Abstract
Summary: High-affinity iron acquisition is mediated by siderophore-dependent pathways in the majority of pathogenic and nonpathogenic bacteria and fungi. Considerable progress has been made in characterizing and understanding mechanisms of siderophore synthesis, secretion, iron scavenging, and siderophore-delivered iron uptake and its release. The regulation of siderophore pathways reveals multilayer networks at the transcriptional and posttranscriptional levels. Due to the key role of many siderophores during virulence, coevolution led to sophisticated strategies of siderophore neutralization by mammals and (re)utilization by bacterial pathogens. Surprisingly, hosts also developed essential siderophore-based iron delivery and cell conversion pathways, which are of interest for diagnostic and therapeutic studies. In the last decades, natural and synthetic compounds have gained attention as potential therapeutics for iron-dependent treatment of infections and further diseases. Promising results for pathogen inhibition were obtained with various siderophore-antibiotic conjugates acting as “Trojan horse” toxins and siderophore pathway inhibitors. In this article, general aspects of siderophore-mediated iron acquisition, recent findings regarding iron-related pathogen-host interactions, and current strategies for iron-dependent pathogen control will be reviewed. Further concepts including the inhibition of novel siderophore pathway targets are discussed.
INTRODUCTION
Most organisms require iron as an essential element in a variety of metabolic and informational cellular pathways. More than 100 enzymes acting in primary and secondary metabolism possess iron-containing cofactors such as iron-sulfur clusters or heme groups. The reversible Fe(II)/Fe(III) redox pair is best suited to catalyze a broad spectrum of redox reactions and to mediate electron chain transfer. Furthermore, several transcriptional (e.g., bacterial Fur and PerR) and posttranscriptional (e.g., mammalian iron regulatory proteins [IRPs]) regulators interact with iron to sense its intracellular level or the current status of oxidative stress in order to efficiently control the expression of a broad array of genes involved mainly in iron acquisition or reactive oxygen species (ROS) protection (131, 167). In special cases, the majority (>80%) of the cellular proteome consists of iron-containing proteins that need iron as a “rivet” for overall structural and functional integrity as found in the archaebacterium Ferroplasma acidiphilum (90). The cellular uptake of iron is restricted to its physiologically most relevant species, Fe(II) (ferrous iron) and Fe(III) (ferric iron). Fe(II) is soluble in aqueous solutions at neutral pH and is hence sufficiently available for living cells if the reductive state is maintained. Generally, Fe(II) can be taken up by ubiquitous divalent metal transporters. Systems for specific Fe(II) uptake are known in bacteria and yeast.
However, in most microbial habitats, Fe(II) is oxidized to Fe(III) either spontaneously by reacting with molecular oxygen or enzymatically during assimilation and circulation in host organisms. In the environment, Fe(III) forms ferric oxide hydrate complexes (Fe2O3 × nH2O) in the presence of oxygen and water at neutral to basic pH. These complexes are very stable, leading to a free Fe(III) concentration of 10−9 to 10−18 M. In mammalian hosts, the assimilated iron is tightly bound to various proteins. Hemoproteins such as hemoglobin contain about two-thirds of the body iron in the heme-bound state. Ferritin, the intracellular iron storage protein, is able to store up to 4,500 Fe(III) ions per oligomer and contains about 30% of the iron pool. The iron of the circulating exchangeable pool that comprises only several milligrams is bound to transport proteins such as transferrin in the plasma delivering iron into the cells via transferrin receptor-mediated endocytosis or innate defense proteins such as lactoferrin in various body fluids. Both transferrin and lactoferrin contain two Fe(III) binding sites per molecule. This strict iron homeostasis leads to a free serum iron concentration of about 10−24 M (272). Thus, a plethora of microorganisms, among them important human and animal pathogens, are severely restricted in iron acquisition. During evolution, this restriction made life advantageous for those microbes that developed skills for highly selective iron uptake, which basically include mechanisms for the utilization of iron sources by either direct or indirect contacts. Direct mechanisms comprise the uptake of various iron sources such as lactoferrin, transferrin, ferritin, heme, and/or hemoproteins (330). The disadvantage of direct uptake is the requirement of a specific receptor for each iron source. Since the composition of iron sources changes between host compartments, those pathogens that use a defined set of iron sources directly are restricted to compartments in which these sources are available. This could be one of the reasons why strategies of iron acquisition that are in contrast based on indirect mechanisms are more broadly distributed. The indirect strategies of iron acquisition are quite diverse. One of them, which is found in gram-negative bacteria, employs specialized secreted proteins called hemophores to acquire heme from different sources. The hxu hemophore system of Haemophilus influenzae uses heme-loaded hemopexin as specific heme/iron source, while the has system of several other gram-negative bacteria uses heme from various sources. However, the hemophore systems are restricted to heme iron sources, making them minimally useful under conditions of low heme availability. In contrast, another indirect strategy is capable of exploiting all available iron sources independent of their nature, thus making it the most widespread and most successful mechanism of high-affinity iron acquisition in the microbial world. In analogy to the hemophore system, it is based on a shuttle mechanism that, however, uses small-molecule compounds called siderophores (generally <1 kDa) as high-affinity ferric iron chelators. Siderophore-dependent iron acquisition pathways can be found among a broad spectrum of prokaryotic and eukaryotic microbes (and even in higher plants) and show a high variety in structure and function of the involved components. The common theme is the production of one or more siderophores by cells during periods of iron starvation (which means that the intracellular iron concentration drops below the threshold of about 10−6 M, which is critical for microbial growth). Secreted siderophores form extracellular Fe(III) complexes with stabilities ranging over about 30 orders of magnitude for different siderophores. Next, either the iron-charged siderophore is taken up by ferric-chelate-specific transporters or siderophore-bound Fe(III) undergoes reduction to Fe(II), which is catalyzed by free extracellular or membrane-standing ferric-chelate reductases. A common advantage for cells is the utilization of xenosiderophores, which means that they possess ferric-chelate reductases and/or uptake systems for siderophores not synthesized by themselves. Baker's yeast, for example, refrains completely from siderophore production but is capable of utilizing several exogenous siderophores as iron sources. If not already released extracytoplasmatically, the iron has to be removed from the Fe-siderophore complex in the cytosol. This is mediated either by intracellular ferric-siderophore reductases or, in a few cases, by ferric-siderophore hydrolases. The following intracellular iron channeling is only partially known. It is uncertain whether iron delivered into the microbial cell could be used immediately for metabolic and regulatory functions such as iron-sulfur cluster assembly and iron-dependent gene expression, respectively, or if intermediate storage has to precede. Several components are involved in iron storage, such as ferritin-like proteins, which either are heme free or, as in the case of bacterioferritins, contain a heme b (47). The DNA-protecting Dps protein that is ubiquitous in prokaryotes also binds iron (29); however, this functional aspect seems to be more related to the prevention of ROS generation than to a particular role in iron storage (357). The binding component of the intermediary mobile Fe(II) pool in Escherichia coli was found to comprise mainly phosphorylated sugar derivatives containing pentose and/or uronic acid as the major fraction (25). The compound was termed ferrochelatin. The suggested oligomeric structure contains monosaccharide building blocks linked by phosphate esters and/or glycosidic bonds. Consistent with the ROS protective function of iron storage proteins, it has been suggested that the sequestration of mobile Fe(II) by the sugar phosphate complex is important to reduce the intracellular level of the Fenton reaction.
SIDEROPHORES: CHEMISTRY AND BIOLOGY
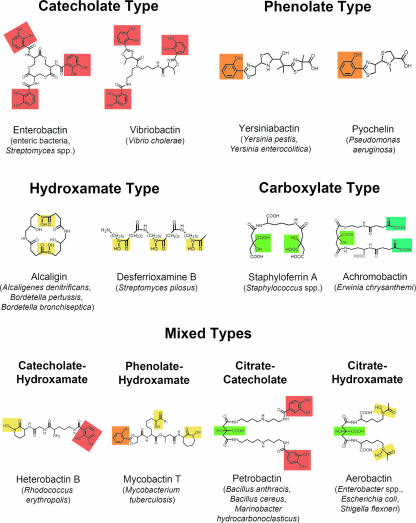
Siderophores can be divided into three main classes depending on the chemical nature of the moieties donating the oxygen ligands for Fe(III) coordination, which are either catecholates (sensu stricto, catecholates and phenolates; better termed as “aryl caps”), hydroxamates, or (α-hydroxy-)carboxylates. However, increasing information about new siderophores led to a more complex classification since many structures that integrate the chemical features of at least two classes into one molecule, resulting in “mixed-type” siderophores, are meanwhile known. Some representative structures of various siderophore types are shown in Fig. 1.
FIG. 1.
Representative examples of different siderophores and their natural producers. Moieties involved in iron coordination are highlighted as follows: catecholates are in red, phenolates are in orange, hydroxamates are in pale yellow, α-hydroxy-carboxylates (deriving from citrate units) are in light green, and α-keto-carboxylates (deriving from 2-oxo-glutarate units) are in blue-green.
Physicochemical Properties of Siderophores
Siderophores are designed to form tight and stable complexes with ferric iron. Generally, the hard Lewis acid Fe(III) is strongly solvated in aqueous solution, forming an octahedral Fe(H2O)63+ complex (73). Due to the gain of entropy, the siderophore donor atoms favorably replace the solvent water and surround Fe(III) in a hexacoordinated state that usually has an octahedral geometry as it is found in the aqueous ion. Provided that the siderophore contains six donor atoms, a 1:1 complex with Fe(III) is generally formed. If there are less than six donor atoms provided by the ligand, the vacancies may be occupied by alternative oxygen donors such as water molecules, or siderophore complexes with higher stoichiometry may be built up as in the cases of rhodoturolic acid that forms Fe2L3 complexes (46), pyochelin that forms both FeL and FeL2 complexes (319), or cepabactin that forms FeL3 complexes (168). Even the formation of mixed complexes was observed for cepabactin and pyochelin, forming 1:1:1 complexes with Fe(III) (168). The diverse formation of iron-siderophore complexes with higher stoichiometry is strongly dependent on ligand concentration and protonation (319). The bound Fe(III) is always found in a high-spin d5 electronic configuration in the siderophore complex. Although there is no ligand field stabilization energy provided by this configuration, the complex is also kinetically stable, since the oxygen donor atoms that are mainly used in siderophores for iron coordination represent hard Lewis bases that allow additional strong ionic interactions between metal and ligand. Thus, siderophores display an enormous affinity towards Fe(III). Fe(III)-siderophore complexes often show characteristic UV-visible and circular dichroism spectra, and as there are no spin-allowed d-d transitions of the metal center in the case of bound Fe(III), the spectra are caused by ligand-to-metal or ligand-to-ligand charge transfer (271). The borderline Lewis acid Fe(II), in contrast, prefers interaction with softer donor atoms such as nitrogen or sulfur, which are only partially employed for iron coordination in natural siderophores. Furthermore, the higher Fe(II) electron density is poorly compensated for in the oxygen-donor atom-dominated siderophore complexes, and the low ratio of charge to ionic radius of Fe(II) compared with that of Fe(III) might be additionally unfavorable to maintain the optimal complex geometry. Thus, the capability of siderophores of forming stable complexes with Fe(II) is rather low.
In the thermodynamics of iron-siderophore binding, formation constants (Kf values) are attractive primarily for comparisons of iron affinities of various siderophores. In general, the overall equilibria of metal-ligand stability constants are expressed by a standard convention as βmlh values for the reaction mM + lL + hH = MmLlHh, where M is metal, L is ligand, and H is proton(s). For the wide variety of siderophores known so far, the corresponding formation constants for iron binding to the fully deprotonated ligand (Kf ☰ β110) ranges over about 30 orders of magnitude. This enormous affinity range is representatively demonstrated by two E. coli siderophores: the mixed citrate-hydroxamate aerobactin as one of the weakest monohexadentatic iron chelators, with a log β110 of 22.5 (133), and the triscatecholate enterobactin as the strongest iron-chelating compound ever found, displaying a log β110 of 49 (198). However, since protonation of the donor atoms is a competitive reaction to metal chelation, the pKa values of the donor groups have to be considered in terms of effectiveness of iron complexation. Catecholate siderophores have pKa values from 6.5 to 8 for the dissociation of the first hydrogen and about 11.5 for the second hydrogen from the catecholic hydroxyl groups. Hydroxamates show pKa values from 8 to 9. The pKa values of carboxylates ranging from 3.5 to 5 make them efficient siderophores under lower-pH conditions at which catecholates and hydroxamates are still fully protonated. Thus, microbes living in acidic habitats, such as many fungi, tend to prefer carboxylate siderophores for iron mobilization, although they could not compete with stronger siderophores such as catecholates at physiological pH (74).
Because proton-independent Kf values do not reflect the real iron binding capacity of the siderophores under physiological conditions for which complete deprotonation is usually not achieved, a more convenient measure for comparing the true relative abilities of different siderophores to bind ferric iron is the pH-analogous pFe value, which gives the negative decadic logarithm of the free iron concentration if, according to a standard convention, the total Fe(III) concentration is 10−6 M and the total ligand concentration is 10−5 M. Since the pH of the medium strongly influences the chelation efficiency, pFe is a pH-dependent value. In this respect, only at above a pH of ∼5.0 is enterobactin significantly more efficient as an iron chelator than aerobactin (322). At a serum pH of 7.4, the free iron concentrations in the presence of enterobactin (pFe[pH 7.4] = 35.5) and aerobactin (pFe[pH 7.4] = 23.4) differ by more than 10 orders of magnitude (73).
Furthermore, the iron affinity of the siderophore determines the redox potential of the ferric iron within the complex. While the standard redox potential (E°) of the Fe(III)/Fe(II) couple in water is +0.77 V, iron chelation can change the E° of the ligand-Fe(III) complex [LFe(III)]/LFe(II) couple dramatically. It may increase for Fe(II) complexes such as Fe-phenanthroline or Fe-bipyridyl above 1 V, meaning that Fe(II) in the complex will not be available any more as a reducing agent, e.g., for the Fenton reaction. On the other hand, it may decrease below −0.4 V in the case of Fe(III) chelates such as ferrioxamine B and Fe-enterobactin, with E° values at pH 7.0 of −0.45 V and −0.75 V, respectively, meaning that the chelated Fe(III) is not accessible to reduction by biological reducing agents such as NAD(P)H or flavins (251). Generally, increasing formation constants of Fe(III) complexes result in decreasing redox potentials of the bound Fe(III). However, the actual redox potential, E, does not necessarily correspond with E°, since it depends furthermore on the ratio of the current LFe(III) and LFe(II) concentrations: E = E° + 0.059 log[LFe(III)]/log[LFe(II)]. Thus, if the concentration of LFe(II) is reduced, which can be achieved in biological systems by Fe(II) sequestration into apoproteins or porphyrins or by transport processes, E is significantly increased, and the reduction of complexed Fe(III) by usual biological reducing agents may become thermodynamically favorable (251). An increase in the Fe(III)-chelate redox potential may additionally occur by reducing the pH (in the periplasm or in vacuoles) or increasing the hydrophobicity (by membrane association) of the reaction environment.
Gene Regulation of Microbial Iron Homeostasis
For the utilization of siderophores, microorganisms have to tightly regulate enzymes and transport systems that allow concerted siderophore biosynthesis, secretion, siderophore-delivered iron uptake, and iron release. In bacteria, gene regulation of siderophore utilization and iron homeostasis in general is mediated mainly at the transcriptional level by the ferric uptake repressor Fur or the diphtheria toxin regulator DtxR (131). While Fur is the global iron regulator in many gram-negative (e.g., enteric bacteria) and low-GC-content gram-positive (e.g., Bacillus spp.) bacteria, DtxR fulfills a comparable role in gram-positive bacteria with a high GC content (streptomycetes, mycobacteria, and corynebacteria). In bacteria that regulate iron homeostasis by Fur, DtxR-like proteins generally regulate manganese transport. Although there are no obvious sequence similarities present, the structural aspects of both regulators are very similar in terms of domain organization and metal binding (253-255). The C-terminal domains involved in homodimer formation contain a structural binding site for Zn(II) and a regulatory binding site for Fe(II). Zn(II) binding was shown to mediate the dimerization of E. coli Fur, although the Zn(II) binding site seems not to be strictly conserved among all Fur orthologues (248). Fe(II) binds to the regulatory site as a corepressor and was shown to enable E. coli Fur to bind to its DNA recognition sites (Fur boxes) (14). The first structural analyses were performed with DtxR from Corynebacterium diphtheriae. An Mn(II)-DtxR structure in the presence of either sulfate or selenate allowed the identification of an anion binding site near metal binding site I, leading to the suggestion that phosphate might act as a “co-corepressor” of DtxR under physiological conditions (261). Crystallization of the DtxR homologue IdeR from Mycobacterium tuberculosis as well as Pseudomonas aeruginosa Fur in the presence of 10 mM Zn-acetate and 10 mM ZnSO4, respectively, led to structures of both regulators in the homodimeric state with the two metal binding sites per monomer fully occupied by Zn(II) (253, 254). Concomitantly, extended X-ray absorption spectroscopy analyses with the Fur protein in solution confirmed the postulated redox state of Fe(II) bound to the regulator, and it was suggested that Zn(II) binding site I observed in the Zn(II)-Fur structure might represent the regulatory Fe(II) binding site in vivo (253). The putative Fe(II) binding site is composed of five amino acid residues (two His residues, two Asp residues, and one Glu residue) and one water molecule, suggesting an octahedral ligand arrangement.
In addition to the global repression systems, various transcriptional regulators of a lower hierarchy are involved in the regulation of siderophore utilization in bacteria. Generally, they act as activators, which are functionalized by direct or indirect sensing of extra- or intracellularly present Fe-siderophores and can be grouped into different classes including (i) alternative sigma factors, (ii) two-component sensory transduction systems, (iii) AraC-type regulators, and (iv) further transcriptional regulator types.
In the first class, the E. coli extracytoplasmic function (ECF) sigma factor FecI regulates the fecABCDE ferric citrate transport genes by the indirect sensing of extracellular ferric dicitrate via its cognate outer membrane (OM) receptor FecA, the N-terminal periplasmic region of which transmits the siderophore binding signal to the C terminus of FecR, the membrane sensor factor located in the cytoplsamic membrane. FecR interacts via its cytoplasmic N-terminal domain with FecI and thus may function as both an FecI chaperone and an anti-sigma factor (88). Homologous systems of FecI-FecR-FecA are PupI-PupR-PupB and FpvI/PvdS-FpvR-FpvA, regulating pseudobactin (type of linear, pyoverdin-like siderophore) and pyoverdin utilization in Pseudomonas putida and P. aeruginosa (31). In these systems, PupR is the anti-sigma factor of PupI, and FpvR is the anti-sigma factor of both ECF sigma factors FpvI and PvdS. The FecI homologue PbrA of Pseudomonas fluorescens activates the transcription of several iron utilization genes encoding pseudobactin biosynthesis and Fe-pseudobactin receptors (65). Several pathogenic Bordetella spp. regulate heme utilization via homologous ECF sigma factor-dependent systems.
The two-component sensory transduction system PfeR-PfeS of P. aeruginosa induces the pfeA Fe-enterobactin receptor by sensing periplasmic Fe-enterobactin (71).
AraC-like transcriptional regulators represent fusion proteins of AraC-type DNA binding domains and various substrate binding domains that bind Fe-siderophores as coregulators. Such intracellular siderophore sensors are found among both pathogenic and nonpathogenic siderophore-utilizing bacteria and may have evolved in terms of fine-tuned siderophore pathway regulation by directly responding to the presence of the iron chelator either before secretion or after uptake as an iron-charged complex. The first described member of this class was the PchR regulator of P. aeruginosa, which induces the pyochelin biosynthesis genes pchDCBA and the Fe-pyochelin receptor ftpA and represses its own gene in the presence of Fe-pyochelin (214). The AlcR regulator of Bordetella pertussis and Bordetella bronchiseptica was found to induce the alcaligin biosynthesis genes alcABCDER and the Fe-alcaligin receptor fauA in response to alcaligin (36). Recently, the Btr (YbbB) regulator of Bacillus subtilis was identified as being another siderophore-binding AraC-type regulator. Btr binds to the promoter of the Fe-bacillibactin uptake operon feuABC ybbA in the absence of bacillibactin (formerly also termed “corynebactin”) but needs Fe-bacillibactin for full induction (A. Gaballa and J. D. Helmann, unpublished data). Very likely, the AraC-type regulator YbtA from the high-pathogenicity island of Yersinia pestis, Yersinia enterocolitica, and Yersinia pseudotuberculosis is also part of this class since it induces the yersiniabactin biosynthesis operon irp21 ybtUTE, the ybtPQXS operon involved in Fe-yersiniabactin uptake and salicylate synthesis, and the Fe-yersiniabactin receptor fyuA (psn) gene and represses its own expression (92). It was found that YbtA binds, possibly as a dimer, to its promoter target sequences independent of yersiniabactin (10). However, as transcriptional activation is yersiniabactin dependent (249), the interaction of the YbtA-DNA complex with Fe-yersiniabactin was proposed to be a prerequisite for the recruitment of the RNA polymerase complex (9).
IrgB, in contrast, is a LysR family-type regulator acting as a transcriptional activator of the Vibrio cholerae IrgA Fe-enterobactin receptor (118). IrgB was not reported to interact with enterobactin or another iron chelator. An unusual type of regulator is the virulence plasmid-encoded Vibrio anguillarum AngR, a Fur-repressed nonribosomal peptide synthetase (NRPS) module-like protein of 120 kDa that acts as an activator of anguibactin biosynthesis and the fatDCBA Fe- anguibactin transport genes at the biosynthetic level and/or as a transcriptional regulator (334). In addition to NRPS condensation, adenylation, and thiolation domains, AngR possesses two helix-turn-helix domains that are preceded by leucine zipper motifs. A point mutation causing a His-to-Asn exchange at position 267, which is located between the first leucine zipper/helix-turn-helix motif, was found to be the cause of anguibactin hyperproduction in the natural V. anguillarum isolate 531A (317). Anguibactin is also involved in the regulation of its uptake since it was found to be an additional inducer of fatDCBA expression (56).
Of these specialized siderophore-dependent regulation systems, the components of ECF sigma factor-dependent regulation, the AraC-type regulators, as well as IrgB and AngR are all regulated by Fur.
Posttranscriptional regulation of bacterial iron homeostasis includes two general mechanisms. In various bacteria, small RNA-targeted mRNA degradation of genes encoding iron-utilizing proteins was observed. The small RNAs involved in this targeting are Fur-regulated antisense RNAs including RhyB in enteric bacteria (205), PrrF1/PrrF2 in P. aeruginosa (335), and, putatively, virulence plasmid pJM1-derived RNAα in V. anguillarum (65). The other mechanism corresponds to posttranscriptional regulation of iron homeostasis in higher eukaryotes. The B. subtilis aconitase CitB is a bifunctional protein with a function analogous to that of mammalian IRP1, a cytosolic aconitase that interacts with regulatory secondary mRNA structures termed iron-responsive elements (IREs) upon losing its [4Fe-4S] cluster during iron deprivation or oxidative stress (283). CitB interacts in an iron-dependent manner with rabbit ferritin IRE as well as IRE-like structures that were found in B. subtilis operons coding for major cytochrome oxidase and Fe-bacillibactin uptake (7).
In yeast, iron homeostasis regulation is also mediated at the transcriptional and posttranscriptional levels. The Saccharomyces cerevisiae transcriptional activator Aft1p and its paralogue Aft2p were shown to bind “iron-responsive” promoter elements during iron starvation (285). Targets of Aft1p/Aft2p regulation include components of the reductive iron assimilatory system such as FET3, FTR1, and the FRE1 to FRE6 genes; the four known Fe-siderophore importers ARN1, TAF1 (ARN2), SIT1 (ARN3), and ENB1 (ARN4); and putative accessory components FIT1 to FIT3 and the low-affinity transporter FET4 (108, 204, 331, 355). Aft1p is constitutively produced, and its function is regulated by its subcellular localization: the protein localizes to the nucleus only if cells are iron depleted (348). Aft1p and Aft2p possess a Cys-X-Cys motif that was suggested to participate in iron-sulfur cluster binding (347) and a nuclear export sequence-like motif. Mutations in either motif cause nuclear retention and constitutive activation of Aft1p (348). Iron-dependent deactivation of Aft1p/Aft2p is abrogated in cells defective for mitochondrial Fe-S cluster biogenesis (55), and iron sensing depends on mitochondrial Fe-S export (286). However, there is no indication that regulation occurs by the direct binding of an Fe-S cluster to the transcription factors, and posttranslational modification might be alternatively involved in Aft1p localization (124). The nuclear monothiol glutaredoxins Grx3 and Grx4 were recently reported to be additional components required for Aft1p iron regulation (238). Further iron-regulatory mechanisms in yeast include the general repressor system Ssn6p-Tup1p, which was shown to regulate the expression of several uptake systems for Fe-siderophores in S. cerevisiae (186), and a global metabolic reprogramming during iron deficiency is achieved by targeted mRNA degradation, which is mediated by Aft1p/Aft2-regulated Cth2p, which is also conserved in plants and mammals (260).
The regulation of siderophore biosynthesis in filamentous fungi was shown to depend on orthologues of the GATA family transcription factor Urbs1, a protein similar to the erythroid transcription factor GATA-1, which was first described for the basidiomycete Ustilago maydis (328). Urbs1 orthologues were also found in ascomycetes such as Sre in Neurospora crassa (358) and SreA in Aspergillus nidulans (126). They are distinguished from all other known fungal GATA factors by the presence of two zinc fingers and a conserved intervening cysteine-rich region, which might represent the most probable binding site for the direct sensing of iron by binding iron or an iron-sulfur cluster (124). While Urbs1 appears to be the exclusive siderophore biosynthesis regulator in U. maydis that represses the siderophore biosynthesis genes sid1 and sid2 during iron repletion (351), additional regulatory factors seem to be present in N. crassa and A. nidulans (124). In A. nidulans, SreA deficiency leads not only to the derepression of siderophore biosynthesis but also to the deregulation of siderophore-mediated iron uptake (236).
In contrast to microorganisms, the regulation of iron homeostasis in mammalians appears to be done exclusively at the posttranscriptional level via the IRP1/IRP2-IRE system (283).
General Steps of Siderophore Pathways
Although the elucidation of new siderophore pathway components has seen much progress during recent years, there is still a substantial discrepancy between information given by the considerable number of well-characterized siderophore biosynthesis systems and the often initially characterized further pathway components. This might be reasonably explained by the fact that siderophore biosynthesis is attractive for semisynthetic drug design combined with various therapeutical applications. However, also, siderophore transport and iron release mechanisms are potentially interesting targets with respect to pathogen control, as discussed below. In this section, siderophore biosynthesis will be treated as a comparative overview of the best-studied systems and their general enzymology. Aspects of siderophore secretion, uptake, and iron release shall then be introduced extensively to the current state of knowledge and with special respect to the latest findings in these growing fields.
Siderophore biosynthesis catalyzed by nonribosomal peptide synthetases.
