Abstract
A mutant of Methylobacterium extorquens AM1 with lesions in genes for three formate dehydrogenase (FDH) enzymes was previously described by us (L. Chistoserdova, M. Laukel, J.-C. Portais, J. A. Vorholt, and M. E. Lidstrom, J. Bacteriol. 186:22-28, 2004). This mutant had lost its ability to grow on formate but still maintained the ability to grow on methanol. In this work, we further investigated the phenotype of this mutant. Nuclear magnetic resonance experiments with [13C]formate, as well as 14C-labeling experiments, demonstrated production of labeled CO2 in the mutant, pointing to the presence of an additional enzyme or a pathway for formate oxidation. The tungsten-sensitive phenotype of the mutant suggested the involvement of a molybdenum-dependent enzyme. Whole-genome array experiments were conducted to test for genes overexpressed in the triple-FDH mutant compared to the wild type, and a gene (fdh4A) was identified whose translated product carried similarity to an uncharacterized putative molybdopterin-binding oxidoreductase-like protein sharing relatively low similarity with known formate dehydrogenase alpha subunits. Mutation of this gene in the triple-FDH mutant background resulted in a methanol-negative phenotype. When the gene was deleted in the wild-type background, the mutant revealed diminished growth on methanol with accumulation of high levels of formate in the medium, pointing to an important role of FDH4 in methanol metabolism. The identity of FDH4 as a novel FDH was also confirmed by labeling experiments that revealed strongly reduced CO2 formation in growing cultures. Mutation of a small open reading frame (fdh4B) downstream of fdh4A resulted in mutant phenotypes similar to the phenotypes of fdh4A mutants, suggesting that fdh4B is also involved in formate oxidation.
Formate oxidation to CO2 has been traditionally considered an important last step in the oxidation of more reduced C1 compounds, such as methane, methanol, or methylamine (1, 9). However, mutant phenotype-based evidence in support of the essential role of this step has been missing. Three sets of nonhomologous formate dehydrogenase (FDH) genes have been reported in Methylobacterium extorquens, two predicted to encode NAD-linked FDHs (FDH1 and FDH2) and one predicted to encode a cytochrome-linked FDH (FDH3) (3). One of the NAD-linked enzymes, FDH1, has been purified and characterized (8). FDH1, FDH2, or FDH3 single mutants revealed no phenotypic defect during growth on methanol, while an FDH1/FDH3 double mutant, as well as the triple mutant, with lesions in all three FDH enzymes, has been shown to transiently excrete formate (3). Only the triple mutant was negative for growth on formate, suggesting functional redundancy of the three FDH enzymes. While the formate-negative phenotype of the triple mutant suggested that no additional functional FDHs were present, the ability of this mutant to grow on methanol with low substoichiometric formate accumulation in the medium was puzzling. Such a phenotype implied that an alternative enzyme or a pathway that converts formate must be present. In this work, we identify genes responsible for formate oxidation in the triple mutant, obtain evidence for a fourth functional FDH in M. extorquens, and confirm the essential role of formate oxidation during growth on methanol.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli strains JM109 (Invitrogen), Top10 (Invitrogen), and S17-1 (19) were grown in Luria-Bertani medium in the presence of appropriate antibiotics as described by Sambrook et al. (18). M. extorquens AM1 was routinely grown in the minimal medium described previously (6). Succinate (20 mM) or methanol (125 mM) was used as a substrate for growth in liquid medium. For growth on solid medium, succinate (20 mM), methanol (125 mM), or formate (25 mM) was used. The following antibiotic concentrations were used: kanamycin, 100 μg/ml; rifamycin, 50 μg/ml; and tetracycline, 10 μg/ml. To test the metal-dependent expression of FDHs, a similar medium was used in which molybdenum was omitted and molybdenum, tungsten, or both were added, as appropriate, at 0.3 to 3 μM. Chemostat cultivation was performed in a bench-top fermentor essentially as previously described (5, 21). The methanol concentration in the feed was 25 mM for the wild type and quadruple mutant A and either 25 or 45 mM for the triple-FDH mutant, and dilution rates were 0.1 h−1 unless otherwise stated. Cultures were maintained at steady state with an optical density at 600 nm (OD600) of 1.07 ± 0.01 and a pH of 6.79 ± 0.01.
