilize the overall structure; the more thermal stable proteases bind four calcium atoms.
Members of the M4 family are secreted bacterial enzymes that are synthesized as preproenzymes. The presequence, or signal peptide, targets the proenzyme to the secretion apparatus of either gram-negative or gram-positive cells. Maturation of the secreted proenzyme to its active mature form is thought to proceed by autocatalytic processing and transient association of the propeptide or prosequence with the active form (19, 26). The primary purpose of the propeptide is to function as an intramolecular chaperone facilitating the proper folding of the active protease (20, 21).
Here we showed that an active gelatinase is required for processing of the propeptide at the amino terminus and cleavage of the C-terminal tail. Mutation of a critical residue in the GelE active site (E137) resulted in secretion of a protein whose molecular weight approximated the molecular weight of the gelatinase proenzyme and that retained the carboxy-terminal histidine tag. This is reminiscent of the previously reported autocatalytic processing of thermolysin (19). However, autoprocessing of GelE may not necessarily be an intramolecular event, as reported for thermolysin (19). In supernatants of strain FA2-2 expressing the GelE E137Q His-tagged protein, we observed disappearance of the histidine tag upon addition of purified wild-type GelE, as determined by Western blot analysis with an anti-C-terminal His tag antibody (see Fig. S4 in the supplemental material). This suggested that at least the C-terminal processing event may not be an intramolecular event.
In addition to the amino-terminal processing, some M4 family members from bacteria belonging to the family Vibrionaceae also require processing at the carboxy terminus to become fully active (15, 23). Here we show that the E. faecalis gelatinase also requires proteolytic processing at the carboxy terminus, which makes it unique among members of the M4 family from gram-positive bacteria. Determination of the precise molecular mass of GelE purified from E. faecalis culture supernatants resulted in a value (33,030 Da) in good agreement with the original estimates of Mäkinen et al. obtained by using SDS-PAGE (33,000 Da), sizing column analysis (32,000 Da), or fast protein liquid chromatography (31,500 Da) (18).
Our investigation of the reason for the approximately 1,500-Da discrepancy with the predicted molecular mass of gelatinase indicated that a processing event takes place at the carboxy-terminal end, most likely between the D304 and I305 residues, and this requires the Q306 residue. As mentioned above, evidence suggested that GelE itself may be involved in the processing. An isoleucine residue is not as common as leucine on the imino side of the scissile bond of known substrates, but such a residue was found for angiotensin and neurotensin, providing further support for the autocatalytic hypothesis (18). Additionally, a hydrophobic amino acid residue has often been found at this position. Furthermore, the molecular weight of the I305P mutant protein suggests that there is an alternative processing event, possibly between Q306 and V307; this could also be a gelatinase processing site as glutamine at the carboxylic side and leucine at the imino side of the bond were found in the gelatinase processing site of the insulin B chain (18).
Nevertheless, additional proteases, such as the coregulated enzyme encoded by sprE, may contribute to the C-terminal processing, as suggested by the molecular weight of the Q306P mutant protein, for which cleavage between the E312 and S313 or S313 and V314 residues must have occurred to explain the molecular weight obtained by mass spectrometry. Singh et al. recently showed that in the absence of a functional fsr system, a basal level of GelE and SprE is produced; thus, we cannot rule out the possibility that a basal level of SprE contributes to GelE maturation in strain FA2-2 (35).
The role of the Q306 residue in the processing of the C-terminal end of GelE is clearly critical for efficient cleavage, as observed with the wild-type protein; in the purification conditions used in this study, this protein is found essentially fully processed, while the Q306P mutant protein was processed at an alternative site, resulting in a protein with a slightly higher molecular weight than the wild-type protein (Fig. 4). Nevertheless, the GelE-CT14 His-tagged variant seemed to be processed to a molecular weight essentially identical to that of the wild-type protein (Fig. 3) despite the lack of the Q306 and I305 residues, which were replaced by Glu and Leu residues (introduced by the XhoI cloning site), respectively, that linked the protease to six histidine residues. However, this processing event was not highly efficient, and its efficiency seemed to increase when the I305 residue alone or the I305 and Q306 residues were still present (pML42 and pML43) (Fig. 3A and B), again suggesting a role for these residues in proper cleavage. Further studies of the biochemical properties of GelE are required to define the mechanism of this autocatalytic event.
The exact role of the GelE C-terminal processing event in the overall activity of the protease appears to be marginal because the Q306P mutant protein was only 25 to 30% less active than the wild-type proteins in the protease assay and it did not have a significant effect on biofilm formation. The C-terminal processing event, however, may delay maturation of the protein, and this may affect protease activity in some specific environmental conditions.
The observation that the Q306P mutant protein had a higher rate of autolysis (Fig. 6) raised the possibility that this GelE variant may remain more tightly associated with the cell membrane or cell wall, perhaps due to the slightly hydrophobic nature of the 14-amino-acid C-terminal tail. This could favor processing and maturation of the autolysin substrate of GelE, muramidase-1 (33), giving rise to the observed phenotype. Thus, processing of the C-terminal tail of gelatinase may have a regulatory role in cell division through the regulation of the autolysin's activity.
It is possible that the C-terminal 14 amino acids have a secondary signaling function. The role of the conjugative peptide pheromones in E. faecalis, which arise from processing of a bacterial signal peptide sequence of surface lipoproteins, is well characterized and provides a precedent for secondary functions for processed peptide sequences. However, so far there is no evidence for such a role for the GelE C-terminal peptide (3, 6). There is also no evidence that the C-terminal 14 amino acids of GelE have a distinct function in the protein other than slightly inhibiting gelatinase activity, as reported for the Vibrio cholerae non-O1 hemagglutinin/protease, whose processing of the C-terminal 2 kDa results in increased protease activity but decreased hemagglutinin activity (23). Thus, the precise role of this domain in the biology of Enterococcus awaits elucidation.