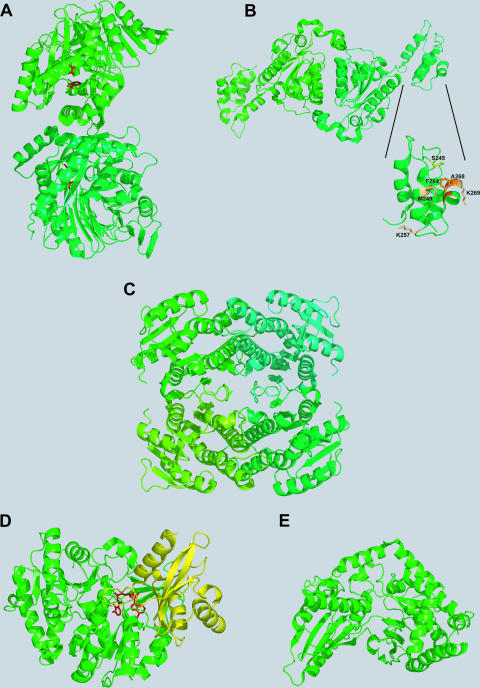
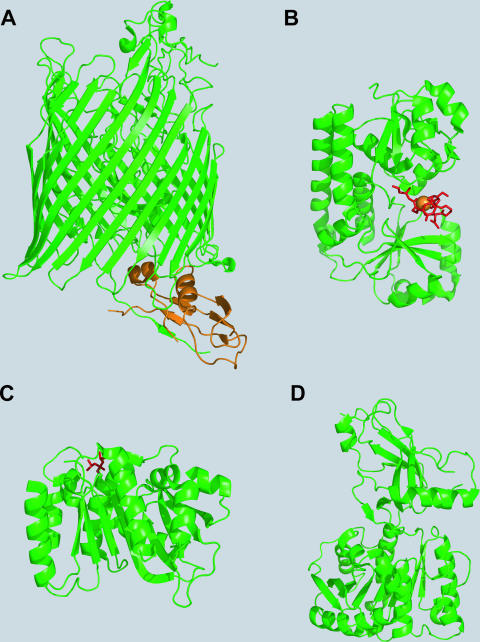
Depending on the chemical nature of the siderophores, their biosynthesis occurs via different mechanisms. In general, the biosynthesis pathways can be distinguished as being either dependent on or independent of an NRPS. NRPSs represent large multienzyme complexes that activate and assemble a broad array of amino, carboxy, and hydroxy acids, leading to a high structural variability of the generally macrocyclic peptidic products (123). This diversity may be enhanced through various substrate modifications occurring during assembly by the action of specialized domains that are integrated into the standard NRPS domain architecture comprising modular sequences of adenylation (A), thiolation (T, or peptidyl carrier protein [PCP]), and condensation (C) domains. In most cases, the peptide chains are released from the synthetase by an intra- or intermolecular cyclization event catalyzed by commonly C-terminally-located thioesterase (TE) domains (171). NRPSs are responsible mainly for the synthesis of aryl-capped siderophores. NRPS-dependent siderophore biosynthesis in several human pathogens has been elucidated in detail, e.g., enterobactin synthesis in enteric bacteria such as E. coli, Salmonella enterica, Klebsiella spp., and Shigella spp.; yersiniabactin synthesis in Yersinia spp.; pyochelin and pyoverdin synthesis in P. aeruginosa; vibriobactin synthesis in V. cholerae; and mycobactin synthesis in M. tuberculosis (66). Prior to NRPS-catalyzed assembly, the aryl acids 2,3-dihydroxybenzoate (DHB) and salicylate, which are generally used as aryl caps, have to be provided by approaching enzymes. In most bacteria, the genes encoding the NRPS and the enzymes for aryl acid synthesis are directly iron regulated via the Fur repressor. Meanwhile, the enzymes for DHB and salicylate formation as well as several NRPS domains involved in catecholate siderophore assembly have been extensively characterized, and crystal structures are available in many cases. Representative structures of enzymes involved in aryl-capped siderophore biosynthesis are shown in Fig. 2. The Y. enterocolitica salicylate synthase Irp9 is a homodimer (Fig. 2A), each protomer of which catalyzes the conversion of chorismate into salicylate via an isochorismate intermediate (162, 163). The X-ray structure of MbtI, the Irp9 homologue in M. tuberculosis and the second known example of a bacterial salicylate synthase, was also recently solved and found to be in a monomeric state (134). Salicylate synthesis in P. aeruginosa was shown to depend on two distinct enzymes, the isochorismate synthase PchA and the isochorismate-pyruvate lyase PchB (104). The activities of both salicylate and isochorismate synthases are highly magnesium dependent, which is also the case for the structurally similar chorismate-utilizing enzymes TrpE, the prototype of anthranilate synthases, and PabB, the aminodeoxychorismate synthase involved in p-aminobenzoate synthesis. In the crystal structure of Irp9 soaked with chorismate, the Mg(II) cofactor was found to be coordinated by two glutamate residues of the active site and the carboxy group salicylate that was found together with pyruvate in the catalytically active crystal, suggesting that this coordination is crucial during catalysis.
FIG. 2.
Structures of proteins involved in siderophore biosynthesis. (A) The Y. enterocolitica salicylate synthase Irp9 homodimer (PDB accession number 2FN1) with reaction products salicylate and pyruvate (shown in red) and the Mg(II) cofactor (shown in yellow). (B) Homodimer of the E. coli isochorismatase-aryl carrier protein EntB (PDB accession number 2FQ1). The enlarged section is the aryl carrier protein domain with Ser245 (yellow) for cofactor modification and residues involved in minor (bright orange) and major (dark orange) interactions with the nonribosomal peptide synthetase EntF. (C) The E. coli 2,3-dihydro-2,3-DHB dehydrogenase EntA in the homotetrameric state (PDB accession number 2FWM). (D) The lone-standing DHB adenylation domain DhbE of B. subtilis cocrystallized with DHB-AMP (shown in red) (PDB accession number 1MDB). (E) The lone-standing condensation domain VibH of V. cholerae (PDB accession number 1L5A) linking VibB-activated DHB and norspermidine via amide bond formation.
The synthesis of DHB from chorismate needs three enzymatic activities. While after isochorismate formation (either as a transient or stable intermediate), during salicylate synthesis, the lyase reaction leads directly to the formation of salicylate when pyruvate is cleaved off, one water molecule is incorporated instead of the pyruvate at the C-3 position during DHB synthesis by the EntB-type isochorismate lyase (or isochorismatase), leading to the formation of a second intermediate, which is 2,3-dihydro-DHB (284). The crystal structure of the E. coli isochorismatase EntB, acting downstream of the PchA-homologous isochorismate synthase EntC (192), was solved recently (81) (Fig. 2B). In fact, it is a bifunctional enzyme that comprises an N-terminal domain harboring the isochorismatase activity and a smaller C-terminal domain that functions as a PCP-homologous ArCP (aryl carrier protein) domain during enterobactin assembly as discussed below. Both domains are connected via a long proline-rich loop that keeps their active sites at a distance of about 45 Å, thus ensuring the independent activity of both domains. EntB forms a homodimer, and the dimerization takes place via the isochorismatase domain. The last step in DHB synthesis is catalyzed by a 2,3-dihydro-DHB dehydrogenase, a member of the short-chain oxidoreductase enzyme family, which oxidizes the EntB product into the aromatic catechol DHB using NAD+ as a cofactor (289). The crystal structure of the E. coli 2,3-dihydro-DHB dehydrogenase EntA revealed a homotetramer formation (Fig. 2C), which, as shown previously for other tetrameric members of this enzyme family, is the result of a tight dimer-to-dimer interaction (312). Once synthesized, the aryl acid is used together with further precursors, usually amino acids or polyamines, for siderophore scaffold assembly by the corresponding NRPS. The usually lone-standing aryl acid A domains catalyze the initial step of aryl-capped siderophore assembly by converting either DHB or salicylate in their acyl adenylates. The crystal structure of the B. subtilis DHB adenylation domain DhbE (Fig. 2D) is an archetype of aryl acid-activating domains (211). Homologous enzymes are the DHB-activating domains EntE and VibE of E. coli and of V. cholerae, respectively, and the salicylate-activating domains YbtE of Yersinia spp., PchD of P. aeruginosa, and MbtA of Mycobacterium species. As a characteristic of NRPS adenylation domains, DhbE possesses a large N-terminal domain that bears the bisubstrate binding site (which is occupied in the depicted structure by DHB-AMP, the native product of catalysis) and a small compact C-terminal “lid” domain that is presented here in superior orientation to the active site, which is the proposed “adenylate-forming conformation.” The N- and C-terminal domains are connected via a short flexible hinge that allows the distal rotation of the lid for substrate binding and product release (“thioester-forming conformation”). Thioester formation occurs upon the adenylation of the aryl acid, which is subsequently tethered to the 4′-phosphopantethein cofactor of an ArCP and is then transferred to the donor position of the first C or Cy (condensation/cyclization) domain. The C-terminal domain of EntB that carries out this function in the enterobactin system bears the covalent attachment site for the cofactor at serine 245, the modification of which converts the domain into its active holo form. Since EntB has to interact with three proteins, which are the phosphopantetheinyl transferase Sfp, the A domain EntE, and the downstream-acting multidomain NRPS EntF, it is a model protein for studying the mechanisms of protein-protein recognition. Mutational studies showed that the interaction with EntE is quite tolerant of a number of point mutations on both the EntB and EntE surfaces (81). The EntB surface for the interaction with EntF, however, was found to comprise three highly conserved hydrophobic residues (M249, F264, and A268) located near the phosphopantetheinylated S245 and in particular on the small helix 3 (Fig. 2B), suggesting that this structural element is important for the EntB-EntF interaction (179). Homologues of the bifunctional EntB protein exist in B. subtilis (DhbB) and V. cholerae (VibB), bacteria that produce triple-DHB-capped siderophores as well. In contrast, in Pseudomonas spp., Yersinia spp., and Mycobacterium spp., which all produce single-salicylate-capped siderophores, the aryl carrier protein is the first domain in a multidomain NRPS complex (262). The C and Cy domains that catalyze the peptide bond formation between the activated substrates and, in the case of Cy domains, also their cyclization are usually integrated into the NRPS multidomain structure. An exception to this rule is the unusual C-domain VibH catalyzing the first condensation step in vibriobactin synthesis (159), which gave the first crystal structure of an NRPS C domain (160) (Fig. 2E). VibH couples the thioester-activated DHB with norspermidine, the remaining two amines of which are acylated during following synthesis steps with DHB-methoxazoline-carbonyl moieties that result form cyclizations-condensations of DHB and threonine catalyzed by the multidomain synthetase VibF (158). While VibH is a monomeric pseudodimer, enzymes that are structurally related to NRPS C domains such as chloramphenicol acetyltransferase and dihydrolipoamide acetyltransferase are known to be homotrimers.
As vibriobactin synthesis demonstrates, the Cy domain-catalyzed formation of intramolecular heterocycles takes place in various aryl-capped siderophores. Anguibactin, pyochelin, and yersiniabactin contain thiazoline rings resulting from the cyclization of cysteine side chains (106, 264). In-trans-acting reductase domains catalyze the subsequent conversion of thiazolines into thiazolidines during pyochelin and yersiniabactin synthesis (217, 243). In mycobactins and structurally related mixed aryl-capped hydroxamate siderophores such as acinetobactin and frankobactin, oxazoline rings are found as a result of serine or threonine cyclization (341), and in the case of heterobactin A, a hydroxybenzoxazole ring is built up during the assembly process (45). In some biosynthesis systems, polyketide synthase (PKS) domains are part of the siderophore assembly lines. High-molecular-weight protein 1, involved in yersiniabactin synthesis, is an NRPS/PKS hybrid synthetase, the PKS domains of which introduce a malonyl moiety into the nascent peptide chain and perform subsequent bismethylation and β-ketoreduction on this building block (217). During mycobactin synthesis, a β-hydroxyacyl is incorporated by the NRPS/PKS hybrid MbtB and the MbtC and MbtD PKSs (263). Deletion of MbtB, which additionally catalyzes phenyloxazoline formation between salicylate and serine, completely disrupts salicylate-derived siderophore synthesis in M. tuberculosis, leading to reduced growth in iron-limited medium and in macrophage-like THP-1 cells (80). Mycobactins are produced by enzymes of the mbt-1 and mbt-2 gene clusters, the latter of which was recently found to comprise the four tailoring enzymes for mycobactin acylation (178). Mycobactins contain a long lipid chain that renders them lipid soluble and favorable for associations with the mycobacterial cell envelope. Pathogenic mycobacteria such as M. tuberculosis and Mycobacterium leprae, however, apparently use the same genes to synthesize mycobactin derivatives called carboxymycobactins, which possess a shorter carboxyalkyl chain that renders them more hydrophilic with respect to their function as extracellular siderophores (268). However, it was recently shown that lipophilic mycobactins can also serve as extracellular siderophores within mycobacterium-colonized macrophages by diffusing through the membrane of the occupied macrophage phagosomes and, after scavenging iron from the intracellular pool, accumulating in intracellular lipid droplets that are delivered to the phagosomes by lipid trafficking (201). Nonpathogenic mycobacteria such as Mycobacterium smegmatis and Mycobacterium neoaurum use, in addition to carboxymycobactins, non-aryl-capped exochelins as extracellular siderophores, which are predicted to be synthesized by the FxbB and FxbC NRPSs (350, 360). Product release from the NRPS multienzyme complex is, in the majority of cases, catalyzed by a C-terminus-standing TE domain, and this final reaction step often leads to the concomitant formation of macrocyclic structures if side chains of the assembled peptide are used for the nucleophilic attack at the TE-bound ester intermediate. NRPS-dependent siderophore macrocyclization has so far been reported for enterobactin and bacillibactin that contain trilactone backbones resulting from iterative cyclization catalyzed by the C-terminal TE domains of the EntF and DhbF assembly lines, respectively (212, 300). During this cyclotrimerization reaction, three peptide chain intermediates, either DHB-seryl or DHB-glycine-threonyl, are successively assembled via ester bond formation between the hydroxy groups of the seryl or threonyl residues of the chain tethered to the TE and the electrophilic thioester of the following chain, which is tethered to the PCP upstream of the TE. NRPS-dependent cyclization reactions are also putatively involved in the synthesis of the cyclopeptide backbones of pyoverdins, alterobactin A, and fungal ferrichrome-type siderophores.
Finally, the optimization of NRPS-dependent peptide siderophore synthesis is mediated in several systems by trans-acting type II thioesterases, which have a general editing function by catalyzing the deacylation of misprimed or misacylated PKS acyl carrier proteins and NRPS ArCPs or PCPs (294). Siderophore biosynthesis proofreading was reported for the type II thioesterases PchC and EntH during pyochelin and enterobactin formation in P. aeruginosa and E. coli, respectively (D. Leduc and E. Bouveret, unpublished data; 274). While PchC proved specific for regenerating the PCP domains of PchE and PchF that were not correctly charged with l-cysteine but were charged with its analogue 2-aminobutyrate, EntH was shown to interact specifically with the ArCP of EntB and to regenerate the domain when mischarged with salicylate. Thioesterase-dependent NRPS (and PKS) editing might be suspected in further siderophore synthesis systems that bear lone-standing thioesterases such as yersiniabactin synthesis cluster-encoded YbtT, which was found not to be necessary for both product release and full in vitro activity of the yersiniabactin assembly machinery (217), or such as Streptomyces coelicolor CchJ, which is required for coelichelin biosynthesis in vivo and hence was provisionally suggested to be a thioesterase for hydrolytic product release rather than for assembly line editing (182).
In some cases, NRPSs are partially involved in the synthesis of hydroxamate and carboxylate siderophores to build a peptidic backbone to which the iron-coordinating residues are attached. This has been reported for coelichelin of S. coelicolor (52, 182), the pyoverdins of fluorescent Pseudomonas spp. (2, 224), the exochelins from nonpathogenic mycobacteria (350, 360), and the ferrichromes/ferricrocins from various fungi (84, 295, 333, 351) and can be anticipated for alterobactin of Alteromonas luteoviolacea, the pseudobactins of Pseudomonas spp., the azotobactins of Azotobacter spp., and the ornibactins of Burkholderia spp.
Siderophore biosynthesis independent of nonribosomal peptide synthetases.
Hydroxamate and carboxylate siderophores are assembled by NRPS-independent mechanisms in the majority of cases. The synthesis of siderophores belonging to these two main classes commonly relies on a diverse spectrum of enzymatic activities such as monooxygenases, decarboxylases, aminotransferases, ac(et)yltransferases, amino acid ligases, and aldolases (51). Siderophores synthesized by NRPS-independent pathways are found as virulence factors in several pathogens, e.g., aerobactin in enteric bacteria (114), alcaligin in B. pertussis and B. bronchiseptica (220, 234), staphylobactin in Staphylococcus aureus (68), and petrobactin (formerly anthrachelin) in Bacillus anthracis (48, 174). Among the NRPS-independently assembled siderophores, petrobactin represents an exception because it is capped with two catecholate moieties for iron coordination (see Fig. 1 for structure). However, the use of 3,4-dihydroxybenzoate moieties is so far unique in the structural world of siderophores, and a possible explanation for their utilization is given in the section dealing with pathogen-host interactions below.
In general, hydroxamate moieties are built in two steps. The first reaction step is an N-hydroxylation catalyzed by reduced flavin adenine dinucleotide (FAD)-dependent monooxygenases that use molecular oxygen and a set of amino acids and polyamines as substrates. In most of the known pathways, one oxygen atom is transferred either to the ɛ-amino group of lysine (aerobactin pathway), to the δ-amino group of ornithine (ferrichrome/ferricrocin, coprogen, rhodotorulic acid, and fusarinine pathways), or to one amino group of the corresponding decarboxylation products cadaverine (desferrioxamine E and, putatively, bisucaberin pathways) and putrescine (alcaligin and, putatively, putrebactin pathways), respectively (51, 341). In the rhizobactin 1021 pathway, the unusual diamine 1,3-diaminopropane is suggested to be the substrate for N-hydroxylation (203). In contrast to the NRPS-independently catalyzed sequence of hydroxamate siderophore assembly, the introduction of hydroxamate functions into the NRPS-derived mycobactin scaffold was concluded to be the final synthesis step due to the detection of didehydroxymycobactins in early-infection cultures of M. tuberculosis (219). In this case, the monooxygenase MbtG catalyzes N6-hydroxylation of two lysines that are already integrated into the complex siderophore scaffold (178).
The formylation (leading to free hydroxamic acid moieties) or acylation of the hydroxylated amine generally represents the second step yielding the functional hydroxamate and is catalyzed in the case of acylation by acyl coenzyme A transferases. Formylated N5-hydroxy-ornithines are present in pyoverdins and in ornibactin. Acylation of hydroxylated amines is much more frequent. As substrates, coenzyme A derivatives of various carboxy acids such as acetate (aerobactin), succinate (desferrioxamine E), β-hydroxybutyrate (pyoverdins and ornibactin C4), or decenoate (rhizobactin 1021) are used. In mycobactin synthesis, however, the acylation is proposed to occur prior to the N-hydroxylation since the cyclization of the C-terminal lysine, which might be catalyzed by some kind of thioesterase or condensation domain (66), was already found in isolated didehydroxymycobactins (219). The free (α-hydroxy-)carboxylate moieties in (α-hydroxy-)carboxylate siderophores may be derived from hydroxy-carboxylic acids such as citrate (staphyloferrin A and vibrioferrin), 2-oxo-glutarate (achromobactin and vibrioferrin), and malonic acid (rhizobactin DM4) or amino-carboxy acids such as alanine in rhizobactin DM4 or 2,3-diaminopropionic acid in staphyloferrin B, which are often (but not always) coupled to the backbone by ester or amide bonds. Generally, the ligation of citrate occurs via one of its prochiral carboxyl groups, while 2-oxo-glutarate is attached via its C-5 carboxyl group (51). Free α-hydroxy-carboxylate moieties may also result from introducing β-hydroxy-aspartates in a peptidic backbone sequence such as in pyoverdins and alterobactin.
The final ligation of the building blocks in NRPS-independent siderophore assembly is catalyzed by NTP-dependent siderophore synthetases of the IucA type and/or the IucC type mainly by amide bond formation and sometimes also via ester linkages such as those in achromobactin or vibrioferrin. IucA and IucC are the prototypes of NRPS-independent siderophore synthetases that catalyze the penultimate and the final steps in aerobactin synthesis, respectively, and all known NRPS-independent siderophore synthesis pathways utilize at least one enzyme with high sequence similarity to the aerobactin synthetases (51).
Siderophore secretion.
Siderophore secretion systems have been identified in only a few microorganisms so far. The exporters that were found or suggested to be involved in siderophore release belong to efflux pumps of the major facilitator superfamily (MFS); the resistance, nodulation, and cell division (RND) superfamily; and the ATP-binding cassette (ABC) superfamily.
The MFS is a large and diverse superfamily of transporters that carry out uniport, solute:cation (proton or sodium) symport and/or solute:proton or solute:solute antiport (287). They exhibit specificity for a broad range of compounds including various primary metabolites, drugs, neurotransmitters, siderophores (efflux), iron-charged siderophores (see “Uptake of siderophore-delivered iron”), and organic and inorganic anions. They possess either 12, 14, or 24 α-helical transmembrane segments (TMSs). In contrast, members of the RND superfamily probably all catalyze substrate efflux via a proton antiport mechanism (320). Generally, they possess a single transmembrane-spanning region at the N terminus that is followed by a large extracytoplasmic region and then six additional transmembrane regions, a second large extracytoplasmic section, and five final transmembrane regions at the C terminus. Substrates of RND efflux pumps are heavy metals, various drugs (e.g., tetracycline, chloramphenicol, fluoroquinolones, and β-lactams), lipooligosaccharides, lipids, pigment(s), siderophore(s), and possibly sterols in eukaryotes. RND members of several families act in conjunction with a membrane fusion protein (MFP) and an OM factor (OMF) to yield efflux across both membranes of the gram-negative bacterial cell envelope in a single energy-coupled step. Both MFS and RND superfamily members are found ubiquitously in all three kingdoms of life.
The MFS involved in enterobactin secretion in E. coli is a 12-TMS protein termed EntS that is encoded in the Fur-regulated ent-fep gene cluster (102). An entS mutant shows significantly reduced secretion of enterobactin but increased release of enterobactin breakdown products, which was explained by Fes-catalyzed intracellular enterobactin hydrolysis and the subsequent diffusion and/or release of the breakdown products by further cytoplasmic export systems. Thus, while export across the cytoplasmic membrane (CM) is mediated primarily (but not exclusively) by EntS, the OM channel protein TolC was shown to be exclusively responsible for enterobactin release across the OM (23). Altogether, there are a couple of possibilities for how enterobactin export is committed, which can be addressed either as a one-step or as a two-step scenario. As a one-step process, enterobactin export may involve a tripartite transport system composed of a CM transporter (primarily EntS but putatively at least one more efflux pump), an MFP (yet unknown), and TolC as an OMF. In a possible two-step process, enterobactin could be released by EntS (and putatively another CM transporter) into the periplasm, from where clearance is mediated by a TolC-comprising RND-type transporter. Such a two-step process was recently shown for heavy metal efflux (228). As a third alternative, a TolC-dependent but EntS-independent export system(s) that transports enterobactin from both the cytoplasm and the periplasm to the environment might exist. Enterobactin breakdown products, however, were also released through the OM in a tolC mutant (23), suggesting that there are further OM gates for clearing the periplasm from catecholate fragments.
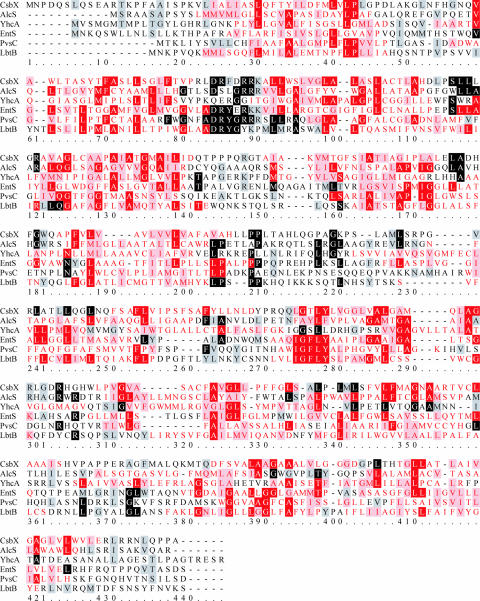
MFS efflux pumps that possess 12 membrane segments are involved in siderophore export in other bacteria. In most cases, they are Fur-dependently regulated. Legiobactin, a siderophore of yet unknown structure produced by Legionella pneumophila, is exported by the MFS exporter LbtB (8). LbtB is encoded by the second gene of the lbt operon that also encodes the IucA-IucC-related siderophore synthetase LbtA and the LbtB-like transmembrane protein LbtC, which, however, is not involved in legiobactin efflux. The Erwinia chrysanthemi and the Vibrio parahaemolyticus hydroxycarboxylate siderophores achromobactin and vibrioferrin are exported via the YhcA and the PvsC MFS exporters encoded within the corresponding siderophore biosynthesis operons, respectively (314). The CsbX MFS exporter encoded upstream of the catecholate siderophore biosynthesis (cdb) operon of Azotobacter vinelandii facilitates the secretion of catecholate siderophores related to protochelin (240). The AlcS MFS exporter of B. pertussis and B. bronchiseptica that mediates the export of the cyclic hydroxamate siderophore alcaligin is, in contrast, constitutively produced and, as it regulates the intracellular alcaligin concentration, is part of the alcaligin/AlcR-dependent regulatory circuit controlling alcaligin gene expression (35). An alignment of the six MFS efflux proteins involved in siderophore secretion identified so far (Fig. 3) reveals low protein sequence identities (below 20%). However, this might be expected since neither the phylogenetic relationship of the bacterial sources (except for E. coli and E. chrysanthemi) nor the structural relationship of the transported siderophores (if the structure is known at all) is very high. Generally, members of the MFS do not possess such well-defined conserved motifs as it is known from other transporter superfamilies, e.g., those of the ABC type. The conserved 13-amino-acid (aa) motif (GX2ADRYGR[R/K] [R/K]X[L/I]; boldface indicates conservation higher than 50% in the alignment shown in Fig. 3) found in the alignment between TMS2 and TMS3 is very similar to the major consensus motif described for the MFS in this sequence region (241). This motif is thought to be involved in promoting conformational changes in the protein upon substrate binding, allowing the trafficking of substrates through the membrane, and may also act as a gate for substrate transport regulation (308). Further studies on the growing number of MFS-type siderophore exporters should provide deeper insights into transport type, affinity, and specificity.
FIG. 3.
Alignment (ClustalW, shown in BOXSHADE format) of MFS-type proteins involved in the secretion of siderophores. Sections of predicted TMSs are shown individually for each protein and are indicated with red letters and/or red shading. Nonshaded letters indicate nonsimilar amino acids; gray- or light red-shaded letters indicate similar amino acids; black- and dark red-shaded letters indicate identical amino acids.
The P. aeruginosa efflux system MexA-MexB-OprM represents a typical RND exporter of gram-negative bacteria consisting of an MFP (MexA), an RND efflux pump with 12 TMSs (MexB), and an OMF (OprM) (189). Since the putative operon of this efflux machinery is iron regulated and since a mutant strain was growth sensitive in dipyridyl-containing iron limitation medium, it was suggested that this exporter is involved in the efflux of pyoverdin, the primarily virulence-associated siderophore of P. aeruginosa (257). Several drugs that are exported by MexA-MexB-OprM have structures related to the pyoverdin chromophore and are capable of binding iron. However, data showing a reduction of pyoverdin secretion in the exporter mutant strain have not been reported so far.
The ABC-type transporter superfamily contains both efflux and uptake transport systems that generally mediate transport via ATP hydrolysis without protein phosphorylation (28, 69, 72, 152). They consist of two integral membrane domains and two cytoplasmic domains (ABC subunits) for NTP (usually ATP) binding and hydrolysis. Both the transmembrane and the cytoplasmic domains may be present as homodimers or heterodimers. While uptake porters generally have their domains as distinct polypeptide chains, efflux systems usually have them fused. ABC-type efflux systems are abundant in both prokaryotes and eukaryotes. The eukaryotic efflux systems often have the four domains (two cytoplasmic domains representing the nucleotide-binding folds [NBFs] and two integral membrane domains) fused into either one or two polypeptide chains, while in prokaryotes, they are generally fused into two chains, both of them containing one integral membrane channel constituent and one cytoplasmic ATP-hydrolyzing constituent. Generally, the integral membrane domains of the ABC-type efflux systems possess six α-helical TMSs each. Representative of the structural topology of bacterial ABC-type exporters are the crystal structures of the open and the closed conformations of the E. coli and V. cholerae MsbA multidrug transporter homologues, respectively (53, 54), and the structure of the S. aureus Sav1866 multidrug transporter (70). Based on different structural aspects, two mechanistic models of substrate translocation, involving either a sideward-directed movement of the subunit halves (275) or domain swapping, which is mediated by twisting the protomer subunits (70), have been proposed. Cross-linking studies with the human multidrug resistance P glycoprotein suggested another model proposing a substrate-induced conformational change in the TMSs of the transporter (145, 197).