The following cloning vectors were used: pCR2.1 (Invitrogen) for cloning PCR fragments, pCM184 (11) as a suicide vector, pCM62 and pCM80 (10) as expression vectors, pCM130 (9) for promoter fusion construction, and pRK2013 (4) as a helper plasmid for conjugation. Plasmids created in this work and oligonucleotide primers used in PCR amplifications are listed in Document S1 in the supplemental material.
DNA manipulations.
Plasmid isolation, E. coli transformation, restriction enzyme digestion, and ligation were carried out as described by Sambrook et al. (18). The chromosomal DNA for PCR amplification was isolated essentially as described by Saito and Miura (17) from 3 ml of culture (OD600, approximately 1.5, resulting in approximately 200 μg of DNA).
Genome analysis.
The complete genome of M. extorquens is now available, but the data are proprietary to the Lidstrom laboratory until publication (pending). An early version of the genome (6.5× sequence coverage) was described earlier (2) and is available at http://www.integratedgenomics.com/genomereleases.html#list4. Gene candidates potentially encoding molybdopterin-linked enzymes were identified using word searches against the automated genome annotation (key words formate, FDH, molybdenum, and molybdopterin were used), as well as BLAST searches using protein sequences for the three previously characterized FDH enzymes from M. extorquens (Fdh1A, Fdh2A, and Fdh3A) (3).
Microarray hybridization and analysis.
Wild-type M. extorquens and the triple-deletion mutant (FDH123) were cultivated in a chemostat as described above with a dilution rate of 0.1 h−1. The concentration of methanol as a growth-limiting nutrient was 0.1% for the wild type and 0.18% for the mutant. RNA and labeled cDNA were prepared and hybridized to the Agilent 60-mer custom oligonucleotide microarray as previously described (16), except that only a single array comparing the wild type to the triple-FDH mutant was analyzed. Microarray data comparing wild-type methanol- and succinate-grown cultures were previously published (16).
Enzyme assays.
Enzyme activities were determined in crude extracts obtained by passing cells through a French pressure cell at 1.2 × 108 Pa, followed by centrifugation for 10 min at approximately 15,000 × g. Measurements were done at room temperature (24 to 25°C) in a total volume of 1 ml. FDH was assayed as described previously (8), and catechol dioxygenase was assayed as described previously (7). Enzyme assays were done in triplicate, and the values obtained agreed within 20%. The protein concentration was assessed spectrophotometrically (23).
Formate detection in culture medium.
Samples (1 ml) were taken from growing cultures and pelleted, and the supernatants were used for formate assays. For these, appropriate volumes of supernatant (0.01 to 0.2 ml) were added to the reaction mixture containing 50 mM Tris HCl buffer, pH 8.0, 2 mM NAD, and yeast FDH (Sigma-Aldrich; 2 U per ml), in a total volume of 1 ml. The reaction mixtures were incubated at 37°C for 60 min, and the resulting NADH was measured spectrophotometrically at 340 nm. All measurements were done in triplicate and agreed within 15%.
Medium acidification as a function of formate concentration.
To determine the correlation between the concentration of excreted formate and medium acidification, we built pH curves as a function of the concentration of formic acid (more than 95% pure; Sigma-Aldrich) added to freshly prepared growth medium and also to spent medium (see Document S2 in the supplemental material.). The latter experiment was important because the medium routinely used to cultivate M. extorquens is weakly buffered (15 mM phosphate buffer) and growth of M. extorquens leads to acidification even in the absence of measurable levels of formate.
Matings.
For mutant selection, biparental matings were performed, and for plasmid transfers, triparental matings were performed overnight at 30°C on nutrient agar (BD Diagnostics, Maryland) as previously described (3). The cells were then washed and plated on selective medium supplemented by succinate.
Mutant generation.
A marker exchange technique described in our previous studies (3, 11) was used to generate insertion mutations in fdh4A and fdh4B, and null mutations resulting from a double-crossover event were confirmed by diagnostic PCR. Details of plasmids used to generate mutations are given in Document S1 in the supplemental material.
13C NMR experiments.
