Abstract
Summary: Actinobacteria constitute one of the largest phyla among Bacteria and represent gram-positive bacteria with a high G+C content in their DNA. This bacterial group includes microorganisms exhibiting a wide spectrum of morphologies, from coccoid to fragmenting hyphal forms, as well as possessing highly variable physiological and metabolic properties. Furthermore, Actinobacteria members have adopted different lifestyles, and can be pathogens (e.g., Corynebacterium, Mycobacterium, Nocardia, Tropheryma, and Propionibacterium), soil inhabitants (Streptomyces), plant commensals (Leifsonia), or gastrointestinal commensals (Bifidobacterium). The divergence of Actinobacteria from other bacteria is ancient, making it impossible to identify the phylogenetically closest bacterial group to Actinobacteria. Genome sequence analysis has revolutionized every aspect of bacterial biology by enhancing the understanding of the genetics, physiology, and evolutionary development of bacteria. Various actinobacterial genomes have been sequenced, revealing a wide genomic heterogeneity probably as a reflection of their biodiversity. This review provides an account of the recent explosion of actinobacterial genomics data and an attempt to place this in a biological and evolutionary context.
INTRODUCTION
General Features of Actinobacteria
In terms of number and variety of identified species, the phylum Actinobacteria represents one of the largest taxonomic units among the 18 major lineages currently recognized within the domain Bacteria (406), including 5 subclasses and 14 suborders (404). It comprises gram-positive bacteria with a high G+C content in their DNA, ranging from 51% in some corynebacteria to more than 70% in Streptomyces and Frankia. An exception to this is the genome of the obligate pathogen Tropheryma whipplei, with less than 50% G+C.
Actinobacteria exhibit a wide variety of morphologies, from coccoid (Micrococcus) or rod-coccoid (e.g., Arthrobacter) to fragmenting hyphal forms (e.g., Nocardia spp.) or permanent and highly differentiated branched mycelium (e.g., Streptomyces spp.) (15). They also exhibit diverse physiological and metabolic properties, such as the production of extracellular enzymes and the formation of a wide variety of secondary metabolites (389). Notably, many such secondary metabolites are potent antibiotics (255), a trait that has turned Streptomyces species into the primary antibiotic-producing organisms exploited by the pharmaceutical industry (29). Furthermore, various different lifestyles are encountered among Actinobacteria, and the phylum includes pathogens (e.g., Mycobacterium spp., Nocardia spp., Tropheryma spp., Corynebacterium spp., and Propionibacterium spp.), soil inhabitants (Streptomyces spp.), plant commensals (Leifsonia spp.), nitrogen-fixing symbionts (Frankia), and gastrointestinal tract (GIT) inhabitants (Bifidobacterium spp.). Unusual developmental features are displayed by many actinobacterial genera, such as formation of sporulating aerial mycelium in Streptomyces species or the persistent nonreplicating state exhibited by certain mycobacteria. Actinobacteria are widely distributed in both terrestrial and aquatic (including marine) ecosystems, especially in soil, where they play a crucial role in the recycling of refractory biomaterials by decomposition and humus formation (152, 403). Furthermore, many bifidobacteria are used as active ingredients in a variety of so-called functional foods due to their perceived health-promoting or probiotic properties, such as protection against pathogens mediated through the process of competitive exclusion, bile salt hydrolase activity, immune modulation, and the ability to adhere to mucus or the intestinal epithelium (273, 329, 407).
The actinobacterial genomes sequenced so far belong to organisms relevant to human and veterinary medicine, biotechnology, and ecology, and the observed genomic heterogeneity is assumed to be a reflection of their biodiversity. This review will give an account of the recent explosion of actinobacterial genomics data and will place this in a biological and evolutionary context.
Evolution and Dynamics of Bacterial Genomes
The principal genetic events that determine genome shape and structure are believed to be gene duplication, horizontal gene transfer (HGT), gene loss, and chromosomal rearrangements. Despite efforts to quantify the relative contribution of each of these processes, no reliable model can yet explain and trace the evolutionary development of bacteria based on their current genome structure (8, 183, 243, 398).
Gene duplications.
It was previously thought that bacterial genomes have evolved from a much smaller ancestral genome through numerous gene duplication events and the consequent generation of paralogs (244). However, an analysis based on the currently available bacterial genome data does not support this theory and shows that gene duplications contribute only modestly to genome evolution (79). Despite this, it has been noted that genes involved in a specific adaptation have been preserved after duplications, suggesting that gene duplication does have an evolutionary role (79). This is nicely illustrated by the mycobacterial paranome, which largely corresponds to a functional class of genes involved in fatty acid metabolism, in agreement with the complex nature of the mycobacterial cell wall and probably reflecting adaptive evolution of this cellular structure (79, 432).
HGT.
The introduction of novel or alien genes by HGT allows for rapid niche-specific adaptation, which in turn may lead to bacterial diversification and speciation (80). Bacterial genome evolution is based on the combined outcome of genes acquired through cell division, i.e., vertically inherited, and by HGT (482). Taking this concept to its extreme, one can claim that two bacterial taxa are more related to each other than to a third one not because they share a more recent ancestor but because they exchange genes more frequently (151). HGT is held responsible for enhancing the competitiveness of bacteria in their natural environments. For example, in some pathogenic bacteria, segments of DNA containing many virulence genes and gene clusters, called pathogenicity islands, appear to have been acquired by HGT (321). Actinobacterial examples of transmission of virulence genes through HGT are rare (376). Of these, the following three cases appear to represent obvious HGT events: (i) phages of Corynebacterium diphtheriae carry the major diphtheria toxin gene, (ii) a linear plasmid carries the genes for the macrolide toxin responsible for the ulceration that gives Mycobacterium ulcerans its name (411), and (iii) a large segment of the chromosome of Streptomyces turgidiscabies concerned with causing potato scab can be transferred by conjugation (280). In addition, it has been argued that the Mycobacterium tuberculosis Rv0986-8 virulence operon, which plays an important role in parasitism of host phagocytic cells by increasing the ecological fitness of the infecting mycobacterium (339), was acquired horizontally by the ancestor of M. tuberculosis, Mycobacterium prototuberculosis. Other genetic studies of the ancestral M. prototuberculosis species have indicated that various HGT events occurred before the evolutionary bottleneck that led to the emergence of the M. tuberculosis complex (167), probably from the Indian subcontinent (124).
Bioinformatic methods to identify HGT events are based principally on the analysis of divergence in the G+C content (GC deviation), dinucleotide differences, four-letter genomic signatures, and/or codon usage, though geneticists would often be satisfied with HGT as the explanation for genes found in only one organism. If the latter is correct, it would mean that HGT frequency is rather low (below 10% of the total gene complement) (243, 398). Interestingly, a recent analysis showed that many of the proteins that appeared to be specific for actinobacteria are also encoded by the genome of an alphaproteobacterium, Magnetospirillum magnetotacticum, but not by any other sequenced alphaproteobacterial genome, leading to the proposal that M. magnetotacticum acquired these genes by HGT from actinobacterial species (137).
Two other interesting cases of HGT between Chlamydia and a subset of Actinobacteria (e.g., Streptomyces, Tropheryma, Bifidobacterium, Leifsonia, Arthrobacter, and Brevibacterium) have recently been described (158). In the enzyme serine hydroxymethyltransferase (GlyA protein), two conserved inserts of 3 and 31 amino acids (aa) are present in various chlamydiae as well as the above-mentioned subset of Actinobacteria. Similarly, these bacteria contain a conserved 16-amino-acid insert in the peptidoglycan biosynthesis enzyme UDP-N-acetylglucosamine enolpyruvyl transferases (MurA). The functional and physiological significance of these apparent HGT events between chlamydiae and Actinobacteria is presently unclear.
Gene decay.
Bacterial genome size is determined by the outcome of several opposing forces. Deletion bias and genetic drift cause genomes to contract, while selection on gene function promotes genomes to preserve DNA. Genome increments depend on both gene duplications and acquisition of alien DNA, coupled to adaptive benefits (293). DNA loss may range from large deletions that span multiple loci to deletions of one or a few nucleotides (7). The influence of these different routes is variable among bacterial lineages (293). Inactivating and deleterious mutations in genes with little contribution to fitness can be transmitted to progeny and accumulate in populations, eventually leading to gene loss; whereas such mutations in genes that are critical will prevent the production of progeny and so will be eliminated from populations, resulting in the preservation of the functional gene (320).
Gene inactivation and loss are particularly apparent in several bacterial groups with a host-associated lifestyle, in which the host supplies many of the metabolic intermediates, thereby obviating the need to maintain many biosynthetic genes. In endosymbiotic bacteria, such as Buchnera and Rickettsia, loss of individual loci or operons is the only source of divergence in the gene inventories between species (289, 419). A clear example of genome degradation is provided by Mycobacterium leprae, which has discarded more than 1,000 genes compared with M. tuberculosis (84). Moreover, the presence of an even larger set of nonfunctional genes, i.e., pseudogenes, in M. leprae indicates that this genome contraction is still in progress. Although the criteria for identifying pseudogenes differ among studies, the overall rationale is identical: the predicted protein must be altered to a degree that abolishes its function. The thresholds applied for pseudogene identification are based on the known size and organization of functional domains within proteins, the observed length variation within individual gene families, and available information on experimentally disrupted proteins (320). Generally, pseudogenes include cases in which a stop codon or deletion has resulted in an encoded protein that is less than 80% of the length of its functional counterpart in the contrasted genome and cases in which a frameshift or insertion has altered more than 20% of the amino acid sequence (263). Most of the pseudogenes so far annotated in bacterial genomes are among the open reading frames (ORFs) whose functions are unknown. The lack of pseudogenes shared among multiple strains of the same species suggests that pseudogenes are generated continually, are eliminated rapidly, and thus only rarely persist in bacterial genomes (320). Other bacteria show a lower level of gene loss: in the obligate intracellular pathogen Rickettsia prowazekii only 76% of the potential coding capacity is used, while just 12 pseudogenes were identified (9); and a recent genome analysis of two Streptococcus thermophilus strains (33) found that 10% of the genes were pseudogenes, perhaps reflecting adaptation of S. thermophilus to its specialized environment, milk (33).
When all bacterial genomes are compared with each other, a set of only 50 to 100 genes, which are called the core genome sequences, appear to be maintained universally (for a review, see reference 147).
Genome rearrangements.
Apart from the events described in the previous sections that affect gene content, the organization of a genome is subject to change through chromosome rearrangements. Synteny, a term used here to indicate the conservation of gene order between genomes, can be applied as a phylogenetic tool to investigate relationships between species, since the degree of genome rearrangements increases linearly in relation to the time of divergence of bacterial taxa (236, 484).
Chromosomal rearrangements are largely dependent on the activity of repeated and mobile elements such as insertion sequences (ISs), transposons, prophage sequences, and plasmids (233). Bacterial genomes containing a higher repeat density have higher rates of rearrangements, leading to an accelerated loss of gene order (371). Homologous recombination events between such repeat sequences catalyze both gene rearrangement and gene loss in the genome, thus leading to diversification of taxa. Such recombination events may have promoted speciation in the T. whipplei taxon (357). Furthermore, chromosome evolution is influenced by large chromosomal rearrangements, e.g., large inversions, roughly symmetrically centered around the replication origin, which lead to the occurrence of X-shaped patterns in the alignments of whole genomes (117).
Taxonomy of Actinobacteria
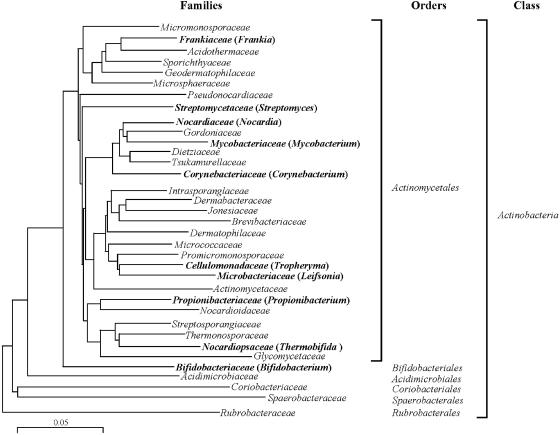
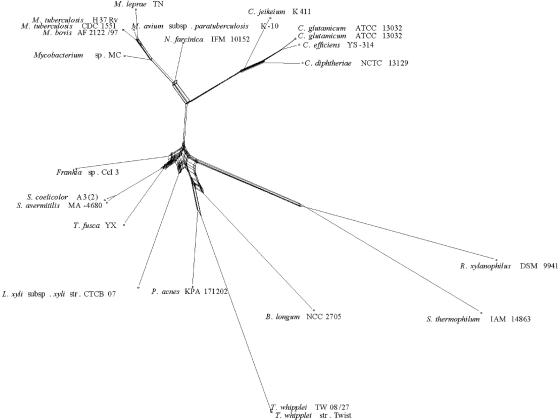
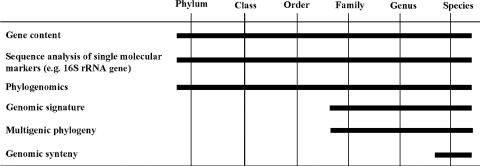
Actinobacteria include many organisms that exhibit, or have a tendency towards, mycelial growth. 16S rRNA gene sequencing has led to the recognition of 39 families and 130 genera, which also include high-G+C gram-positive bacteria with simpler morphology, such as bifidobacteria and micrococci (Fig. 1) (119). The deepest branch separates bifidobacteria from all other known families. The divergence of actinobacteria from other bacteria is so ancient that it is not possible to identify the phylogenetically closest bacterial group to Actinobacteria with confidence (119).
FIG. 1.
Phylogenetic tree of Actinobacteria based on 1,500 nucleotides of 16S rRNA. Scale bar, 5 nucleotides. Families containing members subjected to complete genome sequencing at the time of this writing are depicted in bold. Orders are indicated.
Actinobacteria have a unique molecular synapomorphy, i.e., a shared derived character: a homologous insertion of about 100 nucleotides between helices 54 and 55 of the 23S rRNA gene (375).
