Abstract
Direct bacterial conversion of the hemicellulose fraction of hardwoods and crop residues to biobased products depends upon extracellular depolymerization of methylglucuronoxylan (MeGAXn), followed by assimilation and intracellular conversion of aldouronates and xylooligosaccharides to fermentable xylose. Paenibacillus sp. strain JDR-2, an aggressively xylanolytic bacterium, secretes a multimodular cell-associated GH10 endoxylanase (XynA1) that catalyzes depolymerization of MeGAXn and rapidly assimilates the principal products, β-1,4-xylobiose, β-1,4-xylotriose, and MeGAX3, the aldotetrauronate 4-O-methylglucuronosyl-α-1,2-xylotriose. Genomic libraries derived from this bacterium have now allowed cloning and sequencing of a unique aldouronate utilization gene cluster comprised of genes encoding signal transduction regulatory proteins, ABC transporter proteins, and the enzymes AguA (GH67 α-glucuronidase), XynA2 (GH10 endoxylanase), and XynB (GH43 β-xylosidase/α-arabinofuranosidase). Expression of these genes, as well as xynA1 encoding the secreted GH10 endoxylanase, is induced by growth on MeGAXn and repressed by glucose. Sequences in the yesN, lplA, and xynA2 genes within the cluster and in the distal xynA1 gene show significant similarity to catabolite responsive element (cre) defined in Bacillus subtilis for recognition of the catabolite control protein (CcpA) and consequential repression of catabolic regulons. The aldouronate utilization gene cluster in Paenibacillus sp. strain JDR-2 operates as a regulon, coregulated with the expression of xynA1, conferring the ability for efficient assimilation and catabolism of the aldouronate product generated by a multimodular cell surface-anchored GH10 endoxylanase. This cluster offers a desirable metabolic potential for bacterial conversion of hemicellulose fractions of hardwood and crop residues to biobased products.
Structural polysaccharides comprise up to 90% of plant cell walls, and include cellulose and hemicellulose fractions as prominent resources renewable through photosynthesis. The hemicellulose fractions constitute from 22 to 30% of the dry weight of lignocellulosic biomass derived from wood and agricultural residues (12). The quest for alternatives to petroleum has led to the search for and discovery of microorganisms that can serve as biocatalysts for the production of fuels and chemical feedstocks from renewable resources. The major hemicellulose polymer in hardwoods and crop residues is methylglucuronoxylan (MeGAXn), a linear chain of β-1,4-linked d-xylopyranose residues regularly substituted with α-1,2-linked 4-O-methyl-d-glucuronopyranosyl residues. Variable substitutions on xylose residues may include 2′- and 3′-O-acetyl esters, as well as α-1,2- or α-1,3-linked l-arabinofuranosyl residues (24). Additional substituents may include O-feruloyl and O-p-coumaroyl esters linked to hydroxyl groups on the arabinofuranosyl residues.
Both yeast and bacteria have been developed for the bioconversion of glucose derived from the cellulose fraction, and bacteria have been developed for the bioconversion of pentoses, principally xylose, from the hemicellulose fraction (9, 11, 12). Pretreatment has relied on a combination of chemical and enzymatic hydrolytic procedures to solubilize the hemicellulose fraction and release fermentable xylose and to depolymerize the cellulose to fermentable glucose. Pretreatment protocols are still being developed to provide cost-effective production of ethanol and other biobased products from these resources (13). The use of xylanolytic systems to enhance the extracellular depolymerization of methylglucuronoxylan and to efficiently assimilate and metabolize the products of depolymerization has become a promising route to more complete conversion of cellulosic biomass to alternative fuels and chemicals.
The natural processing of methylglucuronoxylans is catalyzed by the combined action of endoxylanases, α-glucuronidases, arabinosidases, and esterases (7, 18, 24). Xylanolytic bacteria secrete endoxylanases of the glycohydrolase families GH5, GH10, and GH11 that catalyze the depolymerization of the xylan backbone with the generation of different products (4, 18). The GH10 endoxylanases generate xylobiose, xylotriose, and the aldotetrauronate, methylglucuronoxylotriose (MeGAX3), in which β-1,4-linked d-xylotriose is substituted at the nonreducing terminus with α-1,2-linked 4-O-methyl-d-glucuronate. Bacteria that secrete a GH10 endoxylanase may assimilate and metabolize all of the products derived from the depolymerization of MeGAXn. The utilization of the aldouronate requires the expression of genes encoding transporters, α-glucuronidase, and enzymes that convert xylooligosaccharides to xylose and have been found in several gram-positive bacteria (16, 21, 22, 25).
We previously reported on the isolation and characterization of an aggressively xylanolytic gram-positive endospore-forming bacterium, Paenibacillus sp. strain JDR-2, which secretes a multimodular cell-anchored GH10 endoxylanase (23). The rapid and complete utilization of MeGAXn without accumulation of the aldotetrauronate, methylglucuronoxylotriose (MeGAX3), in the medium implicated an efficient system for assimilation and complete metabolism of aldouronates. A structural gene, aguA, has been cloned from genomic DNA of Paenibacillus sp. strain JDR-2 and expressed in Escherichia coli with the formation of a recombinant GH67 α-glucuronidase (AguA) that catalyzes the conversion of MeGAX3 to methylglucuronate and xylotriose. This gene is followed by xynA2, encoding an intracellular GH10 endoxylanase catalytic domain (XynA2) that processes the xylotriose product generated by the action of AguA on MeGAX3 (17). Here we report the identification of an aldouronate-utilization regulon of Paenibacillus sp. strain JDR-2 that includes genes for the extracellular depolymerization of methylglucuronoxylans, the assimilation of aldouronate products, and the intracellular release of monosaccharides. This coordinately regulated process in which xylan depolymerization and product assimilation are coupled in Paenibacillus sp. strain JDR-2 provides a favorable system for the conversion of lignocellulosic biomass to biobased products.