Carbon-13 labeling experiments were carried out as previously described (3). Late-exponential-phase cells (OD578, ∼1.2 to 1.5) of wild-type and FDH mutant strains were centrifuged, washed, and resuspended to a final density of 15 mg (dry weight) ml−1 into 6 ml of fresh medium containing no Mn, Fe, or carbon source. The cell suspension was transferred into an airlift nuclear magnetic resonance (NMR) tube (3). At that time, 0.6 ml of D2O was added for the field-locking signal, and aeration at 38 ml/min was switched on. The airlift NMR tube was placed into the NMR magnet, and the initial spectrum was acquired to test for the naturally abundant signals. The labeled carbon source, i.e., 99.9% [13C]methanol or 99.9% [13C]formate (both purchased from Eurisotop, France), was then added to a final concentration of 120 mM or 20 mM, respectively. Spectra were accumulated in consecutive 5-min blocks of 200 scans each. All 13C NMR spectra were obtained at 30°C in the Fourier transform mode at 125.79 MHz on a Bruker spectrometer (Avance 500 MHz) equipped with a dual 1H/13C 10-mm probe head with a spectral width of 15 kHz (16,000 data points) and a 90° pulse angle with an interpulse delay of 1.5 s. Proton decoupling was applied during acquisition. The free induction decays were exponentially multiplied (3-Hz line broadening) prior to Fourier transformation. Chemical shifts were expressed as parts per million relative to the resonance of tetramethylsilane at 0 ppm.
14C labeling experiments.
The rate of 14CO2 production and assimilation of labeled carbon from [14C]methanol were determined using modifications of the previously described methods (12) as follows: (i) assays were done at 28°C rather than at room temperature, (ii) cell samples were diluted to prevent them from becoming oxygen limited during the 12-minute duration of the assay, and (iii) the assimilation was interrupted by pipetting cells directly onto 0.2-μm polyvinylidene difluoride filters and submerging the filters in scintillation fluid, rather than by adding NaOH. In the wild type and the triple-FDH mutant, approximately 20% of the total 14C was recovered as CO2 or biomass (with the rest remaining as [14C]methanol). In the quadruple mutant A, essentially all [14C]methanol remained in the reaction mixture due to naturally low flux into biomass under the conditions used (growth on succinate) and dramatically decreased flux to CO2 (see Results).
Acid stress assays.
Cells were grown to early exponential phase (OD600, 0.3 to 0.7) in minimal medium supplemented with either methanol or succinate. Fifty microliters of the culture was transferred to 2 ml of minimal medium that had been adjusted to pH 2.5 with HCl (70 mM HCl was required). The same amount of culture was transferred to 2 ml of nonacidified medium, and this culture was used as a control. Both were incubated at 30°C with shaking for 45 to 300 min. The number of CFU ml−1 was then determined by plating serial dilutions (in minimal medium) on minimal medium plates supplemented with succinate. In a separate series of experiments, formic acid was used as a stressor instead of HCl. In these experiments, formic acid (Sigma-Aldrich; more than 95% pure) was added to minimal medium to a 20 mM concentration, resulting in a final pH of 4.0.
Nucleotide sequence accession number.
The sequence of the chromosomal region including fdh4AB has been deposited with GenBank under accession number EU073598.
RESULTS
Further phenotypic characterization of the triple-FDH mutant.
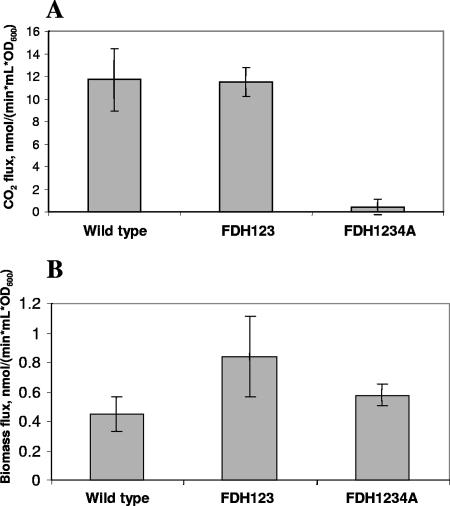
The ability of the mutant in which all three known FDHs had been deleted to grow on methanol suggested the presence of an alternative formate oxidation system. To test this hypothesis, we employed NMR to assess 13CO2/H13CO3− production from [13C]methanol in the triple mutant and found that upon depletion of [3C]methanol, 13CO2/H13CO3− was formed concomitant with the disappearance of accumulated [13C]formate (see Document S3 in the supplemental material.). Carbon flux analysis experiments with 14C labeling revealed that rates of carbon assimilation into biomass and CO2 production were similar in the mutant and wild-type M. extorquens (Fig. 1), supporting the presence of a formate-oxidizing enzyme/pathway in the triple mutant.
FIG. 1.