Actinobacterial Genome Sequencing Projects
The first actinobacterial genome to be sequenced was that of the paradigm strain of the human tuberculosis agent, M. tuberculosis H37Rv (83). In the last few years, genomes of 20 different Actinobacteria (in some cases multiple strains of the same species) have been sequenced to completion (Table 1), while sequencing of genomes from representatives of 43 other high-G+C bacteria are still in progress (Table 1) (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi).
TABLE 1.
Published data for actinobacterial genomes
| Microorganism | Genome size (bp) | No. of ORFs | % G+C content | No. of rRNA operons | No. of tRNAs | No. of pseudogenesa | Reference |
|---|---|---|---|---|---|---|---|
| Bifidobacterium longum biotype longum NCC2705 | 2,266,000 | 1,730 | 60 | 4 | 66 | ND | 384 |
| Corynebacterium diphtheriae NCTC 13129 | 2,488,635 | 2,320 | 53.5 | 5 | 54 | 48 | 60 |
| Corynebacterium efficiens YS-314 | 3,147,090 | 2,950 | 63.4 | 5 | 56 | ND | 316 |
| Corynebacterium glutamicum ATCC 13032 | 3,309,401 | 2,993 | 53.8 | 5 | 60 | ND | 195 |
| Corynebacterium jeikeium K411 | 2,462,499 | 2,104 | 61.4 | 3 | 50 | 68 | 427 |
| Frankia alni ACN14a | 7,497,934 | 6,786 | 72 | 2 | 62 | 12 | 319 |
| Frankia sp. strain Cc13 | 5,433,628 | 4,618 | 70 | 2 | 61 | 50 | NCBI source NC_007777 |
| Leifsonia xyli subsp. xyli CTCB07 | 2,584,158 | 2,351 | 67.7 | 1 | 49 | 307 | 300 |
| Mycobacterium avium subsp. paratuberculosis K-10 | 4,829,781 | 4,350 | 69.3 | 1 | 47 | 0 | 268 |
| Mycobacterium bovis AF2122/97 | 4,345,492 | 3,953 | 65.63 | 1 | 49 | 23 | 139 |
| Mycobacterium leprae TN | 3,268,203 | 1,605 | 57.79 | 1 | 49 | 1116 | 84 |
| Mycobacterium tuberculosis H37Rv | 4,411,532 | 3,994 | 65.61 | 1 | 49 | 6 | 83 |
| Mycobacterium tuberculosis CDC1551 | 4,403,836 | 4,250 | 65.60 | 1 | 49 | ND | 126 |
| Mycobacterium sp. strain MCS | 5,705,448 | 5,391 | 68 | 2 | 59 | 21 | NCBI source NC_008146 |
| Nocardia farcinica IFM10152 | 6,021,225 | 5,674 | 70.8 | 3 | 61 | 0 | 198 |
| Propionibacterium acnes KPA171202 | 2,560,265 | 2,297 | 60 | 3 | 51 | 17 | 46 |
| Streptomyces coelicolor A3 | 8,667,507 | 7,769 | 72 | 6 | 80 | 56 | 26 |
| Streptomyces avermitilis MA-4680 | 9,025,608 | 7,577 | 70 | 6 | 82 | 0 | 194 |
| Thermobifida fusca YX | 3,642,249 | 3,110 | 67 | 4 | 63 | 7 | 281 |
| Tropheryma whipplei TW08/27 | 925,938 | 783 | 46 | 1 | 54 | 1 | 27 |
| Tropheryma whipplei Twist | 927,303 | 808 | 46 | 1 | 54 | 0 | 357 |
ND, not determined.
Although most of the sequenced genomes in Table 1 are circular, like most bacterial genomes, Streptomyces genomes are linear. Using pulsed-field gel electrophoresis, the genomes of some other, still-unsequenced mycelial Actinobacteria taxa, such as Actinomyces, Amycolatopsis, Actinoplanes, Streptoverticillium, and Micromonospora, were also shown to be linear, with sizes ranging from 7.7 Mb (e.g., Micromonospora chalcea) to 9.7 Mb (Streptoverticillium abikoense), while sometimes also harboring large linear plasmids (362). Linear plasmids, typically possessing short inverted repeats at their termini and protein-bound 5′ ends, are often present in Actinobacteria (216).
Below we examine relevant genomic information from some of the best-known actinobacterial taxa (Bifidobacterium, Mycobacterium, Streptomyces, Corynebacterium, Thermobifida, Leifsonia, Frankia, Nocardia, Propionibacterium, and Tropheryma), partially in the light of what is known for Escherichia coli or Bacillus subtilis, as paradigms of gram-negative proteobacteria and gram-positive low-G+C-content bacteria, respectively. We discuss how genomic information can be used to gain insights into the physiology, genetics, and evolution of Actinobacteria.
GENOMICS OF BIFIDOBACTERIUM
General Features
The Bifidobacteriaceae family comprises four genera, Bifidobacterium, Gardnerella, Scardovia, and Parascardovia (404), of which only the first contains more than one species. Bifidobacteria form a deep-branching lineage within the Actinobacteria (136, 137). The Bifidobacterium genus contains six phylogenetic clusters, named B. boum, B. asteroides, B. adolescentis, B. longum, B. pullorum, and B. pseudolongum (448).
Bifidobacteria are nonmotile, nonsporulating, non-gas-producing, anaerobic, and saccharoclastic bacteria. They have been isolated from five different, though somewhat connected, ecological niches: the intestine, the oral cavity, food, the insect gut, and sewage. Those that inhabit the GIT (e.g., B. breve, B. longum biotype longum, and B. longum biotype infantis) have been the subject of growing interest due to their probiotic properties. Bifidobacteria ferment a large variety of oligosaccharides in the GIT, some of which, in particular those that are not digested by their host, are commercially exploited to enhance bifidobacterial numbers (as well as other probiotic bacteria) in situ, a practice that is referred to as the prebiotic concept (146).
Of the currently recognized 29 Bifidobacterium species, three strains that belong to the B. longum and B. adolescentis phylogenetic groups have been sequenced to completion (Table 2), while the sequences of others, e.g., B. dentium Bd1, are at various stages of completion: detailed sequence information for some of these genomes is expected to become publicly available in the near future. Furthermore, genome sequencing of B. breve M-16V, B. breve Yacult, B. animalis subsp. lactis, B. longum biotype longum, and B. longum biotype infantis (276) is under way. These genomes range in size from 1.9 to 2.9 Mb and generally display architectural features of a typical bacterial chromosome. Some of these are the co-orientation of gene transcription and DNA replication (288); a G-rich, C-poor bias in the nucleotide composition of the leading DNA strand (129); and a typical presumptive origin-of-replication region (350), including a gene constellation near the origin (comprising rpmH, dnaA, dnaN, and recF), a particular GC nucleotide skew ([G-C]/[G+C]), and the presence of multiple DnaA boxes and AT-rich sequences immediately upstream of the dnaA gene (77).
TABLE 2.
General features of bifidobacterial genomes
| Microorganism | Statusa | Genome size (bp) | No. of ORFs | % G+C content | No. of rRNA operonsb | Reference |
|---|---|---|---|---|---|---|
| B. longum biotype longum NCC2705 | C | 2,266,000 | 1,730 | 60 | 4 | 384 |
| B. longum biotype longum DJO10A | UF | 2,375,800 | 1,811 | 59 | 4 | NCBI source NZ_AABM00000000 |
| B. adolescentis ATCC15703 | C | 2,084,445 | 1,564 | 59 | 5 | NCBI source NC_008618 |
| B. breve UCC2003 | C | 2,422,668 | 1,868 | 59 | 2 | 254 |
| B. dentium Bd1 | UF | ∼2,600,000 | ∼2,270 | 59.2 | NA | NCBI source (project ID 17583) |
C, finished; UF, unfinished.
NA, not available.
The number of rRNA operons in bifidobacteria varies between one and five (58), perhaps reflecting different ecological strategies (230). The number of tRNA genes in the bifidobacterial genomes sequenced so far is relatively stable, i.e., 54 and 56 in B. breve UCC2003 and B. longum biotype longum NCC2705, respectively. These are representative of all 20 amino acids, with redundant tRNAs for all amino acids except cysteine, histidine, isoleucine, phenylalanine, and tryptophan.
Comparative Bifidobacterial Genome Analysis
Dot plot comparisons (at the nucleotide level) of the fully sequenced bifidobacterial genomes revealed a high degree of conservation and synteny across the entire genomes., i.e., those of B. longum biotype longum NCC2705, B. longum biotype longum DJO10A, B. breve UCC2003, and B. adolescentis ACC15703. Preliminary analysis against the draft genome sequences of B. dentium Bd1 confirmed and extended this result. However, there are also several breakpoint regions that seem to represent inversions or DNA insertion/deletion points (S. Leahy and D. Van Sinderen, unpublished data).
Recently, a B. longum biotype longum NCC2705-based spotted DNA microarray was employed to compare the genomes of 10 bifidobacterial strains, including other B. longum biotype longum strains as well as the closely related B. longum biotype infantis and B. longum biotype suis taxa (232). Results revealed seven large genome regions of variability, the majority of which encompass DNA with a deviating G+C content. These regions correspond to a prophage remnant; a cluster of genes for enzymes involved in sugar metabolism, such as an α-mannosidase; and a capsular polysaccharide biosynthesis gene cluster, which could play a role in host-bacterium interactions (see Fig. S1 in the supplemental material). Though very useful, microarray-based comparative genome analyses suffer from some limitations. It is not possible to identify regions present in the test strains but absent from the strain that was used to construct the array, and it will generally not allow synteny studies.
DNA Regions Acquired by HGT in Bifidobacterial Genomes
It has been suggested that selected genes involved in sugar metabolism as well as in the production of exopolysaccharides in B. longum biotype longum NCC2705 have been acquired via HGT, as part of the adaptation of this organism to a specific ecological niche. For example, a region encoding rhamnosyl transferases seems to have been acquired from streptococci (384), while two other regions that contain genes encoding restriction-modification systems also appear to have been acquired through HGT. Overall, about 5% of the B. longum biotype longum NCC2705 genome content seems to have been recently acquired by this mechanism (384).
Prophage-Like Elements in Bifidobacteria
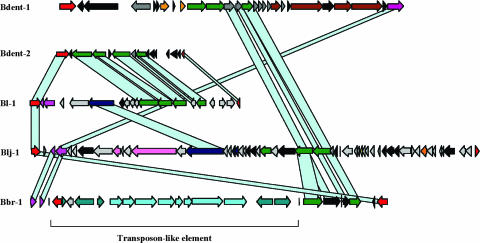
Until recently bifidobacteria were not considered to be suitable targets for phage infection. However, prophage-like elements, designated Bbr-1, Bl-1, and Blj-1, are present in the genomes of B. breve UCC2003, B. longum biotype longum NCC2705, and B. longum biotype longum DJO10A (455). These prophage-like elements display homology to genes of double-stranded DNA phages that infect a broad phylogenetic range of bacteria. Surprisingly, using the proteomic tree method to investigate the evolution of these phages (373), it became clear that the Bbr-1, Bl-1, and Blj-1 prophage-like elements exhibit a close phylogenetic relationship with phages infecting low-G+C bacteria (e.g., lactococcal and staphylococcal phages) (455), perhaps because these bacteria and their phages have shared the same ecological niche (i.e., the animal GIT) during their evolution, thereby allowing DNA exchange. This may therefore point to DNA transfer events between low- and high-G+C bacteria. Perhaps such phages originally infected the ancestor of high-G+C gram-positive bacteria, in line with the concept that high-G+C gram-positive bacteria originated from low-G+C ancestors (455). The unfinished B. dentium Bd1 genome contains at least two prophage-like elements, one of which resembled that of the NCC2705 Bl-1 prophage (Fig. 2). Notably, all three published bifidobacterial prophage-like elements are integrated in a tRNAMet gene, which is the first case of this tRNA gene as a target for phage integration in any gram-positive or gram-negative bacterium (56). Analysis of the distribution of this integration site revealed that the attB sites are well conserved in many bifidobacterial species and in a phylogenetically unrelated bacterium, Thermosynechococcus elongatus BP-1 (306), but surprisingly not in other sequenced Actinobacteria.
FIG. 2.
Comparative genome maps of the prophage-like elements detected in Bifidobacterium genomes. Genes sharing similarity are linked. Probable functions of encoded proteins identified by bioinformatic analysis are indicated. The modular structure is color coded: red, lysogeny; green, DNA packaging and head; blue, tail; mauve, tail fiber; violet, lysis module; yellow, transcriptional regulator; orange, DNA replication; gray, unknown genes; black, genes similar to other functionally unknown bacteriophage genes. Vertical blue lines, tRNA genes.
The 36.9-kb Blj-1 prophage is induced by mitomycin C or hydrogen peroxide and is the first reported inducible and molecularly characterized Bifidobacterium prophage, presenting possibilities for further studies on the biology of bifidophages. Interestingly, the Blj-1 element possesses a putative reverse transcriptase-encoding gene, a homolog of which was shown to represent a diversity-generating retroelement (275, 455).
The Bbr-1 and Bl-1 prophage-like elements appear to be defective prophages, although they may constitute functional satellite phages, whose mobility depends on helper phages in a manner similar to that described for the cryptic mycophages Rv1 and Rv2 (175, 455).
Extrachromosomal DNA Elements
Plasmids are not ubiquitous in bifidobacteria (332, 392), and when present they are small, i.e., ranging from 1.5 kb to 15 kb. Completely sequenced plasmids from different B. longum biotype longum strains include pMB1 (378); pKJ36 and pKJ50 (332, 333); pBLO1 (384); pNAC1, pNAC2, and pNAC3 (95); pDOJH10S and pDOJH10L (260); pTB6 (421); pB44 (GenBank accession number NC004443); and pNAL8 (162). In addition, six plasmids from other bifidobacterial species have been sequenced: pVS809 from Bifidobacterium globosum (285), pCIBb1 from B. breve (327), pNBb1 from B. breve (GenBank accession number E17316), pAP1 from B. asteroides (GenBank accession number Y11549), pBC1 from Bifidobacterium catenulatum (5), and p4M from Bifidobacterium pseudocatenulatum (GenBank accession number NC003527). These plasmids do not encode any obvious phenotypic trait, except for the plasmid isolated from B. bifidum NCFB 1454 (492), which was proposed to encode a bacteriocin, bifidocin B.