MATERIALS AND METHODS
Preparation of a cosmid library of Paenibacillus sp. strain JDR-2.
Culture media (50 ml) were inoculated with 1/250 volume of a starter culture of Paenibacillus sp. strain JDR-2 (23). The culture was grown at 30°C with vigorous shaking. After they reached mid-log phase (i.e., an optical density at 600 nm [OD600] of 0.7 × 109 cells/ml), the cells were collected by centrifugation, resuspended in buffer A (50 mM Tris-HCl [pH 8.0], 1.0 M NaCl), pelleted by centrifugation, and resuspended in buffer A again at 4 × 109 cells/ml. An equal volume of 2% low-melting-point agarose was added to the cell suspension, and the cell-agarose mix was poured into 800-μl plug molds. Plugs were further processed as described previously (1).
Partial digestion of genomic DNA.
To determine the optimal amount of enzyme to use for digestion, 360 mg of plugs was equilibrated two times in 50 ml of TBE (90 mM Tris-borate, 2 mM EDTA [pH 8.0]) for 15 min, rinsed with 15 ml of 0.1% Triton X-100, chopped into a slurry and distributed into six 1.5-ml centrifuge tubes. A brief centrifugation (12,000g, 30 s) packed the agarose slurry and the supernatants were removed by aspiration. To each tube containing approximately 60 μl of packed plugs were added 10 μl of 40 mM spermidine, 10 μl of 10× HindIII reaction buffer, and 1 μl of bovine serum albumin (10 mg/ml). Water was added to adjust the final volume to 100 μl. The mixtures were equilibrated on ice for 30 min, and various amounts (0.05 to 0.7 U) of HindIII were added to each tube, equilibrated further for 15 min, and finally incubated at 37°C for 30 min to allow restriction digestion. The reactions were immediately stopped by adding 11 μl of 0.5 M EDTA (pH 8.0).
Field inversion gel electrophoresis.
Slurries of agarose gel plugs were loaded into the wells of a 1% agarose (Bio-Rad) gel in 0.5× TBE or 1× TAE buffer (40 mM Tris-HCl, 20 mM acetic acid, 1 mM EDTA [pH 8.3]). Electrophoresis was carried out by using the FIGE MAPPER apparatus (Bio-Rad) set at program 8, which separated DNA fragments in the 25- to 150-kb range. The initial current was 47 to 50 mA, and the run lasted 20 h. At completion, 0.5 g of an agarose gel piece containing HindIII-digested DNA ranging in size from 20 to 48 kb was cut out and treated with 6 U of Gelase (Epicenter) at 45°C. The released DNA was ready to be ligated to the cosmid vector.
Construction of cosmid library.
Size-selected HindIII-digested genomic DNA fragments were ligated to the HindIII-digested and dephosphorylated cosmid vector pCC1 (Epicenter). Ligation products were packaged into lambda phage packaging extracts (Epicenter) and electroporated into E. coli EPI300 (Epicenter) according to the manufacturer's protocol. Transformed E. coli was plated onto LB-chloramphenicol plates (LB broth [3] in 1.5% Bacto agar containing chloramphenicol at 12.5 μg/ml), and the colonies were picked and stored individually in wells of 9X12 microtiter plates supplemented with LB-chloramphenicol media.
Screening of cosmid library for aldouronate utilization genes.
Pooled cultures from 300 transformants were screened for the presence of the aguA gene by PCR using the primers PF54 and PR569 (Table 1). Cosmid DNA preparations from positive transformants were sequenced.
TABLE 1.
Nucleotide sequences of primers used in this study
| Primer | Nucleotide sequence |
|---|---|
| PF54 | CGAGAGAGAGACATTCCTTATTACG |
| PR569 | CATCTGGTTGGTATGCTCCATCG |
| rre178f | GTGCTGGACGGATTGGAGCTTA |
| rre459r | CTCCGAGAACTGGCCTTGAACA |
| sbp1081f | AACTCGTATGGCGTAGGCAACC |
| sbp1361r | TGGCCTGTATAGTCGCTCCAGA |
| agua1069f | CGGACGCTTCAAGGACAATGTG |
| agua1354r | GGCCGTAATGCCGCTATGAGTA |
| xyl247f | CATACGCTGGTGTGGCACAATC |
| xyl623r | CCGTGAATCGGCACTTGCTTAG |
| bex948f | GGACAAGTCGGTGACCACCAAG |
| bex1291r | CTTGCGCCATCGCCGTTACAAG |
| xynA1-2237f | GCGTCGGAATGCAAGGCCATTA |
| xynA1-2503r | TCTCGGCTCTCCAGCTTGTGTT |
| amp10f | GATCTGGCAGCTTCCTGCATTC |
| amp204r | TCCAGTCCGCGGCTCTTATCAA |
| oxr535f | TCACGGCGCGAACACTTATCTC |
| oxr774r | GCTCATCACAGGCGGAAGGTAT |
| perm-agua791f | TAACGGCGGTTACGCCAACCTC |
| perm-agua81r | CCAGCCTGCGTATTGCTCCAAG |
Preparation of mRNA.