A mutagenesis study of M. smegmatis led to the identification of the IdeR-regulated exiT locus encoding an ABC-type exporter that is responsible for the secretion of exochelin MS (360), which belongs to the group of water-soluble extracellular hydroxamate siderophores produced by nonpathogenic mycobacteria. ExiT, which is encoded upstream of the exochelin biosynthesis genes fxbB and fxbC, possesses a C-terminally-located NBF and eight proposed TMSs that were suggested to be expanded up to 12 due to the length of three of these segments. Although fusions of two transporter half-segments leading to ABC-type exporters comprised in a single tetradomain polypeptide chain are observed in bacteria, the second NBF is missing in the ExiT sequence. A closer examination reveals that six of the eight proposed TMSs cluster adjacent to the C-terminal ABC, and the two remaining segments are found at the N terminus, leaving a large hydrophilic region of 430 aa between the TMS regions. It was speculated that these features might be an adaptation to the unique architecture of the mycobacterial cell envelope (360). In eukaryotes, the group of multidrug-resistance-related protein-like ABC-type transporters is known to possess an additional trans-membrane-spanning domain of approximately 200 aa at the N terminus of the protein; however, these proteins have a TMSn[TMS6-NBF]2 topology and a characteristic “regulatory” or “connector” domain between their homologous halves (308), both of which are not characteristic for ExiT. Since numerous possible start codons were found in the exiT open reading frame and since the reported protein sequence was based on the farthest upstream-located translation initiation site that was tentatively chosen, the putative N terminus of ExiT might be significantly shorter, thus leading to an ExiT protein with a more common bacterial ABC-type exporter architecture.
Another ABC-type transport system has recently been suggested to function in the secretion of salmochelins (191), which are glucosylated derivatives of enterobactin produced by pathogenic E. coli and Salmonella strains. The iroA gene cluster harbors the gene iroC that encodes a four-domain ABC-type fusion protein with [TMS6-NBF]2 topology showing about 30% identity to eukaryotic P glycoproteins involved in multidrug resistance (15, 16, 132). Genome database searches reveal that there are only few bacterial species harboring four-domain ABC-type fusion proteins, and as in the case of IroC-harboring strains, they often live in close contact to eukaryotes either as pathogens (opportunistic pathogens such as Nocardia farcinica or obligate pathogens such as M. tuberculosis or C. diphtheriae) or as symbionts (such as Frankia alni). Thus, it might be speculated that the prokaryotic development of such fusion types has been advanced by interspecies gene transfer. Although the similarity of IroC to ABC-type efflux proteins is significant, growth promotion studies indicated that IroC might be responsible for the uptake of linearized forms of Fe-salmochelin and Fe-enterobactin (359). Thus, two conflicting models of salmochelin utilization currently exist, one assigning IroC as a salmochelin exporter (95, 191) and the other one assigning IroC as an importer of the iron-charged linearized salmochelin trimer (359). IroC-dependent Fe-salmochelin transport studies or iroC mutant phenotype analyses might help to clarify the opposing interpretations.
Interestingly, during investigations of phenotypes of several exporter mutants, it was observed that they either accumulate (35) or do not accumulate (102, 360) intracellular siderophores. The latter and rather unexpected case occurred if NRPSs were involved in siderophore biosynthesis, which was generally taken as an indication of a tightly coupled process of siderophore synthesis and export, suggesting that a permanent drain of siderophore might be required for continued biosynthesis. Such a coupling would be reasonable in order to avoid the accumulation of nonloaded siderophore molecules in the cell and to prevent the degradation of newly synthesized siderophores, e.g., by esterases that are involved in iron release. In this context, indirect evidence for a membrane association of the enterobactin EntEBF synthetase was provided by the finding that enterobactin synthesis proteins are released from cells by osmotic shock but not by spheroblasting, thus behaving like Beacham group D class proteins that are released from cells in a similar way due to their association (partial, loose, or transient) with the CM (128). EntE, EntB, and EntF polypeptides were also detected in total membrane preparations from cells grown in iron-deficient medium (129). However, the subcellular localization of siderophore biosynthesis enzymes in possible conjunction with membrane-standing exporter systems remains to be investigated in future studies.
Uptake of siderophore-delivered iron.
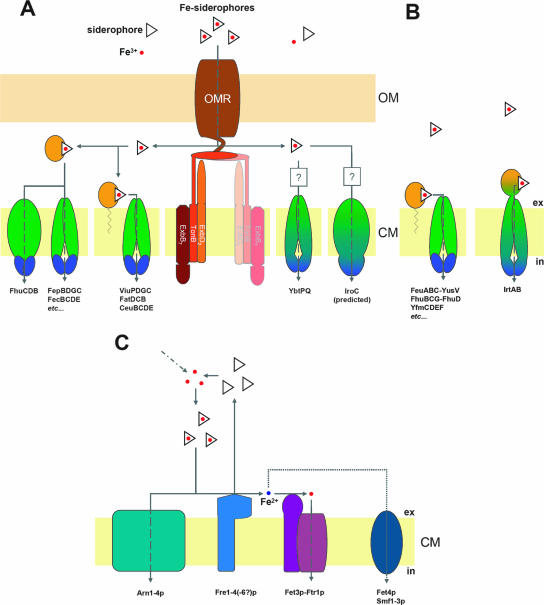
Once Fe(III) is mobilized and captured by the secreted siderophore molecule, it is accessible for cellular uptake. This step is committed in two general ways: either iron is released by reduction from the siderophore at the extracellular surface and is taken up as single ion or the whole Fe(III)-siderophore complex is internalized. Eukaryotes, namely, filamentous fungi and yeast, possess membrane-standing ferrisiderophore reductases and are hence capable of both uptake routes. In bacteria, systems for the extracytosolic reduction of siderophore-bound iron are much less well known (see “Mechanisms of iron release”), and thus, the main uptake route for siderophore-delivered iron in bacteria is represented by the import of iron-siderophore complexes into the cytosol. The uptake systems for Fe-siderophores have been much more intensely studied than siderophore secretion systems, and hence, many importers were identified and partially well characterized in prokaryotes and eukaryotes. In addition to the fact that different routes for the uptake of siderophore-delivered iron can be used, the common presence of a greater battery of Fe-siderophore importers than siderophore exporters in a single organism is owing simply to the possibility of using exogenously derived siderophores as iron sources, which demands the use of further specialized transporters. A general scheme of siderophore-delivered iron uptake routes and the involved types of transporters in bacteria and fungi is given in Fig. 4.
FIG. 4.
Cellular transport systems for uptake of siderophore-delivered iron in bacteria and fungi. (A) Gram-negative bacteria. Fe-siderophore uptake through the OM is mediated by an OM receptor (OMR), which is energized by the proton motive force transduction system ExbB7-ExbD2-TonB (“TonB complex”). The second TonB complex shown in faint colors suggests a possible 2:1 interaction with the OM receptor (see the text for details). Fe-siderophore uptake through the inner membrane depends on ABC-type transporters with different domain arrangements and localizations (see the text for details): periplasmic binding proteins/domains are shown in bright orange, membrane-spanning proteins/domains are shown in green, and cytoplasmic ATP-binding proteins/domains are shown in dark blue. Domain fusions are indicated by corresponding color transitions. Question marks point to the unknown involvement of periplasmic binding proteins. (B) Gram-positive bacteria. Fe-siderophore uptake through the CM by known ABC-type transporters is shown. (C) S. cerevisiae. Fe-siderophore uptake is either mediated by MFS transporters (Arn proteins) or Fe-siderophores are reduced extracytoplasmatically by membrane-standing metalloreductases (Fre proteins). Released ferrous iron either is then reoxidized and taken up by the multicopper ferroxidase-high-affinity uptake complex (Fet3p-Ftr1p) or may be imported by divalent metal transporters such as Smf proteins (low specificity) or Fet4p (low affinity and specificity).
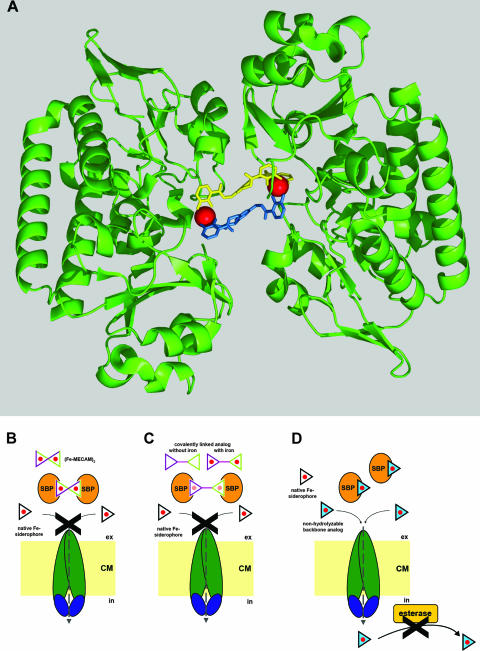
Bacterial uptake of Fe-siderophores depends on ABC-type transporters and, in the case of gram-negative bacteria, OM receptors as the “first gate” of Fe-siderophore recognition. The crystal structures of the OM receptors FepA (40), FhuA (194), and FecA (352) from E. coli and FpvA (60) and FptA (61) from P. aeruginosa for Fe-enterobactin, ferrichrome, ferric dicitrate, Fe-pyoverdin, and Fe-pyochelin uptake, respectively, have recently been solved, with those of the four latter receptors in ligand-bound forms. The overall structures are similar and show the presence of similar domains. All of them consist of a 22-β-strand barrel formed by ca. 600 C-terminal residues, while ca. 150 N-terminal residues fold inside the barrel to form a “plug” or “cork” domain (50). Transport studies upon site-directed mutagenesis of the FepA (49) and FhuA (50) plug domains, also in combination with disulfide tethering in the case of FhuA (85), suggested that conformational rearrangements of this domain are responsible for substrate passage rather than complete displacement of the plug. Energy for the transport is supplied by the TonB complex, consisting of three proteins: the CM-anchored, periplasm-spanning TonB protein and the CM-embedded ExbB and ExbD proteins. It is thought that the TonB complex “transduces” the proton motive force energy to the receptor to allow substrate translocation. TonB makes contacts with both ExbB and ExbD with a stoichiometry of 1 (TonB):7 (ExbB):2 (ExbD) via its N-terminal domain (143). Additionally, TonB interacts via its C-terminal domain with the N terminus of the OM receptor. Interaction studies of FhuA with TonB showed an intermediate 1:1 complex and a stable 1:2 complex, the latter of which increased upon the addition of ferricrocin as an FhuA ligand (164). These findings are supported by the crystal structure of the C-terminal domain of TonB, revealing dimer formation, which, however, seems to depend on the length of the recombinant protein fragments chosen (169). A crystal structure of FhuA in complex with the TonB C-terminal domain was solved recently (245), which showed the interaction between the receptor and the energy transducer in more detail (Fig. 5A). FhuA and TonB interact by forming an interprotein β-sheet between the FhuA-Ton box, a conserved N-terminal region of 8 to 10 aa residues present in all ferrisiderophore receptors, and the β3-sheet of the C-terminal TonB domain. Furthermore, upon this interaction, the TonB α1-helix is positioned proximal to the FhuA plug domain, which results in an electrostatic interaction between TonB Arg166 and FhuA Glu56. It was suggested that Arg166 transfers a mechanical pulling or shearing force from the β3-sheet to the plug, leading to its disruption or to some kind of conformational change that is necessary to permit Fe-siderophore translocation (245).
FIG. 5.
Structures of proteins involved in siderophore uptake and iron release. (A) The E. coli OM ferrichrome transporter FhuA in complex with the periplasmic C-terminal TonB domain (with the latter shown in orange) (PDB accession number 2GRX). (B) The periplasmic ferrihydroxamate binding protein FhuD of E. coli cocrystallized with ferricoprogen (coprogen is shown in red, and iron is shown in orange) (PDB accession number 1ESZ). (C) The E. coli enterobactin and diglucosylenterobactin (salmochelin S4) esterase IroE covalently linked to DFP (DFP is shown in red) at the active-site serine (PDB accession number 2GZS). (D) Ferrienterobactin esterase Fes from Shigella flexneri (PDB accession number 2B20).
The OM receptors for Fe-siderophore uptake display high ligand affinities and specificities. E. coli K-12 possesses at least five OM receptors for the uptake of different siderophores: FepA for the uptake of Fe-enterobactin (250), Cir (also termed CirA) and Fiu for linear Fe-enterobactin degradation products (67, 232), FecA for ferric dicitrate (329), FhuA for ferrichrome (155), and FhuE for Fe-rhodotorulate and Fe-coprogen (130). Although FhuE was also identified as being a receptor for ferrioxamines B and D1 (130), an additional receptor termed FoxB was suggested for ferrioxamine B transport (230). E. coli strains carrying the virulence-associated plasmid CoIV and the iroA gene cluster additionally possess the IutA receptor for Fe-aerobactin uptake (122) and the IroN receptor for Fe-salmochelin uptake (132), respectively. The Cir receptor was additionally found to mediate the uptake of Fe-salmochelins and/or their linear degradation products (132). This is in congruence with another study showing that IroN function seems to be replaceable to a certain extent during virulence development since a Salmonella enterica iroN mutant had a similar 50% lethal dose in mice compared to that of the parent strain upon intraperitoneal infection (344). In several cases, the chirality of the Fe-siderophores was shown to be decisive for uptake by the OM receptors. Stereoselective siderophore recognition was reported for ferrichrome (340), Fe-enterobactin (229), and Fe-rhodotorulate (209) receptors in E. coli and for ferrichrome in Y. enterocolitica (170) and P. putida (154). The affinity of FepA for Fe-enterobactin was found at dissociation constants (KD values) of 20 nM in vitro (246) and <0.2 nM in vivo (231), and the affinity of FhuA for ferrichrome was estimated to have a KD of 50 to 100 nM in vitro (195).
The delivery of the iron-siderophore complex into the cytoplasm depends on ABC-type uptake systems that generally possess, in contrast to efflux systems, extracytoplasmic substrate binding proteins (one or more per system), which are usually located in the periplasm (and are hence also called “periplasmic binding proteins”) in gram-negative bacteria and, in contrast, are present as lipoproteins tethered to the external surface of the CM in gram-positive bacteria (313). In systems with two or more extracytoplasmic substrate binding proteins, the receptors may interact in a cooperative fashion (21). The extracytoplasmic substrate binding proteins scavenge the Fe-siderophore, which, upon interactions with the transmembrane permease unit, is channeled through the membrane. Analogously to ABC-type secretion, the transport is energized by cytoplasmic ABC subunits, whose dimerization and conformational changes upon NTP binding and/or hydrolysis are thought to act as the “power stroke” that pushes the substrate through the transmembrane channel (157). Four subtypes of ABC-type transporters involved in Fe-siderophore uptake in gram-negative bacteria have been described so far, and additionally, a fifth one was suggested to be involved in Fe-salmochelin uptake. The first and most common subtype possesses its domains as distinct polypeptide chains and is thus composed of usually four different proteins: a substrate binding protein located at the extracytoplasmic surface, two integral membrane proteins forming a heterodimeric transmembrane channel, and a cytoplasmic ABC subunit that interacts with the channel subunits at the cytoplasmic site of the membrane. This subtype is exemplified by the E. coli FepBDGC Fe-enterobactin and FecBCDE ferric dicitrate uptake systems. E. coli FhuDCB represents a similar system; however, the transmembrane component is here comprised of one single polypeptide chain of FhuB, which can be regarded as a fusion of two “normal” transmembrane subunits. Thus, FhuDCB represents a second ABC-dependent Fe-siderophore uptake subtype in gram-negative bacteria. As a third subtype, ABC-dependent iron uptake systems that comprise membrane lipoproteins for substrate binding, which was described as being a typical feature of ABC-type transport systems in gram-positive bacteria, were identified in some gram-negative bacteria. Examples for these “unusually” modified substrate binding proteins in gram-negative bacteria are ViuP for Fe-vibriobactin and Fe-enterobactin uptake in V. cholerae (345), FatB for Fe-anguibactin uptake in V. anguillarum (3), and CeuE for Fe-enterobactin uptake in Campylobacter jejuni and Campylobacter coli (242, 276). Since the putative heme binding protein HutB in V. cholerae was also predicted to be a membrane lipoprotein, it was speculated whether this modification type is a common feature of substrate binding proteins in Vibrio spp. (345).
A fourth subtype of ABC-type Fe-siderophore importers was found to be involved in the uptake of Fe-yersiniabactin. YbtP and YbtQ from the high-pathogenicity island of Y. pestis represent domain fusion proteins comprising a TMS domain and an NBF, and hence, the composition of YbtPQ resembles an ABC-type secretion system. Together with the Fe-yersiniabactin OM receptor FyuA, YbtPQ was sufficient to restore Fe-yersiniabactin-supported growth in the enterobactin-deficient strain E. coli H1884 but not in the nonpathogenic strain Y. enterocolitica NF-O, suggesting that there are additional factors needed (e.g., a periplasmic binding protein), which are not provided in every case (33). Thus, the necessity for a periplasmic binding protein for YbtPQ-mediated Fe-yersiniabactin uptake is still in question.
The fifth putative subtype is the four-domain fusion protein IroC, the role of which was discussed above. Since it was observed that E. coli fepB mutants that possess the iroA locus were still able to utilize salmochelin as an iron source, it was concluded that IroC, the only CM permease encoded in the iroA cluster, might be responsible for Fe-salmochelin uptake (359). Direct evidence for IroC function, however, for its role in uptake or in secretion does not exist so far.
In gram-positive bacteria, so far, two subtypes of ATP-dependent transporters are known to be mainly involved in Fe-siderophore import. One is characterized by lipid-modified substrate binding proteins tethered to the outer leaflet of the CM. The most common explanation for the presence of membrane-attached substrate binding proteins in gram-positive bacteria is the lack of a periplasmic space, which forbids free extracytoplasmic diffusion of the binding proteins. Another subtype was recently found to be responsible for Fe-carboxymycobactin uptake in M. tuberculosis (278). The ABC-type uptake components IrtAB display significant similarity in domain organization to the Y. pestis YbtPQ system; however, IrtA possesses an additional 292-aa N-terminal domain that is predicted to be an extracytoplasmic siderophore interaction domain. Thus, IrtA likely represents a three-domain fusion protein of extracellular binding, transmembrane channeling, and cytoplasmic ATP hydrolysis that acts together with its two-domain counterpart IrtB. That such a domain arrangement is possible is supported by the recent finding of two families of ABC-type transporters in which one or two extracytoplasmic substrate binding domains are fused to either the N or C terminus of the translocator protein (323).
Substrate binding proteins are the “second gate” of substrate recognition prior to cytosolic import in gram-negative bacteria, while in gram-positive bacteria, they commonly represent the first and exclusive layer of such recognition processes. The crystal structures of the Fe-hydroxamate binding protein FhuD from E. coli (59) (Fig. 5B) and of the Fe-triscatecholate binding protein CeuE from C. jejuni (225) (see Fig. 10A) in ligand-bound forms have been solved. Both proteins display the overall kidney bean-shaped bilobate structure, which is typical of periplasmic binding proteins in general; however, they show novel folding features that differentiate them from the known groups I and II of periplasmic binding proteins (265). Most importantly, while the two domains of these proteins are usually linked by flexible antiparallel β-strands, which are also found in ferric ion binding proteins such as FbpA (235), SfuA, and YfuA (301), belonging to the transferrin structural superfamily, the bidomains of FhuD and CeuE are connected via α-helical elements, making their backbones more rigid, and abolish broad conformational changes upon ligand binding. However, molecular dynamic simulations suggested a 6° closure of the FhuD C-terminal domain upon ferrichrome release, and this motion might lead to FhuD conformers that could selectively interact with the FhuB transmembrane component of the transporter (177). The ligand binding site is commonly the most variable part in periplasmic binding protein structures and can be used for phylogenetic classification as in the case of periplasmic ferric ion binding proteins that are currently divided into three classes depending on their coordination modes for the Fe(III) ligand (301). The ligand binding site of CeuE is more hydrophilic and more strongly positively charged than that of FhuD, which is owing to the overall chemical nature of the ligands and, in particular, to the geometry and atom composition of their iron binding moieties that are mainly recognized by the proteins. However, while Fe-siderophores that bind to OM receptors such as FhuA and FepA are completely buried in their external cavities prior to transport, there are clearly fewer residues involved in the FhuD- and CeuE-dependent ligand interaction, which is consistent with the higher ligand promiscuity generally observed for substrate binding proteins compared to OM receptors. While many E. coli strains possess several OM receptors for recognition and transport of different Fe-catecholates (Cir, Fiu, and FepA), different Fe-hydroxamates (FhuA, FhuE, and IutA), and ferric dicitrate (FecA), only one periplasmic binding protein for each siderophore class (FepB for Fe-catecholates, FhuD for Fe-hydroxamates, and FecB for ferric dicitrate) is provided for the recognition of the ligands in the periplasm. The KDs of FhuD for coprogen, aerobactin, ferrichrome, and albomycin determined by intrinsic tryptophan fluorescence quenching upon ligand binding were found within 1 order of magnitude at 0.3, 0.4, 1.0, and 5.4 μM, respectively (281). However, the KD for ferrichrome A that possesses a 3-methylvaleryl modification instead of the ferrichrome methyl group at one of the Fe(III)-coordinating hydroxamate moieties was found at 79 μM. This demonstrates that modifications near the iron-coordinating site have a much greater impact on ligand affinity than structural differences in the ligand backbones. This is consistent with the finding that ferrioxamines B and E, which on the one hand display much greater structural differences in their backbones compared to each other than ferrichrome A compared with ferrichrome but on the other hand do not possess modifications at their trishydroxamate iron-coordinating site, had lower KDs with FhuD of 36 and 42 μM, respectively. As FhuD has a 10- to 100-fold-lower affinity to its ligands than FhuA, essentially the same difference was observed for the affinities of FepB and FepA to Fe-enterobactin, although the total affinity of the Fe-catecholate transport systems is about 100-fold higher than the affinity of the Fe-hydroxamate transporters in E. coli.
FIG. 10.
Models for uptake and iron release inhibition. (A) Crystal structure of the C. jejuni Fe-enterobactin periplasmic binding protein CeuE (PDB accession number 2CHU), which was found in dimerization with (Fe-MECAM)2, a synthetic enterobactin analogue with a benzoyl backbone. (B) Model for (Fe-MECAM)2-dependent dimerization of substrate binding proteins (SBPs), leading to Fe-siderophore uptake inhibition. (C) Model for dimerization of substrate binding proteins by covalently linked siderophore analogues, which may induce dimerization in both iron-charged and nonloaded forms. (D) Model for intracellular inhibition of esterase-dependent iron release by siderophore analogues containing nonhydrolyzable backbones.
In contrast to E. coli, B. subtilis uses five lipid-anchored substrate binding proteins for Fe-hydroxamates, Fe-catecholates, and ferric dicitrate as the exclusive gates for extracellular Fe-siderophore recognition prior to cytosolic uptake. Ligand specificity is partially more pronounced in B. subtilis since fungal ferrichromes, bacterial ferrioxamines, and citrate-hydroxamates such as schizokinen and arthobactin have distinct substrate binding proteins (FhuD, YxeB, and YfiY, respectively) (239). Ferric dicitrate and Fe-catecholates are recognized by YfmC and FeuA, respectively. The Fe-bacillibactin binding protein FeuA has ligand specificities and affinities similar to those of E. coli FepB. Recombinant FeuA in its nonacylated form has a KD for Fe-bacillibactin of 57 nM, while Fe-enterobactin and Fe-(DHB)3 are bound with 80% and 0.8% of that affinity, respectively (216). Interesting observations have been made concerning the stereoselectivity of Fe-triscatecholate siderophore binding proteins. The free binding substrates Fe-enterobactin and Fe-bacillibactin have right- and left-handed Δ and Λ configurations, respectively (24), whereas the ferric complex of the synthetic triscatecholate ligand MECAM represents a racemic mixture in solution. Binding of the ferric MECAM complex (as a dimer) to CeuE leads to an induced preference of the Λ configuration, as shown by circular dichroism spectroscopic analysis (225). A similar observation for the preferred binding of the left-handed configuration was made with FepB and Fe-enterobactin. The KD for Fe-enterobactin was found at 30 nM; however, the nonnative Λ stereoisomere Fe-enantioenterobactin displayed a KD of 15 nM (305). This indicates a change of configuration if native Fe-enterobactin is bound to FepB. Such an induced change of configuration has also been observed in the crystal structure of siderocalin with (partially degraded) Fe-enterobactin that shows an Λ configuration in the siderocalin calyx (117, 225) (see also Fig. 7A). In the case of B. subtilis FeuA, the binding of Fe-bacillibactin is preferred over that of Fe-enterobactin, also indicating a preferred binding of the Λ configuration, and uptake of Fe-enantioenterobactin, which was proposed to be FeuA dependent, was also observed (75).
FIG. 7.
Structural comparison of siderocalin (PDB accession number 1L6M) (A) and tear lipocalin (PDB accession number 1XKI) (B) binding pockets. Siderocalin was cocrystallized with Fe-enterobactin (iron is shown in orange, and ligand is shown in red) (the trilactone backbone is partially degraded).
The Fe-hydroxamate uptake system of S. aureus is an example of a cooperative system that involves two FhuD homologues, FhuD1 and FhuD2, which are suggested to interact with both the FhuBGC complex, consisting of the permease, and ABC subunits. While FhuD1 recognizes ferrichrome and ferrioxamine B, FhuD2 mediates the uptake of a broader array of (citrate)-hydroxamate ligands (297). Interestingly, the KDs of FhuD2 for ferrichrome and ferrioxamine B were found to be 20 nM and 50 nM, respectively. The higher ligand affinity of S. aureus FhuD2 than that of E. coli FhuD is due to a ligand binding site with substantially different topology. The superior affinity of FhuD2 might have been enforced upon the lack of substrate concentration in the extracytoplasmic environment of gram-positive bacteria, which is in contrast to the corresponding situation in gram-negative bacteria, in which substrate accumulation is supposed to occur in the periplasm, hence leading to more relaxed affinities of periplasmic binding proteins (298).
Since ABC-type uptake systems are not found in eukaryotes, Fe-siderophore uptake in yeast and filamentous fungi is somewhat different from that of bacteria. Full Fe-siderophore complexes are generally imported by MFS-type transporters (175). In S. cerevisiae, four Fe-siderophore importers of this type have been found: Arn1p for the uptake of ferrichromes that may possess anhydromevalonyl residues (140), Taf1p (Arn2p) for Fe-triacetylfusarinine C (TAFC) (triacetyl fusigen) uptake (141), Sit1p (Arn3p) for preferred ferrioxamine B uptake (354, 355), and Enb1p (Arn4p) for Fe-enterobactin uptake (139). Since the specificities of these MFS-type porters for the tested siderophores were not found to be absolute, a certain redundancy in substrate utilization was noted, especially for Fe-hydroxamate uptake by Arn1p and Sit1p (175). The Km values for the uptake of ferrichrome-bound iron were determined to be 0.9 μM and 2.3 μM for Arn1p and Sit1p, respectively (355). Two ferrichrome binding sites were predicted in Arn1p, one with a higher affinity (KD of 8.1 nM) and one with a lower affinity (KD of 1.2 μM). Although the half-maximal saturation of ferrichrome-delivered iron uptake by Arn1p is similar to the KD of the lower-affinity binding site, which might speak for predominantly low-affinity binding of ferrichrome to Arn1p if substrate binding rather than translocation is supposed as the rate-limiting uptake step (223), high-affinity binding was suggested to occur under normal in vivo conditions since it was observed that under nonoverexpression conditions, Arn1p localizes to the plasma membrane already in the presence of low nanomolar concentrations of ferrichrome but not in the absence of substrate that leads to the sorting of Arn1p to the endosomal compartment (166). Further transport studies revealed a high stereoselectivity of fungal Fe-siderophore uptake systems, as shown in Rhodotorula pilimanae, which preferred Δ rhodotorulic acid (226). For the uptake of ferrichrome by S. cerevisiae and N. crassa, the Λ configuration was preferred (338, 339). Ferrichrome producers such as Aspergillus quadricinctus and Penicillium parvum discriminated more strictly for the Λ isomers than ferrichrome nonproducers. Compared to E. coli, which could still use enantioferrichrome via FhuA with an approximately 50% slower uptake rate than ferrichrome, enantioferrichrome was totally excluded from the P. parvum ferrichrome uptake system (340).