Most of the plasmids contain characteristic genetic features for plasmid replication via a rolling-circle replication system, i.e., repB, traA, and mob genes. In contrast, pDOJH10S from B. longum biotype longum DJO10A and pBC1 from B. catenulatum contain sequences homologous to replication functions of theta-type replicating plasmids (5, 260).
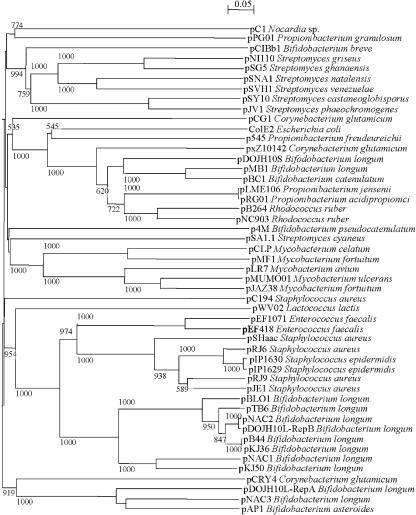
Rep proteins from different bifidobacterial plasmids do not cluster together phylogenetically (110) but resemble replication proteins from different hosts, including gram-negative bacteria such as E. coli (5, 260) (Fig. 3). Horizontal transfer is also indicated for pDOJH10S, which may have been acquired from another Actinobacteria member, possibly Rhodococcus rhodochrous (260).
FIG. 3.
Phylogenetic relationships of Rep proteins from actinobacterial plasmids and several prototype plasmids of different plasmid families from gram-positive and gram-negative bacteria. The phylogenetic tree was calculated by the sequence distance method using the neighbor-joining algorithm.
Bifidobacteria and Carbohydrate Metabolism
Mammalian (including human) biology is partially shaped by the vast community of commensal bacteria that colonize the GIT. Plant-based foods that are commonly consumed by mammals are rich in complex polysaccharides that contain, among others, glucose, fructose, xylan, pectin, and arabinose moieties. Mammalian genomes do not appear to encode the enzymes necessary for degrading most of these glycans (CAZy database; see below), which are supplied instead by the distal GIT microbiome (16). The human GIT microbiome is enriched in genes involved in metabolism of sugars, including glucose, galactose, fructose, arabinose, mannose, and xylose, as well as other sugars that escape digestion by the host's enzymes, including many prebiotic compounds, such as fructooligosaccharides, galactooligosaccharides, glucooligosaccharides, xylooligosaccharides, lactulose, and raffinose (for reviews, see references 148, 161). Gill et al. (148) describe more than 81 different glycoside hydrolase families distributed in a mixture of anaerobic bacteria, i.e., the GIT microbiome, which includes Bifidobacteriales, Clostridiales, Bacteroidales, Enterobacteriales, Fusobacterales, Thermoanaerobacteriales, and Methanobacteria (148, 447. Many of these enzymes are not represented in the human glycobiome. Moreover, GIT mucus provides an abundant reservoir of glycans for microbiota, which serve to reduce the effects of marked changes in the availability of dietary polysaccharides (16).
The type of sugar available is likely to influence the species composition and abundance of the microbiota along the GIT (447). In this context, bacteria such as Lactobacillus are particularly prevalent in the upper GIT, where they mainly ferment relatively simple mono-, di-, and trisaccharides (447). In contrast, bacteria active in the lower parts of the colon, such as bifidobacteria, probably owe their specific ecological success to their capacity to metabolize complex carbohydrates. It therefore comes as no surprise that genes for complex sugar metabolism abound in the genomes of B. breve and B. longum biotype longum. According to the sequence-based classification of carbohydrate-active enzymes (CAZy), over 8% of the annotated genes of these bifidobacterial genomes may encode enzymes involved in the metabolism of carbohydrates, including various glycosyl hydrolases for utilization of diverse, but in most cases not identified, plant-derived dietary fiber or complex carbohydrate structures. Relatively few of these glycosyl hydrolases are predicted to be secreted, including those that are thought to hydrolyze arabinogalactans and arabinoxylans (384). Instead, most of the bifidobacterial glycosyl hydrolases are predicted to be intracellular, and the genes that encode them are almost without exception associated with genes predicted to encode systems for the uptake of structurally diverse carbohydrate substrates (see below). Moreover, carbohydrate-modifying enzymes may also shape the overall metabolic state of the colon to sustain a microbiota that indirectly provides the host with about 10 to 15% of its calories from the degradation of complex carbohydrates through short-chain fatty acids (447).
Bifidobacteria can also utilize sialic acid-containing complex carbohydrates in mucin, glycosphingolipids, and human milk (187, 465). Thus, the mammalian host supplies substrates for intestinal commensals such as bifidobacteria and lactobacilli, in a remarkable symbiotic (or altruistic) relationship (94, 308). Starch and amylopectin are other examples of polysaccharides which may escape digestion in the upper human GIT and which are plant-derived high-molecular-weight carbohydrates. The ability to degrade these sugars appears to be restricted to certain species or to certain strains of a particular species, including B. breve and B. adolescentis (379).
Nearly 10% of the total bifidobacterial gene content is dedicated to sugar internalization, via ABC transporters, permeases, and proton symporters rather than phosphoenolpyruvate-phosphotransferase systems (PEP-PTSs) (384), though a PEP-PTS has been experimentally demonstrated in B. breve to be active for the internalization of glucose (108). The PTS acts through the concomitant internalization and phosphorylation of carbohydrates, in which the transfer of phosphate from PEP to the incoming sugar is mediated via a phosphorylation chain involving enzyme I (EI), histidine-containing protein (HPr), and EII. The B. longum biotype longum NCC2705 genome has a single EII-encoding locus, and that of B. breve UCC2003 has four (287). The latter system was shown to transport fructose, but it appears to transport glucose as well (287). This difference in the number of PEP-PTSs may indicate that B. breve more frequently encounters less complex sugars in its preferred niche, the GITs of infants, than B. longum biotype longum encounters in the GITs of adults, where it is prevalent. Thus, the different diets of infants and adults may affect the compositions of their GIT microbiomes.
Bifidobacteria and Prebiotic Properties
Prebiotics, such as fructo- and galactooligosaccharides, are indigestible food ingredients that beneficially affect the host by selectively stimulating growth of commensal bacteria (36, 370). Bifidobacterial genomes have a rich arsenal of genes for such prebiotic metabolism (116, 176, 218, 259). For example, B. breve UCC2003 has a fos operon encompassing a putative permease-encoding gene, a gene specifying an unknown protein, and a β-fructofuranosidase gene, which has been shown to be involved in fructooligosaccharide degradation (380). Prebiotic oligosaccharides are also provided in human milk (467). These include galactooligosaccharides (325) ranging in degree of polymerization from 3 to over 32 galactose moieties (247). Certain bifidobacteria can also hydrolyze high-molecular-weight prebiotic carbon sources, such as trans-galactooligosaccharides, the latter through an extracellular enzyme encoded by galA (176).
Interaction of Bifidobacteria with the GIT
The molecular basis of interactions with the host epithelium has been investigated in detail for several pathogens such as Listeria monocytogenes and Salmonella spp. (256, 291), but little is known about this for commensal bacteria such as bifidobacteria. Bifidobacterial genome analyses did not reveal clear candidate genes for GIT-bifidobacterial interaction. However, bifidobacteria are predicted to encode cell envelope-associated structures which may play a role in host association. All sequenced bifidobacteria appear to encode an extracellular polysaccharide (EPS) or capsular polysaccharide, and such an extracellular structure may be important in bacterial adherence to host cells, while it could also contribute to resistance to stomach acids and bile salts (338).
The various different EPS clusters present in the commensal microorganism Bacteroides tetaiotamicron help to avoid immune recognition by the host (240). The B. longum biotype longum NCC2705 genome has two regions related to polysaccharide biosynthesis that, like the cps/eps cluster of B. breve UCC2003, are flanked by IS elements and show a strong divergence in G+C content relative to the remainder of the genome. These appear to be a genetic hallmark of cps/eps loci examined thus far (127) and may facilitate inter- and intraspecies transfer of such gene clusters.
Genes predicted to encode glycoprotein-binding fimbria-like structures, which have been identified in the genome sequences of both B. longum biotype longum NCC2705 and B. longum biotype longum DJO10A, may mediate another interaction with the host (232, 384). In addition, B. longum biotype longum NCC2705 encodes a serpin-like protease inhibitor that has been demonstrated to contribute to host interaction in the GIT (199). The NCC2705 serpin is an efficient inhibitor of human neutrophil and pancreatic elastases, whose release by activated neutrophils at the sites of intestinal inflammation represents an interesting mechanism of innate immunity (199).
GENOMICS OF TROPHERYMA
General Features
The only sequenced member of the genus Tropheryma is T. whipplei, the causative agent of Whipple's disease, which is characterized by intestinal malabsorption leading to cachexia and death. T. whipplei isolates are typically found in human intracellular niches, such as inside intestinal macrophages and circulating monocytes (355, 356). However, extracellular and metabolically active T. whipplei cells have been found in the intestinal lumen (130). An environmental reservoir of T. whipplei is also suspected, as PCR experiments revealed its presence in sewage water (283).
Phylogenetic analyses based on the 16S rRNA, 5S rRNA, 23S rRNA, groEL, and rpoB genes placed T. whipplei within the phylum Actinobacteria (282, 479). T. whipplei was difficult to propagate until relatively recently, when cultivation methods using human fibroblasts were established (354). Two T. whipplei strains, TW08/27 and Twist, have been fully sequenced (27, 357). Both strains have a small genome (less than 1 Mb) (Table 1) bearing the traits of strictly host-adapted microorganisms, which include pronounced deficiencies in energy metabolism, dependence on external amino acids, and a lower G+C content (i.e., 46%) than free-living relatives (302).
A large amount of coding capacity is devoted to the biosynthesis of surface-associated features that may sustain the intricate interaction with eukaryotic cells. These surface features include a prominent family of predicted surface proteins termed WiSP (Wnt-induced secreted protein), ranging in size from 103 to 2,308 aa residues. Only a few WiSP family members contain a predicted transmembrane motif near the C terminus that can anchor such proteins to the bacterial membrane. Alignment of all the WiSP members revealed the presence of a single β-strand motif (27).
The two T. whipplei genomes contain many noncoding repetitive DNA regions, which may promote recombination events that allow the bacteria to expose different sets of proteins at their surface, possibly in response to host defense actions and/or specific environmental conditions (27, 357). All these genome characteristics are discussed in detail below. It is noteworthy that the genomes of T. whipplei Twist and TW08/27 contain 808 and 784 coding sequences (CDSs), respectively, with only a small number of pseudogenes. This apparent low degree of gene decay, which contrasts with the conspicuous gene decay of M. leprae (see below), may be related to the complete absence of mobile genetic elements within the genomes, or it may mean that most redundant DNA has already been removed.
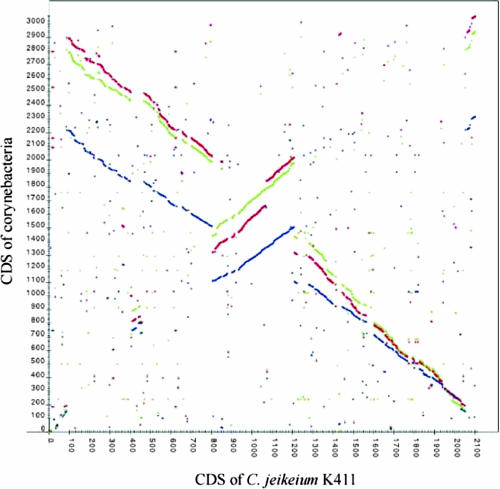
Tropheryma Comparative Genome Analysis
The two available T. whipplei genomic sequences are >99% identical but differ by a large chromosomal inversion (Fig. 4). The extremities of this inversion include two identical nucleotide sequences corresponding to the WND domain of the WiSPs. Consequently, the inversion event caused differences in the WiSPs in the two strains: TW08/27 has eight copies of WND domain sequences that are identical across an 800-bp nucleotide span, whereas the rest of these WiSP genes do not display any DNA similarity. This suggests that WND motifs act both as coding regions and as DNA repeats to promote genome recombination (357).
FIG. 4.
Circular map of genome diversity found in Tropheryma. From inside to outside: ring 1, GC deviation; ring 2, G+C content; ring 3, atlas of T. whipplei strain TW08/27; ring 4, comparison to the genome sequences of T. whipplei Twist. Green indicates homologies of >95%. The synteny plot comparing the order of homologous genes in sequenced genomes of Tropheryma is depicted in the panel inside the circular map.
The comparison of T. whipplei genome sequences with those of other reduced bacterial genomes (less than 1 Mb), such as Mycoplasma species, Ureaplasma, and Buchnera revealed a reduced complement repertoire of genes for most functional categories (357). However, mycoplasmas are genetically better equipped for carbohydrate metabolism and transport, and Buchnera displays a larger gene content devolved to energy production and conversion. This variability shows that small bacterial genomes did not necessarily follow the same reductive evolutionary pathway.
DNA Region Acquired by HGT in T. whipplei Genomes
HGT events appear to be less frequent for intracellular bacteria with small genomes than for free-living bacteria (27, 40, 321, 357). In T. whipplei Twist, only nine genes, about 1% of the entire genome, appear to have been acquired by HGT. These encompass aminoacyl-tRNA synthetase-encoding genes, genes involved in nucleotide metabolism (purB and pyrB), and genes specifying hypothetical proteins (357).
Possibly, the comparatively nonpromiscuous lifestyles of intracellular bacteria do not offer extensive opportunities to exchange DNA with other bacteria. The absence of mobile elements is also consistent with the notion that T. whipplei resides in an isolated niche, being sheltered from foreign bacterial DNA.