Typically, 8 ml of media in 125-ml flasks was each inoculated with a fresh colony of Paenibacillus sp. strain JDR-2, followed by incubation at 30°C with vigorous shaking at 240 rpm. Cells were collected by centrifugation when growth reached an OD600 of 0.6, and RNA was isolated according to the method of Cheung et al. (5). To remove residual genomic DNA in the resultant RNA fraction, DNase (Promega M610A) was added at 20 U/ml and digested at 37°C for 45 min. The DNase was then inactivated by mixing it with 5 volumes of GTC (4 M guanidine thiocyanate, 25 mM sodium acetate [pH 7.0], 0.1 M β-mercaptoethanol, 0.5% Sarkosyl). One volume of 1.0 M sodium acetate (pH 4.4), six volumes of water-saturated phenol, and one volume of chloroform were added and mixed. The mixtures were centrifuged to separate phases, the aqueous phases were transferred to separate tubes, and RNA fractions were precipitated the after addition of equal volumes of isopropanol. The RNA precipitates were further rinsed with 75% ethanol and dissolved in 100 μl of water. The DNase treatment was repeated until there were no significant traces of genomic DNA-directed PCR products in the subsequent reverse transcription-PCRs (RT-PCRs).
RT-PCR.
Real-time RT-PCR was performed in 16-μl reactions, each containing 100 to 200 ng of RNA, 3.2 μl of 0.25 μM primer pair mixtures, and 8 μl of 2× iScript SYBR mix (Bio-Rad iScript). A typical reaction consisted of the following steps: (i) incubation with reverse transcriptase for 10 min at 58°C; (ii) melting for 3 min at 95°C; (iii) 45 cycles of 10 s at 95°C, 20 s at 58°C, and 20 s at 72°C; and (iv) one cycle of melting curve determination. The reactions were conducted in the Bio-Rad iCycler iQ real-time detection system. The primer pairs used for RNA transcript detection were rre178f and rre459r for yesN, sbp1081f and sbp1361r for yesM, agua1069f and agua1354r for aguA, xyl247f and xyl623r for xynA2, bex948f and bex1291r for xynB, and xynA1-2237f and xynA1-2503r for xynA1. The primer pairs for the flanking genes were amp10f and amp204r for the aminopeptidase gene and oxr535f and oxr774r for the oxidoreductase gene. The primer pair used for probing the readthrough transcript from ytcP to aguA were perm-agua791f and perm-agua81r (Table 1). A total of 15,272 bp of cosmid VC2 was sequenced and submitted to GenBank (accession number EU024644).
RESULTS
Cosmid library analysis.
The cosmid library was screened by PCR with degenerate primers PF54 and PR569. Two transformants, VC1 and VC2, each yielded aguA-specific PCR-generated fragments, as confirmed by nucleotide sequencing, and therefore contained the gene encoding α-glucuronidase. VC1 had a 28-kb insert, while VC2 had a 35-kb insert. Analyses by restriction digestion showed that VC1 and VC2 shared a majority of fragments, indicating that they were from the same genomic region.
Sequence organization of the aldouronate gene cluster in cosmid VC2.
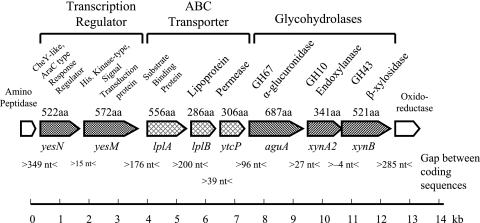
VC2 yielded the 486-bp aguA-specific product by PCR screening with primers PF54 and PR569. These primers were established as specific for the aguA gene identified and sequenced from genomic DNA derived from Paenibacillus sp. strain JDR-2 (17). Cosmid VC2 insert DNA was digested into smaller fragments which were then inserted into pUC19 to facilitate nucleotide sequencing. The results are shown in Fig. 1.
FIG. 1.
Genomic organization of aldouronate-utilization operons in Paenibacillus sp. strain JDR-2.
Genes in this segment were identified by a BLAST search and defined by identification of open reading frames (ORFs). Central in this region is an aguA gene encoding a 687-amino-acid GH67 α-glucuronidase. This aguA gene was followed by a xynA2 gene encoding a 341-amino-acid protein with a GH10 endoxylanase catalytic domain. Following the xynA2 gene was a xynB gene encoding a 521-amino-acid protein, classified as β-xylosidase/arabinofuranosidase in the GH43 family. These three genes constituted a triad of structural genes expected to encode enzymes for the processing of the product generated by the anchored multimodular endoxylanase, XynA1, to xylose after assimilation by the cell.
Immediately 5′ to this triad were three genes that are presumed to encode proteins that comprise an ABC transporter complex. The expected translated products from this triad include a substrate binding protein (556 amino acids), a lipoprotein (286 amino acids), and a permease protein (306 amino acids). Immediately 5′ to the ABC transporter triad were genes capable of encoding a transcription regulation element made up of two proteins: a receiver protein of an AraC-type response regulator of 522 amino acids and a histidine kinase protein of 572 amino acids. An amino peptidase gene was located 349 bp upstream of the transcription regulation unit, while 285 bp downstream of the xynB gene was an NADH-dependent flavin oxidoreductase gene.
Translation start sites of these genes were indicated by the presence of putative ribosome-binding sites (canonical sequence GGAGGG) (14) located 5 to 15 nucleotides 5′ to the translation start sites ATG. These predicted protein products were compared to the archived sequences in GenBank. The genes encoding characterized proteins in the protein databases showing greatest similarity are summarized in Table 2. The average GC content of DNA of Paenibacillus sp. JDR-2 sequenced thus far (∼33 kb) is 52%.
TABLE 2.
