Abstract
Under conditions of nutrient deprivation, Myxococcus xanthus undergoes a developmental process that results in the formation of a fruiting body containing environmentally resistant myxospores. We have shown that myxospores contain two copies of the genome, suggesting that cells must replicate the genome prior to or during development. To further investigate the role of DNA replication in development, a temperature-sensitive dnaB mutant, DnaBA116V, was isolated from M. xanthus. Unlike what happens in Escherichia coli dnaB mutants, where DNA replication immediately halts upon a shift to a nonpermissive temperature, growth and DNA replication of the M. xanthus mutant ceased after one cell doubling at a nonpermissive temperature, 37°C. We demonstrated that at the nonpermissive temperature the DnaBA116V mutant arrested as a population of 1n cells, implying that these cells could complete one round of the cell cycle but did not initiate new rounds of DNA replication. In developmental assays, the DnaBA116V mutant was unable to develop into fruiting bodies and produced fewer myxospores than the wild type at the nonpermissive temperature. However, the mutant was able to undergo development when it was shifted to a permissive temperature, suggesting that cells had the capacity to undergo DNA replication during development and to allow the formation of myxospores.
When deprived of nutrients, the soil bacterium Myxococcus xanthus undergoes a complex developmental program leading to the formation of fruiting bodies containing environmentally resistant myxospores. Development is a social event, requiring approximately 105 cells (9). The initiation of development involves a stringent response and production of the signaling molecule (p)ppGpp (11, 33). At least five different extracellular signals at specific times during development help monitor the status of the starving population and coordinate the regulation of signaling pathways for development (14). During the developmental process, cells aggregate and form mounds, and these mounds eventually form the fruiting body, where rod-shaped cells undergo morphological changes that result in spherical myxospores.
In addition to myxospore formation, M. xanthus cells can have two other fates during development: they may undergo autolysis (approximately 65 to 90% of the cell population) or they may become peripheral rod cells (5 to 30% of the population) (25, 41). It is believed that these processes may ensure that there are sufficient nutrients and/or signals for the developmental process (41). Our laboratory has previously shown that during fruiting body formation, all myxospores contain two copies of the genome, whereas peripheral rod cells contain only one copy of the genome (36; L. Tzeng and M. Singer, unpublished results). We have also shown that in the presence of replication inhibitors, such as nalidixic acid, cells fail to sporulate and fail to form fruiting bodies during development (35). The coordination of DNA replication and development is important during early development, because addition of replication inhibitors within the first 10 to 12 h of development inhibited the developmental process (35). Addition after 12 h had little effect on fruiting body formation and sporulation (35). These results suggest a role for the cell cycle during development.
The cell cycle involves two major events, DNA replication and septation (or cell division). As modeled in Escherichia coli (22), initiation of DNA replication involves the binding of the initiator protein, DnaA, to binding sites at the origin of replication. Along with DnaA, other DNA-associated proteins bind and cause the separation of the double-stranded DNA. The exposed single-stranded DNA allows DnaC to load DnaB, the replicative helicase, onto the DNA. DnaB unwinds the DNA, which allows DnaG, the primase, to bind and synthesize a complementary RNA strand. The DNA-RNA hybrid acts as a site for DNA polymerase to bind and synthesize DNA. A complex containing DnaB, DnaG, and other accessory proteins is associated with DNA polymerase throughout the replication (elongation) process. Upon completion of DNA replication, cell division is initiated by the formation of a septum. The site of the septum is initially determined (localized) by the formation of an FtsZ ring at midcell. The FtsZ ring acts as a site for at least 12 other proteins involved in the formation of the septum, the separation of the duplicated chromosomes, and the generation of two daughter cells (for reviews, see references 10 and 39). DNA replication in M. xanthus has not been investigated in any detail since the initial studies of Zusman, Rosenberg, and their colleagues in the late 1960s and 1970s (15, 27, 42-45), and no work investigating septation in M. xanthus has been described.
In an attempt to study the role of DNA replication during development, we generated a mutation in the M. xanthus dnaB gene by site-directed mutagenesis. In this paper, we describe the isolation and characterization of the temperature-sensitive mutant DnaBA116V. Our studies not only support the model indicating that DNA replication is necessary for development but also demonstrate that cells can resume the developmental program when DNA replication is initiated under permissive conditions.
MATERIALS AND METHODS
Media and strains.
E. coli strain DH5α was grown at 37°C in Luria-Bertani (LB) medium or on plates containing LB medium with 1.5% agar in the presence of antibiotics where indicated. M. xanthus wild-type strain DK1622 was grown in CTTYE broth (1% Casitone [Difco], 10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4, 8 mM MgSO4) or on CTTYE medium plates containing 1.5% agar in the presence of antibiotics where indicated.
Cell growth assays and developmental assays.
Overnight cultures of DK1622 and the DnaBA116V mutant were grown at 28°C in CTTYE broth. Cells were then diluted in fresh CTTYE broth at the indicated temperatures, and cell growth was monitored with a Klett-Summerson photometer. At indicated time points approximately 10 ml of a culture was removed, placed into a new flask, and shifted to a different temperature, after which cell growth was monitored. Viability plating was performed by diluting cell cultures 1/104 to 1/106 in TPM medium (10 mM Tris [pH 7.6], 8 mM MgSO4, 1 mM KH2PO4) and plating 100-μl portions of diluted cells in CTTYE soft agar (1%) in CTTYE agar plates. The plates were incubated at 28°C, and colonies were counted.