14CO2 (A) and 14C-biomass (B) production from [14C]methanol measured in the wild type, the triple-FDH mutant, and the quadruple-FDH4A mutant grown on succinate. The error bars show standard deviations (n = 4). The low flux into biomass is typical of cells grown on succinate (12).
In addition, we carried out further phenotypic characterization of the triple mutant. Chemostat-grown cultures of the triple mutant showed a lower yield on methanol than the wild type. Wild-type M. extorquens is typically maintained in chemostat culture limited by methanol (25 mM) at an OD600 of approximately 1 (21). In contrast, the triple-mutant culture reached an OD600 of only approximately 0.8 under these conditions. Only low levels of formate (approximately 0.5 mM) were detected in the culture supernatant, confirming the previous results from batch cultures and suggesting that most of the formate produced from methanol was oxidized (3).
We also conducted experiments to determine dependence on or inhibition of the triple mutant by Mo or W, the metals known to be essential to the characterized molybdopterin enzymes (22). The triple mutant grew normally in medium with no W or Mo added or in medium to which both W and Mo were added but could not grow in medium supplemented with W in the absence of Mo. A similar phenotype was observed for the FDH1/FDH3 double mutant (L. Chistoserdova and M. E. Lidstrom, unpublished data). In the latter mutant, only one known FDH was functional, FDH2, which is highly similar to the previously characterized Mo-linked FDHs (3). Inhibition of Mo-dependent enzymes by W is a well-characterized phenomenon (14, 20). Thus, the phenotypes of the double FDH1/FDH3 and the triple mutants were consistent with the presence of another Mo-linked enzyme that is important for formate oxidation.
Genome-based predictions and microarray-based transcription analysis.
The genome of M. extorquens (a gapped version is publicly available at http://www.integratedgenomics.com/genomereleases.html#list4; publication of the complete genome sequence is pending) was searched for molybdopterin enzyme candidates other than the three known FDH enzymes, as described in Materials and Methods. A total of 10 candidates were detected. Of these, four were annotated as dehydrogenases of the xanthine dehydrogenase family, with three being parts of gene clusters containing other putative xanthine dehydrogenase genes. Six other candidates were annotated as putative molybdopterin oxidoreductases or putative FDHs (data not shown). Of these, only three were predicted to encode polypeptides of the size typical of molybdopterin binding subunits of known FDHs (over 700 amino acid residues).
Whole-genome transcription analysis was conducted with the triple mutant, in comparison with wild-type M. extorquens, in order to detect promising candidates for mutation tests. cDNAs produced from RNAs isolated from the triple-mutant and wild-type strains growing in a chemostat with a dilution rate of 0.1 h−1 were labeled with Alexa-Fluor 555 and Alexa-Fluor 647, respectively; hybridized to a single whole-genome M. extorquens microarray (16); and analyzed as previously described (16). A list of genes was generated that were differentially expressed in the triple mutant compared to the wild type (see Document S4 in the supplemental material). Only one molybdopterin protein candidate identified via genome searches (see above) was part of this list, represented by two open reading frames (RMQ08549 and RMQ08550) in the incomplete version of the genome, due to a frame shift. The corrected sequence of the region, containing no frame shifts, has been deposited with GenBank (see above). In accordance with COG (clusters of orthologous groups) classification, the peptide translated from this gene carries similarity to uncharacterized putative molybdopterin-binding oxidoreductase-like proteins (COG0243). We tentatively designated this gene fdh4A and chose it as the primary candidate for mutation. At the amino acid level, Fdh4A carries little similarity to known FDH alpha subunits from M. extorquens or from other microorganisms (24 to 36% identity). However, BLAST analysis against the nonredundant database (NCBI) revealed the presence of homologs (up to 72% identity at the protein level) in a variety of proteobacterial species, suggesting that the function encoded by fdh4A is widespread.
Mutant generation and mutant phenotypes.