Tropheryma Genome and Biological Lifestyle
Metabolic reconstruction of T. whipplei from genome data indicates the absence of biosynthetic pathways for arginine, tryptophan, and histidine biosynthesis and incomplete pathways for the synthesis of glycine, serine, leucine, and cysteine. There are also deficiencies in cofactor biosynthesis, energy metabolism, and carbohydrate metabolism. The absence of genes required for prototrophic growth is another indicator of the reliance of these microorganisms on their host for various compounds. Nevertheless, among intracellular microorganisms with reduced genomes, T. whipplei has the most complete biosynthetic pathways for purine and pyrimidine nucleotides, fatty acids, several cofactors, and other small biomolecules. Like Buchnera, T. whipplei genomes have maintained about half of the amino acid biosynthetic pathways. In Buchnera the retained metabolic pathways correspond to those necessary for the synthesis of amino acids essential for their insect hosts. However, since no such symbiotic association is known for T. whipplei, its retained biosynthetic capacity is probably a reflection of the amino acids that are in limited supply in its natural environment or host. All this genome-acquired knowledge allowed the formulation, through computer modeling of the T. whipplei metabolic networks, of a comprehensive culture medium that supported axenic growth of this organism (366). Acquisition of iron is crucially important for bacterial pathogens in the iron-depleted host environment. The genome survey of T. whipplei identified a gene cluster predicted to be involved in ferri-siderophore uptake. However, apart from the presence of a homolog of the gene for the mycobacterial iron dependent regulatory protein IdeR, no genes with clear homology to known siderophore genes have been identified, indicating that T. whipplei might only be able to scavenge xenosiderophores (27).
Interaction of Tropheryma with the Environment
Almost 15% of the T. whipplei predicted proteins and 74% of the hypothetical proteins are expected to be exported from the cell or associated with the cell envelope. The assignment of so many extracellular proteins may be a reflection of the importance of host interactions to the organism. The T. whipplei WiSPs (see above) resemble proteins known to be involved in pathogenesis and host-immune evasion, such as the Staphylococcus aureus Bap protein, a biofilm-associated protein that projects from the cell surface, allowing interactions with adjacent surfaces (100).
GENOMICS OF PROPIONIBACTERIUM
General Features
Currently, only one complete propionibacterial genome sequence, that of Propionibacterium acnes strain KPA171202, is publicly available (46). P. acnes is a non-spore-forming, anaerobic, pleomorphic rod whose end products of fermentation include propionic acid. The organism belongs to the human cutaneous propionibacteria, along with P. avidum, P. granulosum, P. innocuum, and P. propionibacterium. P. acnes is ubiquitous on human skin, preferably within sebaceous follicles, where it is generally a harmless commensal. Nevertheless, P. acnes is an opportunistic pathogen (180, 197). It has been isolated from sites of infection and inflammation ranging from acne to diverse other conditions such as corneal ulcers, endocarditis, synovitis, pulmonary angitis, hyperostosis, endophthalmitis, and osteitis (SAPHO) syndrome (99, 193, 203). P. acnes grows slowly and can resist phagocytosis and persist intracellularly within macrophages (472). This resistance to phagocytosis may be conferred by a complex cell wall structure, which also includes a surface fibrillar layer (301).
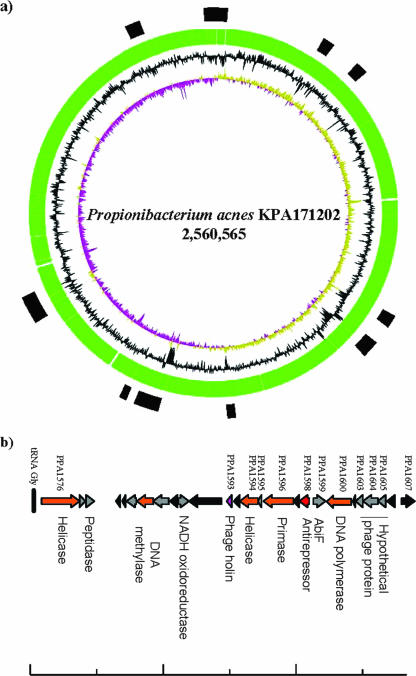
The circular 2.5-Mb chromosome of P. acnes KPA171202 (DSM16379) contains 2,333 predicted genes and 35 pseudogenes (Table 1). A function has been assigned for around 70% of the identified genes.
Data concerning the relatedness of the skin isolate KPA171202 to other P. acnes isolates are limited to just a few genes, including the 16S rRNA, gehA, groEL, and dnaK genes. Such analyses are not very informative, since only limited variability exists between homologs of these genes at the DNA level. Comparative genome analyses with closely related genera, such as Mycobacterium, Streptomyces, and Corynebacterium, revealed that the closest related genome is that of Streptomyces avermitilis. However, genomic synteny between P. acnes and S. avermitilis is limited to just a few gene clusters (46).
Extrachromosomal DNA Elements in Propionibacterium
Endogenous small plasmids, of 6 to 10 kb, have been identified in a few Propionibacterium species, i.e., P. acidipropionici, P. jensenii, P. granulosum, and P. freudenreichii (363). Only four have been sequenced: pRGO1 from P. acidipropionici (224), p545 from P. freudenreichii (211), the cryptic plasmid pPGO1 from P. granulosum (NCBI source NC_004526), and pLME106 from P. jensenii (NCBI source NC_005705). Two genes, repA and repB, encoding putative replication proteins similar to those of ColE theta-type replicating plasmids, were found in all but pPGO1, indicating that a similar replication mechanism is used by many Propionibacterium plasmids (Fig. 3).
DNA Region Acquired by HGT
A survey of the P. acnes genome for DNA regions that show an uncommon codon usage as well as a deviation in G+C content highlighted 10 regions that may have been acquired by HGT (45) (Fig. 5a). These include a cryptic prophage (see below), a cluster of genes predicted to be involved in conjugal DNA transfer, and a putative lanthionine biosynthesis cluster. Various other suspected HGT-acquired DNA regions, apart from those that specify predicted PTS-mediated substrate uptake systems, are assumed to express virulence traits. These include genes specifying factors for iron acquisition and adhesion, as well as hemolysin/cytotoxin factors. Another interesting alien DNA region is a 43-gene cluster predicted to encode a nonribosomal peptide synthetase similar to a nonribosomal peptide synthetase system of a Streptomyces species that enhances the biological fitness of the bacterium (62).
FIG. 5.
(a) Genome map of Propionibacterium acnes KPA171202. The genome variability regions are indicated by black boxes. (b) Genome map of the P. acnes KPA171202 Pro-1 prophage. Probable functions of encoded proteins indicated by bioinformatics analysis are noted.
Prophage-Like Elements in Propionibacterium
P. acnes KPA171202 contains a single, apparently defective, prophage-like element, named Pro-1 (57). The small number of phage-related genes represent functions (e.g., an antirepressor, holin, helicase, DNA polymerase, and primase) found in bacteriophages infecting low-G+C bacteria (Enterococcus, Streptococcus, Clostridium, and Lactobacillus) (Fig. 5b). These phage-related genes are not organized in the modular structure typical of lambdoid phages (35), but are dispersed among genes displaying classical bacterial origins, such as genes encoding an NADH oxidoreductase and a peptidase. Interestingly, Pro-1 carries a gene (abiF) encoding a protein resembling a Lactococcus lactis phage defense system that interferes with intracellular phage multiplication (for a review, see reference 72). The Pro-1 element is flanked on one site by a glycine tRNA gene, although no clear attachment sites can be distinguished in the flanking phage sequences. Based on its genetic structure, it seems that Pro-1 has been subject to intensive genome reshuffling and decay, indicating that the integration of this prophage was not a recent event.
P. acnes Genome and Biological Lifestyle
The genome sequence of P. acnes reflects its presence and activity as a ubiquitous commensal on human skin. Metabolic reconstruction revealed the capacity of P. acnes to cope with varying oxygen availability, in accordance with its growth under microaerobic as well as anaerobic conditions (96, 156). The genome encodes all key components of oxidative phosphorylation, employing two terminal oxidases, a cytochrome aa3 oxidase, a cytochrome d oxidase whose E. coli homolog predominates when cells are grown at low aeration, and an F0F1-type ATP synthase (46). All genes of the Embden-Meyerhof and pentose phosphate pathways are present. Under anaerobic conditions P. acnes grows on different carbon sources, including glucose, ribose, fructose, mannitol, trehalose, mannose, N-acetylglucosamine, and glycerol (46). Fermentation products are short-chain fatty acids, especially propionic acid. P. acnes can also mobilize metabolic capabilities such as nitrate reductase, dimethyl sulfoxide reductase, and fumarate reductase to allow anaerobic respiration.
Various P. acnes gene products can degrade and use host-derived substances. Before the genome decipherment, knowledge of the capacity of P. acnes to use skin tissue was limited to a secreted extracellular triacylglycerol lipase, GehA, isolated from P. acnes P-37 (294). This enzyme degrades skin lipids, such as sebum, which may be a crucial activity for skin colonization. Furthermore, it was proposed that free fatty acids, released by P. acnes lipase activity on sebum, assist bacterial adherence and colonization of the sebaceous follicle (155). As expected, the genome of P. acnes KPA171202 contains a gehA homolog, while it also contains other genes that encode predicted extracellular (secreted and cell wall-bound) lipases.
The degradation of host tissues is also facilitated by hyaluronate lyase, which acts on a key constituent of the extracellular matrix of connective tissues (408). The P. acnes genome encodes such an enzyme, as well as numerous additional enzymatic activities with suspected roles in host tissue degradation, such as two endoglycoceramidases and four sialidases, a putative endo-β-N-acetylglucosaminidase, and various extracellular peptidases. There are also genes specifying homologs of CAMP factors, which are typically found in pathogenic staphylococci (140). The CAMP reaction causes synergistic lysis of erythrocytes due to the interaction of the CAMP factor with the Staphylococcus aureus sphingomyelinase C. CAMP factors can bind to the Fc fragment of immunoglobulins of the immunoglobulin G and immunoglobulin M classes (140).
Interaction of P. acnes with Its Environment
The capacity of P. acnes to modulate the immune system has been thoroughly studied. Increased cellular and humoral immunity to P. acnes has been detected in patients with severe acne (209, 237). The P. acnes genome encodes various cell surface or surface-exposed proteins that may exhibit cell-adherent properties. Many of these possess a C-terminal LPXTG-type cell wall-sorting signal required for binding of surface proteins to the cell wall through the action of a so-called sortase (92). Three genes within the P. acnes genome possess contiguous stretches of 12 to 16 guanine or cytosine residues, distributed either in the promoter or in the coding region. The sequences within these regions were ambiguous with respect to the length of the poly(C/G) stretch (46). Such variable homopolymeric C or G stretches, which are generated by slipped-strand mispairing during replication, have been reported to be involved in phase variation, an adaptive strategy commonly noticed in bacterial pathogens (46). P. acnes has a lipoglycan-based cell wall envelope (474), which may well play a role in adherence to skin and in the formation of a biofilm matrix (53). Furthermore, the genome of P. acnes contains three clusters involved in EPS biosynthesis. All these extracellular structures may modulate immunogenicity towards the microorganism and/or may constitute a barrier against antimicrobial compounds.
P. acnes abundantly produces porphyrins, which might contribute to skin damage (156). The interaction of porphyrins with oxygen is thought to contribute to keratinocyte damage and consequently to have implications regarding the pathogenesis of progressive macular hypomelanosis (473). However, it is known that P. acnes can be eradicated by illumination with intense blue light, which induces photoexcitation of bacterial porphyrins, singlet oxygen production, and thus bacterial destruction (14). The P. acnes genome contains two clusters of 26 and 8 genes, respectively, which are involved in vitamin B12 biosynthesis (46).
GENOMICS OF MYCOBACTERIUM
General Features
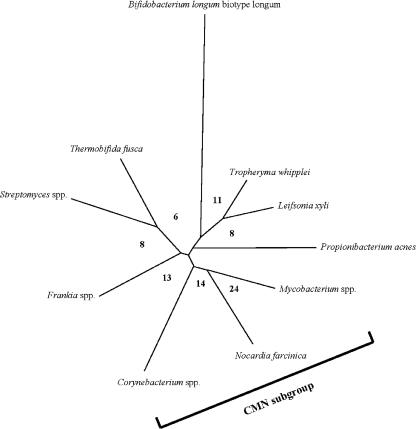
The genera Corynebacterium, Mycobacterium, and Nocardia form a monophyletic taxon, the so-called CMN group, within the Actinobacteria (119). These bacteria share an unusual cell envelope composition, characterized by the presence of a waxy cell envelope containing mycolic acids, conferring alcohol and acid-fast staining properties on these bacteria which distinguish them from other bacteria.
The genus Mycobacterium is highly diverse and comprises 85 different species, which have been identified since the isolation of M. leprae in 1873 (358). In addition, there are a number of Mycobacterium bovis bacille Calmette-Guérin (BCG) vaccine substrains, which are derived from an attenuated M. bovis strain obtained in 1921 (24). Finally, individual Mycobacterium species, such as M. tuberculosis, display great diversity (219). Generally, mycobacteria are free-living saprophytes (121) and are well adapted to different habitats, such as soil (485) and aquatic environments (87). A few species, such as M. bovis and M. tuberculosis, first identified in infected animals, have never been isolated from other environments, suggesting that they are obligate parasites of humans and animals (86). Nevertheless, caution should be taken in drawing such a conclusion: the pathogenic M. ulcerans has also been isolated as a soil inhabitant in symbiosis with roots of certain plants present in tropical rain forests or similar environments (174).
Mycobacteria are the causative agents of a broad epidemiological, clinical, and pathological spectrum of diseases in humans. Mycobacterial diseases are very often associated with immunocompromised patients, especially AIDS patients. M. tuberculosis and related species, such as M. bovis, cause tuberculosis, surviving within macrophages. M. tuberculosis may primarily cause pulmonary disease, although organs other than lungs may be affected. M. leprae causes leprosy, living within Schwann cells and macrophages to give rise to a chronic granulomatous disease of the skin and peripheral nerves (201). M. ulcerans is the third most common mycobacterial disease (443). However, in contrast to the other mycobacteria, M. ulcerans grows outside of its host cells, and its pathogenicity is attributed to the secretion of a toxin. The M. ulcerans-mediated chronic disease results in painless, expanding skin ulcers. Many other environmental mycobacteria (e.g., M. avium) may on occasion cause localized or disseminated clinical illness such as lymphadenitis (121).