The development of fruiting bodies by M. xanthus cultures was examined on TPM agar plates containing 1.5% agar or by using a submerged culture assay as previously described (19, 36). Cells were allowed to develop at 28 or 37°C for 3 to 5 days. Quantification of heat- and sonication-resistant spores was done essentially as previously described; however, strains were incubated at 28°C instead of 33°C for growth (18).
Mutagenesis.
A 1,775-bp DNA fragment containing the M. xanthus dnaB gene was PCR amplified from DK1622 genomic DNA with the upstream primer 5′-AAGGATCCTCAGCCGCACCTGGCGT (underlining indicates a BamHI site) and the downstream primer 5′-CCAAGCTTGGCCACACCGGGCCGCG (underlining indicates a HindIII site). The DNA fragment was digested with the appropriate restriction enzymes and ligated into pBluescript (Gibco-BRL) with the same restriction sites, creating pCJR1. DH5α was transformed with pCJR1 and plated on LB agar containing ampicillin (50 μg/ml). Resulting colonies were isolated, and the plasmid was extracted for DNA sequencing. The DNA sequence of the cloned fragment was confirmed by sequencing at the University of California, Davis sequencing facility.
Mutagenesis was performed with pCJR1 using a GeneEditor mutagenesis kit (Promega) with the primer sequence 5′-GTCAAGGACCAGGTCATCCGCCGCCGC, which changed the alanine at position 116 in the M. xanthus DnaB protein to valine, creating pCJR2. The mutation was confirmed by sequencing. The dnaB gene was digested from pCJR2 with BamHI/HindIII and ligated into the same sites in pBJ113 (13), generating plasmid pCJR3. DK1622 was transformed by electroporation with pCJR3, and transformants were selected on CTTYE agar containing kanamycin (50 μg/ml) as previously described (13). Integration of pCJR3 into the DK1622 genome was confirmed by PCR amplification with primers directed at the dnaB locus, followed by sequencing for the presence of both dnaB alleles (one allele containing the mutation). Counterselection was performed with CTTYE medium containing 2% galactose to screen for loss of the pCJR3 plasmid, leaving only one copy of the dnaB gene. Individual colonies were streaked on CTTYE agar plates and incubated at 28 and 37°C. DNA from colonies that grew at 28°C but failed to grow at 37°C was extracted and used in a PCR to amplify the dnaB gene. The PCR-amplified fragment was purified and used for DNA sequencing to confirm the presence of the mutation.
[3H]thymidine incorporation.
Steady-state incorporation was performed as described by Tzeng and Singer (36) by addition of [6-3H]thymidine (20 to 30 Ci/mmol; catalog no. TRK410; Amersham) at a final concentration of 1 μCi/ml in liquid culture, and the culture was incubated at 28 or 37°C. At various time points, 100 μl of culture was removed and added to 1 ml of ice-cold 5% trichloroacetic acid. Treated cultures were placed at 4°C prior to collection of precipitated counts on glass fibers. Precipitated material was collected on a glass filter and washed with 5% trichloroacetic acid and then with 95% ethanol. The glass filter was counted with a Beckman Coulter LS 6500 scintillation counter. For each time point the experiment was performed in duplicate, and the results were expressed in cpm minus the background level at each time point.
Flow cytometry.
Flow cytometric analysis was performed as described by Tzeng and Singer, with minor changes (36). Approximately 1 ml of cells was removed and fixed in PME buffer [50 mM piperizine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.9), 5 mM EGTA, 1 mM MgSO4] containing 4% paraformaldehyde for 10 min at room temperature, and this was followed by incubation in 95% ethanol for at least 30 min at −20°C. Cells were then treated with RNase (100 μg/ml) overnight at 37°C. After RNase treatment, cells were resuspended in 200 mM Tris (pH 7.6), and propidium iodide was added at a final concentration of 25 μg/ml and incubated for 30 min on ice. Flow cytometry was performed with approximately 10,000 to 20,000 cells using a Becton-Dickinson FACSCalibur instrument system and CellQuestPro software (Becton-Dickinson) for data analysis.
RESULTS
Isolation of the M. xanthus DnaBA116V mutant.
DnaB is a helicase that unwinds double-stranded DNA into single-stranded DNA during initiation and elongation in DNA synthesis. DnaB (length in E. coli, 471 amino acids) is a DNA-dependent ATPase that consists of three functional domains, the α, β, and γ domains, in E. coli (3). Domain α consists of amino acids 1 to 156 and has been shown to be important for protein-protein interactions with DnaA, DnaC, and DnaG (17, 21, 23, 24, 40). Very little is known about which amino acid residues or subdomains in DnaB are involved in these interactions. Domain β (amino acids 157 to 300) contains the ATPase activity (2, 24). The ATPase is characterized by two major motifs, a Walker A motif and a Walker B motif, based on comparative analysis of known ATPases (20, 37). Domain γ (amino acids 310 to 471) contains sites for DNA binding and oligomerization (4). No domain can act alone as a helicase, suggesting that all domains are necessary for this function (3).