The utilization of siderophore-bound iron by reductive iron uptake includes the activities of cell surface metalloreductases and Fe(II)-specific transport systems. In S. cerevisiae, the membrane-bound reductases Fre1p to Fre4p belonging to the flavocytochrome superfamily are involved in Fe-siderophore reduction, while the function of two additional homologues, Fre5p and Fre6p, which are also upregulated upon iron starvation, has not been elucidated yet. The major iron-chelate reductase activity in S. cerevisiae is provided by Fre1p and Fre2p, which reduce Fe(III) bound to citrate, ferrioxamine B, ferrichrome, TAFC, and rhodotorulic acid, while the Fre3p reductase activity for these substrates was found to be 40-fold lower (293, 353). Fre4p could facilitate the utilization of iron bound to rhodotorulic acid only. Furthermore, Fre1p and Fre2p were shown to reduce enterobactin-bound Fe(III), which is remarkable due to its extremely low redox potential of approximately −0.75 V at pH 7. However, since this is equilibrium dependent, the drain of the reduced iron into the cell mediated by the Fe(II) uptake systems might facilitate the reduction of Fe-enterobactin. As a Fre-dependent extracellular acidification was also observed (187), which might be the result of proton gradient generation, the lowered extracellular pH is also favorable for Fe(III)-chelate reduction. Fre1p and Fre2p are also capable of reducing Cu(II) to Cu(I), which is then taken up by the plasma membrane transporters Ctr1 and Ctr3 (109). Further connections of iron and copper metabolism in eukaryotes are exemplified by multicopper-dependent oxidases such as yeast Fet3p or its human homologue ceruloplasmin, which oxidizes Fe(II) coming into circulation for subsequent apotransferrin charging, and the failure of loading ceruloplasmin with copper due to copper starvation or Wilsons disease leads to the development of anemia (12, 306). Fet3p and ceruloplasmin also have cuprous oxidase activity to prevent the accumulation of the prooxidant Cu(I) (309).
The cellular import of Fe(II) that is reductively released from iron-siderophore complexes is mediated mainly by the high-affinity uptake system Fet3p-Ftr1p (306, 310), which is the prototype of the newly classified oxidase-dependent Fe(II) transporter family as part of the iron/lead transporter superfamily (288). The multicopper-dependent ferroxidase Fet3p reoxidizes Fe(II) prior to its uptake by the ferric iron permease Ftr1p. The ferroxidase reaction is coupled to the reduction of oxygen to water and has an in vitro Km of 5.4 μM (315). The crystal structure of the Fet3p glycoprotein reveals the presence of three domains (domains 1 to 3): domain 3 binds one copper atom, while a trinuclear copper cluster is bound at the interface of domains 1 and 3 (315). The physical interaction of Fet3p and Ftr1p was demonstrated by fluorescent resonance energy transfer measurement in S. cerevisiae (302) and cross-linking studies with suspensions of Pichia pastoris membrane fractions (26). The system has an overall Km for iron uptake of 0.2 μM (175). The tight coupling of Fe-siderophore reduction and high-affinity iron uptake also suggests an interaction between the Fre proteins, and the Fet3p-Ftr1p complex (353), however, remains speculative so far. The high-affinity iron transporter Fet5p-Fth1p, which is a Fet3p-Ftr1p homologue, is located at the vacuolar membrane of S. cerevisiae, mediating intracellular iron channeling into the cellular vacuole as a main metal storage compartment (321). In addition to the high-affinity Fe(II) uptake system, S. cerevisiae possesses several divalent metal transporters such as Fet4p and the NRAMP2 homologues Smf1p to Smf3p; however, they display low affinity towards Fe(II) (Km for uptake of >2 μM) and are not suspected to contribute significantly to siderophore-delivered iron uptake. While normal locus concentrations of Fet4p, which has an apparent Km for Fe(II) import of 35 μM, are sufficient to maintain cell growth at medium iron concentrations higher than 1 μM, the overexpression of Smf1p is necessary for growth rescue during iron limitation (175).
Similar to S. cerevisiae, Candida albicans possesses both major uptake routes for the utilization of siderophore-bound iron. The C. albicans siderophore transporter Arn1p (CaArn1p) (CaSit1p), transports various ferrichrome-type siderophores, was found to be required for the invasion and penetration of reconstituted human epithelium, while it is not essential for systemic infection in mice (142). By contrast, CaFTR1 is required for systemic infection in mice (266); however, no change in pathogenicity was observed for a CaFET3 mutant (82). In Ustilago sphaerogena and U. maydis, ferrichrome is taken up as an iron-ligand complex, while iron bound to ferrichrome A is imported via the reductive route (11, 83). A. nidulans, which appears to lack a reductive iron assimilatory system (84), possesses three paralogous iron-regulated importers (MirA, MirB, and MirC), of which MirA and MirB are specific for Fe-enterobactin and Fe-TAFC uptake, respectively, while the function of MirC was not yet elucidated (125).
Mechanisms of iron release.
There are two known general mechanisms that lead to iron release from siderophores. The first comprises the reduction of siderophore-bound Fe(III) to Fe(II) followed by the spontaneous release or competitive sequestration of the reduced species. Most Fe-siderophores are supposed to be discharged in this way, which essentially includes the activity of ferrisiderophore reductases. Although a considerable number of enzymes with ferrisiderophore reductase activity have been found, only a few of them possess significant specificities for their Fe-siderophore substrates, suggesting that reductive iron release has in many cases not led to the development of highly specialized enzymes but has led to the involvement of already established reductase activities. In contrast, specialized enzymes are employed for the second concept of iron release, which is mediated by Fe-siderophore hydrolysis, leading to a dramatic loss of complex stability and hence facilitating the subsequent removal of the iron, e.g., by following reduction or by interaction with other cellular iron-binding components. It is a fairly limited strategy since it is feasible only if the siderophore structure provides a hydrolyzable backbone that allows enzymatic degradation. Since hydrolytic iron release results in the destruction of the siderophore scaffold, it is more cost-intensive to the cell than ferrisiderophore reduction that basically allows siderophore reutilization. In addition to the general mechanisms of iron release, it has to be differentiated whether this pathway step occurs before or after the cytosolic import of the siderophore-delivered iron. As indicated above, iron release may already take place in the extracytoplasmic environment prior to uptake into the cytosol. This scenario has been best studied on the basis of the yeast plasma membrane reductase systems encoded by the FRE genes. The FRE proteins are similar to b-type cytochromes such as the large β-subunit gp91phox of NADPH oxidase from human phagocytes or flavocytochrome b558 of human neutrophils, sharing the FAD and NAD(P)H binding motifs with them (299). Site-directed mutagenesis of Fre1p suggested four histidines as coordinating residues for two heme b groups (93). The hemes are supposed to be involved in transmembrane electron transfer, which is probably coupled with the generation of a proton gradient as shown for gp91phox and indicated by differences in extracellular acidification of wild-type S. cerevisiae and a FRE1 mutant (138, 187). In addition to the generated proton motive force, the acidification might also be for the benefit of Fe-siderophore reduction by increasing the Fe(III) redox potential in the iron-chelate complex. In contrast to the membrane-standing cytochrome b-like reductases in yeast, ferrisiderophore reduction in prokaryotes is in the majority of cases carried out by soluble flavin reductases that catalyze reduction of several flavin-based substrates such as flavin mononucleotide (FMN), FAD, and riboflavin by using NAD(P)H as an electron donor (251). The reduced flavins are released into solution to serve as reducing agents for a variety of compounds including Fe(III)-chelates. The general mechanism of iron release is assumed to proceed by electron transfer from the enzyme-released free reduced flavins to the siderophore-bound Fe(III). Thus, the enzyme is here rather indirectly involved in the reaction, and hence, its ferrisiderophore activity is a nonexclusive function. This is consistent with the general observation that purified bacterial ferrisiderophore reductases are neither substrate specific nor subjected to iron-dependent regulation (127). Both extra- and intracellular assimilatory ferric iron reductases with ferrisiderophore reductase activities in a broad spectrum of bacteria were described (293). The reduction of ferrioxamine B-bound iron by extracellular or periplasmic ferrisiderophore reductases from E. coli, P. aeruginosa, Y. enterocolitica, and Listeria monocytogenes was studied in vitro by using NADH as a coenzyme and FMN as an enzyme substrate (63). While NADH alone was kinetically restricted from iron reduction, the generated reduced FMN reduced ferrichrome-bound iron, and the observed semiquinone intermediate indicated one-electron transfer in two steps (63). The presence of bacterial extracellular ferrisiderophore reductases is consistent with the broad distribution of homologous and/or analogous Fet3p-Ftr1p-like uptake systems in bacteria, such as the YwbLMN uptake system for elemental iron in B. subtilis, indicating high-affinity uptake of the extracellularly released iron (239). The best-studied intracellular flavin reductase is the E. coli Fre reductase that uses both NADH and NADPH as electron donors and a variety of flavin analogues as electron acceptors. The Fre crystal structure revealed extensive similarity to flavoproteins of the ferredoxin reductase family, with the main difference being that it lacks the binding site for AMP of the FAD coenzyme, which is tightly bound to flavoproteins. This explains the relaxed specificity of Fre towards flavin substrates since they are bound only through their isoalloxazine rings via hydrophobic interactions and with very limited contribution to the binding by the ribityl side chain (150). However, proteins containing tightly bound FAD that does not diffuse from the enzyme after reduction are also able to reduce Fe-siderophores, as shown for the hemoglobin-like flavohemoprotein Hmp of E. coli that reduces, in addition to cytochrome c, Fe-hydroxamate K under a nitrogen atmosphere with a Km of 104 μM (258). Since the Km was significantly increased in the presence of carbon monoxide, which completely blocks electron transfer from Fe(II)-heme, it was concluded that electron transfer to the Fe-siderophore proceeds directly via the FAD cofactor. Recently, a new type of Fe-siderophore reductase was discovered with the E. coli FhuF protein that shows for the first time both an iron-dependent regulation mediated by the Fur repressor and substrate specificity to a set of Fe-hydroxamate siderophores including ferrioxamine B, Fe-coprogen, and ferrichrome (206). FhuF binds a [2Fe-2S] cluster via an unusual C-C-x10-C-x-x-C motif located at the very C terminus of the protein, and it is proposed that this redox-active iron-sulfur cluster mediates the electron transfer to the Fe-siderophore substrates (227). The midpoint redox potential of FhuF determined by electron paramagnetic resonance spectroscopy was found to be −310 ± 25 mV, which is near the standard redox potentials of ferrioxamine B, coprogen, and ferrichrome, at −468 ± 15 mV (20, 62), −447 ± 7 mV (342), and −400 ± 10 mV (332), respectively. As discussed above, the standard redox potentials of Fe-siderophores may be increased in the presence of a Fe(II) scavenger, making electron transfer from the reductase to the iron-chelate complex thermodynamically favorable. Homologues of FhuF containing the unusual [2Fe-2S] binding motif are found in Rhizobium spp. and Bacillus halodurans, in which they are encoded downstream of a Fe-hydroxamate and a putative Fe-dicitrate uptake system, respectively. Further homologues are encoded in V. parahaemolyticus and B. pertussis upstream of aerobactin uptake and within alcaligin biosynthesis components, respectively (206). Thus, FhuF could be the prototype of a new class of bacterial Fe-siderophore reductase in addition to unspecific flavin reductase. In addition to FhuF, which is loosely associated with the CM, there are few reports dealing with bacterial membrane-bound Fe-siderophore reductase activities. Such activities were found in E. coli (98) and B. subtilis (105); however, they are again caused by nonspecific flavin reductases.
Interestingly, most reports show the capability of Fe-siderophore reductases to remove iron from Fe-siderophores such as Fe-dicitrate, Fe-(DHB)3, and Fe-hydroxamates, all of which do not display formation constants essentially higher than 1030 M−1 (271). Reductive iron release from the highest-affinity siderophores such as Fe-enterobactin and Fe-bacillibactin, with Kfs of 1049 (198) and 1047.6 (75), respectively, remains an exception consistent with the extremely low redox potential of the Fe(III) bound to these chelators. The reduction of iron bound to triscatecholate siderophores is only known to be catalyzed by Fre1p and Fre2p (see above) and was suggested to occur in E. coli and B. subtilis since their growth was promoted in the presence of iron-charged nonhydrolyzable enterobactin analogues (136, 196). Cell extracts of B. subtilis were shown to catalyze the reductive release of iron from these synthetics in the presence of NADPH as an electron donor; for maximal reaction activity, FMN and Mg(II) were also required (196). However, studies of E. coli and B. subtilis did not exclude the possibility that Fe(III) bound to the enterobactin analogues was partially transferred to the native siderophores enterobactin and bacillibactin, respectively, which would lead to the mobilization of cellular processes. Such a bypassing effect is indicated by the unexpected observation that a mutation in the fes gene, coding for hydrolytic iron release activity from Fe-enterobactin, as discussed below, abolished growth promotion in E. coli (136).
Thus, intracellular reductive iron release from triscatecholate chelators was not clearly demonstrated yet, and it seems not only a lucky coincidence that the iron chelators with the highest known affinities are supplied with hydrolyzable backbones that allow iron release through an esterase-catalyzed reaction. The E. coli ferric enterobactin esterase Fes was the first Fe-siderophore esterase that was discovered quite early and remained the exclusive example of a hydrolytic iron release activity for a long time (37, 237). Fes hydrolyzes the trilactone backbone of Fe-enterobactin, leading to the formation of linear trimers, dimers, and monomers of 2,3-dihydroxybenzoylserine. Meanwhile, there are two additional trilactone hydrolases known in pathogenic enteric bacteria, which are encoded by the iroD and iroE genes of the virulence-associated iroA gene cluster comprising genes for the synthesis and utilization of glucosylated enterobactin derivatives called salmochelins. The three enterobacterial trilactone hydrolases belong to the family of α/β-hydrolases and possess a conserved GxSxG serine esterase motif. They are transcriptionally regulated by Fur and have been studied in detail with regard to substrate specificities and reaction kinetics; however, contrary results were partially reported. While cell extracts containing Fes activity were reported to hydrolyze enterobactin only about 30% as much as Fe-enterobactin in 1 h (180), studies with semipurified and purified Fes showed a 2.5-fold- and 4-fold-higher activity with enterobactin than with Fe-enterobactin, respectively (37, 121). However, a recent study reported catalytic efficiencies (kcat/Km) for Fes (purified to 90%) with enterobactin and Fe-enterobactin of 4.0 min−1 μM−1 and >90 min−1 μM−1, respectively, clearly demonstrating that iron-charged enterobactin is the preferred Fes substrate, displaying a Km that is more than 100-fold lower than that of the deferrated ligand (191). The catalytic efficiencies of IroD and IroE for enterobactin and several salmochelins (mono-, di-, and triglucosylated enterobactins) were furthermore determined during this study. Generally, IroD greatly preferred iron-charged substrates, while essentially the inverse was observed for IroE, which was used in a 30-aa N-terminally-truncated version due to low solubility in the presence of the predicted N-terminal transmembrane region. IroD and IroE hydrolyzed non-, mono-, di-, and triglucosylated enterobactin with decreasing efficiencies; however, the absolute efficiencies for hydrolyzing the glucosylated derivatives were higher than those observed for the corresponding Fes activities, suggesting that IroD and IroE are mainly responsible for salmochelin degradation. While Fes and IroD cleaved their substrates down to the monomeric products, the main products of IroE-mediated hydrolysis were the linear trimers of the cyclic substrates. Thus, it was suggested that the predicted membrane-tethered periplasmic esterase IroE hydrolyzes cyclic salmochelins after their cytosolic export only once in the periplasm, leading to the release of linear salmochelin trimers (191). Indeed, salmochelin S2 that represents linear diglucosylated enterobactin was independently reported to be the main salmochelin fraction in the supernatant of iron-limited salmochelin producers (132). In contrast, the cytosolic esterases Fes and IroD were proposed to mediate iron release from the imported iron-charged enterobactin and salmochelin complexes, respectively. IroD was furthermore reported to catalyze regioselective hydrolysis of nonloaded mono- and diglucosylated enterobactin (191). Significantly, the linear trimeric product resulting from IroD-catalyzed monoglucosylated enterobactin hydrolysis is structurally consistent with the linearized monoglucosylated enterobactin found in posttranslationally modified microcin E492 (MccE492), and thus, a biosynthetic role of the IroD homologue MceD, which is present in the corresponding biosynthesis cluster of the natural MccE492 producer Klebsiella pneumoniae RYC492, was suggested (96, 191). A later study that investigated the activities of IroD and IroE led to partially different results concerning the substrate specificities of the esterases (359). In this case, IroD was reported to greatly prefer deferrated enterobactin and salmochelin substrates over the iron-charged complexes, while IroE was found to hydrolyze both nonloaded and iron-charged diglucosylated enterobactin with essentially the same efficiency. Furthermore, the Km values observed in that study were substantially higher than those reported by Lin et al. Due to their observations, Zhu et al. suggested a model for salmochelin utilization in which IroE preferentially hydrolyzes iron-charged diglucosylated enterobactin after uptake into the periplasm, which supposes that all three esterases function primarily in iron release from iron-charged siderophores. Recently, the crystal structures of Fes (Fig. 5C) and IroE (Fig. 5D) were solved (Protein Data Bank accession number 2B20) (Y. Kim, N. Maltseva, I. Dementieva, P. Quartey, D. Holzle, F. Collart, and A. Joachimiak, unpublished data; 181). Interestingly, the enzymes differ in two important ways. First, Fes possesses, in addition to the catalytic α/β-hydrolase fold, an N-terminal β-sandwich lid region covering the active site, while IroE lacks this domain and hence displays an open catalytic center. Second, Fes possesses a typical catalytic triad that is formed by a histidine, the highly conserved serine of the GxSxG motif, and a glutamate, whereas IroE bears an atypical catalytic diad comprised of the conserved serine and a histidine (181). The lid domain is proposed to confer a higher substrate specificity to the esterase since it participates in binding pocket formation; however, the exact molecular mechanisms that are responsible for the differences observed in specificity, reaction kinetics, and product formation remain speculative so far. Due to sequence alignment analysis, IroD was proposed to be structurally very similar to Fes, including the presence of an N-terminal lid domain and a catalytic triad, and it was suggested that these structural features might favor the complete hydrolysis of iron-charged trilactone siderophores (181). However, this might be qualified by the characterization of a further trilactone hydrolase, which was found to be present in those Bacillus spp. that utilize the enterobactin-related siderophore bacillibactin (see Fig. 6 for structure) for iron acquisition. The Fe-bacillibactin esterase BesA was identified and characterized in B. subtilis and was found to hydrolyze Fe-bacillibactin into monomers without generating detectable amounts of hydrolysis intermediates, while nonloaded bacillibactin used as a substrate led to intermediate accumulation (216). The catalytic efficiency of BesA was 25-fold higher for the three-step hydrolysis with Fe-bacillibactin than for the single-step hydrolysis of nonloaded bacillibactin. Thus, BesA greatly preferred the iron-charged ligand as a substrate and, since it was hydrolyzed completely, very much resembles the enzymology that was reported for Fes and IroD (191). However, BesA is the closest homologue of IroE, with both of them sharing a protein sequence identity of 20% and the presence of the characteristic catalytic diad; hence, its structural topology is predicted to be like that of IroE. BesA was also shown to hydrolyze enterobactin with a threefold-lower efficiency than bacillibactin, which is consistent with the observation that B. subtilis can also use enterobactin as an exogenous iron source (75, 196, 239).
FIG. 6.
Coevolution of siderophore-related defense between the mammalian host and bacterial pathogens.
The trilactone hydrolases for Fe-enterobactin and Fe-salmochelin utilization and the Fe-bacillibactin esterase are the only known bacterial enzymes involved in hydrolytic iron release. Homologues of them are found in various species that use these siderophores as exogenous iron sources or as modification groups for further secondary metabolites. Their exclusiveness among bacteria is consistent with the fact that no further ester-linked bacterial siderophores are known in addition to bacillibactin, enterobactin, and its glucosylated derivatives.
In fungi, trishydroxamate fusarinine C (fusigen) and the related TAFC are further trilactone siderophores consisting of three ester-linked N5-cis-anhydromevalonyl-N5-hydroxyornithine residues, which are additionally N2 acetylated in TAFC. Fusarinine C is produced by different genera such as Fusarium, Giberella, Aspergillus, Agaricus, and Penicillium, while TAFC was isolated from Aspergillus and Penicillium species (341). An esterase activity that hydrolyzes the ornithine ester bonds in the cyclotrimeric scaffold was found to be active on deferrated fusarinine C in Fusarium roseum, while TAFC was found to also be hydrolyzed in the iron-charged form in Penicillium spp. and Mycelia sterilia EP-76 (5, 87). The presence of such an “ornithine esterase” that has the activity under SreA control was also observed in A. nidulans (236). The linear trimeric, dimeric, and monomeric hydroxamate products formed upon TAFC hydrolysis were found to be excreted. Due to the detection of such hydrolytic products in culture filtrates in Aspergillus fumigatus (233), a similar activity was proposed for TAFC utilization in this fungus (236).
Another class of proteins that are putatively involved in iron release is comprised of the homologous siderophore-interacting proteins (SIPs). SIPs are present in various bacteria such as E. coli, Deinococcus radiodurans, M. tuberculosis, P. aeruginosa, Shewanella putrefaciens, and V. cholerae. The first SIP was described as ViuB in V. cholerae, which was shown to be required for Fe-vibriobactin utilization and to be iron-dependently regulated by Fur (43). Interestingly, viuB could complement the function of the fes gene in E. coli, although it has no sequence similarity to Fes, nor is it proposed to code for an esterase. In fact, SIPs seem to represent a group of FAD-binding oxidoreductases, which was revealed by recent crystal structure analysis of the S. putrefaciens SIP that was cocrystallized with a tightly bound FAD (Protein Data Bank accession number 2GPJ) (Joint Center for Structural Genomics and I. A. Wilson, unpublished data). The isoalloxazine ring of FAD is located directly below an open cleft at the concave side of the structure that might represent the binding site for the iron-chelate complex. The bilobate-like overall topology remotely resembles that of substrate binding proteins for Fe-siderophore recognition; however, since SIPs are not known to possess signal sequences for targeted membrane transport, they are proposed to be cytosolic proteins. The fact that Fes could not complement a ViuB deficiency in V. cholerae (43) furthermore points to a function that is not related to hydrolytic iron release from triscatecholate siderophores but that is likely related to reductive release. Since vibriobactin is a triscatecholate siderophore that does not contain a hydrolyzable backbone such as enterobactin and bacillibactin, it is reasonable that reductive iron release has to occur for Fe-vibriobactin utilization. Since no further activity for iron release from Fe-vibriobactin has been reported yet, ViuB might be a new type of Fe-siderophore reductase with specificity for Fe-triscatecholates. In E. coli, however, the chromosomal locus of the endogenous ViuB homologue YqjH, which is also under Fur control (213), apparently does not to complement a Fes deficiency, in contrast to plasmid-provided ViuB. Further studies are necessary to understand the role of SIPs in Fe-siderophore utilization.
Generally, the fate of ligand(s) and iron after successful release is poorly understood. In the case of nondestructive release, it is assumed that the emptied siderophore is used for a new iron acquisition cycle. However, degradation is also possible in this case, as observed for Erwinia chrysanthemi that employs the CbsH peptidase for chrysobactin degradation after the step of reductive iron release (269). CbsH, which is encoded by an iron-regulated gene, shows 45.6% identity to the E. coli Fes esterase; however, it is not required for Fe-enterobactin utilization in E. chrysanthemi. The protein, which has characteristics of the S9 prolyl oligopeptidase family, was rather found to prevent the intracellular accumulation of deferrated chrysobactin. E. chrysanthemi does not possess a further fes-like gene (269), and it could not be explained how Fe-enterobactin is utilized by this bacterium. Interestingly, the genome sequence of its close relative Erwinia carotovora reveals the presence of at least one ViuB homologue, which could complement Fes activity as discussed above. A further fate of catecholate-containing ligands is oxidative inactivation, since catecholates are able to reduce Cu(II) into the prooxidant Cu(I), which in turn oxidizes catechols under conditions of ROS formation (156). The E. coli multicopper oxidase CueO, which is expressed in the presence of copper, oxidizes enterobactin and its degradation products (but also its catecholate precursors), leading to the formation of 2-carboxymuconate if DHB is the substrate (120). The fate of the iron after release may be binding to storage protein like ferritins and bacterioferritins or, if reduced, to ferrochelatin. In A. nidulans, TAFC-bound iron is transferred to cellular storages of desferriferricrocine, revealing iron transfer from an extracellular to an intracellular siderophore (84). A similar mechanism was previously suggested for the utilization of coprogen-bound iron in N. crassa (207). Ferricrocin deficiency in A. nidulans causes an increase in the intracellular labile Fe(II) pool, which is linked with the upregulation of antioxidative systems and an increased sensitivity to paraquat that causes superoxide stress (84).
DEFENSE AND ANTIDEFENSE: IRON-RELATED COEVOLUTION OF PATHOGENS AND MAMMALIAN HOSTS
The struggle for iron between the parasite and the host organism can be regarded as a red queen scenario of coevolution. The pathogen responds to iron scarcity by developing systems for high-affinity iron acquisition. Notably, most pathogens possess an arsenal of systems for iron acquisition via direct and/or indirect contacts with host iron sources, which provides an advantage for multiplication in different compartments with changing iron source composition and pH conditions. The broad spectrum of iron acquisition mechanisms is the result of several constraints made by the host to suppress pathogen multiplication using passive and active strategies. In general, the withholding of iron is mediated by several iron-binding proteins such as ovalbumin, lactoferrin, ferritin, and transferrin. Unoccupied iron binding sites of these proteins serve in a buffer capacity to sequester a surplus of iron entering into circulation, thus keeping levels of free iron constantly low. The labile iron pool is permanently regulated via IRE/IRP-dependent expression of the transferrin receptor and ferritin (283). Furthermore, during the acute-phase reaction occurring upon inflammation, iron storage is enhanced by an interleukin-1- and tumor necrosis factor-dependent increase of the macrophage ferritin pool. Coincidently, the concentration of lactoferrin receptors on macrophages is also increased, while transferrin-mediated iron absorption in the lumen is decreased (153). In addition to these basic strategies of the hypoferremic response, the development of siderophore binding proteins as components of the innate immune system is an intriguing example of a specialized and pathogen-directed host defense. While it has been shown previously that siderophore sequestering can also be mediated by abundant host proteins such as serum albumin, the human variant of which binds enterobactin and Fe-enterobactin with KDs (at 4°C) of 1.6 × 10−4 and 1.0 × 10−5 M, respectively (172), this binding affinity is far too low to outcompete the bacterial Fe-enterobactin uptake system. The E. coli outer membrane transporter FepA displays a KD for Fe-enterobactin lower than 2 × 10−10 M (231), and thus, serum albumin does not efficiently impair enterobactin-mediated iron acquisition. This, in contrast, is ensured by the ligand binding protein siderocalin (also known as uterocalin, lipocalin 2 [Lcn2], NGAL, or 24p3), which is a member of the lipocalin superfamily and is found as an acute-phase protein in mammals (99, 193). Siderocalin was shown to bind Fe-enterobactin with a KD of about 4 × 10−10 M and, with lower affinity, also Fe-(DHB)3 (97, 117). Thus, siderocalin is able to compete successfully with the high-affinity Fe-enterobactin uptake system of enteric bacteria. The employment of innate defense proteins for siderophore neutralization is likely due to the fact that siderophores are hardly recognized as antigens by the acquired immune response if not presented in surface-immobilized forms. Although early studies reported the presence of enterobactin-specific immunoglobulins in normal human serum (222), which were suggested to be primarily of the immunoglobulin A isotype (221), no further observations of siderophores acting as natural immunogens have been made so far.