Due to their clinical importance, genome sequences of several mycobacterial species have been determined (Table 1). These include M. tuberculosis and M. leprae (83, 84, 126); M. bovis, which causes bovine tuberculosis (139); and M. avium subsp. paratuberculosis, the agent of Johne's disease in cattle (271). Additional ongoing mycobacterial genome sequencing projects include those for three undetermined mycobacterial species (NCBI sources NC_008146, NZ_AAQC00000000, and NZ_AAQD00000000), M. flavescens PYR-GCK (NCBI source NZ_AAQ00000000), M. tuberculosis C (NCBI source NZ_AAKR00000000), M. tuberculosis F11 (NCBI source NZ_AAIX00000000), M. tuberculosis strain Haarlem (NCBI source NZ_AASN00000000), and M. vanbaalenii PYR-1 (NCBI source NZ_AAPF00000000). Here we focus on the published and completely sequenced mycobacterial genomes (84, 126, 139, 271).
Genomics of M. tuberculosis
The first mycobacterial genome sequence to be determined, that of M. tuberculosis H37Rv, consists of a 4.41-Mb circular chromosome encoding 3,924 proteins (Table 1) (84). The genome sequence of second strain, M. tuberculosis CDC1551, which causes widespread skin test conversion in rural parts of the United States (441), has also been published (126). This genome, though slightly smaller, is nearly identical (99.94%) to that of H37Rv (Table 1).
In contrast to the situation described for fast-growing bacteria such as B. subtilis, the orientation of genes with respect to the direction of replication is less biased (59% of them are transcribed with the same polarity as the replication forks, instead of 75% in B. subtilis [246]). It is believed that higher expression levels can be achieved by coordinating directions of transcription and replication, and it is therefore assumed that the observed gene orientation in M. tuberculosis is in concordance with its low growth rate.
M. tuberculosis genome architecture.
More than 50% of the identified M. tuberculosis H37Rv genes appear to have been subjected to gene duplication or domain-shuffling (432), while 3.6% of the genome is occupied by IS elements located at 56 loci (153). Furthermore, a novel repeated sequence belonging to the REP13E12 family is present in seven copies on the H37Rv chromosome, and part of this sequence can be used as the integration site of phage phiRvi1. Notably, in M. tuberculosis strains Erdman and CDC1551, this phage is integrated in a different copy of REP13E12 compared to that of strain H37Rv (83). Another IS element that has dramatically affected the genome shape of M. tuberculosis strains is IS6110, an IS3-like element, which has been used as an epidemiological tool due to its nisin-induced transposition frequency, which generates observable genetic variability among isolated strains (e.g., H37Rv possesses 16 copies, while CDC1551 has just 4 copies) (445). Transposition causes both gene inactivation through insertion and genome decay as a consequence of recombination-mediated deletion between two copies of the element (122). M. tuberculosis H37Rv possesses four loci that could potentially be removed by such a mechanism (41). One of these loci was indeed absent from the genomes of some M. tuberculosis strains, thus constituting a hot spot for genome variability (179).
The H37Rv genome also contains about 65 copies of a novel dispersed repeat named the mycobacterial interspersed repetitive unit (MIRU) (131, 412), which ranges in size from 46 to 101 bp. Generally, MIRUs occur at the 5′ ends of genes, whereas MIRU copies located between genes in operons are often predicted to encode peptides and appear to have inserted into such positions so as to allow translational coupling (412).
M. tuberculosis genome and biological lifestyle.
M. tuberculosis contains a remarkable mixture of polyketides, lipoglycans, lipids, and glycolipids in its waxy envelope layers (101). The M. tuberculosis genome sequences revealed genes that sustain the production of all of the above-mentioned molecules. An example of genome adaptation of M. tuberculosis to its ecological niche is the presence of large gene clusters that confer lipolytic functions; in host tissues, lipid substrates are more abundant than carbohydrate substrates. In addition to genes for the prototype β-oxidation cycle, the M. tuberculosis genome contains about 100 genes for enzymes involved in alternative lipid oxidation pathways, in which exogenous lipids are metabolized following the degradation of host cell membranes (83). The derived acetyl coenzyme A (acetyl-CoA) can then be used for the synthesis of mycobacterial cell wall components or utilized for other metabolic pathways (e.g., the Krebs cycle or glyoxylate shunt).
In addition to lipolysis, energy may also be generated through hydrolysis of a wide variety of carbohydrates, alcohols, ketones, and other hydrocarbon compounds. Genome pathway reconstruction has identified all genes necessary for the glycolytic and pentose phosphate pathways, as well as a large arsenal of genes for putative oxidoreductases, oxygenases, and dehydrogenases, thereby allowing the metabolism of other carbon sources. Notably, the H37Rv genome contains a gene for a cytochrome family member that may catalyze the introduction of oxygen groups into organic molecules, perhaps as a mechanism to degrade organic matter (11). This enzyme is commonly found in the genomes of bacterial soil inhabitants, and its presence in the M. tuberculosis genome indicates that, like many other mycobacteria, the tubercle bacillus may have occupied this niche before evolving as an obligate pathogen.
The genome of M. tuberculosis contains all the genes required for oxidative phosphorylation under aerobic conditions. However, analysis of the genome suggests that M. tuberculosis can also respire anaerobically using alternative terminal electron acceptors (e.g., fumarate or nitrite) (471). This may allow growth of M. tuberculosis under conditions of limited oxygen availability in abscesses and granulomas.
About 10% of the predicted coding capacity of the genome of M. tuberculosis is dedicated to two large and unrelated novel families of acidic, glycine-rich proteins, the PE and PPE families, whose genes are clustered and often present in multiple copies (82, 83). Of particular interest are the PE proteins and PPE proteins belonging to the major polymorphic tandem repeat (MPTR) subfamily or the polymorphic G+C-rich sequence (PGRS) subfamily (Fig. 6). PGRS proteins contain almost 50% glycine, in tandem repetitions of the motif Asn-Gly-Gly-Ala-Gly-Gly-Ala or variants thereof. Some MPTR proteins display multiple repetition of the motif Asn-X-Gly-X-Gly-Asn-X-Gly. Both the PGRS and MPTR subfamilies include surface proteins, some of which may also possess antigen properties (74, 109). The PPE-PPW subfamily (3) has a characteristic conserved 44-amino-acid region in the C terminus containing Gly-Phe-X-Gly-Thr and Pro-X-X-Trp motifs (Fig. 6). Members of the last PPE subfamily, the PPE-SVP proteins, have low homology at the C terminus and are characterized by the motif Gly-X-X-Ser-Val-Pro-X-X-Trp (446) (Fig. 6). Several members of these subclasses appear to be involved in pathogenesis (55, 271, 353), while others act as adhesins and influence phagocytosis (83).
FIG. 6.
(a and b) Diagrammatic representation of the gene structures of the members of the PE and PPE gene family, displaying conserved 5′-end domains, motif positions, and differences between different subfamilies found in the two families modified by van Pittius et al. (446). (c) Alignment of the region surrounding the SVP motif Gly-XXSer-Val-Pro-XX-Trp in the members of the PPE-SVP subfamily. (d) Alignment of the region surrounding the GFGT motif (Gly-Phe-X-Gly-Thr) and the PPW motif (Pro-XX-Pro-XX-Trp) in the members of the PPE-PPW subfamily. (Modified from reference 446 with permission from BioMed Central.)
Comparative genomics within the M. tuberculosis complex.
The H37Rv and CDC1551 sequences revealed an unexpectedly high level of polymorphism (126): the H37Rv genome contains 37 insertions (greater than 10 bp) relative to strain CDC1551. Moreover, a different repertoire of IS elements was identified in these genomes, particularly for IS6110 (see above) (122). While H37Rv contained several deletions associated with a possible IS6110 recombination mechanism, none of the deletions described for CDC1551 appeared to be the result of such mechanism.
Whole-genome comparison between CDC1551 and H37Rv also highlights the presence of large sequence variation in several genes, such as those encoding a phospholipase C, a membrane lipoprotein, and clustered members of an adenylate cyclase gene family, which in the H37Rv and CDC1551 genomes include three and four genes, respectively (see Fig. S2 in the supplemental material). One of the clustered adenylate cyclase-encoding genes in H37Rv is an in-frame chimera of the 3′ and 5′ ends of two adjacent genes in the CDC1551 genome, generated by a deletion-fusion event, thus suggesting an ancestral structure of four tandemly organized genes (126).
Several genes, including members of the PE/PPE gene family, have significantly higher synonymous and nonsynonymous substitution frequencies than the genome as a whole. Notably, the ratio of nonsynonymous to synonymous substitutions in M. tuberculosis is approximately 1.6, much lower than those in housekeeping genes in E. coli and Salmonella enterica, in which they range from 4 to 17 (37, 466).
The low ratio observed in M. tuberculosis indicates either additional selective pressure in favor of synonymous substitutions or decreased selection against nonsynonymous substitutions (126).
The genomes of 100 epidemiologically well-characterized M. tuberculosis clinical isolates have been investigated with DNA microarrays (438). This analysis highlighted 68 different large sequence polymorphisms, representing 4.2% of the genome, which are present in H37Rv but absent from one or more clinical isolates. Deletions were found to be clustered in the genome, and further analysis suggested two distinct causes. Some deletion clusters appeared to be specific to a single mycobacterial lineage, whereas other clusters seemed to indicate the presence of regions of genomic vulnerability throughout the species. Some deletions may offer short-term advantages, such as escape from the host immune system, whereas others could reduce the load of mobile elements, such as prophages; others again could offer strong advantages such as antibiotic resistance.
Interestingly, genes encoding proteins associated with pathogenesis in M. tuberculosis, such as the genes coding the Snm protein secretion system, are critical for mediating interactions during infection to allow pathogen survival in the hostile environment and are conserved among all mycobacteria, including the nonpathogenic saprophyte Mycobacterium smegmatis (93). Furthermore, only a few genes that make up the pathogenicity island (Rv0298 to Rv0303, Rv0323c to Rv0331, Rv0656c to Rv0666, Rv1041 to Rv1055c, Rv2302 to Rv2312, Rv2801c to Rv2824c, Rv2954c to Rv2961, Rv3108 to Rv3126c, and Rv3173c to Rv3191c) are present in the genomes of members of the M. tuberculosis complex or in a few phylogenetically closely related groups (e.g., Corynebacterium or Rhodococcus) (22).
Prophage-like elements in M. tuberculosis.
The two sequenced M. tuberculosis strains contain two prophage-like elements (each approximately 10 kb), phiRv1 and phiRv2. The phiRv1 element is predicted to encode three head proteins, a primase, and an integrase of a serine recombinase family that catalyzes integration and excision (an adjacent small ORF controls the directionality of such a recombination event). The phiRv2 element is similarly organized, except for the presence of an IS element (175).
Much more is known about inducible prophages from nonsequenced M. tuberculosis strains. This knowledge has been used to develop genetic tools, e.g., integration vectors, for the manipulation of mycobacteria (261). The most intensely studied mycobacterial phage is L5. The genome of this temperate phage consists of a left part, containing a structural gene cluster, which shares similarity with many dairy phages and lambdoid coliphages (172). The right part of the L5 genome contains the lysogeny module. Notably, in this phage lysogeny is established by a phage repressor that requires stabilization by a protein encoded by an adjacent ORF (310). The L5 repressor regulates transcription at an early lytic promoter, but it also affects gene expression of the phage genome through binding to several so-called stoperator sites located within short intergenic spaces in both early and late lytic operons (44). Other well-studied temperate mycobacteriophages include phages D29 and Bxb1, whose overall genome organizations are very similar, though sequence similarity is patchy and in general does not exceed 60% amino acid identity. Interestingly, Bxb1 specifies many enzymes that could degrade or modify the mycobacterial cell wall (290).
Genomics of M. bovis
The only sequenced M. bovis genome, that of strain AF2122/97, is a circular 4.4-Mb chromosome encoding 3,952 proteins (Table 1) (139). Although its genome shares over 99.9% DNA sequence identity with the other members of the M. tuberculosis complex, M. bovis can be differentiated on the basis of its different host range and virulence and a few other distinctive phenotypes.
M. bovis genome architecture.
Deletion of genetic information is the dominant trend in M. bovis, which has many pseudogenes (139). These pseudogenes resemble intact genes that are involved in transport and cell surface structures (e.g., pstB, ugpA, mce3A to -F, lppO, lpqG, lprM, pks6, mmpL1, and mmpL9), detoxification (e.g., ephA, ephF, and alkA), intermediary metabolism (e.g., epiA and gmdA), fatty acid metabolism (e.g., fadE22 and echA1), and cofactor biosynthesis (e.g., moaE and moaC2). M. bovis (and M. leprae) also lacks the AtsA system for the hydrolysis of sulfate esters to recycle sulfate (190).
M. bovis genome and biological lifestyle.
The M. bovis cell wall contains phenolic glycolipids that are absent from M. tuberculosis. Consistent with this, the M. bovis genome contains a TbD1 locus consisting of the mmp genes, which specify a family of membrane-spanning proteins involved in the export of the phenolic cell wall glycolipids.
Another key feature of M. bovis is a requirement for pyruvate when glycerol is the sole carbon source (468). In M. bovis, glpK, which encodes a glycerol kinase, is a pseudogene, preventing the phosphorylation of glycerol and therefore its use as a carbon source (139). Other genes that may influence the biological lifestyle of M. bovis are those encoding antigens. The cell surface-located antigen repertoire specified by a (pathogenic) microorganism may reflect its immune modulatory strategy. The M. bovis genome encodes a group of antigens, the ESAT-6 family, which were originally described as T-cell antigens secreted by M. tuberculosis (401) and belong to a large family that contains other T-cell antigens such as CPF-7 and CPF-10 (432). The demonstration of an interaction between ESAT-6 and CPF-10 suggests that other members of the family may also act in pairs, possibly in a mix-and-match arrangement (367). However, six members of the ESAT-6 family in M. tuberculosis are absent from the genome of M. bovis. The effects of the absence of these proteins are difficult to predict, although it may affect the function of other members if they act in combination (139).