The M. xanthus dnaB gene was identified by a BLAST search for the M. xanthus DK1622 protein sequence (http://tigrblast.tigr.org/cmr-BLAST/) with the sequence from the E. coli DnaB protein. MXAN5084 coded for a protein that was 456 amino acids long with a level of amino acid sequence identity of 43.6% to the 471-amino-acid sequence of E. coli DnaB. M. xanthus DnaB characteristically has the conserved residues for its numerous activities. A Walker A box, which is characterized by the consensus sequence G/AXXXXGKT (amino acids 213 to 220 in M. xanthus), and a Walker B site, which contains the catalytic aspartate residue (amino acid 324), are present. The C-terminal region also contains a conserved DNA-binding site, RAKARR (amino acids 305 to 310), and the dimerization domain, a leucine zipper motif (amino acids 342 to 370) (4). Interestingly, the N-terminal 14 amino acids of the E. coli DnaB protein, which is important for maintaining the activity of the protein (24), is not present at the M. xanthus DnaB N terminus.
In an attempt to isolate temperature-sensitive dnaB mutants of M. xanthus, site-directed mutagenesis was performed based on known mutations in the E. coli dnaB gene (5, 7, 29, 30). For this study, we created the dnaB mutant DnaBA116V, whose mutation corresponds to the A130V mutation in the E. coli DnaB mutant (29). The DnaBA116V mutant had a temperature-sensitive phenotype and was able to grow on CTTYE agar plates at 28 and 33°C but did not grow at 37°C. While 37°C is an unusual temperature for M. xanthus growth, previous studies have shown that cells grow at wild-type rates at this elevated temperature (16).
Growth phenotype of the M. xanthus DnaBA116V mutant.
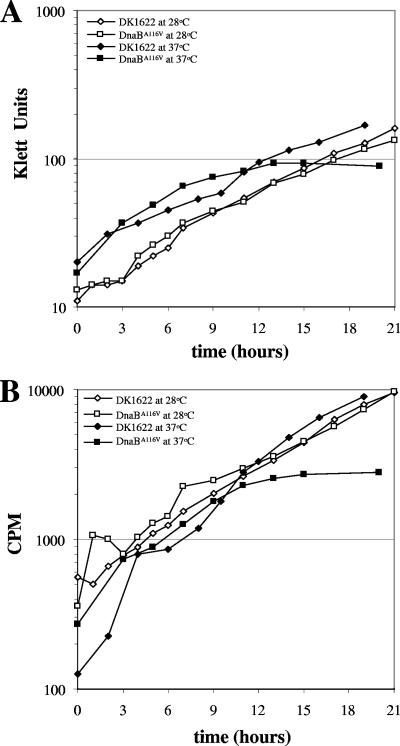
To further demonstrate that the M. xanthus DnaBA116V mutant had a growth defect, growth assays were performed in liquid CTTYE medium. As shown in Fig. 1A, wild-type DK1622 and the DnaBA116V mutant displayed similar growth rates at 28°C, with doubling times of 5.1 and 5.6 h, respectively. At 37°C, DK1622 and the DnaBA116V mutant initially displayed similar growth rates, with doubling times of 5.2 and 5.0 h, respectively, for the first 6 h of growth (Fig. 1A). However, by 6 h the growth rate of the DnaBA116V mutant began to decrease dramatically and by 12 h growth halted altogether at 37°C (Fig. 1A). The cell density decreased after 12 h (Fig. 1A and 2A), suggesting that cell death was occurring. Interestingly, the M. xanthus DnaBA116V mutant consistently showed this growth phenotype. The corresponding E. coli DnaB mutant stopped growing immediately when a culture was shifted to the nonpermissive temperature (7).
FIG. 1.
Growth of DK1622 and the DnaBA116V mutant and steady-state [3H]thymidine incorporation into DNA of these strains at 28 and 37°C. DK1622 and the DnaBA116V mutant were initially grown overnight at 28°C. The cells were diluted to the indicated starting concentrations and incubated at 28 or 37°C in the presence of 1 μCi/ml [3H]thymidine. At the indicated times, cell density was monitored using a Klett-Summerson colorimeter (A), and 100 μl of culture was removed for scintillation counting (B), as described in Materials and Methods. The values for cpm are averages of duplicate samples at each time point. In addition, each experiment was performed at least three times, and the results of a representative experiment are shown.
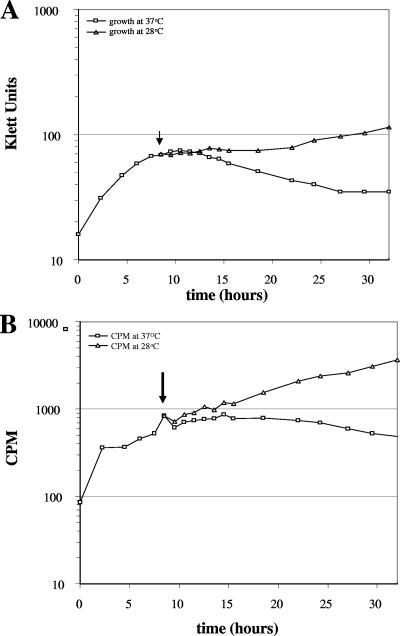
FIG. 2.