Siderocalin-dependent binding outrules enterobactin as a virulence factor, which was shown by comparing the mortality rates of Lcn2+/+ and Lcn2−/− mice after intraperitoneal infection with the enterobactin-producing E. coli strain H9049 (97). The same study showed that the introduction of the iroA gene cluster from the uropathogenic E. coli strain CFT073 into E. coli H9049 rendered the nonvirulent strain highly virulent. Most Salmonella enterica serotypes and some E. coli strains such as the uropathogenic strain CFT073 harbor the iroA gene cluster. The cluster comprises the Fur-regulated genes iroNEDCB, whose functions were in part elucidated (see above). Homologues of IroBD and IroNE, termed MceCD and PfeAE, are also present in K. pneumoniae RYC492 and P. aeruginosa PAO1, in which they serve in posttranslational microcin modification (G. Vassiliadis, G., J. Peduzzi, S. Zirah, X. Thomas, S. Rebuffat, and D. Destoumieux-Garzon, unpublished data) and suggested exogenous siderophore utilization, respectively (95). The C-glycosyltransferase IroB was shown to function as an enterobactin-tailoring enzyme that attaches glucose moieties to the C-5 position of the enterobactin DHB moieties via an unusual C-C-bond formation (96). The glucosylated variants of enterobactin that were called salmochelins had been isolated and structurally characterized previously (22, 132). The main salmochelin component, S2, isolated from culture supernatant of S. enterica, was found to be a diglucosylated linear enterobactin, while the cyclic diglucosylated derivative S4 was found in only minor amounts. The reason for opening the trilactone cycle might be due to the lower membrane partitioning coefficient of the linear enterobactin and salmochelin trimers (202). The glucosylation of enterobactin at two catechol C-5 positions efficiently impairs the sequestration of the siderophore by siderocalin and serum albumin, thus representing an efficient antidefense of the pathogen as the latest layer of this pathogen-host coevolution (22, 97). While siderocalin is quite flexible in binding diverse enterobactin-metal complexes and enterobactin analogues with different backbones (such as tripodal TREN- and benzoyl-based scaffolds), the methylation of the critical C-5 aryl cap positions is already sufficient to sterically hinder siderocalin binding (1, 144).
In addition to Fe-enterobactin, siderocalin also binds the ferric forms of bacillibactin, carboxymycobactins, and parabactin (1, 144). The pathogens that are constrained by siderocalin-mediated defense use different strategies to circumvent sequestration of their high-affinity iron scavengers (Fig. 6). Mycobacteria that produce carboxymycobactins as extracellular siderophores colonize the intracellular niche of macrophage phagosomes, thereby blocking their maturation and exploiting the iron-transferrin uptake system of the early phagosomes (290). By choosing this compartment, mycobacteria ensure that the carboxymycobactins, which are involved in the virulence process, are protected against siderocalin-dependent sequestration. V. cholerae produces the catechol siderophore vibriobactin, which is structurally highly similar to parabactin (produced by Paracoccus denitrificans) with three minor alterations: (i) the polyamine backbone is norspermidine in the case of vibriobactin and spermidine in the case of parabactin; (ii) vibriobactin contains two l-threonines per molecule, forming two oxazoline rings, while parabactin contains one; and (iii) vibriobactin contains three DHB moieties as aryl caps, whereas parabactin has two DHB caps and one salicylate cap that is tethered to the oxazoline ring. Although V. cholerae can use heme directly as an iron source, it has been shown that vibriobactin is also an important virulence factor (137). A vibriobactin biosynthesis mutant showed attenuated virulence in mice that were challenged with both the mutant and the vibriobactin producer by intestinal infection. The infection localization, however, appears to be decisive for virulence development with respect to siderocalin-mediated defense, which is illustrated by the fact that enteric bacteria with a mutagenized iroA cluster show impaired virulence upon infection of serum-covered compartments such as the peritoneum but not after oral infection (16, 97). These examples suggest that as long as pathogens do not refrain from using siderophores as virulence factors that could be sequestered by siderocalin, they are forced to occupy niches that either are not accessed by the secreted defense protein or provide only remote possibilities of protein-ligand interactions. As an alternative to siderophore modification and self-restriction to certain compartments for multiplication, the establishment of completely novel siderophores as virulence factors is a third strategy that allows the circumvention of siderophore sequestration. Examples for this strategy can be found in gram-negative (e.g., enteric bacteria) and gram-positive (e.g., Bacillus spp.) pathogens. Aerobactin is a mixed citrate-hydroxamate siderophore of many pathogenic E. coli, Shigella flexneri, and K. pneumoniae strains and is not sequestered by siderocalin (144). Since aerobactin is not produced by most Salmonella spp. that predominantly use salmochelins, the utilization of aerobactin seems to be an alternative strategy that is distributed among those enteric bacteria that do not use enterobactin modification to prevent siderocalin-dependent sequestration. In terms of iron binding efficiency, however, the usage of aerobactin as a virulence-related siderophore seems to be less advantageous, since its pFe at physiological pH is about 10 orders of magnitude lower than that of enterobactin and its derivatives. Thus, salmochelin-producing Salmonella strains are capable of exploiting a broad spectrum of host iron sources in all accessible compartments, while for aerobactin-dependent strains, some iron sources are less accessible, such as Fe-transferrin (38), which has a KD for Fe(III) of 10−22 M that is comparable to the equivalent value of Fe-aerobactin (6, 133) and which furthermore regulates the free serum Fe(III) concentration just below the range of the Fe-aerobactin dissociation constant. Among gram-positive bacteria, many Bacillus species, including nonpathogenic B. subtilis or B. licheniformis as well as pathogenic B. anthracis, B. cereus, and B. thuringiensis, produce the triscatecholate siderophore bacillibactin. Bacillibactin is a triscatecholate trilactone siderophore and hence is structurally related to enterobactin. Structural differences include glycine spacers between the aryl caps and the trilactone backbone that, in contrast to enterobactin, is formed by three threonine moieties instead of serines. Furthermore, the chirality of the Fe-bacillibactin complex is Λ and not Δ as in the case of Fe-enterobactin (24, 212). Fe-bacillibactin was shown to bind at pH 7.4 with a KD of 0.49 nM almost as tight to siderocalin as to Fe-enterobactin (1). Consequently, the mammalian pathogens B. anthracis and B. cereus produce a second siderophore that is structurally identical to the siderophore petrobactin, which was isolated from cultures of the oil-degrading ocean bacterium Marinobacter hydrocarbonoclasticus (174). Phenotypic analyses of biosynthesis mutants revealed that petrobactin (formerly termed anthrachelinin in B. anthracis), and not bacillibactin (formerly termed anthrabactinin in B. anthracis), is the virulence-related siderophore in B. anthracis (48). The utilization strategy of the bacillibactin/petrobactin siderophore pair in pathogenic bacilli is analogous to that of enterobactin/aerobactin utilization in pathogenic enteric bacteria. Petrobactin is built on a citrate bis-spermidine backbone that is structurally related to the citrate bis-lysine backbone of aerobactin, and the NRPS-independent biosynthesis of B. anthracis petrobactin was proposed to be analogous to aerobactin synthesis (185). However, while the lysyl moieties of aerobactin are both N6 hydroxylated and N6 acetylated, which yields four of the six oxygen donor atoms for Fe(III) coordination (the other two are provided by the citrate backbone as in petrobactin), petrobactin possesses two 3,4-dihydroxybenzoate caps linked to the spermidinyl moieties to provide the full number of oxygen donor atoms. The use of aryl caps generally includes the possibility for the ligand to get trapped in the siderocalin calyx via cation-π interactions; however, the unusual C-3 and C-4 hydroxylations of the catecholate moieties apparently prevent petrobactin capturing. This was comparatively demonstrated by binding studies with siderocalin and the ferric complexes of DHB and 3,4-dihydroxybenzoate. The binding of iron-charged 3,4-dihydroxybenzoate was not observed, and in silico modeling revealed considerable steric clashes in at least two pockets of the siderocalin cavity due to the altered positioning of the carboxylate functions in the Fe-(3,4-dihydroxybenzoate)3 complex (1). The possibility that petrobactin production is not a general requirement for virulence development in bacilli but is directly related with the mammal-specific siderocalin defense is furthermore supported by the finding that entomopathogenic B. thuringiensis does not produce petrobactin (337).
In addition to its role in siderophore neutralization, it was shown that siderocalin delivers scavenged iron-chelator complexes (Fe-enterobactin was used in essentially all studies) by several endocytic pathways into the liver, the proximal tubules of the kidney, and the placenta by using cellular receptors such as megalin (149) and 24p3R (78) for endosome internalization. It was shown that the uptake of holo-siderocalin via 24p3R increases the intracellular iron pool, while the uptake of apo-siderocalin, in contrast, diminishes intracellular iron levels, leading to cellular apoptosis (78, 79). The extent of siderocalin-mediated body iron trafficking is remarkable. It can sufficiently complement transferrin-mediated iron delivery as observed in the metanephric mesenchym and has several important physiological consequences such as protection of epithelial progenitors from apoptosis (349). Thus, it has been proposed that mammals possess endogenous iron chelators similar to enterobactin, which may reside intracellularly (78) and/or are secreted in order to charge siderocalin extracellularly with iron. Siderophore-like activities were observed in the urine of human, dog, and mouse, and a catechol-like structure was anticipated for the responsible molecule(s) (292). In this context, it is remarkable that the catecholamine hormone l-norepinephrine (NE) that is substantially produced by mesenteric organs (86) was found to scavenge iron from transferrin and lactoferrin and to support the growth of pathogens that were supplied with these iron sources in vitro (101). Infection model studies in mice showed that NE can complement enterobactin- and salmochelin-mediated iron acquisition in an S. enterica serovar Typhimurium iroN fepA double mutant; however, it was not able to support the growth of an iroN fepA cir triple mutant that is not capable of catecholate siderophore uptake (336). It was not shown whether siderocalin is capable of NE binding; however, the general impact of the sequestration of host-derived bacterial iron sources such as NE is illustrated by frequently occurring intra-abdominal sepsis caused by commensal bacteria in trauma and burn patients. Severe tissue damage is associated with a massive systemic release of NE and likely spillover into the intestine (336). Further studies will be necessary to show whether iron-catecholamines might associate with host binding proteins and, if so, whether this complex formation could be connected with iron-depleting pathogen defense and/or siderocalin-dependent iron delivery pathways in the host.
Another human lipocalin was found to act as a siderophore binding protein. Since it was first discovered in human tear fluid, it was termed tear lipocalin (Tlc, also known as lipocalin 1 or von Ebners gland protein), although it was found later on to be produced by many other secretory tissues and glands, thus being present in fluids covering most of the epithelial surfaces. Tear lipocalin exhibits highly promiscuous binding of hydrophobic ligands such as fatty acids, phospholipids, glycolipids, cholesterol, retinol, arachidonic acid, and other lipid peroxidation products and a variety of siderophores (100, 115, 183, 273). The cross sections of the binding cavities of siderocalin and tear lipocalin are shown in Fig. 7. While the siderocalin binding pocket is perfectly tailored in shape, polarity, and charge for binding triscatecholate or structurally similar ring systems mainly via hybrid electrostatic/cation-π interactions, the pocket of tear lipocalin is deeper (≈15 Å) and more hydrophobic, enabling the binding of a broader ligand spectrum (34, 117). Siderophores that bind to tear lipocalin comprise catecholates (enterobactin), hydroxamates (desferrioxamine B and the fungal siderophores rhodotorulic acid, TAFC, coprogen, ferrichrome, and ferricrocin), and mixed citrate-hydroxamates (aerobactin). The Pseudomonas sp. siderophore pyoverdin was not bound, very likely due to steric hindrances caused by its bulky chromophoric group. In contrast to siderocalin, which binds iron-charged enterobactin about 1 order of magnitude more strongly than nonloaded enterobactin (1), ligand binding to tear lipocalin was shown to be independent of iron charging (100). The KD of tear lipocalin for TAFC was in the range of 0.1 to 0.5 μM, while affinities for ferrichrome, coprogen, and rhodotorulic acid were slightly higher. Since tear lipocalin was not found to bind bacterial siderophores such as enterobactin, aerobactin, and ferrioxamine B with significantly stronger affinity, it was suspected that it might not be able to compete with high-affinity bacterial siderophore uptake systems such as those for Fe-enterobactin. However, it could be sufficient to prevent the uptake of fungal Fe-hydroxamate siderophores since both bacterial and, partially, fungal uptake systems display dissociation constants for these iron sources that are comparable to their tear lipocalin affinities. Thus, tear lipocalin might fulfill its role in sufficiently neutralizing a broad array of fungal hydroxamate siderophores, which would partially complement the specialized siderocalin-mediated defense against triscatecholates and carboxymycobactins. Indeed, it was shown that there is no antifungal activity of tear lipocalin except for siderophore sequestration (100).
NATURAL AND SYNTHETIC COMPOUNDS FOR IRON-DEPENDENT PATHOGEN CONTROL
There are several possibilities for how the need for iron can be exploited to affect pathogen multiplication and hence virulence development. Three basic concepts of iron-dependent pathogen control are introduced in this section. The “Trojan horse” concept employs siderophores as mediators to facilitate the cellular uptake of antibiotic compounds. A further strategy provides artificial iron depletion by applying siderophores that cannot be utilized as an iron source by the pathogen. The third strategy strives to prevent endogenous siderophore utilization by the inhibition of siderophore pathways. These concepts show varying degrees of efficacy for different sets of pathogens. Many of the natural and synthetic compounds that were found or rationally designed to encounter microbes in an iron-dependent manner have been only initially tested, and thus, current research focuses on their further validation and improvement with the aim of therapeutic applications.
“Trojan Horse” Antibiotics
A strategy for infection treatment is inspired by natural compounds that integrate two functional molecule parts into one structural scaffold. They are called siderophore-antibiotic conjugates (or sideromycins) since the siderophore part of these compounds is able to scavenge iron and is recognized by cellular Fe-siderophore uptake systems, while the other part bears an antibiotic activity that uses the siderophore as a “Trojan horse” and exploits the Fe-siderophore uptake system as a cellular entry gate. Examples of natural siderophore antibiotic conjugates are albomycins, ferrimycins, danomycins, and salmycins, which were isolated mainly from streptomycetes or actinomycetes, and several microcins from enteric bacteria. Albomycins are composed of a tris(N5-acetyl, N5-hydroxyornithine) peptide and a nucleoside-analogous thioribosyl pyrimidine moiety linked by a serine spacer (17). Because of its similarity to fungal ferrichromes, albomycin is recognized by the FhuA receptor and the FhuD binding protein, which was shown by both mutagenesis (259) and crystallization studies (58). After hydrolysis from the siderophore carrier, the seryl-thioribosyl pyrimidine moiety blocks protein biosynthesis by inhibiting seryl-tRNA synthetases (307). Danomycins and salmycins produced by actinomycetes contain the trishydroxamate danoxamine as a siderophore unit that is connected with an aminoglycoside moiety (147, 326). The compounds are highly active protein synthesis inhibitors in gram-positive bacteria such as staphylococci and streptococci. Ferrimycins are hybrids of ferrioxamine B and an antibiotically active group. Their transport is antagonized by ferrioxamines (356), and their activity is directed against gram-positive bacteria such as S. aureus and Bacillus spp. by altering their protein biosynthesis (30). Microcins (Mcc) are ribosomally synthesized antibiotics that are either plasmid or chromosome encoded. Chromosome-encoded MccH47, MccI47, and MccM from certain E. coli strains and MccE492 from K. pneumoniae RYC492 undergo a similar posttranslational maturation process that includes the addition of a siderophore moiety. Synthesis of MccE492 and MccH47 was shown to depend on enterobactin production, and the linkage of monoglucosylated linear enterobactin to the Ser84 residue of MccE492 via an O-glycosidic bond was demonstrated (13, 316). In addition to enterobactin synthesis, the modification requires genes of the iroA cluster for salmochelin synthesis, essentially iroB and iroD for glucosylation and backbone linearization, respectively. A similar posttranslational processing is anticipated for MccI47 and MccM (244, 252). The modified microcins are recognized and taken up by the catecholate siderophore receptors FepA, Cir, and Fiu of E. coli and FepA, Cir, and IroN of S. enterica in a cooperative and TonB-dependent manner (77, 244). The uptake of MccE492 was found to be inhibited by 2,3-dihydroxybenzoylserine dimers and trimers (311). Although both modified and unmodified forms of MccE492 were found to be transported by catecholate receptors, modified MccE492 displayed a two- to eightfold-higher activity against enterobacterial strains, suggesting that a salmochelin-like modification enhances the efficacy of MccE492 by improving its channeling to the CM as one (possible) mode-of-action target (76).
Although inhibition concentrations for some sideromycins are remarkably low, as found for the protein biosynthesis inhibitor albomycin acting effectively against E. coli at 10−8 M (341), the main drawback for therapeutical use is the general high frequency of resistant mutants that have gained mutations in the Fe-siderophore-specific receptors or the TonB complex proteins. In this respect, synthetic development of sideromycins has proven to be an outgrowing field during the last three decades with the aim to generate a broad array of new antibiotic compounds (Fig. 8). Several studies showed that synthetic siderophore-drug conjugation is feasible for creating antibiotics with improved cell permeability and reduced susceptibility to resistance mechanisms such as secretion or enzymatic inactivation. The compounds can generally be produced by derivatizing siderophores and/or antibiotics isolated from cell cultures (semisynthesis) or by total synthesis. Semisynthesis may be advantageous if suitable functional groups are provided by the natural compound, as was shown for P. aeruginosa pyoverdin that was coupled to various β-lactam antibiotics (41). However, total synthesis is often preferred since it allows the rational introduction of useful functional groups at appropriate positions, preferably in the periphery of the siderophore scaffold. In principle, each structural type of siderophore may be coupled with each type of antibiotic, yielding a broad spectrum of sideromycin variants. The siderophore part of the synthesized compounds comprises in most cases hydroxamate, catecholate, or mixed hydroxamate-catecholate functions tethered to various backbones. The use of mixed ligand-siderophore that can be taken up via several pathways is seen as advantage since it reduces the frequency of resistant mutants and, if occurring, makes them more susceptible to iron starvation since the transport systems were found to be the main targets for resistance formation rather than the antibiotic targets themselves. The advantage of mixed-ligand siderophore conjugates was proven with various antibiotic functions, and activity of a biscatechol-hydroxamate-carbacephem conjugate that was more than 2,000-fold higher than that of to the parent drug loracarbef (Lorabid) could be reached in Acinetobacter (MIC = 0.03 μg/ml) (110, 113). The antibiotic functions of the conjugates tested so far are quite diverse. While at the beginning, conservative drugs such as sulfonamides and a number of β-lactam antibiotics, especially cephalosporins, were used, since their synthesis was well established earlier (282), later studies included antibiotics from various classes, such as spiramycin, vancomycin, erythromycylamine, nalidixic acid, and norfloxacin. In many cases, the activity of the siderophore-conjugated antibiotics increased compared to those of the nonmodified antibiotics during conditions of iron starvation-mimicking growth conditions in various mammalian body fluids, thus proving the general potential of siderophore-dependent “Trojan horse” antibiotics for therapeutic applications. Not only antibacterial but also antifungal activities were provided by using nucleoside analogues such as 5-flourouridine (218) and neoenactin as well as derivatives such as norneoenactin and desketoneoenactin (18, 112). Most of these compounds gave considerable inhibition of C. albicans (MIC < 1 μg/ml) and other fungi (282). The mechanisms of drug uptake into the pathogens are still under investigation, and it is remarkable that broadly modified compounds can be transported especially by highly selective outer membrane receptors. This implies that the siderophore part might behave like a key that unlocks the receptor by specific recognition and that transport itself is less specific and might essentially comprise the pumping of the successfully recognized compound through the membrane channel. Crystal structure analysis with FhuA and several ligands revealed that the Fe-hydroxamate part of albomycin binds to the same site and interacts with the same residues as the native substrate ferrichrome (89). The antibiotic part of albomycin is bound in the external cavity of the pocket by five residues that are not involved in ferrichrome binding. Two different conformations of FhuA-bound albomycin have been observed in the crystal structure, both fitting into the recognition site and the external pocket by interacting with seven different amino acid residues. The overall transport rate of albomycin is half the transport rate of ferrichrome; however, it was not distinguished between outer and inner membrane transport mediated by FhuA and FhuDBC, respectively (30). The possibility of a high-affinity interaction between receptors, especially for Fe-siderophore uptake and noncognate substrates, is also demonstrated by the outer membrane association of various phages and colicins that use the receptors as docking stations (32, 188).
FIG. 8.
Representative iron-chelating compounds involved in natural microbial defense and considered as potential therapeutics for treatment of infection, iron overload, and tumorigenesis. PIHs, pyridoxal isonicotinoyl hydrazones.
Antimalarial Agents
While the development and therapeutical application of sideromycins are one of the basic strategies for siderophore-dependent bacterial and fungal infection treatment, some iron chelators have also been found to be of value as antimalarial agents. Desferrioxamine B (deferoxamine [DFO]) (see Fig. 1 for structure) is a linear trishydroxamate siderophore that is conventionally produced by large-scale fermentation of Streptomyces pilosus A21784 as methane sulfonate salt (Desferal; Norvatis Inc.) but is now also available via chemical synthesis (341). Iron-charged DFO displays a Kf of 1030.6 and a pFe[pH 7.4] of 26.6 (271). The use of DFO for in vitro and in vivo treatment of Plasmodium falciparum infection was shown early (256, 270). Mode-of-action studies suggested that DFO enters the parasite for intracellular iron depletion, displaying an in vitro activity of 30.8 μM (199, 200, 296). DFO conjugated with methylanthranilic acid showed 10-fold-higher in vitro activity against P. falciparum (50% inhibitory concentration [IC50] of 3 to 4 μM) (116, 200), which could even be improved by using nalidixic acid as a conjugate, displaying an IC50 of 0.6 μg/ml (≈0.7 μM) against multidrug-resistant P. falciparum (111). The mode of action in the latter case was suggested to involve metal-catalyzed oxidative DNA damage upon intercalation of the drug. Salicylaldehyde isonicotinoyl hydrazone and benzoyl hydrazone derivatives were found to be active at similar low micromolar concentrations in vitro (318). Salicylaldehyde isonicotinoyl hydrazone together with DFO significantly protected mice from infection with Plasmodium vinckei; however, they also inhibited the in vitro proliferation of B and T cells and allogeneic mixed-lymphocyte reactions to a certain extent (119).
In addition to infection treatment, there is an important therapeutic application for DFO and synthetic iron chelators such as deferiprone (1,2-dimethyl-3-hydroxypyrid-4-one) (L1, Ferriprox; Apotex Inc.), deferitrin (a dihydroxyphenyl-methylthiazolecarboxylate derivative developed by Genzyme Corp.), or deferasirox [Exjade, a bis(hydroxyphenyl)benzoate-triazole derivative developed by Novartis Inc.] in iron overload therapy (173). Furthermore, DFO and mycobacterial desferriexochelins as well as synthetic iron chelators such as dexrazoxane (Savene [Europe] and Totect [United States]) and isonicotinoyl hydrazone derivatives are also used in cancer therapy (Fig. 8) (42, 57).
Siderophore Pathway Inhibitors
Another strategy for therapeutic iron-dependent pathogen control is the employment of iron limitation itself as a bacteriostatic condition that prevents pathogen multiplication in the host. This condition can be achieved by suppressing siderophore utilization. In the early 1970s, it was already reported that siderophore-mediated iron acquisition can be inhibited with small-molecule compounds. The antitubercular drug p-aminosalicylate (PAS) was found to interfere with iron metabolism in Mycobacterium smegmatis and M. bovis by causing the inhibition of mycobactin formation (39). Iron uptake was also inhibited, and the activity of several iron-containing enzymes declined, which can be regarded as putative consequences of a mycobactin deficiency. Further studies supported this effect by detecting a 40 to 80% decrease in mycobactin production under conditions of iron starvation in the presence of PAS in M. smegmatis, while exochelin synthesis was unaffected (4). Although a concomitant increase of 50 to 55% in the accumulation of carboxymycobactin was observed, it was noted that there was a diminished flux of salicylate along the mycobactin and carboxymycobactin biosynthesis pathway and that salicylate accumulated up to 10-fold if PAS was added to the cultures (4). Thus, the salicylate adenylation domain MbtA catalyzing the first step in mycobactin and carboxymycobactin synthesis was suggested to be a target for PAS. However, since growth inhibition was also observed under conditions of iron repletion and a mycobactin-requiring auxotroph of M. smegmatis also retained this high sensitivity to PAS, even in the presence of mycobactin, PAS seems to act at additional sites other than the conversion of salicylate to mycobactin (4, 39). Salicylate itself can be used to inhibit the production of enterobactin in E. coli (Leduc and Bouveret, unpublished). In vitro reconstitution of enterobactin biosynthesis showed that salicylate can be covalently linked to the EntF PCP domain, but this intermediate does not lead to product formation (possibly by negatively affecting iterative cyclization) and hence may block the biosynthesis machinery (107). However, since siderophore biosynthesis systems are often provided with type II thioesterases that regenerate such misacylated PCPs, this mode of biosynthesis inhibition is not as effective as blocking the initial biosynthesis step to prevent the accumulation of iron-chelating siderophore intermediates.