Identification of the relevant xylanolytic genes in the 15-kb genomic segment
| ORFa | Protein | COG no. | Function | E value(s)b | Similar protein, identification no. | % Identityc |
|---|---|---|---|---|---|---|
| yesN* | YesN | 4753 | Response regulator containing CheY-like receiver domain and AraC-type DNA-binding domain (signal transduction mechanisms) | 3e-51 | B. subtilis subsp. subtilis 168, GeneID 938764 | 40 (64/159) |
| yesM* | YesM | 2972 | Predicted signal transduction protein with a C-terminal ATPase domain (signal transduction mechanisms) | 1e-48 | B. subtilis subsp. subtilis 168, GeneID 936078 | 23 (138/592) |
| lplA* | UgpB | 1653 | ABC-type sugar transport system, periplasmic component (carbohydrate transport and metabolism) | 4e-10 | B. subtilis subsp. subtilis 168, GeneID 936079 | 25 (49/191) |
| lplB* | LplB | 4209 | ABC-type polysaccharide transport system, permease component (carbohydrate transport and metabolism) | 3e-92 | B. subtilis subsp. subtilis 168, GeneID 936088 | 39 (112/285) |
| ytcP* | UgpE | 0395 | ABC-type sugar transport system, permease component (carbohydrate transport and metabolism) | 9e-41 | B. subtilis subsp. subtilis 168, GeneID 938095 | 34 (102/294) |
| aguA | AguA | pfam03648 | Glycosyl hydrolase family 67; family of α-glucuronidase. | 0.0 | G. stearothermophilus T-6, α-glucuronidase chain A, gi:37926810 | 67 (422/680) |
| xynA2 | XynA2 | smart00633 | Endoxylanase, glycosyl hydrolase family 10 | 8e-90 | G. stearothermophilus, intracellular xylanase IXT6, gi:114054545 | 60 (199/327) |
| xynB* | XynB | pfam04616/3507 | Arabinofuranosidase glycohydrolase family 43/β-xylosidase. | 2e-74/8e-86 | G. stearothermophilus, intracellular xylanase IXT6, XynB, gi:114054567 | 32 (178/540) |
*, The gene name is assigned to that closest in homology found in B. subtilis subsp. subtilis 168.
Similarity to the functional protein family assignment as determined by CD SEARCH (National Center for Biotechnology Information).
The percent identity is the number of residues of the major portion of the query protein identical to those of the subject protein as determined by the BLAST program (National Center for Biotechnology Information).
Identification of transcriptional regulatory elements in genes associated with utilization of methylglucuronoxylan.
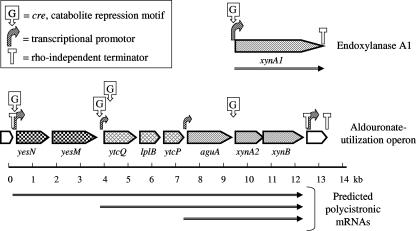
The BPROM program (Softberry) was used to locate bacterial promoters, and the high scoring transcription start sites were located at the 5′ termini of genes potentially encoding YesN, the receiver protein of the response regulator; UgpB, the substrate binding protein of the ABC transporter; and AguA, the α-glucuronidase protein of the glycohydrolase triad in the aldouronate-gene cluster. The promoter 5′ to YesN was identified as having the greatest potential of the cluster. The FindTerm program (Softberry) was used to locate the rho-independent transcriptional stop sites. The transcriptional terminator upstream of yesN was located at 14 bp after the termination codon of the preceding amino peptidase gene and consisted of a 14/20-bp stem-9-bp loop, followed by a 7/8 AT stretch, and punctuated the beginning of the aldouronate utilization gene cluster. The site found downstream from xynB, consisting of a 13/14-bp stem-7-bp loop, followed by an 8/9 AT stretch, was located 16 bp after the termination codon for XynB and punctuated the end of the aldouronate utilization gene cluster. The same analyses applied to the xynA1 gene (23) identified a potential promoter immediately upstream. We also identified a stop site downstream from xynA1 located 1 bp after the termination codon and consisting of a 18/23-bp stem-4-bp loop, followed by a 4/7 AT stretch. The positions for different promoters and terminating sites are presented in Fig. 2.
FIG. 2.
Transcriptional regulation and gene expression of the aldouronate utilization genes in Paenibacillus sp. strain JDR-2.
Transcriptional regulatory genes.
The yesN and yesM genes together made up a two-component transcriptional regulation unit. Sequence homology analysis by CDD Search (www.ncbi.nlm.nih.gov/Structure/cdd) of yesN indicated that it coded for a response regulator protein containing a CheY-like receiver domain at the initial 121 amino acids at the amino terminus with Asp55 as the phosphorylated residue and an AraC-type DNA-binding domain spanning residues 432 to 515 at the carboxyl terminus. We have designated this yesN since it was similar to the yesN gene of the gram-positive prototype organism, Bacillus subtilis subsp. subtilis 168. yesM, on the other hand, contained a HAMP (for histidine kinase/adenyl cyclase/metal-binding proteins/phosphatases) domain at residues 275 to 344, a histidine kinase domain at residues 367 to 450 with His378 as the phosphorylated residue, and an ATPase domain at residues 461 to 558. Again, we designated this yesM for the same reason above. Analysis of the two-component unit yesN-yesM by CDD Search identified loci 2109 and 2110 in Bacillus halodurans C-125 to be most similar in amino acid content (42% [221 of 524] and 45% [261 of 580] identities, respectively) and the above-described domain architecture. Another similar loci pair identified was Clostridium cellulolyticum H10 Draft 2754 and 2755.
ABC-type transporter.