Growth and [H3]thymidine incorporation into DNA during a temperature shift from the nonpermissive temperature (37°C) to the permissive temperature (28°C). An overnight culture of the DnaBA116V mutant was grown at 28°C. The culture was diluted to 20 Klett units in the presence of 1 μCi/ml [3H]thymidine and incubated at 37°C. At various time points, growth was monitored, and 100 μl of cells (in duplicate) was removed for scintillation counting. At 9 h during growth at 37°C, 10 ml of the culture was shifted to 28°C (indicated by the arrow). At the indicated time points, growth was monitored (A), and 100 μl of cells (in duplicate) was removed for scintillation counting (B) from the cultures at 28 and 37°C, as described in the legend to Fig. 1.
The DnaBA116V mutant also showed a partial phenotype when it was incubated at an intermediate temperature (35°C) (data not shown). The DnaBA116V mutant had a growth rate similar to that of DK1622 when cells were diluted from an overnight culture grown at 28°C and then incubated at 35°C. However, when overnight cultures were grown at 35°C, the DnaBA116V mutant grew at a much lower rate, with a doubling time of 12 h. Unlike what occurred at 37°C, growth did not cease and cells continued to grow at this rate for at least 3 days (data not shown).
One explanation for the increase in turbidity of the DnaBA116V mutant for the first 12 h of incubation at the nonpermissive temperature could be that there was an increase in cell mass without cell division. To address this possibility, we examined the cell morphology and cell viability of DK1622 and the DnaBA116V mutant. At 37°C the cells of both DK1622 and the DnaBA116V mutant were three to four times longer than cells that were incubated at 28°C (data not shown). Incidentally, the amount of cell debris increased as the DnaBA116V mutant grew at 37°C, suggesting that cells were lysing. The numbers of viable cells of DK1622 and the DnaBA116V mutant were similar for the first 3 h of growth at 37°C. At later time points, the viability of the DnaBA116V mutant cells decreased compared to DK1622 cells. At 6 h, 55% of the DnaBA116V mutant cells were viable compared to DK1622; by 9 h, only 18% of the DnaBA116V mutant cells were viable; and at 12 h, 10% of the DnaBA116V mutant cells were viable. These results suggest that the DnaBA116V mutant underwent a single round of the cell cycle and that the increase in the turbidity of the culture (from 6 to 12 h) observed was due to an increase in the cell mass of the surviving cells.
To test whether the growth defect could be rescued when cells were shifted back to a permissive temperature, cell cultures that were grown at 37°C for 9 h (the point at which cell growth ceased) were shifted to a permissive temperature, 28°C. As shown in Fig. 2A, the DnaBA116V mutant was able to resume growth at 28°C. This resumption of growth usually followed a lag of 3 to 8 h (Fig. 2A). Furthermore, cells were able to resume growth when cultures were shifted after 15 and 18 h from the nonpermissive temperature (37°C) to 28°C, but the lag phase was longer than that of cells that were shifted at 9 h (data not shown).
DnaBA116V mutant is defective in DNA synthesis at the nonpermissive temperature.
Because DnaB is involved in both initiation and elongation during DNA replication, we wanted to confirm that the defect in growth was a result of a defect in DNA synthesis. To test this, we monitored the steady-state incorporation of [3H]thymidine into DNA at 28 and 37°C. DK1622 and the DnaBA116V mutant had similar rates of [3H]thymidine incorporation when they were grown at 28°C (Fig. 1B). At 37°C, DK1622 and the DnaBA116V mutant had similar rates of [3H]thymidine incorporation for the first 9 h of growth. However, at later time points, the rate of [3H]thymidine incorporation decreased in the DnaBA116V mutant and incorporation stopped by 12 h, suggesting that the defect in growth at 37°C is a result in a defect in DNA synthesis (Fig. 1B). When the DnaBA116V mutant was shifted from 37°C (after 12 h of growth) back to 28°C, cells were able to resume incorporation of [3H]thymidine into DNA after a short lag (Fig. 2B). These data imply that the mutant is capable of synthesizing DNA when it is shifted back to the permissive temperature. Interestingly, [3H]thymidine incorporation always preceded cell growth (Fig. 2).
M. xanthus DnaBA116V mutant is a 1n population at the nonpermissive temperature.
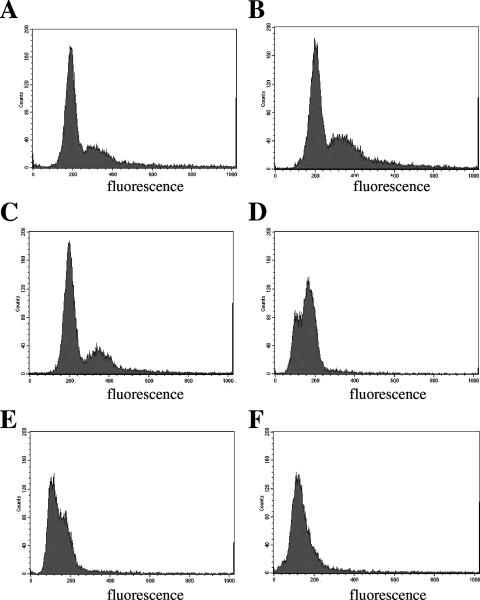
The ability of the DnaBA116V mutant to grow and synthesize DNA at the nonpermissive temperature for at least one cell doubling suggests that the DnaB complexes formed by the mutant were competent to complete replication of the entire genome before cessation of both growth and DNA synthesis. If this is true, we would predict that cells should arrest after cell division as a homogeneous cell population. To test this hypothesis, we determined the DNA contents of DK1622 and the DnaBA116V mutant using cells grown at 28 and 37°C by flow cytometry. Flow cytometric analysis of DK1622 revealed two distinct peaks when cells were grown at 28 and 37°C (a representative profile is shown in Fig. 3A for DK1622). These peaks represented the chromosomal content of individual cells. As previously reported for wild-type DK101 cells grown at 33°C (36), the patterns for DK1622 at 28 and 37°C represent cells that contain one copy of the chromosome (1n; the first peak at ∼200 fluorescence units), cells that contain two copies of the chromosome (2n; the second peak and the shoulder at ∼350 to 400 fluorescence units), and a population in the process of replication (the shoulder between 200 and 400 fluorescence units).