Comparative Genomics of M. bovis and M. tuberculosis
The deletion of segments ranging from 1 to 12.7 kb compared with other members of the M. tuberculosis complex affects a wide range of metabolic functions and putative virulence factors (24, 153). For example, the loss of a DNA region encompassing three phospholipase C-encoding genes, which are known virulence factors in Clostridium and Listeria species (434), is expected to affect pathogenesis.
There are 2,437 single-nucleotide polymorphisms (SNPs) between the genomes of M. bovis and M. tuberculosis H37Rv and 2,423 SNPs when the M. bovis genome is compared with that of M. tuberculosis CDC1551 (139). Some of these are in genes that may have a crucial role in the biology of the organism. For example, the SNP in the pncA gene in M. bovis confers resistance to the key antituberculosis drug pyrazinamide and prevents accumulation of niacin as observed in M. tuberculosis (498). Moreover, a single base change in the principal sigma factor gene may also be sufficient to attenuate M. bovis (88).
Genomic deletions have significantly contributed to the evolution of other, relatively recently clonal organisms (322), in particular those showing a species-specific host dependence (231). The analysis of genome variability within M. bovis and M. bovis-like mycobacteria (e.g., M. pinnipedii, M. caprae, and oryx bacillus) (305) revealed many sequence polymorphisms in predicted virulence genes. In the case of the M. tuberculosis complex, these regions have proven to be very informative both for the identification of genes that vary between strains (438) and for the identification of molecular signatures that differentiate members of the complex (41, 305). From these investigations emerged the notion of geographically defined and host-restricted forms of the M. tuberculosis complex (178, 305). Specific genomic deletion profiles appear to be restricted to well-defined host types. Since host adaptation is not uncommon for pathogens (486), it is plausible that genomic deletions are responsible for the observed differences in host range among members of the M. tuberculosis complex. Genomic analyses have consistently described M. bovis as the furthest derived M. tuberculosis complex member (41, 305). Moreover, based on the deleted regions within the genomes of M. bovis and M. tuberculosis-like organisms, it has been possible to obtain insights into the evolutionary development of the M. tuberculosis complex (Fig. 7) (305).
FIG. 7.
Phylogeny of the M. tuberculosis complex, based on deleted regions as indicated by genomic analysis. Clustered along the vertical axis are organisms for which one or more genomic deletions specific for this evolutionary branch have been observed. The functions of the genes comprising the deleted regions are provided. (Modified from reference 305 with permission.)
Genomics of M. leprae
The genome of M. leprae is the smallest sequenced among mycobacteria (Table 1). Its coding capacity is restricted to only 49.5% of its genome, while recognizable pseudogenes occupy 27% (85). The pseudogenes appear to have lost their function as a result of one or more mutations, including in-frame stop codons, frameshifts, deletions, and insertions.
Pseudogenes appear to be randomly distributed within the M. leprae genome, whereas the identified 1,605 functional genes are in clusters surrounded by long noncoding regions. The process of gene deletion and decay is accompanied by a reduction in the percent G+C content: M. leprae has the lowest G+C content of all mycobacteria. The G+C content of the intact genes (60.1%) is higher than that of the pseudogenes (56.5%), which in turn is higher than that of intergenic regions (54.5%). These findings indicate that the high G+C content of M. leprae, and more generally those of the other mycobacteria, is due to codon preference of the protein-specifying DNA regions. Deamination of cytosine residues in the DNA to thymidine may account for the trend of decaying genomes towards lower G+C content. It is generally true that bacterial genomes that have undergone massive gene decay are richer in adenine and thymine (420). Assuming that the genome of M. leprae was once topologically equivalent and similar in size to those of other mycobacteria (e.g., around 4.4 Mb), then extensive downsizing and rearrangement must have occurred during evolution of this species. Such reductive evolution is most probably associated with the obligate intracellular habitat of M. leprae: reductive evolution has been documented in several obligate intracellular pathogens and in some endosymbionts (see the introduction). It is thought that genes become inactivated once their functions are no longer required in highly specialized niches. This process may have naturally defined the minimal gene set for a pathogenic mycobacterium. Until the genome sequence of M. leprae became available, the most extensive genome degradation reported in a pathogen was in Rickettsia prowazekii, which causes the typhus Brill's disease. In this organism, only 76% of the potential coding capacity is used (9). However, compared to that in M. leprae, the level of gene loss in R. prowazekii is modest. It is remarkable that elimination of pseudogenes by deletion lags far behind gene inactivation in both pathogens, in contrast to what has been found in the aphid endosymbionts Buchnera spp. (394). The mechanism driving pseudogene formation in M. leprae is unknown, although the loss of dnaQ-mediated proofreading activities of DNA polymerase III must have contributed to this (85). Furthermore, genome analyses revealed convincing cases of large-scale rearrangements and deletions that would have arisen from homologous recombination events. For example, 2% of the M. leprae genome is composed of repetitive DNA that is believed to have promoted such DNA rearrangements (85). Notably, comparative genome analysis of M. leprae and M. tuberculosis produced 65 DNA regions that indicated synteny. Breaks in synteny generally correspond to dispersed repeats, tRNAs, or gene-poor regions (85). Copies of all four principal repeats, RLEP (37 copies), REPLEP (15 copies), LEPREP (8 copies), and LEPRPT (5 copies), occur at the junctions of discontinuity, suggesting that the mosaic arrangement of the M. leprae genome reflects multiple recombination events between related repetitive sequences. Notably, although there is very little similarity to known transposable elements, RLEPs occur mainly at the 3′ ends of genes and in several cases within pseudogenes, suggesting that this element was once capable of transposition.
The organism with which M. leprae could possibly exchange DNA is its human (or another ancestral mammalian) host. Interestingly, and in contrast to all other analyzed mycobacteria, the domain structure of M. leprae prolyl-tRNA synthetase is eukaryotic-like, and the corresponding M. leprae proS gene is both displaced and inverted with respect to the M. tuberculosis genome, consistent with a recent acquisition through HGT (85).
Other mobile agents of HGT are prophages. M. leprae possesses just three prophage-like genes, all with M. tuberculosis orthologs (57), whereas the M. tuberculosis complex contains two prophage-like elements (see above). M. leprae has discarded genes that can be compensated for by the assumption of a host-dependent parasitic lifestyle, while preserving or acquiring genes required for host transmission, establishment, and survival. The availability of multiple mycobacterial genome sequences has allowed the genetic dissection of conserved and dissimilar pathways, such as those for lipolysis and those sustaining pathogenesis.
As in M. tuberculosis, the largest protein family in M. leprae represents enzymes involved in polyketide synthesis and fatty acid metabolism. However, this enzyme repertoire is much less extensive than that found in M. tuberculosis, which possesses a cell envelope displaying a greater diversity of lipids, glycolipids, and carbohydrates (101). The M. leprae genome encodes only 12 members belonging to the PE or PPE family, all of which lack the C-terminal repetitions that are implicated in antigenic variation (38, 402). Entire metabolic pathways together with their regulatory circuits and accessory functions appear to have been eliminated, particularly those involved in catabolism. Even though intracellular mycobacteria are thought to derive much of their energy from the degradation of host-derived lipids (85), M. leprae possesses just two lipase-encoding genes, of which lipG clusters with mmaA genes and might, therefore, affect fatty acid reshuffling (85). This is in contrast to the 22 lip genes of M. tuberculosis, which also possesses a much higher number of fatty acid degradation pathways.
Recently, the genome variability of M. leprae was analyzed (83). The carbon source utilization systems that were lost from the M. leprae genome seem to specifically relate to acetate and galactose metabolism. This implies that M. leprae can grow only on a very restricted or even specialized combination of carbon sources. Moreover, presumed deletions have caused the loss of many crucial metabolic activities, including siderophore production, part of the oxidative as well as most of the microaerophilic and anaerobic respiratory chains, and several catabolic systems and their regulatory circuits.
A DNA microarray-based approach with seven M. leprae strains from different geographical areas (299) provided further information regarding gene loss and also revealed rare SNPs between various M. leprae isolates. Such data have provided clues for general evolutionary drift of M. leprae and could correlate the spread of leprosy with an associated M. leprae genotype(s).
Genomics of M. avium subsp. paratuberculosis
M. avium subsp. paratuberculosis is an extremely slow-growing, acid-fast, mycobactin-dependent multispecies pathogen that causes Johne's disease, a chronic granulomatous enteritis in cattle and other wild and domestic ruminants (171). The K-10 strain has a circular 4.8-Mb chromosome with 69.3% G+C (Table 1) (271). About 1.5% of the K-10 genome is repetitive DNA, such as ISs, multigene families, and duplicated housekeeping genes.
The most obvious physiological difference between M. avium subsp. paratuberculosis K-10 and other mycobacteria is the inability of K-10 to produce the iron siderophore mycobactin in laboratory culture. The biosynthetic gene cluster for mycobactin, a cluster of 10 genes (mbtA to -J), is present in the K-10 genome, but the first gene of the operon (mbtA) appears to be truncated, which would predictably impair mycobactin synthesis at its inception and potentially explains the strict dependence of M. avium subsp. paratuberculosis on this siderophore for in vitro growth.
Genes encoding members of the PE/PPE family comprise just 1% of the K-10 genome, as opposed to ∼10% in the genomes of the M. tuberculosis complex (see above). This observation suggests that antimicrobial agents and vaccines directed against members of the PE/PPE protein family should be more effective in M. avium subsp. paratuberculosis than in M. tuberculosis (271).
Many mycobacteria are facultative pathogens and produce specific proteins to allow survival inside the host's macrophages. Emphasis has therefore been placed on identifying virulence genes that are important for entry and persistence in the host (169). In M. tuberculosis and Nocardia farcinica, one such gene, the mammalian cell entry (mce) gene, has been found to actively enhance macrophage survival of an E. coli strain expressing it (13). The K-10 genome contains eight mce homologs. However, mce is also present in apparently nonpathogenic mycobacteria (e.g., M. smegmatis), so its presence does not necessarily endow a microorganism with the ability to cause disease. The study of its expression under specific conditions will be useful to investigate the role of the mce gene(s) in virulence (242).
Approximately 75% of the predicted K-10 proteome is homologous to that of M. tuberculosis (83), but 39 predicted proteins appear to be unique to K-10 (271). These can be used to develop sensitive molecular and immunoassay-based diagnostic detection tests (17). Bioinformatic analysis identified 185 di- and trinucleotide repeat sequences dispersed throughout the K-10 genome, of which 78 are perfect repeats (6). Sequence analysis of these loci in different M. avium subsp. paratuberculosis isolates from different host species as well as geographic locations identified a subset of polymorphic short sequence repeats, which have been used to develop a highly discriminatory typing technique (145).
Extrachromosomal DNA Elements in Mycobacterium
Most of the mycobacterial plasmids used for gene cloning and manipulation are based on the low-copy-number M. fortuitum plasmid pAL5000. Other sequenced mycobacterial plasmids, such as pMSC262 from M. scrofulaceum (349), pLR7 from M. avium (23), pVT2 from M. avium (229), pCLP from M. celatum (257), and pJAZ38 from M. fortuitum (142), have been assigned to one family, the pMSC262 family (229). Among these, pCLP is linear (257). Linear plasmids have also been found in M. xenopi and M. branderi (343). A large DNA region of pCLP has high sequence identity with a region in the genome of M. tuberculosis H37Rv that is located near ISs and contains a transposase-encoding gene (153). Recently, an in vivo transposition system was used to rescue a relatively rare circular DNA species, a cryptic plasmid named pVT2, from M. avium MD22 (229). Remarkably, pVT2 contains a large ORF encoding a protein with significant amino acid similarity to the DNA relaxases of conjugative plasmids, which introduce the specific nick at oriT to facilitate the unwinding of plasmid DNA needed to initiate DNA transfer (252).
Plasmids also mediate virulence in some mycobacteria. For example, the 174-kb plasmid pMUM001 from M. ulcerans contains a cluster of genes encoding very large polyketide synthases and polyketide-modifying enzymes, which are necessary for mycolactone synthesis (411). Mycolactone is a macrolide with cytotoxic, analgesic, and immunosuppressive activities and thus plays a key role in pathogenesis (144).
GENOMICS OF NOCARDIA
General Features
Nocardia spp. are filamentous soil saprophytes but also include pathogenic agents that cause nocardiosis in humans and animals in the lung, central nervous system, brain, and skin (42), while another species, N. asteroides, is suspected to contribute to Parkinson's disease (189). Moreover, Nocardia species can produce industrially important bioactive molecules such as antibiotics and enzymes (78, 393, 422).
The only fully sequenced nocardial genome, that of Nocardia farcinica IFM10152, comprises a circular chromosome of 6,021 kb (Table 1) (198). The 5,674 putative CDSs include many candidate genes for virulence and multidrug resistance as well as secondary metabolism (198).
A codon usage-based computational approach identified 571 putatively highly expressed genes, including some involved in primary metabolism (housekeeping functions) and 25 genes that may be involved in virulence and survival in host cells (487).
Nocardia Comparative Genome Analysis
Consistent with broader phylogenetic analysis, the genomes of M. tuberculosis and Corynebacterium glutamicum bear the closest relationship to that of N. farcinica. Large and frequent inversions have occurred within these genomes, as is obvious from the broken X pattern generated by dot plot analyses. The N. farcinica genome content reflects specific adaptations to accommodate growth in both soil environments and animal tissues (198). In the N. farcinica genome, 36% of the complete gene content belongs to 552 paralogous families. A large paranome set is a common feature of large bacterial genomes. Paralogous gene families found in N. farcinica include those coding for ABC transporters, two-component system proteins, extracytoplasmatic function σ factors, mammalian cell entry family proteins, fibronectin-binding proteins, short-chain dehydrogenases, acyl-CoA hydratase/isomerase family proteins, and lipase/esterase family proteins.