The genes encoding the ABC-type transporter are found in the operon as a cassette of three ORFs. The first ORF in this cassette, lplA, encodes a protein similar to the substrate binding periplasmic component, UgpB, and identified by CDD Search to be at residues 38 to 384. The second coding sequence, lplB, codes for the transmembrane permease component, LplB, which spans the entire length of 286 residues and contains the sequence motif EAA-X3-G-X9-I-X-LP (residues 179 to 198), located in a cytoplasmic loop at a distance of ∼100 residues from the C terminus (20). The third coding sequence, ytcP, encodes a protein with another permease component, spanning residues 16 to 305. Analyses of similarities in sequence showed this ABC transporter to be most similar to B. halodurans C-125 loci 2111 to 2113 (49% [272 of 555], 73% [210 of 287], and 65% [193 of 293] identities in amino acids, respectively). Another similar ABC transporter identified was in C. cellulolyticum H10 Draft, loci 2757 to 2759.
Aldouronate processing functions.
An aguA gene was identified encoding a GH67 α-glucuronidase with a calculated molecular mass of 77,876 Da and a pI of 5.4. This was the same aguA gene cloned and sequenced from genomic DNA and shown to encode a functional α-glucuronidase when expressed in E. coli (17). The identities derived from GenBank entries of AguA proteins were as follows: 63% to Aeromonas punctata, 62% to Geobacillus stearothermophilus T-6, 61% to B. halodurans C-125, and 57% to C. cellulolyticum H10. This protein is highly conserved with respect to catalytic sites. Based upon alignment with the two bacterial GH67 α-glucuronidases of G. stearothermophilus T-6 and Cellvibrio japonicus, for which catalytic mechanisms have been elucidated (10, 15), glutamate and aspartate residues that participate in the acid/base-catalyzed reactions can be discerned. In AguA from Paenibacillus sp. strain JDR-2 Glu401 and Asp373 are homologs of residues Glu392 and Asp364 in G. stearothermophilus T-6 and Glu393 and Asp365 in C. japonicus, which together with a water molecule constitute the catalytic general base. Similarly, Glu294 in Paenibacillus sp. strain JDR-2 probably corresponds to Glu285 in G. stearothermophilus T-6 and Glu292 in C. japonicus as the catalytic general acid. Catalysis results in the hydrolysis of the α-1,2-glycosidic bond between the 4-O-methylglucuronic acid residue and the xylose residue in the aldo-oligouronate substrate by an inverting mechanism.
A xynA2 gene follows aguA. The encoded XynA2 protein contains the catalytic domain of a GH10 endoxylanase spanning residues 10 to 332 of the 341 amino acids. Neither a signal sequence (determined by SignalP [2]) nor any carbohydrate binding module (CBM) was found in the protein sequence. XynA2 has a calculated molecular mass of 39,457 Da and a calculated pI of 5.3 and showed 60 to 61% identity to catalytic domains of GH10 xylanases presumed to function as intracellular enzymes in other bacteria, e.g., G. stearothermophilus T-6 and Thermotoga maritima MSB8.
The last gene in this triad and the last gene in this aldouronate utilization cluster, xynB, encodes a protein of 521 amino acids with an internal family GH43 β-xylosidase/α-arabinofuranosidase defined within residues 11 to 288. It did not have a signal peptide, had a calculated molecular mass of 57,783 Da, and had a calculated pI of 4.9. The encoded XynB protein sequence showed 40% identity to the XynB found in both Bacillus clausii KSM-K16 (gi:56962920) and Geobacillus thermoleovorans (gi:85717961).
Effects of xylan, glucose, and xylose on the relative levels of expression of aldouronate utilization genes.
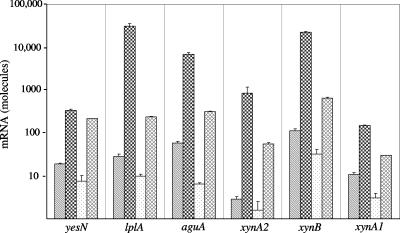
Analysis by real-time RT-PCR of the amount of mRNA produced under different growth conditions (Fig. 3) indicated coordinate induction and repression of expression of genes in this cluster, as well as expression of xynA1 encoding the secreted multimodular GH10 endoxylanase. When growing in only 0.5% yeast extract, all six genes (yesN, lplA, aguA, xynA2, xynB, and xynA1) were expressed, with aguA and xynB mRNAs being slightly more abundant. The levels of mRNA determined in cultures grown in 0.5% yeast extract-containing medium served as points of reference. When 0.5% xylan was added to the yeast extract-containing medium, expressions of the six genes were dramatically enhanced, from 18-fold in the case of the response regulator yesN to more than 200-fold in the case of the substrate binding protein lplA and the β-xylosidase xynB. In contrast, when 0.5% glucose was added to supplement yeast extract in the media instead of xylan, the relative levels of mRNA transcribed by the six monitored genes were all variously reduced to about two-thirds (67% for xynA2) and to as much as one-tenth (10% for aguA) of basal levels. The addition of 0.5% xylose, on the other hand, slightly induced expression, resulting in a 2.7-fold increase (xynA1) to a 19-fold increase (xynA2) over basal levels. In addition, by performing real-time RT-PCR with the primer pair perm-agua791f and perm-agua81r, readthrough transcripts from ytcP to aguA were also identified.
FIG. 3.