FIG. 3.
Flow cytometric profiles of DK1622 and the DnaBA116V mutant grown at 28 and 37°C: flow cytometric profiles of DK1622 incubated at 28°C (A), the DnaBA116V mutant incubated at 28°C prior to a shift to 37°C (B), and the DnaBA116V mutant after 3 h (C), 6 h (D), 9 h (E), and 12 h (F) of incubation at 37°C. Cells were grown in CTTYE medium and fixed in paraformaldehyde as described in Materials and Methods prior to flow cytometric analysis. The y axis represents the number of cells counted for each fluorescence unit, and the x axis represents arbitrary fluorescence units ranging from 0 to 1,000 on a linear scale.
DnaBA116V mutant cells grown at 28°C showed a pattern similar to that of DK1622, with two peaks at positions similar to those observed for DK1622 (Fig. 3A). Flow cytometric analysis of the mutant characteristically revealed two peaks for cell cultures throughout logarithmic growth at 28°C (data not shown), suggesting that the processes for growth and DNA replication in the mutant are similar to those in wild-type cells. At 37°C, the DnaBA116V mutant progressively showed a decrease in the 1n and 2n peaks with an increase in a new peak located between 50 to 100 fluorescence units during growth at 37°C (Fig. 3C to 3E). By 12 h, a single peak was seen (Fig. 3F). We presume that the appearance of this peak during incubation at 37°C may have been due to nonviable cells and that the DNA may have been partially degraded. The remaining viable cells at 12 h (approximately 10 to 30%) appeared to lie within the range for 1n cells (the small shoulder at ∼200 fluorescence units in Fig. 3F), and few or no cells were detected in the range for 2n cells (∼350 to 400 fluorescence units), suggesting that the remaining viable cells largely consisted of a population of 1n cells. These results are consistent with the results obtained from viability plating of the DnaBA116V mutant.
DnaBA116V mutant is defective in development at the nonpermissive temperature.
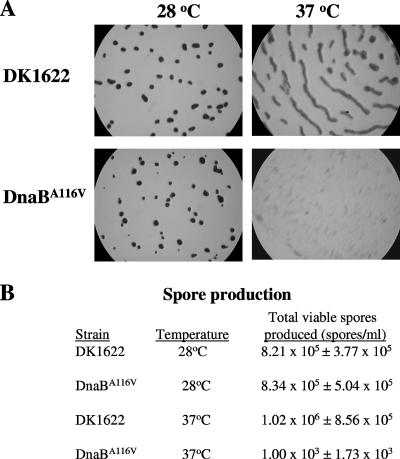
Developmental assays were performed with DK1622 and the DnaBA116V mutant to examine the role of DNA replication during development. As shown in Fig. 4A, after 3 days of development at 28°C, DK1622 and the DnaBA116V mutant both displayed normal developmental phenotypes. Furthermore, the sporulation efficiency for the DnaBA116V mutant was nearly the same (102%) as that for DK1622. At 37°C, DK1622 did not form the typical darkened fruiting bodies normally observed at 33°C; instead, DK1622 formed elongated, compact, darkened structures (Fig. 4A). Nevertheless, DK1622 did form viable spores, with an efficiency of 124% compared to DK1622 cells incubated at 28°C. No fruiting bodies were formed by the DnaBA116V mutant when cells were grown for 12 h at 37°C prior to plating for development at 37°C; however, the DnaBA116V mutant did appear to form translucent cell aggregates (Fig. 4A). In addition to the defect in fruiting body formation, spore production was reduced over 800-fold compared to that in DK1622 (Fig. 4B). This is consistent with the findings of previous work with DNA replication inhibitors, where DK101 failed to develop into fruiting bodies and appeared to be blocked at the aggregation stage (35). Interestingly, when the DnaBA116V mutant was grown at 28°C and then plated and incubated at 37°C for development, the cells were able to form wild-type fruiting bodies with sporulation efficiencies near wild-type levels. Furthermore, when the DnaBA116V mutant was grown in liquid medium at 37°C, cells that were spotted onto developmental plates during periods of active growth (prior to 12 h) were able to undergo normal development (data not shown).
FIG. 4.