The strategy of target-specific siderophore biosynthesis inhibition proved to be an upcoming research field in the last few years due to the availability of crystal structures that allow the rational design of catalytic site inhibitors. So far, most studies have focused on the inhibition of aryl-capped siderophore biosynthesis enzymes including aryl acid adenylation domains from NRPS systems and enzymes for precursor synthesis (Fig. 9). Since the formation of salicylate and DHB as aryl acid precursors is a prerequisite for aryl-capped siderophore assembly, it is reasonable to consider enzymes involved in aryl acid formation as potential inhibition targets. Both salicylate and DHB are derived from chorismate, which is a central precursor for aromatic amino acid, folate, and quinone biosynthesis in primary metabolism. Thus, inhibition has to occur downstream of chorismate biosynthesis to avoid large unspecific side effects. Salicylate synthase and isochorismate synthase catalyzing the formation of salicylate and isochorismate from chorismate, respectively, have been studied as potential inhibition targets. Inhibition studies with the Y. enterocolitica Irp9 salicylate synthase and the E. coli EntC isochorismate synthase were performed (176, 247); however, those studies were done before the final crystal structures were known. A model of salicylate synthase as a rationale for inhibitor design was constructed using four crystal structures of closely related anthranilate synthases as initial templates (247). Among several chorismate and isochorismate analogues tested with Irp9, the compounds shown in Fig. 9 gave the best inhibition results but with inhibition constants (Ki values) in the micromolar range, even higher than the Km value of Irp9 for chorismate. In contrast, the reduced chorismate transition-state analogues tested with EntC proved to be much more effective. Although the C-4,6-diol and the C-4-ol-C-6-amine compounds shown in Fig. 9 as well as a further C-4-amine-C-6-ol analogue were designed as preferential inhibitors for the related enzymes isochorismate synthase, anthranilate synthase, and aminodeoxychorismate synthase (involved in p-aminobenzoate synthesis), respectively, all three analogues proved to be the most potent inhibitors of isochorismate synthase, leading to a reevaluation of the suggested reaction mechanism, especially for aminodeoxychorismate synthase (135, 161). An important step for the improvement of biosynthesis inhibitors was the resolution of the DhbE crystal structure as an archetype of aryl acid-activating domains (211), which is still the only available structural template for the rational design of aryl acid A domain inhibitors. DhbE was cocrystallized with DHB, AMP, and DHB-AMP, which provided insights into demands for conformation and interactions of a synthetic ligand in the active side. The use of aryl acid A domains for aryl-capped siderophore biosynthesis bears several advantages. Aryl acid A domains are highly homologous enzymes catalyzing the initial step of siderophore formation in all organisms that synthesize aryl-capped siderophores. The aryl acid substrates are generally either salicylate or DHB, which are activated via adenylation. Thus, the enzymes possess two substrate binding pockets in close proximity at the active site: one pocket is conserved for ATP binding in virtually all A domains, and one is conserved for salicylate binding in salicylate-activating domains such as PchD, MbtA, and YbtE or DHB binding in DHB-activating domains such as DhbE and EntE. Potential inhibitors are therefore designed as bisubstrate analogues, which may possess slightly varied aryl acid and adenosine moieties and in every case possess a nonhydrolyzable linker that reflects the highest degree of structural variation. Due to the highly similar binding pockets of all known aryl acid A domains, it is potentially possible to achieve their inhibition among a broad range of microorganisms with one potent analogue. Furthermore, arylic acyl adenylate analogues are not expected to cross-react with the analogous amino acid activation system for ribosomal protein synthesis provided by aminoacyl tRNA synthetases. The first aryl acid A domain bisubstrate analogue synthesized and tested was a salicyl sulfamoyl adenosine (SAL-AMS), which inhibited PchD, MbtA, and YbtE in the low nanomolar range and provoked growth inhibition of iron-limited M. tuberculosis and Y. pestis cultures with MIC50s in the micromolar range (91) (Fig. 9). Related aminoacyl sulfamoyl adenosines were shown in previous studies to act as potent inhibitors of NRPS and non-NRPS A domains (94, 210). The in vitro and in-culture inhibition efficacy of SAL-AMS was confirmed in further studies (215, 303, 304, 324). Due to the in vitro IC50 values, which were in the range of the enzyme concentrations used, and the observed noncompetitive inhibition of the analogue with respect to salicylate, SAL-AMS was suggested to be a tight binding inhibitor (91). Similar observations regarding submicromolar enzyme inhibition were made during further studies with the same compound and several derivatives thereof; however, the compounds were always found to be fully competitive for both ATP and the corresponding aryl acid (44, 215, 304). Further sulfamate-linked analogues have been derivatized mainly in their aryl and glycosyl domains (Fig. 9). Changing the salicyl moiety to DHB yielded DHB-AMS, which showed a preferential inhibitory effect on DhbE compared with SAL-AMS (215). The molecular basis for this effect was given by a comparative analysis of the aryl acid binding pockets of salicylate and DHB A domains, revealing a space constriction at the bottom of salicylate-specific pockets in which the conserved valine and serine residues present at the corresponding positions in DHB-specific pockets are replaced by a bulkier leucine/isoleucine and cysteine, respectively (211, 215). Consequently, SAL-AMS was found to inhibit YbtE with a higher preference (1.8-fold) than DHB-AMS, and DHB-AMS inhibited DhbE preferentially better (1.25-fold) than SAL-AMS (215). A systematic investigation of structure-activity relationships of the glycosyl domain in sulfamate-bridged bisubstrate analogues resulted in the identification of a carbocyclic compound that inhibited MbtA in vitro and M. tuberculosis in iron-deficient cultures with an efficacy comparable to that observed for SAL-AMS (304). It was furthermore suggested that the bisubstrate inhibitors utilize a mycobacterial transporter for crossing the cell envelope. The compounds that displayed a remarkable activity against M. tuberculosis had only slight activities against Y. pseudotuberculosis, which was suggested to be the result of different transport mechanisms (304).
FIG. 9.
Representative inhibitors of aryl-capped siderophore biosynthesis and modification. s.a., see above; obs., observed; norm., normalized; A domain, adenylation domain.
Further studies aimed to improve the overall stability and the anticipated ADMET (i.e., absorption, distribution, metabolism, elimination, and toxicology) profile (228a) of the SAL-AMS parent compound. The initially preferred sulfamate linker was identified as being potentially unstable due to the expulsion of the 5′-O-sulfamoyl moiety through spontaneous hydrolysis or cyclonucleoside formation (303, 324). The introduction of an acylsulfamide function led to an analogue with the highest in-culture activity against M. tuberculosis that has been reported so far for an aryl acid A domain inhibitor (303) (Fig. 9). The replacement of the sulfamoyl group by a β-ketosulfonamide linkage, which was intended to prevent hydrolysis leading to the possible generation of cytotoxic sulfamoyladenosines, had an impaired effect on activity both in vitro and in vivo (324). Subsequent studies of docking to the MbtA active site (based on the DhbE X-ray structure) revealed a preference for analogues that are able to adopt a planar geometry of aryl acid and linking group as observed for the native acyl phosphate in the DhbE structure. The planarity can be readily adopted by internal hydrogen bond formation between the free salicyl hydroxyl group and the α-oxygen of the native acyl phosphate linker or the α-nitrogen atom of the synthetic sulfamate or sulfamide linkers but not in compounds that lack such a hydrogen-accepting α-atom, which is the case for β-ketosulfonamides or β-ketophosphonates (324). Cytotoxic sulfamoyladenosine was furthermore shown to inhibit M. tuberculosis with an MIC99 of 50 μM, which is substantially higher than all MICs reported for efficient M. tuberculosis bisubstrate inhibitors including sulfamate-based analogues. For the inhibition of the E. coli EntE domain, another DHB-containing analogue was prepared as a hydroxyamoyl phosphate compound (44) (Fig. 9). The activity of this compound with EntE was higher than the activity of the DHB-AMS compound with DhbE. However, since DHB-AMS also had a higher level activity with YbtE than with DhbE (Fig. 9), this might further indicate enzyme-specific differences of inhibitory susceptibility.
A new class of inhibitory compounds was presented in a study dealing with the inhibition of the first step in iroA-dependent enterobactin tailoring, which is benzoyl-C-5-directed C-glucosylation catalyzed by IroB (see above). Successive mono-, di-, and tri-C-glucosylation of enterobactin that takes place in vitro (although diglucosyl enterobactin is the predominantly generated form in vivo) could be inhibited by introducing bromo substituents at the C-5 position of the enterobactin catecholate moieties (190). The inhibitory activity of the brominated enterobactin analogues increased with the number of introduced bromines, hence revealing the tribrominated analogue to be the best compound (Fig. 9). Surprisingly, mono- and dibrominated enterobactins were not accepted as substrates by IroB at all. Since the KD values for the inhibitors bound to IroB were found to be in the same nanomolar range as the Ki values, high-affinity binding was suggested to be involved in the mechanism of IroB inhibition, which was competitive for all three analogues tested (190). With respect to potential therapeutic applications, however, it should be considered that these analogues are potential binding substrates for siderocalin, which might have yet unpredictable consequences for their ADMET profiles.
In addition to recently accumulating data from studies of siderophore biosynthesis and tailoring inhibition, there are still only a few reports that deal with rational approaches to the inhibition of further siderophore pathway components. One reason, as mentioned above, might be the discrepancy between the detailed characterization of siderophore biosynthesis and the partially rather sketchy knowledge about downstream-acting pathway components. This, however, is currently changing, at least in the field of Fe-siderophore uptake, and future concepts for uptake inhibition may already be anticipated. One concept of earlier studies was the use of siderophores in which Fe(III) was replaced by a nonmetabolizable metal ion, thus leading to the competitive inhibition of siderophore-mediated iron transport by these antimetabolites. In initial studies with enterobactin, a broad array of group III and transition metal complexes was tested, revealing only those containing either indium or scandium to inhibit growth of K. pneumoniae and pathogenic E. coli strains by competitive Fe-enterobactin uptake inhibition (279, 280). The Sc-enterobactin complex exerted further therapeutic effects on K. pneumoniae and E. coli infections in mice. However, the strategy was abandoned due to the toxic effects of scandium, which was easily released in vivo from the metabolically unstable enterobatin scaffold. In similar studies, metal analogues of mycobactin and exochelin failed to act as effective antitubercular agents (268). However, interesting observations have been made concerning the use of heterologous mycobactins as growth inhibition factors for mycobacteria. Mycobacteria produce species-specific mycobactin variants that vary essentially in five scaffold substituents, leaving the central core of the siderophore unchanged (268). When M. aurum, which produces mycobactin A, was supplied with heterologous mycobactins J and S from M. paratuberculosis and M. smegmatis, respectively, these heterologous siderophores promoted growth only up to a concentration of 5 μM and were highly inhibitory for growth above this concentration (27). Essentially the same effect was observed with the synthetic catecholate-spermidine compound FR160. Following the establishment of its total chemical synthesis, synthetic mycobactin S was found to be a potent growth inhibitor of M. tuberculosis (MIC99 = 12.5 μg/ml), although it differs only in one stereogenic center located in the alkenyl side chain from mycobactin T, the endogenously produced mycobactin of M. tuberculosis (146). Testing of further synthetic mycobactin analogues as antitubercular agents revealed one compound, which carried a Boc-protecting group, with an MIC98 of 0.2 μg/ml (346). The molecular mechanism of the inhibitory effect of heterologous mycobactins in mycobacteria awaits further characterization, and it can be assumed that the mycobactin analogues interfere with the uptake of iron delivered by endogenously derived mycobactins. In vivo studies are necessary to evaluate the use of mycobactin analogues as potential drugs, since it has to be anticipated that the targeting of these bulky compounds into macrophage phagosomes, where the mycobacteria reside, will be problematic.
In addition to mycobactins, several other siderophores are known to possess considerable antibiotic activities. Desferri-TAFC from Penicillium and Aspergillus strains and desferrioxamine E (“nocardamine”) from streptomycetes and Pseudomonas stutzeri are compounds that are active against a broad spectrum of bacteria. Also ferrocins from P. fluorescens YK-310 representing cyclic decapeptides with three hydroxamate residues are active against E. coli and P. aeruginosa (341). As in the case of heterologous mycobactins, the modes of action are still under investigation; however, it can be speculated that there is interference with siderophore pathway steps including extracellular complex formation, uptake, and iron release.
CONCLUSIONS AND FUTURE CONCEPTS
Due to the tight relationship between siderophore utilization and virulence, the strategy of iron-dependent pathogen control is a promising field for future investigations and offers a broad array of possible therapeutic applications. While the siderophore-antibiotic strategy uses Fe-siderophore uptake systems as gateways for cellular infiltration with established antibiotics, the siderophore pathway inhibition strategy tries to abolish siderophore utilization in order to starve the pathogens out for iron. Both concepts proved to be successful with in vitro and ex vivo culture model systems. However, most of these compounds have to be evaluated in appropriate in vivo systems. Nevertheless, definition of further targets for directed pathogen control during iron limitation is indispensable to broaden the spectrum of potential drugs. The current status of siderophore pathway inhibition has by far not exhausted all possible mechanisms of iron-dependent control. Inhibition of Fe-siderophore uptake and iron release has been scarcely studied. However, recent findings point to a new, mechanistically sophisticated mode of inhibition. The crystal structure of the Fe-enterobactin binding protein CeuE was found in a status of ligand-dependent dimerization, which was observed upon cocrystallization with the synthetic enterobactin analogue MECAM, possessing an aromatic backbone (225) (Fig. 10A). The ligand dimerization that led to the face-to-face joining of two periplasmic binding proteins was dependent on the iron bound to the ligand complex and the π-stacking interactions between the aromatic backbones of the ligand molecules. Since it was suggested that this dimerization is also possible in solution, this would lead to an intriguing model of dimerization-dependent blocking of substrate binding proteins and hence Fe-siderophore uptake as depicted in Fig. 10B. Thus, in the proposed mode of its action, MECAM resembles those ligands that were previously described as being chemical inducers of dimerization, which are used to study interactions and oligomerization effects between macromolecules (64). Since substrate binding proteins as inhibition targets are located exocytosolically, the synthetic ligand must not cross the CM, which abolishes the permeation problem that appears as one major drawback of siderophore biosynthesis inhibition. That MECAM indeed acts very differently from enterobactin after uptake into the periplasm of E. coli was observed in previous studies that found MECAM-delivered iron in the periplasm; however, only little iron was further delivered into the cytoplasm, suggesting its periplasmic accumulation (208). However, since MECAM-induced dimerization is iron dependent, and the iron-charged compound was also found as an iron source for E. coli and B. subtilis (136, 196), it would be desirable if the ligand-induced dimerization occurred iron independently. This could be achieved if the approach utilizing chemical inducers of dimerization was fully applied to the system, leading to the design of covalently linked triscatecholate units (Fig. 10C). It has been shown that the affinity of nonloaded triscatecholates such as enterobactin and bacillibactin for their substrate binding proteins lies essentially within the same range as that observed for the iron-charged complexes. Furthermore, the ligand-protein oligomerization is expected to confer additional stabilization energy to the complexes. If this approach proved successful, it could be further applied to uptake systems with specificities for hydroxamates, carboxylates, or mixed siderophore ligands. Initial studies of the chemistry of such “sandwich-siderophore”-like compounds and their protein-ligand complexes are in progress (M. Miethke et al., unpublished data). Furthermore, it is conceivable to design inhibitors for iron release enzymes. This might encounter more difficulties since bacterial iron release systems are located mainly in the cytosol and, in the case of Fe-siderophore reductases, are often rather unspecific for their iron-chelate substrates. However, the enterobactin and salmochelin esterases in enteric bacteria were shown to be specific for their cognate substrates, and thus, nonhydrolyzable analogues that are transported into the cytoplasm could be applied for iron release inhibition as depicted in Fig. 10D. An anticipated analogue with high structural similarity to native enterobactin may possess a trilactame backbone conferring resistance against hydrolytical cleavage instead of a trilactone. The chemical synthesis of a nonsubstituted trilactame ring has already been reported (267); however, the cyclization in the substituted state was not achieved during initial studies and needs further effort. Altogether, it seems worth pursuing the concept of inhibiting siderophore pathway components that act downstream of siderophore biosynthesis. Phenotypic comparisons of biosynthesis, uptake, and iron release mutants in the bacillibactin pathway of B. subtilis revealed more severe growth restriction during iron starvation if uptake or iron release rather than siderophore biosynthesis was blocked, which is likely caused by further starvation effects due to ongoing siderophore synthesis and subsequent self-poisoning with the nonconsumable iron chelator (216). Similar observations regarding the essential involvement of downstream-acting siderophore pathway components in cellular growth were made in other studies. An E. coli fur tonB double mutant, which is derepressed in enterobactin synthesis but unable to take up Fe-enterobactin, reached only one-third of the dry weight of an enterobactin-deficient fur entC double mutant (23). Another study reported normal growth rates for an E. coli entB mutant and fes entB double mutant, both defective in enterobactin biosynthesis, but very poor growth of fes and fep mutants defective in Fe-enterobactin uptake and hydrolysis, respectively, under the same iron-depleted conditions (327). Furthermore, the iron-dependent growth defect of Bordetella sp. alcaligin export mutants was found to be suppressed by a defect in alcaligin biosynthesis (35). Since heterologous mycobactins as putative inhibitors of mycobactin-mediated iron uptake were already found to be efficient growth inhibitors of M. tuberculosis, the blocking of Fe-siderophore components of transport and release might generally prove to be a considerable alternative to siderophore biosynthesis inhibition.
Beyond the strategies that employ synthetic small-molecule compounds as working tools, systems of natural defense can be considered for protection against those pathogens that are provided with siderophore utilization mechanisms representing the latest layer of coevolution. Innate defense of mammals has developed siderophore-binding lipocalins to counteract iron-dependent virulence development. In reply, pathogens were forced to develop new iron acquisition strategies by either evolving tailoring enzymes for existing siderophores, evolving new siderophore synthesis pathways, or adopting their colonization to shielded compartments. Thus, the current state of coevolution represents a severe drawback for pathogens that cannot cope with lipocalin-mediated defense but gives an advantage to those that have altered or renewed their siderophore arsenal. Thus, the next logical step is the development of a novel defense layer that should neutralize tailored and newly developed siderophores such as salmochelins and aerobactin from pathogenic enteric bacteria and petrobactin from pathogenic bacilli, respectively. Since nature proved that high-affinity sequestration of (Fe)-siderophores is an efficient concept to abolish or impair iron-dependent pathogen multiplication, it should be challenging to apply this strategy to existing lipocalins by using rational approaches such as directed protein evolution. The plasticity of lipocalin binding pockets for molecular tailoring was proven in studies of directed evolution that reshaped the binding pocket of the bilin-binding protein, a lipocalin from Pieris brassicae, with respect to the high-affinity binding of nonanalogous ligands such as fluorescein, which was achieved with KDs in the low nanomolar range (19). Initial studies revealed a similar capacity for a modification of the human siderocalin (291). If high-affinity sequestration of virulence-associated siderophores by rationally designed binding proteins was achievable, the tailored scavengers could be applied to several therapeutic applications including gene therapy as a long-term goal. The neutralization of virulence-associated siderophores would provide a further example of virulence-specific pathogen control, which has been recently demonstrated for selective targeting of V. cholerae virulence factors (148) and was also suggested for inhibiting components of iroA-dependent enterobactin modification in order to spare the commensal flora from antibiotic treatment (97, 190).
Finally, protection against siderophore-utilizing pathogens via the acquired immune defense system has been envisaged in several studies and seems worthy of further investigation. Since convalescent-phase sera often contain antibodies that are directed against iron-repressible outer membrane receptors of gram-negative bacterial pathogens, these transport proteins were suggested to be candidate immunogens for immunoprophylactic therapy (343). However, a drawback of this approach is the impaired accessibility of antibodies to bacterial cell surface targets as observed in several E. coli isolates producing lipopolysaccharide O antigens (277). Thus, a recent vaccination concept suggested that not Fe-siderophore receptors but rather whole-pathogen cells should be used as immunoglobulin targets. These cells are derived from attenuated strains lacking their virulence-associated siderophore uptake systems. It was demonstrated that an S. enterica serovar Typhimurium iroN fepA cir triple mutant may serve as a potential immunization strain since it protected mice against infection with the fully virulent parental strain (336). These studies will be extended with equivalent mutants of chick-virulent strains in a chicken infection model. If the approach proves to be of general advantage in conferring resistance to siderophore-dependent pathogens, such attenuated strains will be of future interest in clinical and veterinary settings.
Acknowledgments
We thank all members of our group for helpful discussion.
Work conducted in our group was supported by grants from the Deutsche Forschungsgemeinschaft, by EC grant LSHG-CT-2004-503468, and by the Fonds der Chemischen Industrie. M. M. also thanks the Graduiertenkolleg “Protein function at the atomic level” for additional support.
REFERENCES
- 1.Abergel, R. J., M. K. Wilson, J. E. Arceneaux, T. M. Hoette, R. K. Strong, B. R. Byers, and K. N. Raymond. 2006. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc. Natl. Acad. Sci. USA 103:18499-18503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerley, D. F., T. T. Caradoc-Davies, and I. L. Lamont. 2003. Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 185:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1995. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17:197-204. [DOI] [PubMed] [Google Scholar]
- 4.Adilakshmi, T., P. D. Ayling, and C. Ratledge. 2000. Mutational analysis of a role for salicylic acid in iron metabolism of Mycobacterium smegmatis. J. Bacteriol. 182:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adjimani, J. P., and T. Emery. 1987. Iron uptake in Mycelia sterilia EP-76. J. Bacteriol. 169:3664-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aisen, P., A. Leibman, and J. Zweier. 1978. Stoichiometric and site characteristics of the binding of iron to human transferrin. J. Biol. Chem. 253:1930-1937. [PubMed] [Google Scholar]
- 7.Alen, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anisimov, R., D. Brem, J. Heesemann, and A. Rakin. 2005. Molecular mechanism of YbtA-mediated transcriptional regulation of divergent overlapping promoters ybtA and irp6 of Yersinia enterocolitica. FEMS Microbiol. Lett. 250:27-32. [DOI] [PubMed] [Google Scholar]
- 10.Anisimov, R., D. Brem, J. Heesemann, and A. Rakin. 2005. Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int. J. Med. Microbiol. 295:19-28. [DOI] [PubMed] [Google Scholar]
- 11.Ardon, O., R. Nudelman, C. Caris, J. Libman, A. Shanzer, Y. Chen, and Y. Hadar. 1998. Iron uptake in Ustilago maydis: tracking the iron path. J. Bacteriol. 180:2021-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Askwith, C., and J. Kaplan. 1998. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23:135-138. [DOI] [PubMed] [Google Scholar]
- 13.Azpiroz, M. F., and M. Lavina. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 48:1235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 15.Baumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 17.Benz, G., T. Schröder, J. Kurz, C. K. W. Wünsche, G. Steffens, J. Pfitzner, and D. Schmidt. 1982. Konstitution der Desferriform der Albomycine δ1, δ2, ɛ. Angew. Chemie 94:552-553. [Google Scholar]
- 18.Bernier, G., V. Girijavallabhan, A. Murray, N. Niyaz, P. Ding, M. J. Miller, and F. Malouin. 2005. Desketoneoenactin-siderophore conjugates for Candida: evidence of iron transport-dependent species selectivity. Antimicrob. Agents Chemother. 49:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beste, G., F. S. Schmidt, T. Stibora, and A. Skerra. 1999. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc. Natl. Acad. Sci. USA 96:1898-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bickel, H., G. E. Hall, W. Keller-Schierlein, V. Prelog, E. Vischer, and A. Wettstein. 1960. Stoffwechselprodukte von Actinomyceten, 27. Mitt. Ueber die Konstitution von Ferrioxamin B. Helv. Chim. Acta 43:2129-2138. [Google Scholar]
- 21.Biemans-Oldehinkel, E., and B. Poolman. 2003. On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 22:5983-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Sussmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471-481. [DOI] [PubMed] [Google Scholar]
- 23.Bleuel, C., C. Grosse, N. Taudte, J. Scherer, D. Wesenberg, G. J. Krauss, D. H. Nies, and G. Grass. 2005. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187:6701-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bluhm, M. E., S. S. Kim, E. A. Dertz, and K. M. Raymond. 2001. Corynebactin and enterobactin: related siderophores of opposite chirality. J. Am. Chem. Soc. 124:2436-2437. [DOI] [PubMed] [Google Scholar]
- 25.Bohnke, R., and B. F. Matzanke. 1995. The mobile ferrous iron pool in Escherichia coli is bound to a phosphorylated sugar derivative. Biometals 8:223-230. [DOI] [PubMed] [Google Scholar]
- 26.Bonaccorsi di Patti, M. C., R. Miele, M. E. Schinina, and D. Barra. 2005. The yeast multicopper oxidase Fet3p and the iron permease Ftr1p physically interact. Biochem. Biophys. Res. Commun. 333:432-437. [DOI] [PubMed] [Google Scholar]
- 27.Bosne-David, S., L. Bricard, F. Ramiandrasoa, A. DeRoussent, G. Kunesch, and A. Andremont. 1997. Evaluation of growth promotion and inhibition from mycobactins and nonmycobacterial siderophores (desferrioxamine and FR160) in Mycobacterium aurum. Antimicrob. Agents Chemother. 41:1837-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouige, P., D. Laurent, L. Piloyan, and E. Dassa. 2002. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3:541-559. [DOI] [PubMed] [Google Scholar]
- 29.Bozzi, M., G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, and E. Chiancone. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272:3259-3265. [DOI] [PubMed] [Google Scholar]
- 30.Braun, V. 1999. Active transport of siderophore-mimicking antibacterials across the outer membrane. Drug Resist. Updat. 2:363-369. [DOI] [PubMed] [Google Scholar]
- 31.Braun, V., S. Mahren, and M. Ogierman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 32.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 33.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 34.Breustedt, D. A., I. P. Korndorfer, B. Redl, and A. Skerra. 2005. The 1.8-A crystal structure of human tear lipocalin reveals an extended branched cavity with capacity for multiple ligands. J. Biol. Chem. 280:484-493. [DOI] [PubMed] [Google Scholar]
- 35.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brickman, T. J., and M. A. McIntosh. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267:12350-12355. [PubMed] [Google Scholar]
- 38.Brock, J. H., P. H. Williams, J. Liceaga, and K. G. Wooldridge. 1991. Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun. 59:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown, K. A., and C. Ratledge. 1975. The effect of p-aminosalicyclic acid on iron transport and assimilation in mycobacteria. Biochim. Biophys. Acta 385:207-220. [DOI] [PubMed] [Google Scholar]
- 40.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 41.Budzikiewicz, H. 2001. Siderophore-antibiotic conjugates used as trojan horses against Pseudomonas aeruginosa. Curr. Top. Med. Chem. 1:73-82. [DOI] [PubMed] [Google Scholar]
- 42.Buss, J. L., B. T. Greene, J. Turner, F. M. Torti, and S. V. Torti. 2004. Iron chelators in cancer chemotherapy. Curr. Top. Med. Chem. 4:1623-1635. [DOI] [PubMed] [Google Scholar]
- 43.Butterton, J. R., and S. B. Calderwood. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callahan, B. P., J. V. Lomino, and R. Wolfenden. 2006. Nanomolar inhibition of the enterobactin biosynthesis enzyme, EntE: synthesis, substituent effects, and additivity. Bioorg. Med. Chem. Lett. 16:3802-3805. [DOI] [PubMed] [Google Scholar]
- 45.Carrano, C. J., M. Jordan, H. Drechsel, D. G. Schmid, and G. Winkelmann. 2001. Heterobactins: a new class of siderophores from Rhodococcus erythropolis IGTS8 containing both hydroxamate and catecholate donor groups. Biometals 14:119-125. [DOI] [PubMed] [Google Scholar]
- 46.Carrano, C. J., and K. N. Raymond. 1978. Coordination chemistry of microbial iron transport compounds: rhodotorulic acid and iron uptake in Rhodotorula pilimanae. J. Bacteriol. 136:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 49.Chakraborty, R., E. A. Lemke, Z. Cao, P. E. Klebba, and D. van der Helm. 2003. Identification and mutational studies of conserved amino acids in the outer membrane receptor protein, FepA, which affect transport but not binding of ferric-enterobactin in Escherichia coli. Biometals 16:507-518. [DOI] [PubMed] [Google Scholar]
- 50.Chakraborty, R., E. Storey, and D. van der Helm. 2007. Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli. Biometals 20:263-274. [DOI] [PubMed] [Google Scholar]
- 51.Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601-611. [DOI] [PubMed] [Google Scholar]
- 52.Challis, G. L., and J. Ravel. 2000. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol. Lett. 187:111-114. [DOI] [PubMed] [Google Scholar]
- 53.Chang, G. 2003. Structure of MsbA from Vibrio cholera: a multidrug resistance ABC transporter homolog in a closed conformation. J. Mol. Biol. 330:419-430. [DOI] [PubMed] [Google Scholar]
- 54.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 55.Chen, O. S., R. J. Crisp, M. Valachovic, M. Bard, D. R. Winge, and J. Kaplan. 2004. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279:29513-29518. [DOI] [PubMed] [Google Scholar]
- 56.Chen, Q., A. M. Wertheimer, M. E. Tolmasky, and J. H. Crosa. 1996. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol. Microbiol. 22:127-134. [DOI] [PubMed] [Google Scholar]
- 57.Chong, T. W., L. D. Horwitz, J. W. Moore, H. M. Sowter, and A. L. Harris. 2002. A mycobacterial iron chelator, desferri-exochelin, induces hypoxia-inducible factors 1 and 2, NIP3, and vascular endothelial growth factor in cancer cell lines. Cancer Res. 62:6924-6927. [PubMed] [Google Scholar]
- 58.Clarke, T. E., V. Braun, G. Winkelmann, L. W. Tari, and H. J. Vogel. 2002. X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J. Biol. Chem. 277:13966-13972. [DOI] [PubMed] [Google Scholar]
- 59.Clarke, T. E., S. Y. Ku, D. R. Dougan, H. J. Vogel, and L. W. Tari. 2000. The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat. Struct. Biol. 7:287-291. [DOI] [PubMed] [Google Scholar]
- 60.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol. 347:121-134. [DOI] [PubMed] [Google Scholar]
- 61.Cobessi, D., H. Celia, and F. Pattus. 2005. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 352:893-904. [DOI] [PubMed] [Google Scholar]
- 62.Cooper, S. R., J. V. McArdle, and K. N. Raymond. 1978. Siderophore electrochemistry: relation to intracellular iron release mechanism. Proc. Natl. Acad. Sci. USA 75:3551-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cowart, R. E. 2002. Reduction of iron by extracellular iron reductases: implications for microbial iron acquisition. Arch. Biochem. Biophys. 400:273-281. [DOI] [PubMed] [Google Scholar]
- 64.Crabtree, G. R., and S. L. Schreiber. 1996. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem. Sci. 21:418-422. [DOI] [PubMed] [Google Scholar]
- 65.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Curtis, N. A., R. L. Eisenstadt, S. J. East, R. J. Cornford, L. A. Walker, and A. J. White. 1988. Iron-regulated outer membrane proteins of Escherichia coli K-12 and mechanism of action of catechol-substituted cephalosporins. Antimicrob. Agents Chemother. 32:1879-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dale, S. E., M. T. Sebulsky, and D. E. Heinrichs. 2004. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 186:8356-8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 70.Dawson, R. J., and K. P. Locher. 2006. Structure of a bacterial multidrug ABC transporter. Nature 443:180-185. [DOI] [PubMed] [Google Scholar]
- 71.Dean, C. R., S. Neshat, and K. Poole. 1996. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dean, M., and T. Annilo. 2005. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6:123-142. [DOI] [PubMed] [Google Scholar]
- 73.Dertz, E. A., and K. N. Raymond. 2004. Biochemical and physical properties of siderophores, p. 3-17. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 74.Dertz, E. A., and K. N. Raymond. 2003. Siderophores and transferrins, p. 141-168. In L. Que, Jr., and W. B. Tolman (ed.), Comprehensive coordination chemistry II, vol. 8. Elsevier, Ltd., Philadelphia, PA. [Google Scholar]
- 75.Dertz, E. A., J. Xu, A. Stintzi, and K. N. Raymond. 2006. Bacillibactin-mediated iron transport in Bacillus subtilis. J. Am. Chem. Soc. 128:22-23. [DOI] [PubMed] [Google Scholar]
- 76.Destoumieux-Garzon, D., J. Peduzzi, X. Thomas, C. Djediat, and S. Rebuffat. 2006. Parasitism of iron-siderophore receptors of Escherichia coli by the siderophore-peptide microcin E492m and its unmodified counterpart. Biometals 19:181-191. [DOI] [PubMed] [Google Scholar]
- 77.Destoumieux-Garzon, D., X. Thomas, M. Santamaria, C. Goulard, M. Barthelemy, B. Boscher, Y. Bessin, G. Molle, A. M. Pons, L. Letellier, J. Peduzzi, and S. Rebuffat. 2003. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49:1031-1041. [DOI] [PubMed] [Google Scholar]
- 78.Devireddy, L. R., C. Gazin, X. Zhu, and M. R. Green. 2005. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123:1293-1305. [DOI] [PubMed] [Google Scholar]
- 79.Devireddy, L. R., J. G. Teodoro, F. A. Richard, and M. R. Green. 2001. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 293:829-834. [DOI] [PubMed] [Google Scholar]
- 80.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Drake, E. J., D. A. Nicolai, and A. M. Gulick. 2006. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem. Biol. 13:409-419. [DOI] [PubMed] [Google Scholar]
- 82.Eck, R., S. Hundt, A. Hartl, E. Roemer, and W. Kunkel. 1999. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 145:2415-2422. [DOI] [PubMed] [Google Scholar]
- 83.Ecker, D. J., and T. Emery. 1983. Iron uptake from ferrichrome A and iron citrate in Ustilago sphaerogena. J. Bacteriol. 155:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eisendle, M., H. Oberegger, I. Zadra, and H. Haas. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 49:359-375. [DOI] [PubMed] [Google Scholar]
- 85.Eisenhauer, H. A., S. Shames, P. D. Pawelek, and J. W. Coulton. 2005. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J. Biol. Chem. 280:30574-30580. [DOI] [PubMed] [Google Scholar]
- 86.Eisenhofer, G., A. Aneman, D. Hooper, B. Rundqvist, and P. Friberg. 1996. Mesenteric organ production, hepatic metabolism, and renal elimination of norepinephrine and its metabolites in humans. J. Neurochem. 66:1565-1573. [DOI] [PubMed] [Google Scholar]
- 87.Emery, T. 1976. Fungal ornithine esterases: relationship to iron transport. Biochemistry 15:2723-2728. [DOI] [PubMed] [Google Scholar]
- 88.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferguson, A. D., V. Braun, H. P. Fiedler, J. W. Coulton, K. Diederichs, and W. Welte. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferrer, M., O. V. Golyshina, A. Beloqui, P. N. Golyshin, and K. N. Timmis. 2007. The cellular machinery of Ferroplasma acidiphilum is iron-protein-dominated. Nature 445:91-94. [DOI] [PubMed] [Google Scholar]
- 91.Ferreras, J. A., J. S. Ryu, F. Di Lello, D. S. Tan, and L. E. Quadri. 2005. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1:29-32. [DOI] [PubMed] [Google Scholar]
- 92.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 93.Finegold, A. A., K. P. Shatwell, A. W. Segal, R. D. Klausner, and A. Dancis. 1996. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271:31021-31024. [DOI] [PubMed] [Google Scholar]
- 94.Finking, R., A. Neumüller, J. Solsbacher, D. Konz, G. Kretzschmar, M. Schweitzer, T. Krumm, and M. A. Marahiel. 2003. Aminoacyl adenylate substrate analogues for the inhibition of adenylation domains of nonribosomal peptide synthetases. Chembiochem 4:903-906. [DOI] [PubMed] [Google Scholar]
- 95.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2006. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2:132-138. [DOI] [PubMed] [Google Scholar]
- 96.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102:571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fischbach, M. A., H. Lin, L. Zhou, Y. Yu, R. J. Abergel, D. R. Liu, K. N. Raymond, B. L. Wanner, R. K. Strong, C. T. Walsh, A. Aderem, and K. D. Smith. 2006. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. USA 103:16502-16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fischer, E., B. Strehlow, D. Hartz, and V. Braun. 1990. Soluble and membrane-bound ferrisiderophore reductases of Escherichia coli K-12. Arch. Microbiol. 153:329-336. [DOI] [PubMed] [Google Scholar]
- 99.Flo, T. H., K. D. Smith, S. Sato, D. J. Rodriguez, M. A. Holmes, R. K. Strong, S. Akira, and A. Aderem. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917-921. [DOI] [PubMed] [Google Scholar]
- 100.Fluckinger, M., H. Haas, P. Merschak, B. J. Glasgow, and B. Redl. 2004. Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother. 48:3367-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Freestone, P. P., M. Lyte, C. P. Neal, A. F. Maggs, R. D. Haigh, and P. H. Williams. 2000. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 182:6091-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 103.Reference deleted.