Aldouronate utilization gene expression in Paenibacillus sp. strain JDR-2 grown under different nutrient conditions. A colony of Paenibacillus sp. strain JDR-2 was dispersed in 420 μl of Zucker-Hankin (26) salts medium, and 100 μl of this suspension was added to each of four 8-ml portions of culture media in 250-ml baffle flasks containing 1× Zucker-Hankin, 0.5% yeast extract, and either no additional substrate (▧), 0.5% oat spelt xylan ( ), 0.5% glucose (□), or 0.5% xylose (▩). The cultures were incubated at 30°C at 225 rpm for 9 h to an OD600 of 0.6. The cells were harvested, and RNA was prepared from each cell pellet. RNA (100 ng) was added to each 16-μl real-time RT-PCR. At the end of the reaction, threshold cycle levels were converted to mRNA abundance by predetermined standardization of RT-PCR threshold cycles using genomic DNA concentration as a standard.
In separate experiments, analysis of the relative expressed amounts of the 5′ gene (encoding a putative aminopeptidase) and the 3′ gene (encoding a putative oxidoreductase) that flank the eight ORFs comprising the aldouronate utilization gene cluster showed modest variations in response to growth on different substrates, fluctuating from a threefold increase to a threefold decrease of transcripts.
Glucose repression and CcpA binding sites.
In the real-time RT-PCR analyses, expressions of aldouronate utilization genes were reduced up to 10-fold for genes within the cluster and >3-fold for xynA1 outside this cluster when glucose was added to the culture medium containing yeast extract. Glucose catabolite repression in G. stearothermophilus (6) led to the identification of a 14-base canonical sequence (cre motif) within or immediately preceding genes responsive to such transcription repression. With visual inspection of Paenibacillus sp. strain JDR-2 sequences and analysis with the Prokaryotic Promoter Prediction program, at least five such sequences were detected: a sequence 5′ to the response regulator yesN, a second sequence 5′ to lplA, a third sequence about 100 bp 3′ from the translation start site of lplA, a fourth sequence 5′ to xynA2, and a fifth sequence 5′ to xynA1, the endoxylanase gene located distal to this cluster (Table 3). The distribution of the CcpA binding sites, identified as cre sequences, were found in genes comprising the alduronate utilization cluster, as well as in xynA1, the gene encoding the secreted multimodular GH10 endoylanase (Fig. 2).
TABLE 3.
Candidate CcpA binding sites (cre sequences) and their locations 5′ of aldouronate utilization genes in Paenibacillus sp. strain JDR-2
| Binding site sequence and distance from translation start sitea | Similarity to canonical sequenceb | Gene |
|---|---|---|
| 5′-TGWAANCGNTNWCA | 14/14 | crec |
| 5′-TGAAATCGCTTACA*---145 nt---ATG-- | 14/14 | yesN |
| 5′-TGAAAGTGCTTACA*---81 nt---ATG-- | 13/14 | lplA |
| 5′-ATG---104 nt---TGAAGCGGATGACA†-- | 12/14 | lplA |
| 5′-TGAACCGCTGGCAG†---183 nt---ATG-- | 12/14 | xynA2 |
| 5′-TGTAAGCGCTTAAT†---30 nty---ATG-- | 12/14 | xynA1 |
*, Identified by the PPP (Prokaryotic Promoter Prediction) program (Groningen Biomolecular Sciences and Biotechnology Institute, Haren, The Netherlands [http://bioinformatics.biol.rug.nl/websoftware/ppp/ppp_start.php]); †, identified by manual scanning of the upstream region of these genes.
According to S.-G. Cho and Y.-J. Choi (6). Similarity is indicated as the number of matching residues/total number of residues in the sequence.
The canonical sequence, as defined by S.-G. Cho and Y.-J. Choi (6).
DISCUSSION
Genomic organization and regulation of gene expression.
Based upon similarities to orthologs defined in other bacteria, the ABC transporter located 5′ to the aguA gene is most likely concerned with the import of oligoaldouronate substrate for intracellular degradation. Conners et al. (8) in a recent study of the ABC transporters in T. maritima concluded that ABC transporters for carbohydrate uptake are probably controlled by local regulators responsive to the transport substrate or a key metabolic degradation product. Shulami et al. (22) reported that the two-component response regulator and ABC transporter found upstream of the glycohydrolases (GH67 and GH52) in G. stearothermophilus T-6 regulated the expression of this cluster. The organization of the genes in Paenibacillus sp. strain JDR-2 encoding putative transcriptional regulators, transporter proteins, and glycohydrolases, as well as their coordinate regulation, is consistent with these interpretations. The identification of cre motifs within selected genes within each triad further defines the basis for this regulation.
Data from real-time RT-PCR analyses indicated the genes within the aldouronate utilization cluster in Paenibacillus sp. strain JDR-2 were regulated as a unit by the same transcription signals and were differentially expressed compared to the flanking genes encoding amino peptidase and oxidoreductase. The coordinate expression of the aldouronate utilization gene cluster, along with the expression of the xynA1 gene encoding the multimodular and cell anchored GH10 endoxylanase, supports the case made earlier for the coupling of the depolymerization of methylglucuronoxylan with assimilation and processing of the product, MeGAX3 (23). The aldouronate utilization gene cluster, itself comprised of three potential operons coordinately responding to induction or repression (Fig. 2), may thus be considered a regulon. The coordinate response of these genes with xynA1 expands the scope of this regulon to the function of methylglucuronoxylan or xylan utilization. Further definition of these processes awaits the development of transformation systems in Paenibacillus sp. strain JDR-2 or the expression of these systems in Bacillus spp. amenable to transformation.
Comparative genomic organizations of aldouronate utilization clusters.