Development of DK1622 and the DnaBA116V mutant at 28 and 37°C. (A) Light micrographs of developmental morphology of DK1622 and the DnaBA116V mutant. Cells were grown at either 28 or 37°C in liquid CTTYE medium to 80 to 100 Klett units. For the DnaBA116V mutant at 37°C, cells were grown for 12 h to 80 Klett units. Cells were spotted on TPM medium starvation plates and incubated for 3 days at the respective temperatures. (B) Viable spore production by each strain during development at 28 or 37°C. The total number of viable spores was determined for each strain at 28 and 37°C by performing at least three biological assays, and the results are expressed as means ± standard deviations.
DK1622 can resume development following relief from inhibition of DNA replication.
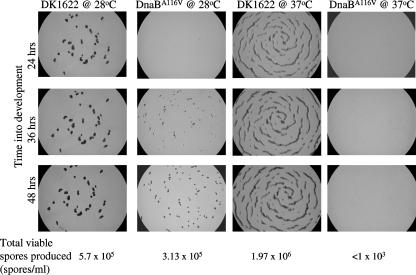
Because the DnaBA116V mutant lost viability in liquid medium at 37°C, one explanation for our results is that there may not have been a sufficient number of viable cells for the developmental process to proceed. Experiments were performed to address this possibility. In these experiments cells that were growing at the nonpermissive temperature for 12 h were spotted onto starvation media and incubated at 28 and 37°C. As shown in Fig. 5, DK1622 formed the previously described elongated structures at 37°C and wild-type fruiting bodies at 28°C between 24 and 36 h. These data imply that a certain function(s) necessary for the formation of compact discrete fruiting bodies, but not myxospores, is temperature sensitive, unrelated to DNA replication. The DnaBA116V mutant was able to form fruiting bodies when it was incubated at 28°C, while no fruiting bodies were formed at 37°C (Fig. 5). Although the fruiting bodies did appear to be smaller than those of DK1622 at the same temperature, the fruiting bodies were more numerous. Furthermore, the production of viable spores by the DnaBA116V mutant was nearly as efficient as that by DK1622 under these conditions (Fig. 5).
FIG. 5.
Fruiting body formation in cultures after a shift from the nonpermissive temperature (37°C) to the permissive temperature (28°C) or in cultures remaining at the nonpermissive temperature: light micrographs of the developmental morphology of DK1622 and the DnaBA116V mutant. Cells were grown in liquid CTTYE medium at 37°C, spotted on TPM starvation medium, and incubated at the indicated temperatures. Micrographs were taken at the indicated times. The total numbers of viable spores recovered in the assay are indicated at the bottom.
To further test whether the developmental program can be delayed when DNA replication in cells is inhibited, we treated DK1622 cells with the DNA gyrase inhibitor nalidixic acid. We have previously shown that M. xanthus cells fail to progress past the aggregation phase of development in the presence of this DNA replication inhibitor (36). However, when nalidixic acid was removed from the culture medium, we were able to observe that M. xanthus could resume the developmental program, even when we inhibited DNA replication and development for 22 h (data not shown). Like the results observed with the DnaBA116V mutant shown in Fig. 5, development resumed 8 to 12 h after removal of the inhibitor (data not shown). These results suggest that when the block in DNA replication during development is relieved, M. xanthus can resume the developmental program.
DISCUSSION
In this paper, we describe the isolation and characterization of a temperature-sensitive dnaB mutant of M. xanthus. The mutation which changes the amino acid at position 116 from alanine to valine corresponds to the mutation in E. coli dnaB which changes the amino acid at position 130 from alanine to valine. We predicted that this change should result in a similar growth and replication phenotype; however, we show here that the analogous change in M. xanthus results in a distinctively different phenotype. Many E. coli dnaB mutants, as well as mutants with mutations involved in elongation during DNA replication, immediately cease growth and DNA synthesis, referred to as a “quick” stop phenotype (1, 7, 32, 38). The M. xanthus DnaBA116V mutant showed a “slow” stop phenotype, where both growth and DNA synthesis eventually stopped during incubation at the nonpermissive temperature. “Slow” stop mutants have been identified in E. coli, many of which were characterized for the initiator protein for DNA replication, DnaA (1, 32, 38), and E. coli dnaB mutants dnaB125, an amber nonsense mutant, and dnaB252, a change at amino acid 299 from a glycine to an aspartate (31, 46). It has been proposed that “slow” stop mutants are able to complete DNA replication but fail to initiate new cycles of DNA replication during incubation at nonpermissive temperatures. This suggests that the M. xanthus DnaBA116V mutant does retain a functional enzyme that sustains activity throughout the replication cycle at the nonpermissive temperature. A DnaB protein in a complex as a hexamer and/or with other proteins (e.g., DnaC) could be protected from inducing changes in protein structure and/or function during incubation at the nonpermissive temperature. As the complex is dissembled, DnaB activity would become compromised and therefore could not initiate new rounds of DNA replication. Sclafani and Wechsler demonstrated that dnaB252 could be suppressed when the helicase loader, DnaC, was overexpressed (30). The dnaB252 mutant does retain all of the activities of a helicase in vitro, suggesting that the defect in vivo may involve an interaction between DnaB and DnaC (29).
This model thus predicts that cells can complete only one round of the cell cycle, resulting in a population of cells with only one copy of the chromosome. Data from flow cytometry (Fig. 3) support this model. The population of viable dnaB mutant cells at the nonpermissive temperature consists largely of cells arrested in the 1n state. This population appears to lack any measurable 2n cells. When the cells were shifted to the permissive temperature, we observed that DNA replication usually preceded cell growth (Fig. 2). We hypothesized that the delay in growth may involve the completion of DNA replication. It is unclear whether the M. xanthus DnaBA116V mutant can become active when it is shifted to permissive conditions, and this requires biochemical characterization.