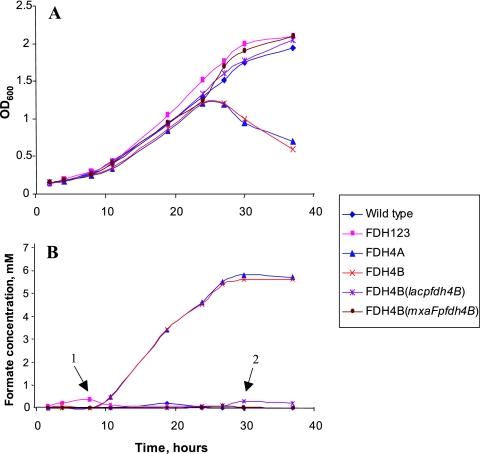
Mutations in fdh4A were generated in the triple-mutant background, as well as in the wild-type background, and growth phenotypes were tested on methanol and formate plates. The fdh4A mutant generated in the triple-mutant background (quadruple mutant A, or FDH1234A) was unable to grow on methanol. As expected, it also did not grow on formate. When succinate-grown cells of this mutant were incubated with methanol, large amounts of formate were excreted into the medium (up to 7 mM), accompanied by medium acidification to pH 4.7. Mutation in the wild-type background (the FDH4A mutant) resulted in diminished growth on methanol-containing plates, while no growth defect was observed on formate plates. When grown in liquid culture on methanol, the FDH4A mutant revealed a dramatic phenotype. In the mid-exponential phase of growth, it accumulated formate up to 6 mM concentration, followed by rapid medium acidification, growth arrest, and lysis (Fig. 2). Perfect correlation was observed between the concentration of excreted formate and medium acidification (see Document S2 in the supplemental material). In a pH-controlled (via automated NaOH additions) chemostat, the mutant grew to the OD typical of the wild type (OD600, 1) only if twice the standard concentration of Fe (i.e., 7.2 instead of 3.6 μM) was added to the medium.
FIG. 2.
Growth of the wild type, the triple-FDH mutant, and the quadruple mutants FDH4A and FDH4B on methanol (A) and formate accumulation in the medium (B). Arrows 1 and 2 indicate low levels of formate transiently accumulated by the triple mutant and by the FDH4B (lacp fdh4B) construct.
A small open reading frame was identified downstream of fdh4A, whose start codon overlapped by 1 nucleotide with the stop codon of fdh4A, suggesting that the two may be cotranscribed. The translated polypeptide is 107 amino acids long and reveals no recognizable motifs in its sequence. Homologs of this gene (sharing 40 to 45% identity at the amino acid level) are present in the genomes of Agrobacterium, Rhizobium, and Brucella species, and in these genomes, they are also located immediately downstream of fdh4A homologs. We tentatively designated the small open reading frame fdh4B and constructed fdh4B mutants in both wild-type and triple-mutant backgrounds. The resulting mutant phenotypes were similar to those for fdh4A mutants. Adding the fdh4B mutation to the triple-mutant background (quadruple mutant B) caused a methanol-negative phenotype, while the fdh4B mutation in a wild-type background (mutant FDH4B) caused formate excretion during growth on methanol, acidification of the medium, and growth arrest in late exponential phase (Fig. 2). These data suggest that fdh4B is involved in oxidation of formate, together with fdh4A. No other genes appeared to be associated with fdh4AB in terms of transcription or predicted function. The DNA region containing fdh4AB and surrounding genes is depicted in Document S5 in the supplemental material.
Acid stress response tests.
Fdh4A shares significant amino acid identity with the YdeP proteins of E. coli and Shigella flexneri (48%), which have been shown to be involved in acid stress response in the respective organisms (13, 15). Mutants of E. coli and S. flexneri defective in ydeP showed a distinctive acid-sensitive phenotype (13, 15). We tested whether the fdh4A mutant might have an acid-sensitive phenotype as well. A protocol similar to the one used for enterobacteria (medium adjusted to pH 2.5 by HCl) was employed, but no difference in survival was observed between wild-type M. extorquens and the fdh4A mutant (data not shown). In addition, we tested formic acid as a stressor (see Materials and Methods), but again, we did not observe any difference in survival between wild-type M. extorquens and the fdh4A mutant.
Labeling experiments with the FDH quadruple mutant.
Formation of 13CO2/H13CO3− from [13C]formate was observed by NMR in suspensions of cells that were grown in the presence of both succinate and methanol (to induce formate oxidation systems). The triple mutant, as well as the wild type, showed the formation of 13CO2/H13CO3− at the first data acquisition point (12 min) after [13C]formate addition, whereas no 13CO2/H13CO3− was detectable in quadruple mutant A (data not shown). However, after 40 min, 13CO2/H13CO3− production was also observable in the quadruple mutant, albeit at a lower concentration than in the triple mutant. This indicates that the quadruple-FDH mutant had a decreased ability to convert formate to CO2; however, CO2 formation from formate was still possible. 13C labeling of CO2/HCO3− was determined qualitatively rather than quantitatively, since CO2 could escape with the air stream that was used for aeration of the cell suspension. Therefore, we performed experiments to follow carbon fluxes from [14C]methanol into CO2/HCO3− and into biomass (12). Levels of CO2 production were found to be similar in the wild type and the triple mutant, while flux into CO2/HCO3− in the quadruple mutant was at background level (Fig. 1A). In contrast, carbon flux into biomass was not affected in either the triple or the quadruple mutant (Fig. 1B).