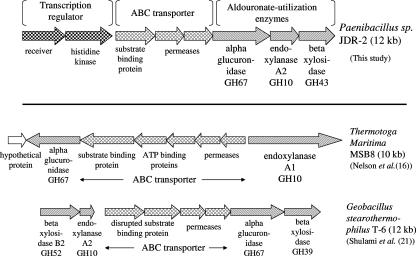
The organization of the aldouronate utilization gene clusters in bacteria that have been studied for this function is presented in Fig. 4. Although there are parallels, as noted above, there are also salient differences. There were no nucleotide-binding domains identified in this Paenibacillus ABC transporter cassette. Neither were genes encoding ATP-binding proteins detected in the four ORFs that precede the aguA gene in G. stearothermophilus T-6, where the first two ORFs were identified as an interrupted substrate binding protein and the last two ORFs were identified as permeases (21). It has been noted in different gram-positive bacteria that a single ATPase may serve more than one set of substrate-binding and membrane-associated proteins that comprise typical ABC transporter systems (19, 20). In the case of T. maritima MSB8 (16), where five ORFs were located adjacent to aguA, two ORFs were identified as genes encoding ATP-binding proteins and were located between genes encoding a substrate binding protein and two permeases.
FIG. 4.
Comparison of aldouronate utilization gene organizations in bacteria in which evidence supports relationships of gene function to substrate utilization.
Another feature distinguishing each of these aldouronate utilization clusters is the relationship to the secreted GH10 endoxylanase, as well as the structural properties of the GH10 endoxylanase itself. Both Paenibacillus sp. strain JDR-2 and T. maritima MSB8 secrete large multimodular enzymes that include CBMs, as well as the GH10 catalytic domain that is distinctive for its generation of the aldotetrauronate MeGAX3. Paenibacillus sp. strain JDR-2 secretes a 1,467-amino-acid endoxylanase comprised of three family 22 CBMs, followed by a GH10 catalytic domain, followed by a single family 9 CBM and triplicate surface layer homology domains (SLH) on the C terminus. This enzyme is cell bound, presumably anchored by the C-terminal SLH modules, and the MeGAX3 and xylooligosaccharides are rapidly assimilated as they are released during depolymerization of methylglucuronoxylan (23). The xynA1 gene encoding this enzyme is located distal from the aldouronate utilization gene cluster, since it is not found in cosmids containing 35-kb inserts that include the 14-kb aldouronate utilization cluster itself. T. maritima secretes a multimodular 1,059-amino-acid GH10 endoxylanase that contains two family 4 CBMs, followed by a GH10 catalytic domain and another two family 9 CBMs, but lacks SLH domains and has not been shown to be cell associated. The xynA1 gene encoding this enzyme is found adjacent to the permease gene for the ABC transporter, and its transcription is in a direction opposite for the genes encoding ABC transporter proteins and AguA. G. stearothermophilus T6 secretes a 407-amino-acid GH10 endoxylanase comprised of a catalytic domain and a 28-amino-acid signal peptide, lacking modules to associate with glucan or xylan polymers or to anchor the enzyme to the cell surface. The xynA1 gene encoding this enzyme is located near the aldouronate utilization cluster, separated by 10 genes most of which encode enzymes involved in glucuronate metabolism (21). Evidence for coordinate expression of aldouronate utilization genes has been demonstrated but not for the expression of the xynA1 gene that encodes the secreted GH10 endoxylanase.
An ortholog of the Paenibacillus xynA2, the third gene in the aldouronate utilization triad, is found also in the vicinity of gene clusters in T. maritima MSB8 and G. stearothermophilus that include orthologs of aguA. In G. stearothermophilus T6, the corresponding gene, xynA2, is located immediately upstream of the genes encoding the ABC transporter and AguA (Fig. 4). This encodes a protein of 339 amino acids and is transcribed in the same direction as the transporter genes. However, a major transcriptional termination site functionally delineates this gene from transporter genes and aguA located immediately downstream (21). An ortholog, xynB (TM0070), encoding a protein of 347 amino acids, is located in the genome of T. maritima MSB8 at a position 17.4 kb from aguA (TM0055), separated by 14 intervening genes. The xynA2 of Paenibacillus sp. strain JDR encodes a GH10 endoxylanase that cleaves the xylotriose molecule to xylose and xylobiose, where xylotriose is a product of the hydrolysis of MeGAX3 by α-glucuronidase encoded by the adjacent aguA gene (17; unpublished data).
Development of bacteria for bioconversion of methylglucuronoxylan.
The rapid and complete utilization of methylglucuronoxylan, along with the synchronized induction and repression of the genes comprising the xylan utilization regulon, supports further development of Paenibacillus sp. strain JDR-2 for the direct conversion of methylglucuronoxylan to biobased products. Growth under conditions of oxygen limitation allows formation of acetate, ethanol, lactate, and succinate. Paenibacillus sp. strain JDR-2 isolate has been approved for complete genome sequencing by the DOE Joint Genome Institute. From this we expect to gain further insight into the metabolic potential of this bacterium, both for the depolymerization and assimilation of polysaccharides comprising cellulosic biomass and for the conversion of carbohydrate components to specific products.
The compact configuration of the aldouronate utilization gene cluster from Paenibacillus sp. strain JDR-2 and its coordinate control recommend it as a cassette for the transformation of other gram-positive bacteria that have been or may be developed for efficient fermentation of xylose. Additional transformation with the xynA1 gene encoding the multimodular GH10 endoxylanase may provide the products for assimilation and subsequent metabolism. The presence of CBMs for interaction with cellulosic polysaccharides and surface layer homology domains that anchor the catalytic domain and associated substrates to the surface of the cell generate products that are in turn rapidly assimilated into the cell. The collective properties that allow extracellular depolymerization, assimilation, and metabolism are presumably the basis for the aggressive xylanolytic activity of Paenibacillus sp. strain JDR-2. Through genetic engineering, gram-positive bacterial biocatalysts may then be developed for the digestion and vectorial conversion of the hemicellulose fraction of cellulosic resources to renewable fuels and chemicals.