Extrachromosomal DNA Elements in Nocardia
Two circular plasmids, pNF1 (184 kb) and pNF2 (87 kb), were identified in N. farcinica IFM10152. Many other indigenous circular plasmids, ranging from 8 kb to 50 kb, from Nocardia spp. have been identified and sequenced (346). However, the mechanisms of plasmid replication, inheritance, and host ranges have not yet received much attention. The two replication proteins (RepA and RepB) found in most small plasmids of Nocardia, as well as those of the closely related Rhodococcus, resemble replication proteins of the theta-type replicating Mycobacterium plasmid pAL5000 (111, 177, 241). The replication protein of Nocardia plasmid pNI100 (308a) resembles the rolling-circle replication proteins encoded by the Streptomyces plasmids pIJ101 and pSG5 (238, 277).
N. farcinica Genome and Biological Lifestyle
As mentioned above, N. farcinica can grow in human as well as soil niches. Thus, a survey of the N. farcinica genome revealed many genes predicted to specify virulence factors. Among these were six so-called mce operons resembling mycobacterial virulence operons (see below) (1). Other candidate virulence genes include those encoding adherence factors similar to mycobacterial invasion proteins (248) and a heparin-binding hemagglutin protein, all of which are expected to play a role in extrapulmonary dissemination. Like many pathogens of mammalian tissue, N. farcinica has a suspected siderophore biosynthetic cluster similar to that responsible for mycobactin production (198), encompassing genes for two polyketide synthases, three nonribosomal peptide synthetases, two lysine modification proteins, and a mycobactin receptor protein.
Another genetic trait that may aid in pathogenic activities is provided by the determinants of a significant number of enzymes (i.e., four catalases, two superoxide dismutases, and an alkylhydroperoxidase) that may protect N. farcinica cells against reactive oxygen species produced by phagocytes. This suggests that Nocardia may also survive under low-oxygen conditions such as in stimulated macrophages (469).
The ecological fitness and virulence of Nocardia may also be linked to the presence of drug resistance genes. Notably, N. farcinica has two genes for the β subunit of RNA polymerase; one encodes a protein, RpoB1, that is sensitive to rifampin, while the second specifies RpoB2, which contains amino acid substitutions at positions expected to confer rifampin resistance (391). Moreover, specific drug resistance genes, such as the one that specifies resistance to aminoglycosides (kanamycin, gentamicin, and streptomycin), are also present. It is not known how and why N. farcinica evolved to contain so many drug resistance genes apparently without needing to resort to HGT; perhaps gene duplication events were instrumental in the emergence of drug resistance, as appears to be the case for the rpoB gene.
GENOMICS OF CORYNEBACTERIUM
General Features
The genus Corynebacterium was originally delineated in 1896 to accommodate primarily pathogenic species that showed morphological similarity to the diphtheroid bacillus (18). Consequently, the genus comprised, for several decades, an extremely diverse collection of morphologically similar gram-positive microorganisms, including nonpathogenic soil bacteria (89). More recently, chemotaxonomic studies and 16S rRNA gene sequence analysis defined the borderline of the genus Corynebacterium, clearly demonstrating that the species assigned to this genus form a monophyletic group (335), even though they exhibit considerable heterogeneity (for instance in mycolic acid content and in DNA base composition, which ranges from 46 to 74% G+C) (134).
Currently, there are almost 70 recognized Corynebacterium species, including many new species isolated from human clinical samples (328, 368), wild animals (90, 123), saline soil (71), or the surface of smear-ripened cheese (39). Four corynebacterial genomes have been completely sequenced and published so far, representing two species of biotechnological relevance and two important human pathogens. Some prominent features of these species are briefly summarized below.
(i) C. glutamicum is widely used in the industrial production of amino acids, especially l-glutamic acid and l-lysine, which are important in human and animal nutrition, respectively (267). Both l-glutamic acid and l-lysine are produced on a large scale by genetically modified high-performance strains of C. glutamicum. Natural habitats of C. glutamicum include soil contaminated with bird feces, sewage, manure, vegetables, and fruits (272). The sequenced type strain (ATCC 13032), which originated from a soil sample from the Ueno Zoo in Tokyo, Japan, is a natural producer of l-glutamic acid (228, 439).
(ii) Three strains of C. efficiens (formerly “C. thermoaminogenes”) were validly published as corynebacterial species in 2002 (132). They were originally obtained from onion bulbs and soil samples during a systematic search for new glutamic acid-producing bacteria that can grow at a higher temperature than C. glutamicum; such strains might reduce the need for cooling during fermentation (132). C. efficiens has 95.3% 16S rRNA gene sequence identity with its closest relative, C. glutamicum. The sequenced type strain of the species (YS-314) was isolated from soil samples collected in Kanagawa, Japan (132).
(iii) C. diphtheriae is a strictly human-adapted species and the causative agent of diphtheria (168). The disease is generally caused by exotoxin-producing C. diphtheriae strains and is characterized by local growth of the bacterium in the pharynx with pseudomembrane formation. Diphtheria toxin, the major virulence factor of C. diphtheriae, is a typical AB toxin that inhibits protein synthesis and kills susceptible host cells (181). The toxin structural gene (tox) is carried by some closely related corynephages and is therefore produced only by C. diphtheriae strains that harbor tox+ prophages. The sequenced strain (NCTC 13129) was isolated in 1997 from a pharyngeal membrane of a diphtheria patient (60).
(iv) C. jeikeium (formerly “Corynebacterium group JK”) was isolated from human blood and was associated with bacterial endocarditis following cardiac surgery (200). C. jeikeium is part of the normal human skin flora, particularly in the axillary, inguinal, and rectal areas (98). It is implicated in a variety of nosocomial infections, most frequently associated with immunocompromised patients, and is typically multiresistant to clinically relevant antibiotics, with the exception of glycopeptides (135). Its growth depends on the addition of lipids to culture media (135). The sequenced isolate (K411) was recovered from the axilla of a bone marrow transplant patient who received immunosuppressive therapy and broad-spectrum antibiotics (222).
Corynebacterium Genome Architecture
The general features of available corynebacterial genome sequences are summarized in Table 1. The first corynebacterial genome to be published, that of C. glutamicum ATCC 13032, was independently determined by two groups (195, 214), whose results were slightly different. In particular, the smaller genome lacked a putative prophage island (CGP4) but contained three additional ISs (213, 214). One of the additional ISs (of the ISCg1 type) disrupted groEL1, whereas the additional ISCg2 elements were located in distinct intergenic regions of the chromosome (20). Highly active ISs and corynephages apparently contribute to the rapid divergence of C. glutamicum genomes.
The 3.3-Mbp wild-type C. glutamicum R chromosome contains 2,990 predicted coding regions (details yet to be published) (414, 463). Remarkably, C. glutamicum R has 11 strain-specific islands (SSIs) larger than 10 kb that are absent from C. glutamicum ATCC 13032 (416) (see below). Overall, the ATCC 13032 and R strains have about 97% sequence identity.
The genome of the C. efficiens type strain YS-314 has a 3.1-Mbp circular chromosome with a 63.4% G+C content and containing 2,950 predicted coding regions. C. diphtheriae NCTC 13129 has a 2.4-Mb circular chromosome (53.5% G+C) containing 2,320 predicted coding regions, of which 45 are annotated as pseudogenes (60).
The 2.4-Mbp circular chromosome of C. jeikeium K411 (61.4% G+C) contains 2,104 predicted coding regions, of which 68 are apparently pseudogenes (427).
The larger genomes of environmental corynebacteria presumably reflect the greater metabolic diversity that is necessary in the changing environmental conditions of soil; fewer metabolic functions may be required in the specialized ecological niche of the corynebacterial pathogen. Reciprocal best-hit BLAST analysis revealed that gene loss has played a major role in the evolution of the C. diphtheriae genome (317).
GC skew profiles of the corynebacterial chromosomes (159, 278) indicate bidirectional replication from an origin of replication (oriC) close to the dnaA gene, though the GC skew plot of C. efficiens is a bit ambiguous (316). Many bacterial chromosomes tend toward an A+T enrichment near the replication terminus (105). This may reflect a higher proportion of horizontally acquired DNA in this region or structural constraints associated with the resolution of chromosome dimers at the end of replication (105). However, the skew of Rag (RGNAGGGS) motifs is maintained in the C. diphtheriae chromosome, even near the replication terminus (60). Rag motifs are G-rich DNA elements whose skew strongly shifts near the origin and the terminus of replication (278, 381). It has been proposed that the polarized Rag motifs, in conjunction with a polarity-reading factor, most probably the septum-anchored FtsK protein, are involved in preventing the capture of the dif region by the septum and thus facilitate the resolution of a chromosome dimer by site-specific recombination (278).
The C. jeikeium genome has 61 iap repeats (427). These highly conserved 29-bp elements contain an imperfect inverted repeat of 7 bp and form a tandem array with 32-bp spacers in a small, low-G+C region that is flanked by a tRNAMet gene at the 5′ junction. The iap repeats are members of the short regularly spaced repeat family of repetitive sequences (297), also referred to as the clustered regularly interspaced short palindromic repeat (CRISPR) family (207). CRISPRs have been implicated, for instance, in large-scale chromosome rearrangements and DNA condensation, but the mechanisms involved in their spread are obscure (107, 337). Recently, a new role has been proposed for CRISPRs, which includes providing immunity against foreign genetic elements via a mechanism based on RNA interference (19).
Corynebacterium Comparative Genome Analysis
Bidirectional best BLASTP match analysis (427) detected 1,089 genes that are considered orthologous in C. glutamicum, C. efficiens, C. diphtheriae, and C. jeikeium and apparently represent the conserved genetic backbone of the four species. These genes make up 52% of all C. jeikeium K411 genes and 36% of the C. glutamicum ATCC 13032 gene complement. A similar approach revealed 748 to 773 orthologous genes in cross comparisons using the three corynebacteria together with M. tuberculosis and Streptomyces coelicolor (when C. diphtheriae was excluded, this number went up to 831 [317]), implying that C. diphtheriae has lost many genes that were present in the common ancestor of actinobacteria (317). A comparison between the gene distributions in corynebacterial genomes was performed by classifying the predicted protein products into major functional categories, according to the COG (clusters of orthologous groups of proteins) protein classification scheme (213). The numbers of proteins within each COG category are generally higher in the environmental species than in the pathogenic species, indicating that bacterial genome size and gene content are largely dictated by environmental pressures (235).
Using two minitransposons based on either the corynebacterial IS IS31831 (462) or the Ez::Tn5 transposome complex, 2,332 C. glutamicum R genes were disrupted, representing 78.1% of the predicted coding regions (414). The average number of hits per coding region was 3.45. Since each of the selected transposon mutants survived on complex medium under normal growth conditions, the disrupted genes are apparently nonessential under these circumstances. The 658 undisrupted genes are therefore candidate essential genes. (Note, however, that experimental strategies based on transposon mutagenesis systems tend to overestimate this set of genes [147]. Blast comparison indicated that 251 C. glutamicum genes have clear orthologs among known E. coli or B. subtilis essential genes. Of these, 221 are among the candidate essential genes of C. glutamicum R [414].)
Although members of the same bacterial genus usually share highly parallel distributions of transporter families, the distribution of transporter families in C. glutamicum, C. efficiens, and C. diphtheriae is an exception. There are eight transporter families specific to C. diphtheriae, while only one is specific for C. glutamicum and three for C. efficiens. For those transporter families with orthologs in C. glutamicum and C. efficiens, but not in C. diphtheriae, orthologs were also identified in the majority of actinobacteria. In contrast, transporter families specific to C. diphtheriae tend to have either no apparent orthologs or only distantly related homologs in other sequenced bacterial species, suggestive of horizontal gene acquisition events (365). Phylogenetic analyses suggest that certain transporter families in C. efficiens are missing because of specific gene deletion occurrences (365).
Brune et al. (47) found genes for 127 DNA-binding transcriptional regulators in C. glutamicum, 103 in C. efficiens, 63 in C. diphtheriae, and only 55 in C. jeikeium, consistent with the general observation that larger genomes require more complex regulation of gene expression by an increased number of regulatory genes (59). The common set of 28 DNA-binding regulators in C. glutamicum, C. efficiens, C. diphtheriae, and C. jeikeium consists of apparent regulators of cell division and septation, SOS and stress responses, carbohydrate metabolism, and macroelement and metal homeostasis (47).
The C. glutamicum, C. efficiens, and C. diphtheriae chromosomes are highly syntenous across their entire lengths (Fig. 8), except for a large putative prophage region (genomic islands CGP3 and CGP4) in C. glutamicum (195, 213, 214). The C. jeikeium chromosome differs by 10 apparent breakpoints of synteny (427). These breakpoints are suggestive of the insertion of a DNA fragment that is flanked by clustered ISs, the translocation of a small DNA region, and two distinct inversion events. One breakpoint of synteny attributed to DNA inversion was localized close to clustered ISs and copy A of the rRNA operons. Since no clearly defined boundaries of the rearranged DNA fragments could be deduced from the genome sequence, the actual molecular mechanisms responsible for the genome rearrangement in C. jeikeium are unclear.
FIG. 8.
Synteny plot comparing the order of homologous genes (through their encoded proteins) in sequenced genomes of corynebacteria. The conservation between the C. jeikeium K411 genome and the genomes of C. glutamicum ATCC 13032, C. efficiens YS-314, and C. diphtheriae NCTC 13129 is shown by x-y plots of dots forming syntenic regions between the corynebacterial genomes. Each dot represents a CDS of C. jeikeium having a homolog in another genome, with coordinates corresponding to the CDS number in each genome. Homologs were identified by best BLASTP matches of amino acid sequences deduced from the 2,104 CDSs of C. jeikeium K411 with proteins encoded by C. glutamicum (3,002 CDSs; red dots), C. efficiens (2,950 CDSs; green), and C. diphtheriae (2,320 CDSs; blue).
In contrast, extensive rearrangements have occurred in mycobacteria throughout their evolution, and the order of orthologous genes between corynebacteria and mycobacteria is considerably disrupted (307). The absence of the recBCD genes from corynebacteria may have suppressed genome shuffling, leading to exceptionally stable genome structures in these species (307): mutations of the recBCD genes of E. coli are known to reduce the frequency of chromosomal inversions (239). The different types of genome rearrangements detected in C. jeikeium indicate, however, that a moderate level of chromosomal reorganization can still occur in corynebacteria by recBCD-independent routes. The apparent stability of corynebacterial gene order indicates that speciation has involved gene gain and gene loss, HGT, and nucleotide substitutions rather than genome shuffling.