- 104.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 105.Gaines, C. G., J. S. Lodge, J. E. Arceneaux, and B. R. Byers. 1981. Ferrisiderophore reductase activity associated with an aromatic biosynthetic enzyme complex in Bacillus subtilis. J. Bacteriol. 148:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gehring, A. M., I. Mori, R. D. Perry, and C. T. Walsh. 1998. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry 37:11637-11650. [DOI] [PubMed] [Google Scholar]
- 107.Gehring, A. M., I. Mori, and C. T. Walsh. 1998. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37:2648-2659. [DOI] [PubMed] [Google Scholar]
- 108.Georgatsou, E., and D. Alexandraki. 1999. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15:573-584. [DOI] [PubMed] [Google Scholar]
- 109.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272:13786-13792. [DOI] [PubMed] [Google Scholar]
- 110.Ghosh, A., M. Ghosh, C. Niu, F. Malouin, U. Moellmann, and M. J. Miller. 1996. Iron transport-mediated drug delivery using mixed-ligand siderophore-beta-lactam conjugates. Chem. Biol. 3:1011-1019. [DOI] [PubMed] [Google Scholar]
- 111.Ghosh, M., L. J. Lambert, P. W. Huber, and M. J. Miller. 1995. Synthesis, bioactivity, and DNA-cleaving ability of of desferrioxamine B-nalidixic acid and anthraquinone carboxylic acid conjugates. Bioorg. Med. Chem. Lett. 5:2337-2340. [Google Scholar]
- 112.Ghosh, M., and M. J. Miller. 1995. Design, synthesis, and biological evaluation of isocyanurate-based antifungal and macrolide antibiotic conjugates: iron transport-mediated drug delivery. Bioorg. Med. Chem. 3:1519-1525. [DOI] [PubMed] [Google Scholar]
- 113.Ghosh, M., and M. J. Miller. 1996. Synthesis and in vitro antibacterial activity of spermidine-based mixed catechol- and hydroxamate-containing siderophore-vancomycin conjugates. Bioorg. Med. Chem. 4:43-48. [DOI] [PubMed] [Google Scholar]
- 114.Gibson, F., and D. I. Magrath. 1969. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta 192:175-184. [DOI] [PubMed] [Google Scholar]
- 115.Glasgow, B. J., A. R. Abduragimov, Z. T. Farahbakhsh, K. F. Faull, and W. L. Hubbell. 1995. Tear lipocalins bind a broad array of lipid ligands. Curr. Eye Res. 14:363-372. [DOI] [PubMed] [Google Scholar]
- 116.Glickstein, H., W. Breuer, M. Loyevsky, A. M. Konijn, A. Shanzer, and Z. I. Cabantchik. 1996. Differential cytotoxicity of iron chelators on malaria-infected cells versus mammalian cells. Blood 87:4871-4878. [PubMed] [Google Scholar]
- 117.Goetz, D. H., M. A. Holmes, N. Borregaard, M. E. Bluhm, K. N. Raymond, and R. K. Strong. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10:1033-1043. [DOI] [PubMed] [Google Scholar]
- 118.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1991. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1125-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Golenser, J., A. Domb, T. Mordechai-Daniel, B. Leshem, A. Luty, and P. Kremsner. 2006. Iron chelators: correlation between effects on Plasmodium spp. and immune functions. J. Parasitol. 92:170-177. [DOI] [PubMed] [Google Scholar]
- 120.Grass, G., K. Thakali, P. E. Klebba, D. Thieme, A. Müller, G. F. Wildner, and C. Rensing. 2004. Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Greenwood, K. T., and R. K. Luke. 1978. Enzymatic hydrolysis of enterochelin and its iron complex in Escherichia coli K-12. Properties of enterochelin esterase. Biochim. Biophys. Acta 525:209-218. [DOI] [PubMed] [Google Scholar]
- 122.Grewal, K. K., P. J. Warner, and P. H. Williams. 1982. An inducible outer membrane protein involved in aerobactin-mediated iron transport by co1V strains of Escherichia coli. FEBS Lett. 140:27-30. [DOI] [PubMed] [Google Scholar]
- 123.Grünewald, J., and M. A. Marahiel. 2006. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70:121-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Haas, H. 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62:316-330. [DOI] [PubMed] [Google Scholar]
- 125.Haas, H., M. Schoeser, E. Lesuisse, J. F. Ernst, W. Parson, B. Abt, G. Winkelmann, and H. Oberegger. 2003. Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J. 371:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haas, H., I. Zadra, G. Stoffler, and K. Angermayr. 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274:4613-4619. [DOI] [PubMed] [Google Scholar]
- 127.Halle, F., and J. M. Meyer. 1992. Iron release from ferrisiderophores. A multi-step mechanism involving a NADH/FMN oxidoreductase and a chemical reduction by FMNH2. Eur. J. Biochem. 209:621-627. [DOI] [PubMed] [Google Scholar]
- 128.Hantash, F. M., M. Ammerlaan, and C. F. Earhart. 1997. Enterobactin synthase polypeptides of Escherichia coli are present in an osmotic-shock-sensitive cytoplasmic locality. Microbiology 143:147-156. [DOI] [PubMed] [Google Scholar]
- 129.Hantash, F. M., and C. F. Earhart. 2000. Membrane association of the Escherichia coli enterobactin synthase proteins EntB/G, EntE, and EntF. J. Bacteriol. 182:1768-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hantke, K. 1983. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet. 191:301-306. [DOI] [PubMed] [Google Scholar]
- 131.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 132.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harris, W. R., C. J. Carrano, and K. N. Raymond. 1979. Coordination chemistry of microbial iron transport compounds. 16. Isolation, characterization and formation constants of ferric aerobactin. J. Am. Chem. Soc. 101:2722-2727. [Google Scholar]
- 134.Harrison, A. J., M. Yu, T. Gardenborg, M. Middleditch, R. J. Ramsay, E. N. Baker, and J. S. Lott. 2006. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J. Bacteriol. 188:6081-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He, Z., K. D. S. Lavoie, P. A. Bartlett, and M. D. Toney. 2004. Conservation of mechanism in three chorismate-utilizing enzymes. J. Am. Chem. Soc. 126:2378-2385. [DOI] [PubMed] [Google Scholar]
- 136.Heidinger, S., V. Braun, V. L. Pecoraro, and K. N. Raymond. 1983. Iron supply to Escherichia coli by synthetic analogs of enterochelin. J. Bacteriol. 153:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Henderson, L. M., G. Banting, and J. B. Chappell. 1995. The arachidonate-activable, NADPH oxidase-associated H+ channel. Evidence that gp91-phox functions as an essential part of the channel. J. Biol. Chem. 270:5909-5916. [PubMed] [Google Scholar]
- 139.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1p) in Saccharomyces cerevisiae. Biometals 13:65-72. [DOI] [PubMed] [Google Scholar]
- 140.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186:221-227. [DOI] [PubMed] [Google Scholar]
- 141.Heymann, P., J. F. Ernst, and G. Winkelmann. 1999. Identification of a fungal triacetylfusarinine C siderophore transport gene (TAF1) in Saccharomyces cerevisiae as a member of the major facilitator superfamily. Biometals 12:301-306. [DOI] [PubMed] [Google Scholar]
- 142.Heymann, P., M. Gerads, M. Schaller, F. Dromer, G. Winkelmann, and J. F. Ernst. 2002. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect. Immun. 70:5246-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 144.Holmes, M. A., W. Paulsene, X. Jide, C. Ratledge, and R. K. Strong. 2005. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure 13:29-41. [DOI] [PubMed] [Google Scholar]
- 145.Hou, Y. X., J. R. Riordan, and X. B. Chang. 2003. ATP binding, not hydrolysis, at the first nucleotide-binding domain of multidrug resistance-associated protein MRP1 enhances ADP.Vi trapping at the second domain. J. Biol. Chem. 278:3599-3605. [DOI] [PubMed] [Google Scholar]
- 146.Hu, J., and M. J. Miller. 1997. Total synthesis of a mycobactin S, a siderophore and growth promoter of Mycobacterium smegmatis, and determination of its growth inhibitory activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 119:3462-3468. [Google Scholar]
- 147.Huber, P., H. Leuenberger, and W. Keller-Schierlein. 1986. Danoxamin, der eisenbindende Teil des Sideromycin-Antibioticums Danomycin. Helv. Chim. Acta 69:236-245. [Google Scholar]
- 148.Hung, D. T., E. A. Shakhnovich, E. Pierson, and J. J. Mekalanos. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670-674. [DOI] [PubMed] [Google Scholar]
- 149.Hvidberg, V., C. Jacobsen, R. K. Strong, J. B. Cowland, S. K. Moestrup, and N. Borregaard. 2005. The endocytic receptor megalin binds the iron transporting neutrophil-gelatinase-associated lipocalin with high affinity and mediates its cellular uptake. FEBS Lett. 579:773-777. [DOI] [PubMed] [Google Scholar]
- 150.Ingelman, M., S. Ramaswamy, V. Niviere, M. Fontecave, and H. Eklund. 1999. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry 38:7040-7049. [DOI] [PubMed] [Google Scholar]
- 151.Reference deleted.
- 152.Jones, P. M., and A. M. George. 2004. The ABC transporter structure and mechanism: perspectives on recent research. Cell. Mol. Life Sci. 61:682-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jurado, R. L. 1997. Iron, infections, and anemia of inflammation. Clin. Infect. Dis. 25:888-895. [DOI] [PubMed] [Google Scholar]
- 154.Jurkevitch, E., Y. Hadar, Y. Chen, J. Libman, and A. Shanzer. 1992. Iron uptake and molecular recognition in Pseudomonas putida: receptor mapping with ferrichrome and its biomimetic analogs. J. Bacteriol. 174:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kadner, R. J., K. Heller, J. W. Coulton, and V. Braun. 1980. Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J. Bacteriol. 143:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kamau, P., and R. B. Jordan. 2002. Kinetic study of the oxidation of catechol by aqueous copper(II). Inorg. Chem. 41:3076-3083. [DOI] [PubMed] [Google Scholar]
- 157.Karpowich, N., O. Martsinkevich, L. Millen, Y. R. Yuan, P. L. Dai, K. MacVey, P. J. Thomas, and J. F. Hunt. 2001. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure 9:571-586. [DOI] [PubMed] [Google Scholar]
- 158.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522-15530. [DOI] [PubMed] [Google Scholar]
- 159.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 160.Keating, T. A., C. G. Marshall, C. T. Walsh, and A. E. Keating. 2002. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat. Struct. Biol. 9:522-526. [DOI] [PubMed] [Google Scholar]
- 161.Kerbarh, O., E. M. Bulloch, R. J. Payne, T. Sahr, F. Rebeille, and C. Abell. 2005. Mechanistic and inhibition studies of chorismate-utilizing enzymes. Biochem. Soc. Trans. 33:763-766. [DOI] [PubMed] [Google Scholar]
- 162.Kerbarh, O., D. Y. Chirgadze, T. L. Blundell, and C. Abell. 2006. Crystal structures of Yersinia enterocolitica salicylate synthase and its complex with the reaction products salicylate and pyruvate. J. Mol. Biol. 357:524-534. [DOI] [PubMed] [Google Scholar]
- 163.Kerbarh, O., A. Ciulli, N. I. Howard, and C. Abell. 2005. Salicylate biosynthesis: overexpression, purification, and characterization of Irp9, a bifunctional salicylate synthase from Yersinia enterocolitica. J. Bacteriol. 187:5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2005. Kinetic analyses reveal multiple steps in forming TonB-FhuA complexes from Escherichia coli. Biochemistry 44:3441-3453. [DOI] [PubMed] [Google Scholar]
- 165.Reference deleted.
- 166.Kim, Y., C. W. Yun, and C. C. Philpott. 2002. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 21:3632-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Klausner, R. D., T. A. Rouault, and J. B. Harford. 1993. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72:19-28. [DOI] [PubMed] [Google Scholar]
- 168.Klumpp, C., A. Burger, G. L. Mislin, and M. A. Abdallah. 2005. From a total synthesis of cepabactin and its 3:1 ferric complex to the isolation of a 1:1:1 mixed complex between iron (III), cepabactin and pyochelin. Bioorg. Med. Chem. Lett. 15:1721-1724. [DOI] [PubMed] [Google Scholar]
- 169.Kodding, J., F. Killig, P. Polzer, S. P. Howard, K. Diederichs, and W. Welte. 2005. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 280:3022-3028. [DOI] [PubMed] [Google Scholar]
- 170.Koebnik, R., K. Hantke, and V. Braun. 1993. The TonB-dependent ferrichrome receptor FcuA of Yersinia enterocolitica: evidence against a strict co-evolution of receptor structure and substrate specificity. Mol. Microbiol. 7:383-393. [DOI] [PubMed] [Google Scholar]
- 171.Kohli, R. M., J. W. Trauger, D. Schwarzer, M. A. Marahiel, and C. T. Walsh. 2001. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 40:7099-7108. [DOI] [PubMed] [Google Scholar]
- 172.Konopka, K., and J. B. Neilands. 1984. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 23:2122-2127. [DOI] [PubMed] [Google Scholar]
- 173.Kontoghiorghes, G. J., E. Eracleous, C. Economides, and A. Kolnagou. 2005. Advances in iron overload therapies. Prospects for effective use of deferiprone (L1), deferoxamine, the new experimental chelators ICL670, GT56-252, L1NA11 and their combinations. Curr. Med. Chem. 12:2663-2681. [DOI] [PubMed] [Google Scholar]
- 174.Koppisch, A. T., C. C. Browder, A. L. Moe, J. T. Shelley, B. A. Kinkel, L. E. Hersman, S. Iyer, and C. E. Ruggiero. 2005. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals 18:577-585. [DOI] [PubMed] [Google Scholar]
- 175.Kosman, D. J. 2003. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47:1185-1197. [DOI] [PubMed] [Google Scholar]
- 176.Kozlowski, M. C., N. J. Tom, C. T. Seto, A. M. Sefler, and P. A. Bartlett. 1995. Chorismate-utilizing enzymes isochorismate synthase, anthranilate synthase, and p-aminobenzoate synthase: mechanistic insight through inhibitor design. J. Am. Chem. Soc. 117:2128-2140. [Google Scholar]
- 177.Krewulak, K. D., C. M. Shepherd, and H. J. Vogel. 2005. Molecular dynamics simulations of the periplasmic ferric-hydroxamate binding protein FhuD. Biometals 18:375-386. [DOI] [PubMed] [Google Scholar]
- 178.Krithika, R., U. Marathe, P. Saxena, M. Z. Ansari, D. Mohanty, and R. S. Gokhale. 2006. A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lai, J. R., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2006. Localized protein interaction surfaces on the EntB carrier protein revealed by combinatorial mutagenesis and selection. J. Am. Chem. Soc. 128:11002-11003. [DOI] [PubMed] [Google Scholar]
- 180.Langman, L., I. G. Young, G. E. Frost, H. Rosenberg, and F. Gibson. 1972. Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. J. Bacteriol. 112:1142-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Larsen, N. A., H. Lin, R. Wei, M. A. Fischbach, and C. T. Walsh. 2006. Structural characterization of enterobactin hydrolase IroE. Biochemistry 45:10184-10190. [DOI] [PubMed] [Google Scholar]
- 182.Lautru, S., R. J. Deeth, L. M. Bailey, and G. L. Challis. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat. Chem. Biol. 1:265-269. [DOI] [PubMed] [Google Scholar]
- 183.Lechner, M., P. Wojnar, and B. Redl. 2001. Human tear lipocalin acts as an oxidative-stress-induced scavenger of potentially harmful lipid peroxidation products in a cell culture system. Biochem. J. 356:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Reference deleted.
- 185.Lee, J. Y., B. K. Janes, K. D. Passalacqua, B. F. Pfleger, N. H. Bergman, H. Liu, K. Hakansson, R. V. Somu, C. C. Aldrich, S. Cendrowski, P. C. Hanna, and D. H. Sherman. 2007. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J. Bacteriol. 189:1698-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lesuisse, E., P. L. Blaiseau, A. Dancis, and J. M. Camadro. 2001. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology 147:289-298. [DOI] [PubMed] [Google Scholar]
- 187.Lesuisse, E., M. Casteras-Simon, and P. Labbe. 1995. Ferrireductase activity in Saccharomyces cerevisiae and other fungi: colorimetric assays on agar plates. Anal. Biochem. 226:375-377. [DOI] [PubMed] [Google Scholar]
- 188.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154:283-287. [DOI] [PubMed] [Google Scholar]
- 189.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lin, H., M. A. Fischbach, G. J. Gatto, Jr., D. R. Liu, and C. T. Walsh. 2006. Bromoenterobactins as potent inhibitors of a pathogen-associated, siderophore-modifying C-glycosyltransferase. J. Am. Chem. Soc. 128:9324-9325. [DOI] [PubMed] [Google Scholar]
- 191.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu, J., N. Quinn, G. A. Berchtold, and C. T. Walsh. 1990. Overexpression, purification, and characterization of isochorismate synthase (EntC), the first enzyme involved in the biosynthesis of enterobactin from chorismate. Biochemistry 29:1417-1425. [DOI] [PubMed] [Google Scholar]
- 193.Liu, Q., J. Ryon, and M. Nilsen-Hamilton. 1997. Uterocalin: a mouse acute phase protein expressed in the uterus around birth. Mol. Reprod. Dev. 46:507-514. [DOI] [PubMed] [Google Scholar]
- 194.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 195.Locher, K. P., and J. P. Rosenbusch. 1997. Oligomeric states and siderophore binding of the ligand-gated FhuA protein that forms channels across Escherichia coli outer membranes. Eur. J. Biochem. 247:770-775. [DOI] [PubMed] [Google Scholar]
- 196.Lodge, J. S., C. G. Gaines, J. E. Arceneaux, and B. R. Byers. 1980. Non-hydrolytic release of iron from ferrienterobactin analogs by extracts of Bacillus subtilis. Biochem. Biophys. Res. Commun. 97:1291-1295. [DOI] [PubMed] [Google Scholar]
- 197.Loo, T. W., M. C. Bartlett, and D. M. Clarke. 2003. Substrate-induced conformational changes in the transmembrane segments of human P-glycoprotein. Direct evidence for the substrate-induced fit mechanism for drug binding. J. Biol. Chem. 278:13603-13606. [DOI] [PubMed] [Google Scholar]
- 198.Loomis, L. D., and K. N. Raymond. 1991. Solution equilibria of enterobactin and metal-enterobactin complexes. Inorg. Chem. 30:906-911. [Google Scholar]
- 199.Loyevsky, M., C. John, B. Dickens, V. Hu, J. H. Miller, and V. R. Gordeuk. 1999. Chelation of iron within the erythrocytic Plasmodium falciparum parasite by iron chelators. Mol. Biochem. Parasitol. 101:43-59. [DOI] [PubMed] [Google Scholar]
- 200.Loyevsky, M., S. D. Lytton, B. Mester, J. Libman, A. Shanzer, and Z. I. Cabantchik. 1993. The antimalarial action of desferal involves a direct access route to erythrocytic (Plasmodium falciparum) parasites. J. Clin. Investig. 91:218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Luo, M., E. A. Fadeev, and J. T. Groves. 2005. Mycobactin-mediated iron acquisition within macrophages. Nat. Chem. Biol. 1:149-153. [DOI] [PubMed] [Google Scholar]
- 202.Luo, M., H. Lin, M. A. Fischbach, D. R. Liu, C. T. Walsh, and J. T. Groves. 2006. Enzymatic tailoring of enterobactin alters membrane partitioning and iron acquisition. ACS Chem. Biol. 1:29-32. [DOI] [PubMed] [Google Scholar]
- 203.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Martins, L. J., L. T. Jensen, J. R. Simon, G. L. Keller, and D. R. Winge. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 205.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Matzanke, B. F., S. Anemüller, V. Schünemann, A. X. Trautwein, and K. Hantke. 2004. FhuF, part of a siderophore-reductase system. Biochemistry 43:1386-1392. [DOI] [PubMed] [Google Scholar]
- 207.Matzanke, B. F., E. Bill, G. I. Müller, A. X. Trautwein, and G. Winkelmann. 1987. Metabolic utilization of 57Fe-labeled coprogen in Neurospora crassa. An in vivo Mössbauer study. Eur. J. Biochem. 162:643-650. [DOI] [PubMed] [Google Scholar]
- 208.Matzanke, B. F., D. J. Ecker, T. S. Yang, B. H. Huynh, G. Müller, and K. N. Raymond. 1986. Escherichia coli iron enterobactin uptake monitored by Mössbauer spectroscopy. J. Bacteriol. 167:674-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Matzanke, B. F., G. I. Müller, and K. N. Raymond. 1984. Hydroxamate siderophore mediated iron uptake in E. coli: stereospecific recognition of ferric rhodotorulic acid. Biochem. Biophys. Res. Commun. 121:922-930. [DOI] [PubMed] [Google Scholar]
- 210.May, J. J., R. Finking, F. Wiegeshoff, T. T. Weber, N. Bandur, U. Koert, and M. A. Marahiel. 2005. Inhibition of the d-alanine:d-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall. FEBS J. 272:2993-3003. [DOI] [PubMed] [Google Scholar]
- 211.May, J. J., N. Kessler, M. A. Marahiel, and M. T. Stubbs. 2002. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 99:12120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 213.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 214.Michel, L., N. Gonzalez, S. Jagdeep, T. Nguyen-Ngoc, and C. Reimmann. 2005. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58:495-509. [DOI] [PubMed] [Google Scholar]
- 215.Miethke, M., P. Bisseret, C. L. Beckering, D. Vignard, J. Eustache, and M. A. Marahiel. 2006. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. FEBS J. 273:409-419. [DOI] [PubMed] [Google Scholar]
- 216.Miethke, M., O. Klotz, U. Linne, J. J. May, C. L. Beckering, and M. A. Marahiel. 2006. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61:1413-1427. [DOI] [PubMed] [Google Scholar]
- 217.Miller, D. A., L. Luo, N. Hillson, T. A. Keating, and C. T. Walsh. 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9:333-344. [DOI] [PubMed] [Google Scholar]
- 218.Miller, M. J., I. Darwish, A. Ghosh, M. Ghosh, J.-G. Hansel, J. Hu, C. Niu, A. Ritter, K. Scheidt, C. Suling, S. Sun, D. Zhang, A. Budde, E. De Clerq, S. Leong, F. Malouin, and U. Moellmann. 1997. Design, syntheses and studies of new antibacterial, antifungal and antiviral agents, p. 116-138. In P. H. Bentley and P. J. O'Hanlon (ed.), Anti-infectives: recent advances in chemistry and structure-activity relationships. Royal Society of Chemistry, Cambridge, United Kingdom.