Previous work in our lab showed that replication inhibitors, including nalidixic acid, caused a defect in the developmental program (35). Moreover, we have demonstrated by use of replication inhibitors that DNA replication is necessary for cells to progress through the early stages (aggregation) of development (35). Development of the DnaBA116V mutant during incubation at the nonpermissive temperature also failed to progress past the aggregation phase (Fig. 5). However, we noted that some viable spores were detectable (<1% compared to DK1622) (Fig. 4). In this work, developmental assays were performed with cultures that were incubated at 37°C for 12 h, the point when cells first appeared to stop growing and synthesizing DNA (Fig. 1B). It is possible that there was a small subpopulation of cells (not detectable by our flow cytometric analysis) which were still undergoing DNA replication and therefore could have contributed to the small amount of sporulation observed.
Results presented here demonstrate that an arrested 1n population of dnaB mutant cells cannot undergo normal development under conditions that are nonpermissive for DNA replication. More importantly, we show that the developmental program can be delayed when certain criteria during the cell cycle are not met. When cells are shifted to permissive conditions that allow DNA replication (e.g., a shift to the permissive temperature or removal of replication inhibitors), cells can proceed with the developmental program, despite experiencing starvation conditions (up to 24 h) during which developmental signals are present. Our results suggest that cells delay the developmental program until the requirement for completion of DNA replication is fulfilled and imply that a regulatory circuit monitoring DNA replication controls progression of the developmental program. In Bacillus subtilis, sporulation is characterized by an asymmetric cell division that forms the endospore and the mother cell (34). The coordination of spore development and DNA replication is regulated by the histidine kinase inhibitor, Sda (6). Sda binds and inhibits phosphorylation of KinA and possibly KinB (6, 28). KinA and KinB are histidine kinases involved in the phosphorelay system for sporulation (6, 28). Rowland et al. demonstrated that Sda inhibits KinA activity by binding to the C-terminal autokinase domain of KinaA and suggested that Sda prevents the interaction between the ATP-binding site and the phosphorylatable His residue in KinA (28).
In Caulobacter crescentus, only nonmotile stalk cells can undergo DNA replication, while motile swarmer cells cannot. It has been shown that when swarmer cells differentiate into stalk cells, DNA replication can proceed. The coordination of differentiation and DNA replication is controlled by CtrA (8, 26). CtrA is a transcriptional regulator and has been shown to bind to the origin of replication site (26). It is believed that this binding may inhibit initiation of DNA replication (26). Domian et al. (8) have shown that the regulation of CtrA protein levels controls the ability of the cells to differentiate and undergo DNA replication. No homologues of Sda or CtrA have been identified in M. xanthus, suggesting that M. xanthus may utilize a novel mechanism to coordinate the cell cycle with development.
Our results suggest that cells have the capacity to undergo DNA replication during development. This implies that cells do not necessarily have to be in a 2n state (or have actively replicating genomes) prior to development. If a monitoring system does exist, at what level does development control DNA replication, and vice versa? We can only speculate that this control could involve regulation at the transcriptional and/or translational level or, in the case of the temperature-sensitive mutant, refolding or renaturation of preexisting proteins. Jakobsen et al. reported that dnaA mRNA expression increases during development in M. xanthus (at 12 h) based on DNA microarray analysis (12), and this was confirmed by Bragg and Singer (J. Bragg and M. Singer, unpublished results). We have also observed an increase in the expression of dnaA during the early stages of development (4 to 5 h) as determined by quantitative PCR analysis (C. J. Rosario and M. Singer, unpublished data), suggesting that the developmental program could feed into the DNA replication pathway, or vice versa. Regulation of dnaB, transcriptionally or posttranscriptionally, has not been extensively studied in any microorganism. Studies to investigate the regulation of the DNA replication machinery during development are currently being performed.