Acknowledgments
This research was supported in part by Public Health Service grant AI052289 from the National Institute of Allergy and Infectious Diseases. The Stein Beneficial Trust partially supported oligonucleotide synthesis and DNA sequencing.
We acknowledge Paula Oliviera for her technical contributions.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
Manuscript 19055 from The Scripps Research Institute.
REFERENCES
- 1.Bonten, M. J., M. K. Hayden, C. Nathan, J. van Voorhis, M. Matushek, S. Slaughter, T. Rice, and R. A. Weinstein. 1996. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 348:1615-1619. [DOI] [PubMed] [Google Scholar]
- 2.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188:2875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 25:1377-1388. [DOI] [PubMed] [Google Scholar]
- 4.Chang, P. C., and Y. H. Lee. 1992. Extracellular autoprocessing of a metalloprotease from Streptomyces cacaoi. J. Biol. Chem. 267:3952-3958. [PubMed] [Google Scholar]
- 5.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 6.Dunny, G. M. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos. Trans. R. Soc. Lond. B 362:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelbert, M., E. Mylonakis, F. M. Ausubel, S. B. Calderwood, and M. S. Gilmore. 2004. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 72:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franco, A. A., S. L. Buckwold, J. W. Shin, M. Ascon, and C. L. Sears. 2005. Mutation of the zinc-binding metalloprotease motif affects Bacteroides fragilis toxin activity but does not affect propeptide processing. Infect. Immun. 73:5273-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmore, M. S., D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.). 2002. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 10.Gustin, J. K., E. Kessler, and D. E. Ohman. 1996. A substitution at His-120 in the LasA protease of Pseudomonas aeruginosa blocks enzymatic activity without affecting propeptide processing or extracellular secretion. J. Bacteriol. 178:6608-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, L. E., and M. Perego. 2004. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J. Bacteriol. 186:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häse, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kothary, M. H., and A. S. Kreger. 1987. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J. Gen. Microbiol. 133:1783-1791. [DOI] [PubMed] [Google Scholar]
- 16.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay, D., and A. von Holy. 2006. Bacterial biofilms within the clinical setting: what healthcare professionals should know. J. Hosp. Infect. 64:313-325. [DOI] [PubMed] [Google Scholar]
- 18.Mäkinen, P.-L., D. B. Clewell, F. An, and K. K. Mäkinen. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 264:3325-3334. [PubMed] [Google Scholar]
- 19.Marie-Claire, C., B. P. Roques, and A. Beaumont. 1998. Intramolecular processing of prothermolysin. J. Biol. Chem. 273:5697-5701. [DOI] [PubMed] [Google Scholar]
- 20.Marie-Claire, C., E. Ruffet, A. Beaumont, and B. P. Roques. 1999. The prosequence of thermolysin acts as an intramolecular chaperone when expressed in trans with the mature sequence in Escherichia coli. J. Mol. Biol. 285:1911-1915. [DOI] [PubMed] [Google Scholar]
- 21.McIver, K. S., E. Kessler, J. C. Olson, and D. E. Ohman. 1995. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol. 18:877-889. [DOI] [PubMed] [Google Scholar]
- 22.Mohamed, J. A., and B. E. Murray. 2005. Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. J. Clin. Microbiol. 43:5405-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naka, A., K. Yamamoto, T. Miwatani, and T. Honda. 1992. Characterization of two forms of hemagglutinin/protease produced by Vibrio cholerae non-O1. FEMS Microbiol. Lett. 77:197-200. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. L. Akkermans, W. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, J., S. Chen, N. Oyama, K. Nishiguchi, E. A. Azab, E. Tanaka, R. Kariyama, and K. Sonomoto. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 188:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donohue, M. J., and A. Beaumont. 1996. The roles of the prosequence of thermolysin in enzyme inhibition and folding in vitro. J. Biol. Chem. 271:26477-26481. [DOI] [PubMed] [Google Scholar]
- 27.O'Donohue, M. J., B. P. Roques, and A. Beaumont. 1994. Cloning and expression in Bacillus subtilis of the npr gene from Bacillus thermoproteolyticus Rokko coding for the thermostable metalloprotease thermolysin. Biochem. J. 300:599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 30.Sabat, A., K. Kosowska, K. Poulsen, A. Kasprowicz, A. Sekowska, B. B. van Den, J. Travis, and J. Potempa. 2000. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect. Immun. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 32.Schmidtchen, A., I. M. Frick, and L. Bjorck. 2001. Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol. Microbiol. 39:708-713. [DOI] [PubMed] [Google Scholar]
- 33.Shockman, G. D., and M. C. Cheney. 2007. Autolytic enzyme system of Streptococcus faecalis. V. Nature of the autolysin-cell wall complex and its relationship to properties of the autolytic enzyme of Streptococcus faecalis. J. Bacteriol. 98:1199-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, K. V., S. R. Nallapareddy, E. C. Nannini, and B. E. Murray. 2005. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect. Immun. 73:4888-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su, Y. A., M. C. Sulavik, P. He, K. K. Mäkinen, P.-L. Mäkinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannock, G. W., and G. Cook. 2002. Enterococci as members of the intestinal microflora of humans, p. 101-132. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 38.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5′-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 41.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng, J., F. Teng, and B. E. Murray. 2005. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect. Immun. 73:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]