Induction of FDH4.
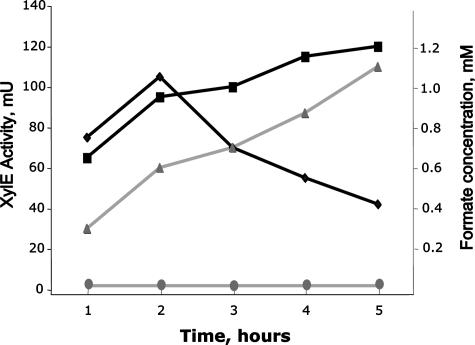
The formate excretion phenotypes of the fdh4A and fdh4B mutants suggested that FDH4 may be important for formate consumption during the late exponential and stationary growth phases of M. extorquens (Fig. 2). In order to obtain insights into the mechanisms of FDH4 induction, we cloned the putative promoter region upstream of fdh4A into a promoter-probe vector, pCM130 (see Documents S1 and S5 in the supplementary material), and followed the activity of the reporter enzyme, catechol dioxygenase (XylE) (7), over different stages of growth of wild-type M. extorquens with methanol as a substrate. However, we did not observe dramatic changes in the expression of fdh4A with the growth phase. It was somewhat higher in the early exponential phase (30 ± 5 nmol min−1 mg protein−1) and somewhat lower in the late exponential phase (10 ± 2 nmol min−1 mg protein−1). We also tested methanol, formate, and HCl as inducers of fdh4A in a wild-type background. For these experiments, cells were grown on succinate to mid-exponential phase, pelleted, and resuspended in medium containing methanol (250 mM), sodium formate (20 mM), or HCl (70 mM; pH 2.5), followed by incubation for 1 h at 30°C with shaking. While no XylE activity above vector background was found in succinate-grown cells, all of the tested inducers resulted in comparable levels of XylE activity: 30 to 50 nmol min−1 mg protein−1. To test whether induction by methanol was due to formate production, we introduced the fdh4Ap-xylE fusion into the quadruple mutant, which excretes large amounts of formate when exposed to methanol. Cultures were grown in succinate-containing medium to stationary phase, and cells were harvested and transferred into fresh medium containing 187.5 mM methanol. Samples were taken every hour over a 5-hour incubation and tested for XylE activity and formate excretion (Fig. 3). In wild-type cells that were used as a control, a spike of XylE activity was followed by gradual decline, and no excreted formate was measured in the medium. In contrast, formate was steadily accumulated by the quadruple mutant, and the activity of XylE increased with time.
FIG. 3.
Induction of fdh4Ap measured via XylE levels (squares, FDH1234A mutant; diamonds, wild type) as a function of formate accumulation (triangles, FDH1234A mutant; circles, wild type).
Expression of fdh4A and fdh4B.
We performed expression experiments in order to attempt to measure the activity of FDH4 and in order to assess whether FDH4 expressed at a higher level could support the growth of M. extorquens on formate, to provide further insights into the formate-negative growth phenotype of the triple mutant. We expressed fdh4A and fdh4B under the mxaFp promoter (with high strength in M. extorquens [10]) and also under the lacp promoter (with low strength in M. extorquens [10]). We also expressed both genes simultaneously under both mxaFp and lacp. Our attempts to measure the activity of FDH4 using standard conditions, aerobically or anaerobically, in the triple mutant grown on methanol or in genetic constructs in which fdh4AB were transcribed from mxaFp were all negative. However, given the novel nature of this enzyme, these results are not surprising. The location of the fdh4AB pair on the chromosome (see Document S5 in the supplemental material) does not suggest a natural electron acceptor (such as a cytochrome), and the divergent natures of Fdh4A and Fdh4B do not suggest any additional cofactors or effectors.