- 219.Moody, D. B., D. C. Young, T. Y. Cheng, J. P. Rosat, C. Roura-Mir, P. B. O'Connor, D. M. Zajonc, A. Walz, M. J. Miller, S. B. Levery, I. A. Wilson, C. E. Costello, and M. B. Brenner. 2004. T cell activation by lipopeptide antigens. Science 303:527-531. [DOI] [PubMed] [Google Scholar]
- 220.Moore, C. H., L. A. Foster, D. G. Gerbig, Jr., D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Moore, D. G., and C. F. Earhart. 1981. Specific inhibition of Escherichia coli ferrienterochelin uptake by a normal human serum immunoglobulin. Infect. Immun. 31:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Moore, D. G., R. J. Yancey, C. E. Lankford, and C. F. Earhart. 1980. Bacteriostatic enterochelin-specific immunoglobulin from normal human serum. Infect. Immun. 27:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Moore, R. E., Y. Kim, and C. C. Philpott. 2003. The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100:5664-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J. P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernandez, M. Schafer, J. Ravel, and P. Cornelis. 2002. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673-1685. [DOI] [PubMed] [Google Scholar]
- 225.Müller, A., A. J. Wilkinson, K. S. Wilson, and A. K. Duhme-Klair. 2006. An [{Fe(mecam)}2]6− bridge in the crystal structure of a ferric enterobactin binding protein. Angew. Chem. Int. 45:5132-5136. [DOI] [PubMed] [Google Scholar]
- 226.Müller, G., Y. Isowa, and K. N. Raymond. 1985. Stereospecificity of siderophore-mediated iron uptake in Rhodotorula pilimanae as probed by enantiorhodotorulic acid and isomers of chromic rhodotorulate. J. Biol. Chem. 260:13921-13926. [PubMed] [Google Scholar]
- 227.Müller, K., B. F. Matzanke, V. Schünemann, A. X. Trautwein, and K. Hantke. 1998. FhuF, an iron-regulated protein of Escherichia coli with a new type of [2Fe-2S] center. Eur. J. Biochem. 258:1001-1008. [DOI] [PubMed] [Google Scholar]
- 228.Munkelt, D., G. Grass, and D. H. Nies. 2004. The chromosomally encoded cation diffusion facilitator proteins DmeF and FieF from Wautersia metallidurans CH34 are transporters of broad metal specificity. J. Bacteriol. 186:8036-8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228a.Nassar, A. E., R. E. Talaat, and A. M. Kamel. 2006. The impact of recent innovations in the use of liquid chromatography-mass spectrometry in support of drug metabolism studies: are we all the way there yet? Curr. Opin. Drug Discov. Devel. 9:61-74. [PubMed] [Google Scholar]
- 229.Neilands, J. B., T. J. Erickson, and W. H. Rastetter. 1981. Stereospecificity of the ferric enterobactin receptor of Escherichia coli K-12. J. Biol. Chem. 256:3831-3832. [PubMed] [Google Scholar]
- 230.Nelson, M., C. J. Carrano, and P. J. Szaniszlo. 1992. Identification of the ferrioxamine B receptor, FoxB, in Escherichia coli K12. Biometals 5:37-46. [DOI] [PubMed] [Google Scholar]
- 231.Newton, S. M., J. D. Igo, D. C. Scott, and P. E. Klebba. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol. Microbiol. 32:1153-1165. [DOI] [PubMed] [Google Scholar]
- 232.Nikaido, H., and E. Y. Rosenberg. 1990. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with β-lactam antibiotics containing catechol and analogous groups. J. Bacteriol. 172:1361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Nilius, A. M., and S. G. Farmer. 1990. Identification of extracellular siderophores of pathogenic strains of Aspergillus fumigatus. J. Med. Vet. Mycol. 28:395-403. [DOI] [PubMed] [Google Scholar]
- 234.Nishio, T., N. Tanaka, J. Hiratake, Y. Katsube, Y. Ishida, and J. Oda. 1988. Isolation and structure of the novel dihydroxamate siderophore alcaligin. J. Am. Chem. Soc. 110:8733-8734. [Google Scholar]
- 235.Nowalk, A. J., S. B. Tencza, and T. A. Mietzner. 1994. Coordination of iron by the ferric iron-binding protein of pathogenic Neisseria is homologous to the transferrins. Biochemistry 33:12769-12775. [DOI] [PubMed] [Google Scholar]
- 236.Oberegger, H., M. Schoeser, I. Zadra, B. Abt, and H. Haas. 2001. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41:1077-1089. [DOI] [PubMed] [Google Scholar]
- 237.O'Brien, I. G., G. B. Cox, and F. Gibson. 1971. Enterochelin hydrolysis and iron metabolism in Escherichia coli. Biochim. Biophys. Acta 237:537-549. [DOI] [PubMed] [Google Scholar]
- 238.Ojeda, L., G. Keller, U. Muhlenhoff, J. C. Rutherford, R. Lill, and D. R. Winge. 2006. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281:17661-17669. [DOI] [PubMed] [Google Scholar]
- 239.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Page, W. J., E. Kwon, A. S. Cornish, and A. E. Tindale. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol. Lett. 228:211-216. [DOI] [PubMed] [Google Scholar]
- 241.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Park, S. F., and P. T. Richardson. 1995. Molecular characterization of a Campylobacter jejuni lipoprotein with homology to periplasmic siderophore-binding proteins. J. Bacteriol. 177:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Patel, H. M., and C. T. Walsh. 2001. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry 40:9023-9031. [DOI] [PubMed] [Google Scholar]
- 244.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557-2570. [DOI] [PubMed] [Google Scholar]
- 245.Pawelek, P. D., N. Croteau, C. Ng-Thow-Hing, C. M. Khursigara, N. Moiseeva, M. Allaire, and J. W. Coulton. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399-1402. [DOI] [PubMed] [Google Scholar]
- 246.Payne, M. A., J. D. Igo, Z. Cao, S. B. Foster, S. M. Newton, and P. E. Klebba. 1997. Biphasic binding kinetics between FepA and its ligands. J. Biol. Chem. 272:21950-21955. [DOI] [PubMed] [Google Scholar]
- 247.Payne, R. J., O. Kerbarh, R. N. Miguel, A. D. Abell, and C. Abell. 2005. Inhibition studies on salicylate synthase. Org. Biomol. Chem. 3:1825-1827. [DOI] [PubMed] [Google Scholar]
- 248.Pecqueur, L., B. D'Autreaux, J. Dupuy, Y. Nicolet, L. Jacquamet, B. Brutscher, I. Michaud-Soret, and B. Bersch. 2006. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J. Biol. Chem. 281:21286-21295. [DOI] [PubMed] [Google Scholar]
- 249.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181-1190. [DOI] [PubMed] [Google Scholar]
- 250.Pierce, J. R., C. L. Pickett, and C. F. Earhart. 1983. Two fep genes are required for ferrienterochelin uptake in Escherichia coli K-12. J. Bacteriol. 155:330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pierre, J. L., M. Fontecave, and R. R. Crichton. 2002. Chemistry for an essential biological process: the reduction of ferric iron. Biometals 15:341-346. [DOI] [PubMed] [Google Scholar]
- 252.Poey, M. E., M. F. Azpiroz, and M. Lavina. 2006. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 50:1411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 254.Pohl, E., R. K. Holmes, and W. G. Hol. 1999. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J. Mol. Biol. 285:1145-1156. [DOI] [PubMed] [Google Scholar]
- 255.Pohl, E., R. K. Holmes, and W. G. Hol. 1998. Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR. J. Biol. Chem. 273:22420-22427. [DOI] [PubMed] [Google Scholar]
- 256.Pollack, S., R. N. Rossan, D. E. Davidson, and A. Escajadillo. 1987. Desferrioxamine suppresses Plasmodium falciparum in Aotus monkeys. Proc. Soc. Exp. Biol. Med. 184:162-164. [DOI] [PubMed] [Google Scholar]
- 257.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 258.Poole, R. K., N. J. Rogers, R. A. D'Mello, M. N. Hughes, and Y. Orii. 1997. Escherichia coli flavohaemoglobin (Hmp) reduces cytochrome c and Fe(III)-hydroxamate K by electron transfer from NADH via FAD: sensitivity of oxidoreductase activity to haem-bound dioxygen. Microbiology 143:1557-1565. [DOI] [PubMed] [Google Scholar]
- 259.Pramanik, A., and V. Braun. 2006. Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J. Bacteriol. 188:3878-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Puig, S., E. Askeland, and D. J. Thiele. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99-110. [DOI] [PubMed] [Google Scholar]
- 261.Qiu, X., E. Pohl, R. K. Holmes, and W. G. J. Hol. 1996. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry 35:12292-12302. [DOI] [PubMed] [Google Scholar]
- 262.Quadri, L. E. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol. Microbiol. 37:1-12. [DOI] [PubMed] [Google Scholar]
- 263.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 264.Quadri, L. E., T. A. Keating, H. M. Patel, and C. T. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 265.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17-25. [DOI] [PubMed] [Google Scholar]
- 266.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 288:1062-1064. [DOI] [PubMed] [Google Scholar]
- 267.Rastetter, W. H., T. J. Erickson, and M. C. Venuti. 1981. Synthesis of iron chelators. Enterobactin, enantioenterobactin, and a chiral analogue. J. Org. Chem. 46:3579-3590. [Google Scholar]
- 268.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 269.Rauscher, L., D. Expert, B. F. Matzanke, and A. X. Trautwein. 2002. Chrysobactin-dependent iron acquisition in Erwinia chrysanthemi. Functional study of a homolog of the Escherichia coli ferric enterobactin esterase. J. Biol. Chem. 277:2385-2395. [DOI] [PubMed] [Google Scholar]
- 270.Raventos-Suarez, C., S. Pollack, and R. L. Nagel. 1982. Plasmodium falciparum: inhibition of in vitro growth by desferrioxamine. Am. J. Trop. Med. Hyg. 31:919-922. [DOI] [PubMed] [Google Scholar]
- 271.Raymond, K. N., and C. J. Carrano. 1979. Coordination chemistry and microbial iron transport. J. Am. Chem. Soc. 12:183-190. [Google Scholar]
- 272.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 100:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Redl, B., P. Holzfeind, and F. Lottspeich. 1992. cDNA cloning and sequencing reveals human tear prealbumin to be a member of the lipophilic-ligand carrier protein superfamily. J. Biol. Chem. 267:20282-20287. [PubMed] [Google Scholar]
- 274.Reimmann, C., H. M. Patel, C. T. Walsh, and D. Haas. 2004. PchC thioesterase optimizes nonribosomal biosynthesis of the peptide siderophore pyochelin in Pseudomonas aeruginosa. J. Bacteriol. 186:6367-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Reyes, C. L., A. Ward, J. Yu, and G. Chang. 2006. The structures of MsbA: insight into ABC transporter-mediated multidrug efflux. FEBS Lett. 580:1042-1048. [DOI] [PubMed] [Google Scholar]
- 276.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141:3181-3191. [DOI] [PubMed] [Google Scholar]
- 277.Roberts, M., K. G. Wooldridge, H. Gavine, S. I. Kuswandi, and P. H. Williams. 1989. Inhibition of biological activities of the aerobactin receptor protein in rough strains of Escherichia coli by polyclonal antiserum raised against native protein. J. Gen. Microbiol. 135:2387-2398. [DOI] [PubMed] [Google Scholar]
- 278.Rodriguez, G. M., and I. Smith. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188:424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Rogers, H. J., C. Synge, and V. E. Woods. 1980. Antibacterial effect of scandium and indium complexes of enterochelin on Klebsiella pneumoniae. Antimicrob. Agents Chemother. 18:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rogers, H. J., V. E. Woods, and C. Synge. 1982. Antibacterial effect of the scandium and indium complexes of enterochelin on Escherichia coli. J. Gen. Microbiol. 128:2389-2394. [DOI] [PubMed] [Google Scholar]
- 281.Rohrbach, M. R., V. Braun, and W. Koster. 1995. Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J. Bacteriol. 177:7186-7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Roosenberg, J. M., II, Y. M. Lin, Y. Lu, and M. J. Miller. 2000. Studies and syntheses of siderophores, microbial iron chelators, and analogs as potential drug delivery agents. Curr. Med. Chem. 7:159-197. [DOI] [PubMed] [Google Scholar]
- 283.Rouault, T. A. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2:406-414. [DOI] [PubMed] [Google Scholar]
- 284.Rusnak, F., J. Liu, N. Quinn, G. A. Berchtold, and C. T. Walsh. 1990. Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and substrate specificity of isochorismatase. Biochemistry 29:1425-1435. [DOI] [PubMed] [Google Scholar]
- 285.Rutherford, J. C., S. Jaron, and D. R. Winge. 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636-27643. [DOI] [PubMed] [Google Scholar]
- 286.Rutherford, J. C., L. Ojeda, J. Balk, U. Muhlenhoff, R. Lill, and D. R. Winge. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135-10140. [DOI] [PubMed] [Google Scholar]
- 287.Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 288.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sakaitani, M., F. Rusnak, N. R. Quinn, C. Tu, T. B. Frigo, G. A. Berchtold, and C. T. Walsh. 1990. Mechanistic studies on trans-2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EntA) in the biosynthesis of the iron chelator enterobactin. Biochemistry 29:6789-6798. [DOI] [PubMed] [Google Scholar]
- 290.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 291.Schlehuber, S., and A. Skerra. 2005. Lipocalins in drug discovery: from natural ligand-binding proteins to “anticalins.” Drug. Discov. Today 10:23-33. [DOI] [PubMed] [Google Scholar]
- 292.Schmidt-Ott, K. M., K. Mori, A. Kalandadze, J. Y. Li, N. Paragas, T. Nicholas, P. Devarajan, and J. Barasch. 2006. Neutrophil gelatinase-associated lipocalin-mediated iron traffic in kidney epithelia. Curr. Opin. Nephrol. Hypertens. 15:442-449. [DOI] [PubMed] [Google Scholar]
- 293.Schroder, I., E. Johnson, and S. de Vries. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427-447. [DOI] [PubMed] [Google Scholar]
- 294.Schwarzer, D., H. D. Mootz, U. Linne, and M. A. Marahiel. 2002. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. USA 99:14083-14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schwecke, T., K. Gottling, P. Durek, I. Duenas, N. F. Kaufer, S. Zock-Emmenthal, E. Staub, T. Neuhof, R. Dieckmann, and H. von Dohren. 2006. Nonribosomal peptide synthesis in Schizosaccharomyces pombe and the architectures of ferrichrome-type siderophore synthetases in fungi. Chembiochem 7:612-622. [DOI] [PubMed] [Google Scholar]
- 296.Scott, M. D., A. Ranz, F. A. Kuypers, B. H. Lubin, and S. R. Meshnick. 1990. Parasite uptake of desferroxamine: a prerequisite for antimalarial activity. Br. J. Haematol. 75:598-602. [DOI] [PubMed] [Google Scholar]
- 297.Sebulsky, M. T., and D. E. Heinrichs. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Sebulsky, M. T., B. H. Shilton, C. D. Speziali, and D. E. Heinrichs. 2003. The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J. Biol. Chem. 278:49890-49900. [DOI] [PubMed] [Google Scholar]
- 299.Shatwell, K. P., A. Dancis, A. R. Cross, R. D. Klausner, and A. W. Segal. 1996. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J. Biol. Chem. 271:14240-14244. [DOI] [PubMed] [Google Scholar]
- 300.Shaw-Reid, C. A., N. L. Kelleher, H. C. Losey, A. M. Gehring, C. Berg, and C. T. Walsh. 1999. Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chem. Biol. 6:385-400. [DOI] [PubMed] [Google Scholar]
- 301.Shouldice, S. R., D. E. McRee, D. R. Dougan, L. W. Tari, and A. B. Schryvers. 2005. Novel anion-independent iron coordination by members of a third class of bacterial periplasmic ferric ion-binding proteins. J. Biol. Chem. 280:5820-5827. [DOI] [PubMed] [Google Scholar]
- 302.Singh, A., S. Severance, N. Kaur, W. Wiltsie, and D. J. Kosman. 2006. Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J. Biol. Chem. 281:13355-13364. [DOI] [PubMed] [Google Scholar]
- 303.Somu, R. V., H. Boshoff, C. Qiao, E. M. Bennett, C. E. Barry III, and C. C. Aldrich. 2006. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 49:31-34. [DOI] [PubMed] [Google Scholar]
- 304.Somu, R. V., D. J. Wilson, E. M. Bennett, H. I. Boshoff, L. Celia, B. J. Beck, C. E. Barry III, and C. C. Aldrich. 2006. Antitubercular nucleosides that inhibit siderophore biosynthesis: SAR of the glycosyl domain. J. Med. Chem. 49:7623-7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sprencel, C., Z. Cao, Z. Qi, D. C. Scott, M. A. Montague, N. Ivanoff, J. Xu, K. M. Raymond, S. M. Newton, and P. E. Klebba. 2000. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182:5359-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 307.Stefanska, A. L., M. Fulston, C. S. Houge-Frydrych, J. J. Jones, and S. R. Warr. 2000. A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot. (Tokyo) 53:1346-1353. [DOI] [PubMed] [Google Scholar]
- 308.Stergiopoulos, I., L.-H. Zwiers, and M. A. De Waard. 2002. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 108:719-734. [Google Scholar]
- 309.Stoj, C., and D. J. Kosman. 2003. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 554:422-426. [DOI] [PubMed] [Google Scholar]
- 310.Stoj, C. S., A. J. Augustine, L. Zeigler, E. I. Solomon, and D. J. Kosman. 2006. Structural basis of the ferrous iron specificity of the yeast ferroxidase, Fet3p. Biochemistry 45:12741-12749. [DOI] [PubMed] [Google Scholar]
- 311.Strahsburger, E., M. Baeza, O. Monasterio, and R. Lagos. 2005. Cooperative uptake of microcin E492 by receptors FepA, Fiu, and Cir and inhibition by the siderophore enterochelin and its dimeric and trimeric hydrolysis products. Antimicrob. Agents Chemother. 49:3083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sundlov, J. A., J. A. Garringer, J. M. Carney, A. S. Reger, E. J. Drake, W. L. Duax, and A. M. Gulick. 2006. Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 62:734-740. [DOI] [PubMed] [Google Scholar]
- 313.Sutcliffe, I. C., and R. R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tanabe, T., H. Nakao, T. Kuroda, T. Tsuchiya, and S. Yamamoto. 2006. Involvement of the Vibrio parahaemolyticus pvsC gene in export of the siderophore vibrioferrin. Microbiol. Immunol. 50:871-876. [PubMed] [Google Scholar]
- 315.Taylor, A. B., C. S. Stoj, L. Ziegler, D. J. Kosman, and P. J. Hart. 2005. The copper-iron connection in biology: structure of the metallo-oxidase Fet3p. Proc. Natl. Acad. Sci. USA 102:15459-15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Thomas, X., D. Destoumieux-Garzon, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 317.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1993. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect. Immun. 61:3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tsafack, A., M. Loyevsky, P. Ponka, and Z. I. Cabantchik. 1996. Mode of action of iron (III) chelators as antimalarials. IV. Potentiation of desferal action by benzoyl and isonicotinoyl hydrazone derivatives. J. Lab. Clin. Med. 127:574-582. [DOI] [PubMed] [Google Scholar]
- 319.Tseng, C. F., A. Burger, G. L. Mislin, I. J. Schalk, S. S. Yu, S. I. Chan, and M. A. Abdallah. 2006. Bacterial siderophores: the solution stoichiometry and coordination of the Fe(III) complexes of pyochelin and related compounds. J. Biol. Inorg. Chem. 11:419-432. [DOI] [PubMed] [Google Scholar]
- 320.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 321.Urbanowski, J. L., and R. C. Piper. 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274:38061-38070. [DOI] [PubMed] [Google Scholar]
- 322.Valdebenito, M., A. L. Crumbliss, G. Winkelmann, and K. Hantke. 2006. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 296:513-520. [DOI] [PubMed] [Google Scholar]
- 323.van der Heide, T., and B. Poolman. 2002. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 3:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Vannada, J., E. M. Bennett, D. J. Wilson, H. I. Boshoff, C. E. Barry III, and C. C. Aldrich. 2006. Design, synthesis, and biological evaluation of beta-ketosulfonamide adenylation inhibitors as potential antitubercular agents. Org. Lett. 8:4707-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Reference deleted.
- 326.Vertesy, L., W. Aretz, H.-W. Fehlhaber, and H. Kogler. 1995. Salmycin A-D, Antibiotika aus Streptomyces violaceus, DSM 8286, mit Siderophor-Aminoglycosid-Struktur. Helv. Chim. Acta 78:46-60. [Google Scholar]
- 327.Vinella, D., C. Albrecht, M. Cashel, and R. D'Ari. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56:958-970. [DOI] [PubMed] [Google Scholar]
- 328.Voisard, C., J. Wang, J. L. McEvoy, P. Xu, and S. A. Leong. 1993. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wagegg, W., and V. Braun. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J. Bacteriol. 145:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 331.Waters, B. M., and D. J. Eide. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277:33749-33757. [DOI] [PubMed] [Google Scholar]
- 332.Wawrousek, E. F., and J. V. McArdle. 1982. Spectrochemistry of ferrioxamine B, ferrichrome, and ferrichrome A. J. Inorg. Biochem. 17:169-183. [Google Scholar]
- 333.Welzel, K., K. Eisfeld, L. Antelo, T. Anke, and H. Anke. 2005. Characterization of the ferrichrome A biosynthetic gene cluster in the homobasidiomycete Omphalotus olearius. FEMS Microbiol. Lett. 249:157-163. [DOI] [PubMed] [Google Scholar]
- 334.Wertheimer, A. M., W. Verweij, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Williams, P. H., W. Rabsch, U. Methner, W. Voigt, H. Tschape, and R. Reissbrodt. 2006. Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 24:3840-3844. [DOI] [PubMed] [Google Scholar]
- 337.Wilson, M. K., R. J. Abergel, K. N. Raymond, J. E. Arceneaux, and B. R. Byers. 2006. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 348:320-325. [DOI] [PubMed] [Google Scholar]
- 338.Winkelmann, G. 1979. Evidence for stereospecific uptake of iron chelates in fungi. FEBS Lett. 97:43-46. [Google Scholar]
- 339.Winkelmann, G. 2001. Siderophore transport in fungi, p. 463-479. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH, Weinheim, Germany.
- 340.Winkelmann, G., and V. Braun. 1981. Stereoselective recognition of ferrichrome by fungi and bacteria. FEMS Microbiol. Lett. 11:237-241. [Google Scholar]
- 341.Winkelmann, G., and H. Drechsel. 1997. Microbial siderophores, p. 200-246. In H. Kleinkauf and H. von Döhren (ed.), Products of secondary metabolism. Wiley-VCH, Weinheim, Germany.
- 342.Wong, G. B., M. J. Kappel, K. M. Raymond, B. F. Matzanke, and G. Winkelmann. 1983. Coordination chemistry of microbial iron transport compounds. 24. Characterization of coprogen and ferricrocin, two ferric hydroxamate siderophores. J. Am. Chem. Soc. 105:810. [Google Scholar]
- 343.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12:325-348. [DOI] [PubMed] [Google Scholar]
- 344.Wu, W. S., P. C. Hsieh, T. M. Huang, Y. F. Chang, and C. F. Chang. 2002. Cloning and characterization of an iron regulated locus, iroA, in Salmonella enterica serovar Choleraesuis. DNA Seq. 13:333-341. [DOI] [PubMed] [Google Scholar]
- 345.Wyckoff, E. E., A. M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Xu, Y., and M. J. Miller. 1998. Total syntheses of mycobactin analogues as potent antimycobacterial agents using a minimal protecting group strategy. J. Org. Chem. 63:4314-4322. [Google Scholar]
- 347.Yamaguchi-Iwai, Y., A. Dancis, and R. D. Klausner. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Yamaguchi-Iwai, Y., R. Ueta, A. Fukunaka, and R. Sasaki. 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277:18914-18918. [DOI] [PubMed] [Google Scholar]
- 349.Yang, J., K. Mori, J. Y. Li, and J. Barasch. 2003. Iron, lipocalin, and kidney epithelia. Am. J. Physiol. Renal Physiol. 285:9-18. [DOI] [PubMed] [Google Scholar]
- 350.Yu, S., E. Fiss, and W. R. Jacobs, Jr. 1998. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180:4676-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Yuan, W. M., G. D. Gentil, A. D. Budde, and S. A. Leong. 2001. Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. J. Bacteriol. 183:4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]
- 353.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218-10223. [DOI] [PubMed] [Google Scholar]
- 354.Yun, C. W., T. Ferea, J. Rashford, O. Ardon, P. O. Brown, D. Botstein, J. Kaplan, and C. C. Philpott. 2000. Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275:10709-10715. [DOI] [PubMed] [Google Scholar]
- 355.Yun, C. W., J. S. Tiedeman, R. E. Moore, and C. C. Philpott. 2000. Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 275:16354-16359. [DOI] [PubMed] [Google Scholar]
- 356.Zähner, H., H. Diddens, W. Keller-Schierlein, and H. U. Nageli. 1977. Some experiments with semisynthetic sideromycins. Jpn. J. Antibiot. 30:201-206. [PubMed] [Google Scholar]
- 357.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]
- 358.Zhou, L. W., H. Haas, and G. A. Marzluf. 1998. Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol. Gen. Genet. 259:532-540. [DOI] [PubMed] [Google Scholar]
- 359.Zhu, M., M. Valdebenito, G. Winkelmann, and K. Hantke. 2005. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 151:2363-2372. [DOI] [PubMed] [Google Scholar]
- 360.Zhu, W., J. E. Arceneaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]