ADDENDUM IN PROOF
The position of the start site for the lplA gene may be moved upstream to include 14 amino acids from the start site as designated in Fig. 1 and thus provide a putative signal sequence. This annotation has been included in the GenBank submission.
Acknowledgments
We thank John D. Rice for assistance with the preparation of substrates and Adnan Hosana for assisting in the preparation of the cosmid library. We also thank K. T. Shanmugam and L. O. Ingram for helpful discussions and review of the manuscript.
This research was supported by U.S. Department of Energy grants DE FC36-99GO10476 and DE FC36-00GO10594, the Consortium for Plant Biotechnology Research Project GO12026-198 (DE FG36-02GO12026), and the Institute of Food and Agricultural Sciences, University of Florida Experiment Station, as CRIS Project MCS 3763.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Bell, K. S., A. O. Avrova, M. C. Holeva, L. Cardle, W. Morris, W. DeJong, I. K. Toth, R. Waugh, G. J. Bryan, and P. R. J. Birch. 2002. Sample sequencing of a selected region of the genome of Erwinia carotovora subsp. atroseptica reveals candidate phytopathogenicity genes and allows comparison with Escherichia coli. Microbiology 148:1367-1378. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides-SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biely, P., J. Hirsch, D. C. la Grange, W. H. van Zyl, and B. A. Prior. 2000. A chromogenic substrate for a beta-xylosidase-coupled assay of alpha-glucuronidase. Anal. Biochem. 286:289-294. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 6.Cho, S.-G., and Y.-J. Choi. 1999. Catabolite repression of the xylanase gene (xynA) expression in Bacillus stearothermophilus no. 236 and B. subtilis. Biosci. Biotechnol. Biochem. 63:2053-2058. [DOI] [PubMed] [Google Scholar]
- 7.Collins, T., C. Gerday, and G. Feller. 2005. Xylanases, xylanase families, and extremophilic xylanases. FEMS Microbiol. Rev. 29:3-23. [DOI] [PubMed] [Google Scholar]
- 8.Conners, S. B., C. I. Montero, D. A. Comfort, K. R. Shockley, M. R. Johnson, S. R. Chhabra, and R. M. Kelly. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 187:7267-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dien, B. S., M. A. Cotta, and T. W. Jeffries. 2003. Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63:258-266. [DOI] [PubMed] [Google Scholar]
- 10.Golan, G., D. Shallom, A. Teplitsky, G. Zaide, S. Shulami, T. Baasov, V. Stojanoff, A. Thompson, Y. Shoham, and G. Shoham. 2004. Crystal structures of Geobacillus stearothermophilus α-glucuronidase complexed with its substrate and products. J. Biol. Chem. 279:3014-3024. [DOI] [PubMed] [Google Scholar]
- 11.Ingram, L. O., H. C. Aldrich, A. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Underwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 12.Kuhad, R. C., A. Singh, and K. E. Eriksson. 1997. Microorganisms and enzymes involved in the degradation of plant fiber cell walls. Adv. Biochem. Eng. Biotechnol. 57:45-125. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd, T. A., and C. E. Wyman. 2005. Combined sugar yield for dilute sulfuric acid pretreatment of corn stover followed by enzymatic hydrolysis of the remaining solids. Biores. Technol. 96:1967-1977. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin, J. R., C. L. Murray, and J. C. Rabinowitz. 1981. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus β-lactamase gene. J. Biol. Chem. 256:11283-11291. [PubMed] [Google Scholar]
- 15.Nagy, T., D. Nurizzo, G. J. Davies, P. Biely, J. H. Lakey, D. N. Bolam, and H. Gilbert. 2003. The α-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J. Biol. Chem. 278:20286-20292. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 17.Nong, G., V. Chow, J. Rice, F. St. John, and J. Preston. 2005. An aldouronic acid-utilization operon in a Paenibacillus sp. encodes an alpha-glucuronidase with activity on aldouronic acids generated by acid and enzyme mediated digestion of methyglucuronoxylan, abstr. O-055. Abstr. 105th Natl. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 18.Preston, J. F., J. C. Hurlbert, J. D. Rice, A. Ragunathan, and F. J. St. John. 2003. Microbial strategies for the depolymerization of glucuronoxylan: leads to biotechnological applications of endoxylanases, p. 191-210. In S. D. Mansfield and J. N. Sadler (ed.), Applications of enzymes to lignocellulosics. American Chemical Society, Washington, DC.
- 19.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 20.Schneider, E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303-310. [DOI] [PubMed] [Google Scholar]
- 21.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid-utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulami, S., G. Zaide, G. Zolotnitsky, Y. Langut, G. Feld, A. L. Sonenshein, and Y. Shoham. 2007. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl. Environ. Microbiol. 73:874-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St. John, F., J. D. Rice, and J. F. Preston. 2006. Paenibacillus sp. strain JDR-2 and xynA1: a novel system for methylglucuronoxylan utilization. Appl. Environ. Microbiol. 72:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunna, A., and G. Antranikian. 1997. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 17:39-67. [DOI] [PubMed] [Google Scholar]
- 25.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zucker, M., and L. Hankin. 1970. Regulation of pectate lyase synthesis in Pseudomonas fluorescens and Erwinia carotovora. J. Bacteriol. 104:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]