However, we were able to demonstrate that plasmid constructs carrying fdh4A or fdh4B transcribed at appropriate levels from either lac or mxaF promoters complemented the respective mutants for growth on methanol and also for formate excretion, as exemplified in Fig. 2 for the Fdh4B mutant. Similar data were obtained for the Fdh4A mutant. These data indicate that the severe phenotypes observed were due to single specific mutations and not to any polar effects. Interestingly, different levels of expression were required for the respective genes to complement mutations in fdh4A and fdh4B. As shown in Table 1, high expression of fdh4A in quadruple mutant A resulted in restoration of growth on both methanol and formate, while low expression did not. In contrast, either high or low expression of fdh4B restored growth of quadruple mutant B on methanol, but none restored growth on formate (Table 1). Constructs carrying mxaFp fdh4AB showed only weak growth on methanol and no growth on formate. In wild-type M. extorquens, this construct caused a severe inhibitory effect, resulting in complete growth arrest with methanol as a substrate, while a negligible inhibitory effect was observed on formate and expressing either gene separately did not affect growth. An inhibitory effect was also apparent in all of the mutants, although to a lesser degree (Table 1). The triple (FDH123) mutant was not complemented for growth on formate by either of the constructs described above (Table 1).
TABLE 1.
Plate tests for growth on methanol and formate for wild-type M. extorquens and mutants described in this work and effects of low versus high expression of fdh4A and fdh4Ba
| Plasmid | Wild type
|
FDH123
|
FDH4A
|
FDH4B
|
FDH1234A
|
FDH1234B
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | M | F | M | F | M | F | M | F | M | F | |
| No plasmid | + | + | + | − | + | + | + | + | − | − | − | − |
| lacpfdh4A | + | + | + | − | + | + | + | + | − | − | − | − |
| lacpfdh4B | + | + | + | − | + | + | + | + | − | − | + | − |
| lacpfdgh4AB | + | + | + | − | + | + | + | + | − | − | + | − |
| mxaFpfdh4A | + | + | + | − | + | + | + | + | + | + | − | − |
| mxaFpfdh4B | + | + | + | − | + | + | + | + | − | − | + | − |
| mxaFpfdh4AB | − | + | −/+ | − | −/+ | + | −/+ | + | −/+ | − | −/+ | − |
M, methanol; F, formate. +, strains grew on plates and also in liquid media with wild-type characteristics; −, strains were negative for growth. Growth on formate was tested only on plates because of extremely poor growth in liquid media. Growth on methanol was tested on plates and in liquid media. −/+, strains grew poorly on plates and showed diminished growth in liquid media.
DISCUSSION
In this work, we further investigated the phenotype of the mutant of M. extorquens deficient in the three previously detected FDH enzyme systems and established that a fourth FDH enzyme system, previously unrecognized as an FDH, is present that allows this mutant to grow on methanol. Mutation of this fourth system in the triple-mutant background resulted in a methanol-negative phenotype concomitant with loss of normal CO2 production from methanol, demonstrating that formate oxidation is indeed an essential step in the methylotrophic growth of M. extorquens. While we could not demonstrate in vitro activity of this enzyme using standard assay conditions, the function of FDH4 in the oxidation of formate was supported by data on formate excretion and by NMR and carbon flux analyses. The appearance of low levels of CO2 in the quadruple mutant after a long lag suggests a fifth alternate for formate consumption, albeit at low levels and not allowing growth on methanol. The possibilities include, besides a fifth, unrecognized FDH present at low levels, a fortuitous set of metabolic pathways that involve a decarboxylation step. Further studies will be necessary to distinguish between these possibilities.
Mutations in both fdh4A and fdh4B caused a dramatic phenotype in M. extorquens involving formate excretion and growth arrest in batch cultures, despite the presence of intact FDH1, FDH2, and FDH3. This phenotype suggests that both genes are important for the function of FDH4 and that the function of FDH4 is nonredundant. However, complementation experiments demonstrated different patterns for fdh4A and fdh4B mutants, suggesting that Fdh4B is unlikely to be a subunit of FDH4 and that both Fdh4A and Fdh4B may have additional functions. Homologs of Fdh4A have been implicated in acid stress response in E. coli and S. flexneri via an unknown mechanism (13, 15). While we were not able to observe any difference between the wild type and the fdh4A mutant in survival tests, the induction pattern for fdh4A was consistent with its being involved in acid stress response. Other data, such as the dramatic growth phenotype, Fe dependence, and differential complementation patterns, point to the possible involvement of either fdh4A, fdh4B, or both in a regulatory mechanism that may or may not be connected to acid stress. An inhibitory effect of YdeP, the E. coli Fdh4A homolog, on wild-type E. coli has been reported, but the mechanism remains unknown (13). However, overexpression of either fdh4A or fdh4B did not inhibit growth of M. extorquens on methanol, while expressing both caused severe inhibition, suggesting that Fdh4AB form a functional couple. The fact that the inhibitory effect was either not present or less pronounced on formate points to different roles for the couple during growth on methanol versus formate. This hypothesis would also be in agreement with the remarkable phenotype of the triple mutant, which grows well on methanol but is unable to grow on formate. The observation that an overexpressed Fdh4A is able to complement the quadruple mutant but not the triple mutant suggests that the ratio of Fdh4A to Fdh4B may be important for the function of Fdh4AB, as genetically, these mutants differ only in expression levels for Fdh4AB. The complementation data also agree with this hypothesis.