Acknowledgments
This work was supported in part by Public Health Service grant GM54592 (to M.S.) from the National Institutes of Health.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Beyersmann, D., W. Messer, and M. Schlicht. 1974. Mutants of Escherichia coli B/r defective in deoxyribonucleic acid initiation: dnaI, a new gene for replication. J. Bacteriol. 118:783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas, E. E., P. H. Chen, and S. B. Biswas. 1994. Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry 33:11307-11314. [DOI] [PubMed] [Google Scholar]
- 3.Biswas, E. E., and S. B. Biswas. 1999. Mechanism of DNA binding by the DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding, and oligomerization. Biochemistry 38:10919-10928. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, E. E., and S. B. Biswas. 1999. Mechanism of DNA binding by the DnaB helicase of Escherichia coli: analysis of the roles of domain γ in DNA biding. Biochemistry 38:10929-10939. [DOI] [PubMed] [Google Scholar]
- 5.Bonhoeffer, F., and H. Schaller. 1965. A method for selective enrichment of mutants based on the high UV sensitivity of DNA containing 5-bromouracil. Biochem. Res. Commun. 109:93-97. [PubMed] [Google Scholar]
- 6.Burkholder, W. F., I. Kuster, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 7.Carl, P. L. 1970. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet. 109:107-122. [DOI] [PubMed] [Google Scholar]
- 8.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin, M., and D. Kaiser. 1985. Cell interactions in myxobacterial growth and development. Science 230:18-24. [DOI] [PubMed] [Google Scholar]
- 10.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobsen, J. S., L. Jelsbak, R. D. Welch, C. Cummings, B. Goldman, E. Stark, and D. Kaiser. 2004. σ54 enhancer binding proteins and Myxococcus development. J. Bacteriol. 186:4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien, B., D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 15.Kimchi, A., and E. Rosenberg. 1976. Linkages between deoxyribonucleic acid synthesis and cell division in Myxococcus xanthus. J. Bacteriol. 128:69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura, Y., H. Nakato, K. Ishibashi, and S. Kobayashi. 2005. A Myxococcus xanthus CbpB containing two cAMP-binding domains is involved in temperature and osmotic tolerances. FEMS Microbiol. Lett. 244:75-83. [DOI] [PubMed] [Google Scholar]
- 17.Kobori, J. A., and A. Kornberg. 1982. The Escherichia coli dnaC gene product. III. Properties of the dnaB-dnaC protein complex. J. Biol. Chem. 257:13770-13775. [PubMed] [Google Scholar]
- 18.Kroos, L., and D. Kasier. 1987. Expression of many developmentally regulated genes in Myxoccocus xanthus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 19.Kuner, J. M., and D. Kaiser. 1982. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J. Bacteriol. 151:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leipe, D. D., L. Aravind, N. V. Grishin, and E. V. Koonin. 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 10:5-16. [PubMed] [Google Scholar]
- 21.Marszalek, J., and J. M. Kaguni. 1994. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269:4883-4890. [PubMed] [Google Scholar]
- 22.Messer, W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26:355-374. [DOI] [PubMed] [Google Scholar]
- 23.Mitkova, A. V., S. M. Khopde, and S. B. Biswas. 2003. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J. Biol. Chem. 278:52253-52261. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama, N., N. Arai, Y. Kaziro, and K.-I. Arai. 1984. Structural and functional studies of the dnaB protein using limited proteolysis. J. Biol. Chem. 259:88-96. [PubMed] [Google Scholar]
- 25.O'Connor, K., and D. R. Zusman. 1991. Development of Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg, E., M. Katarski., and P. Gottlieb. 1967. Deoxyribonucleic acid synthesis during exponential growth and microcyst formation in Myxococcus xanthus. J. Bacteriol. 93:1402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejerski, A. D. Gorssman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinase that regulates initiation of sporulation in Bacillus subtilis. Mol. Cell 13:689-701. [DOI] [PubMed] [Google Scholar]
- 29.Saluja, D., and G. N. Godson. 1995. Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutant dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. J. Bacteriol. 177:1104-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sclafani, R. A., and J. A. Wechsler. 1981. Deoxyribonucleic acid initiation mutation dnaB252 is suppressed by elevated dnaC+ gene dosage. J. Bacteriol. 146:418-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sclafani, R. A., and J. A. Wechsler. 1981. dnaB125, a dnaB nonsense mutation J. Bacteriol. 146:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sevastopoulos, C. G., C. T. Wehr, and D. A. Glaser. 1977. Large-scale automated isolation of Escherichia coli mutants with thermosensitive DNA replication. Proc. Natl. Acad. Sci. USA 8:3485-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer, M., and D. Kaiser. 1995. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633-1644. [DOI] [PubMed] [Google Scholar]
- 34.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet 30:297-341. [DOI] [PubMed] [Google Scholar]
- 35.Tzeng, L., T. N. Ellis, and M. Singer. 2006. DNA replication during aggregation phase is essential for Myxococcus xanthus development. J. Bacteriol. 188:2774-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzeng, L., and M. Singer. 2005. DNA replication during sporulation in Myxococcus xanthus fruiting bodies. Proc. Natl. Acad. Sci. USA 102:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 8:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wechsler, J. A., and J. A. Gross. 1971. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol. Gen. Genet. 113:273-284. [DOI] [PubMed] [Google Scholar]
- 39.Weiss, D. S. 2004. Bacterial cell division and septal ring formation. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 40.Wickner, S., and J. Hurwitz. 1975. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc. Natl. Acad. Sci. USA 72:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zusman, D., and E. Rosenburg. 1968. Deoxyribonucleic acid synthesis during microcyst germination in Myxococcus xanthus. J. Bacteriol. 96:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zusman, D., and E. Rosenberg. 1970. DNA cycle of Myxococcus xanthus. J. Mol. Biol. 49:609-619. [DOI] [PubMed] [Google Scholar]
- 44.Zusman, D., P. Gottlieb, and E. Rosenberg. 1971. Division cycle of Myxococcus xanthus. III. Kinetics of cell growth and protein synthesis. J. Bacteriol. 105:811-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zusman, D. R., D. M. Krotoski, and M. Cumsky. 1978. Chromosome replication in Myxococcus xanthus. J. Bacteriol. 133:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zyskind, J. W., and D. W. Smith. 1977. Novel Escherichia coli dnaB mutant: direct involvement of the dnaB252 gene product in the synthesis of an origin-ribonucleic acid species during initiation of a round of deoxyribonucleic acid replication. J. Bacteriol. 125:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]