Determining the mechanism of Fdh4B or the Fdh4AB couple will require further investigation. At this point, we can only hypothesize that the mechanism will likely involve parts of metabolism that are principally different during growth on methanol versus growth on formate (1, 2, 9). Possibilities include the main pathways for electron transfer and for NADPH regeneration.
Acknowledgments
We are grateful to Jonathan Miller and Yoko Okubo for help with the microarray experiment.
This work was supported by a grant from the NIH (GM36296) to M.E.L.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Chistoserdova, L., S. W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistoserdova, L., M. Laukel, J.-C. Portais, J. A. Vorholt, and M. E. Lidstrom. 2004. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 186:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ditta, G., T. Schmidhauser, F. Yakobson, P. Lu, X. Liang, D. Finlay, D. Guinley, and D. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 5.Guo, X., and M. E. Lidstrom. 2006. Physiological analysis of Methylobacterium extorquens AM1 grown in continuous and batch cultures. Arch. Microbiol. 186:139-149. [DOI] [PubMed] [Google Scholar]
- 6.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 7.Kataeva, I. A., and L. A. Golovleva. 1990. Catechol 2,3-dioxygenase from Pseudomonas aeruginosa 2x. Methods Enzymol. 188:115-121. [DOI] [PubMed] [Google Scholar]
- 8.Laukel, M., L. Chistoserdova, M. E. Lidstrom, and J. A. Vorholt. 2003. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270:325-333. [DOI] [PubMed] [Google Scholar]
- 9.Lidstrom, M. E. 2001. Aerobic methylotrophic prokaryotes, p. 431-445. In E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY.
- 10.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 11.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 12.Marx, C. J., S. J. Van Dien, and M. E. Lidstrom. 2005. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 3:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 14.Notton, B. A., and E. J. Hewitt. 1971. The role of tungsten in the inhibition of nitrate reductase activity in spinach (Spinacea oleracea L.) leaves. Biochem. Biophys. Res. Commun. 44:702-710. [DOI] [PubMed] [Google Scholar]
- 15.Oglesby, A. G., E. R. Murphy, V. R. Lyer, and S. M. Payne. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and YdeP. Mol. Microbiol. 58:1354-1367. [DOI] [PubMed] [Google Scholar]
- 16.Okubo, Y., E. Skovran, X. Guo, D. Sivam, and M. E. Lidstrom. Implementation of microarrays for Methylobacterium extorquens AM1. OMICS, in press. [DOI] [PubMed]
- 17.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 71:619-629. [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmid for in vivo manipulations of Gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interactions. Springer, Berlin, Germany.
- 20.Takahashi, H., and A. Nason. 1957. Tungstate as a competitive inhibitor of molybdate in nitrate assimilation and in N2 fixation by Azotobacter. Biochim. Biophys. Acta 23:433-435. [DOI] [PubMed] [Google Scholar]
- 21.Van Dien, S. J., T. Strovas, and M. E. Lidstrom. 2003. Quantification of central metabolic fluxes in the facultative methylotroph Methylobacterium extorquens AM1 using 13C-label tracing and mass spectrometry. Biotechnol. Bioeng. 84:45-55. [DOI] [PubMed] [Google Scholar]
- 22.Vorholt, J. A., and R. K. Thauer. 2003. Molybdenum and tungsten enzymes in C1 metabolism. Met. Ions Biol. Syst. 39:571-619. [PubMed] [Google Scholar]
- 23.Whitaker, J. R., and P. E. Granum. 1980. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal. Biochem. 109:156-159. [DOI] [PubMed] [Google Scholar]