The 11 SSIs larger than 10 kb in the C. glutamicum R genome mentioned earlier are located in the highly conserved common backbone (416). The SSIs are loaded mainly with transposable elements and genes of unknown function, although several enzymes and proteins of a diverse functional context are also encoded on the islands (415, 416). SSI-8 includes a gene region with deduced amino acid sequence similarity to proteins of the lytic corynephage BFK20 from the industrial amino acid producer “Brevibacterium flavum” (52). By means of a precise genome excision method based on the Cre/loxP recombination system, the 11 SSIs were individually deleted from the C. glutamicum R genome (415, 416). All resulting mutants exhibited normal growth under standard laboratory conditions, indicating that a total of 250 kb (representing 7.5% of the R genome or 233 genes) was dispensable for cell survival. Likewise, no obvious phenotypic changes resulted for multiple deletions of SSIs, though no competition experiments with the C. glutamicum R wild-type strain were conducted (415, 416). Nevertheless, under the conditions tested the SSIs appear to contribute only marginally, if at all, to fitness.
The sequenced type strain C. glutamicum ATCC 13032 is unusual among corynebacteria in lacking an S-layer (surface protein) lattice (170). It lacks a 6-kb DNA region that includes the cspB gene, encoding the S-layer protomer PS2, which is present in all analyzed C. glutamicum wild-type strains capable of S-layer formation. The S-layer gene region of the ATCC 14067 wild-type strain is flanked by a 7-bp direct repeat element that is present only once in the ATCC 13032 genome, suggesting that recombination between these elements has been responsible for gene loss in the ATCC 13032 type strain (170). The loss of the S-layer gene region might reflect a mechanism for deactivating, under favorable culture conditions, a superfluous surface structure that is expressed in considerable amounts (up to 15% of the total protein content).
DNA Regions in C. glutamicum Acquired by HGT
A search of the C. glutamicum ATCC 13032 genome for atypical GC islands (213, 214) revealed the 26.9-kb LCG1 island, whose 25 predicted coding regions had G+C contents ranging from 41% to 50%. The 25 genes include murA and murB, both involved in murein formation, and additional genes that are predicted to be involved in surface polysaccharide biosynthesis. The C. glutamicum genome contains additional copies of the murAB genes (murA2 and murB2), with a more typical G+C content. Another island, HGC1, of 20.6 kb, has a 69% G+C content. HGC1 is flanked on one side by a defective copy of ISCg5, and on the other by the nonfunctional DNA element ISCg19. The products of HGC1 genes include a putative cation-transporting P-type ATPase, a two-component system, and a putative multicopper oxidase (213). Highly similar DNA sequences (up to 99% identity) were localized on SSI-3 of C. glutamicum R (416) and on plasmid pLEW279b from Corynebacterium sp. strain L2-79-05 (477), suggesting the recent plasmid-mediated HGT of the HGC1 island to the C. glutamicum chromosome. A highly similar DNA region in C. diphtheriae also has an exceptionally high G+C content (213).
C. glutamicum ATCC 13032 has 24 different ISs belonging to nine different families (214). Most of the ISs appear to have functional copies in the genome, although 11 IS elements are partially deleted and presumably are defective. The two available C. glutamicum ATCC 13032 genome sequences differ in the number of ISCg1 and ISCg2 copies (214) (see above). The ISs ISCg15a and ISCg15b are both members of the IS6 family. They form terminal parts of the 3,148-bp cryptic, composite transposon TnCg1 (Table 3; Fig. 9), which encodes a putative membrane protein of unknown function (214).
TABLE 3.
General features of transposons detected in C. glutamicum and C. jeikeium
| Transposon | Transposon size (bp) | Target site duplication (bp) | % G+C content | CDS at:
|
Prominent features of the central region | Reference | |
|---|---|---|---|---|---|---|---|
| Left end | Right end | ||||||
| TnCg1 | 3,148 | 8 | 61.0 | cg2756 | cg2758 | Cryptic; putative membrane protein of unknown function | 214 |
| Tn3595 | 2,463 | 6 | 64.2 | jk0815 | jk0817 | Putative glyoxylase/bleomycin resistance protein and AraC-type transcriptional regulator | 427 |
| Tn3596 | 33,607 | 8 | 60.1 | jk1767 | jk1791 | Siderophore biosynthesis genes and iron acquisition system | 427 |
| Tn3597a | 6,543 | 3 | 58.4 | jk0015 | jk0021 | NRAMP manganese uptake system MntH1, Nudix hydrolase Utp1, and TetR-type transcriptional regulator | 427 |
| Tn3597b | 6,541 | 3 | 58.4 | jk0621 | jk0627 | NRAMP manganese uptake system MntH2, Nudix hydrolase Utp2, and TetR-type transcriptional regulator | 427 |
| Tn3598 | 12,021 | 8 | 65.2 | jk0360 | jk0370 | TetAB-type ABC transport system and undecaprenyl pyrophosphate phosphatase UppP2 | 427 |
| Tn3599 | 3,749 | 6 | 59.1 | jk1402 | jk1405 | Chloramphenicol exporter (MFS type) | 427 |
FIG. 9.
Genetic organization of transposons localized in the genome of C. jeikeium K411. The detected transposons are arranged according to size. Predicted coding regions are shown as arrows. Different colors indicate transposase genes of ISs (blue), antibiotic resistance determinants (red), genes potentially involved in protective functions against environmental stress (orange), predicted transcriptional regulators (green), and genes involved in iron acquisition (magenta). Genes with other predicted functions are shown in yellow, and genes encoding hypothetical proteins are depicted in gray.
Prophage-Like Elements in the C. glutamicum Genome
Four putative prophage islands have been recognized in the C. glutamicum ATCC 13032 genome, based on deviations in base composition and the presence of phage integrase genes (213, 495). Prophage island CGP1 (12.7 kb) is integrated at a tRNALeu gene. Prophage island CGP2 (3.9 kb) is apparently a highly degenerated prophage remnant. CGP3 (185.8 kb, 48.4% G+C) is adjacent to a cluster of seven tRNA genes. The few recognizable CGP3 functions include the cglIM, cglIR, and cglIIR genes of the C. glutamicum stress-sensitive restriction-modification system, which is the major barrier to efficient gene transfer into this strain (382, 383). The cglIM-specified 5-cytosine methyltransferase apparently methylates the sequence GCSGC (206, 382, 383). Hybridization experiments revealed that the cgl gene region is absent from other C. glutamicum strains and closely related corynebacterial species (383). The CGP3 island therefore contributes specifically to the evolution of C. glutamicum ATCC 13032 by coupling the efficiency of HGT to unfavorable environmental conditions.
The putative prophage island CGP4 represents one of the major differences between the C. glutamicum ATCC 13032 genome sequences determined at Bielefeld University and Kitasato University. The 23.5-kb CGP4 island is flanked by a 4.4-kb duplication (213). CGP4 is located within CGP3, indicating that the 4.4-kb duplication is related to the integration process. CGP3 is surrounded by an imperfect direct repeat element of approximately 500 bp (213, 495).
DNA Acquired by HGT in the C. efficiens Genome
Four low-GC genomic islands of 32.3 to 70.5 kb were found in the genome of C. efficiens. The largest, CEGI-1, is integrated at a tRNAPro locus. It encodes various enzymes, including a DNA repair protein of the RecF pathway, aspartate kinase, aspartate-semialdehyde dehydrogenase, and isopropylmalate synthase. No further genes encoding these amino acid biosynthesis enzymes are present in the C. efficiens genome. Aspartate kinase catalyzes the first step of the lysine biosynthesis pathway and is a key enzyme in the design of industrial lysine producers (215, 323). The aspartate kinase of C. efficiens has a lower thermal stability than the aspartate kinase of C. glutamicum, whereas other amino acid biosynthesis enzymes and enzymes of central metabolism have a higher thermal stability in the heat-tolerant C. efficiens (316). Presumably, the adaptive mutations necessary for thermal stability have not occurred extensively due to the recent horizontal acquisition of CEGI-1. Genomic island CEGI-2 contains genes that may be involved in capsular polysaccharide biosynthesis, and CEGI-4 harbors transposase genes and IS-related genes. Both genomic islands are flanked at one end by ISs.
Genomic island CEGI-3 (40.4 kb) is flanked by a 26-bp direct repeat element and encodes an integrase at the 5′ junction (494). Seven of the 50 predicted genes of CEGI-3 encode proteins similar to late proteins of the lytic bacteriophage BFK20 (52). A high content of genes coding for hypothetical proteins and the similarity to the genome of bacteriophage BFK20 indicate that CEGI-3 is a likely prophage island.
Prophage-Like Element in the Genome of C. diphtheriae
The genome sequence of C. diphtheriae NCTC 13129 provided the first complete nucleotide sequence of a tox+ corynephage (60). The 36.5-kb prophage genome has a G+C content of 52.2% and encodes 43 predicted proteins. Its closest sequence matches at the amino acid level were with deduced structural proteins of the lytic phage BFK20 of “Brevibacterium flavum” (52). However, its genome organization resembles that of phages infecting low-G+C gram-positive bacteria, and several of its proteins show weak sequence similarities with these phages (50). The prophage is integrated at a tRNAArg locus (359). The tox gene, encoding diphtheria toxin, is situated at the right end of the prophage genome, next to the attachment site and within a region of low G+C content. The predicted coding regions dip0180 and dip0181, both encoding toxin proteins, were detected in a low-G+C region at the left end between the integrase gene and the attachment site (60). The specific location of tox, dip0180, and dip0181 in the prophage genome, in conjunction with the deviating G+C content, led to the hypothesis that these prophage genes actually represent bacterial genes that were acquired from a previous host and that they may be considered lysogenic conversion genes that are not required for the phage life cycle but may affect the phenotype or fitness of the lysogen (50).
The toxin is the major virulence factor of C. diphtheriae, so the phage is pivotal in pathogenicity. Transcription of tox is regulated by the host-encoded diphtheria toxin represssor DtxR and becomes derepressed during iron depletion (181). Iron-dependent regulators of the DtxR family have been detected in many corynebacteria (326) and described as global regulatory proteins in C. diphtheriae and C. glutamicum (48, 245). DtxR obviously links expression of the phage-encoded tox gene with the control of iron homeostasis in the bacterial cell, resulting in a complex host-pathogen-phage interaction. Iron limitation also enhances the interaction of C. diphtheriae with erythrocytes and HEp-2 cells (303). Whether this apparent effect on host-pathogen interaction is under transcriptional control by the DtxR regulator remains to be investigated.
DNA Regions in the C. diphtheriae Genome Acquired by HGT
In addition to the integrated tox+ corynephage, 12 genomic regions with local anomalies in the nucleotide composition were detected. Seven are flanked by tRNA genes, and none is present in the C. glutamicum and C. efficiens genomes. Five contain phage-related genes and may be prophage remnants. Several pathogenicity-related functions are encoded in the genomic islands, including a siderophore biosynthesis and export system and a potential lantibiotic biosynthesis system, as well as three sortase-related fimbrial systems (60, 141, 417, 436). Each fimbrial gene cluster is composed of at least one pilin-specific sortase gene (srtA to -E) and three sortase-mediated pilus assembly genes (spaA to -I). Morphological and genetic studies have clearly demonstrated that each pilus of C. diphtheriae is composed of three pilin subunits and that the pilus assembly process requires pilin-specific sortases (60, 141, 417, 435, 436). Thus, the genomic islands contribute substantially to the structure of the cell surface.
DNA Regions in the C. jeikeium Genome Acquired by HGT
Five genomic islands, of 15.6 to 44.9 kb, have been found in the C. jeikeium K411 chromosome (Table 4). The islands JKGI-1 and JKGI-2 encode different types of restriction-modification systems, including an mrr-like restriction system as well as type I and type II restriction-modification systems (31). These islands may therefore prevent the expression or incorporation of incoming foreign DNA. Island JKGI-3 adds an extra pathway for molybdenum cofactor synthesis to the C. jeikeium genome. Islands JKGI-4 and JKGI-5 are separated only by a small region encoding ribosomal proteins. Both islands harbor genes for siderophore biosynthesis systems and iron ABC transporters and are thus apparently involved in iron uptake. Since iron homeostasis plays a critical role in the survival of a bacterial cell, genomic islands contributing to iron supply can be regarded as potential pathogenicity islands. However, the pathogenicity of C. jeikeium appears to be determined by a variety of factors that are encoded throughout the genome in small islets (427). Thus, clearly defined pathogenicity islands seem to be absent, in contrast to the situation described for C. diphtheriae (60). Interestingly, all genomic islands of C. jeikeium are flanked on both sides by ISs, which may have played a decisive role in integrating the islands into the C. jeikeium genome (427).
TABLE 4.
Genomic islands localized in the C. jeikeium chromosome
| Genomic islanda | CDS at:
|
Size (kb) | No. of genes | % G+C content | Prominent features | Reference | |
|---|---|---|---|---|---|---|---|
| Left end | Right end | ||||||
| JKGI-1 | jk0522 | jk0530 | 15.6 | 9 | 54.9 | Type II DNA restriction-modification system; IS3510 at 5′ junction and IS3511 and tRNAArg at 3′ junction | 427 |
| JKGI-2 | jk1226 | jk1264 | 44.9 | 39 | 55.2 | Type I and type II DNA restriction-modification systems, mrr-like restriction system; IS3503 at 5′ junction and IS3523 and tRNAGlu at 3′ junction | 427 |
| JKGI-3 | jk1579 | jk1602 | 24.3 | 24 | 55.8 | Molybdate ABC-type transport system and molybdenum cofactor biosynthesis genes; IS3506 at 5′ junction and IS3519 and tRNAMet at 3′ junction | 427 |
| JKGI-4a | jk1767 | jk1791 | 33.6 | 25 | 60.1 | Siderophore biosynthesis genes and iron ABC-type transport system; IS3503 at 5′ junction and IS3503 at 3′ junction | 427 |
| JKGI-5 | jk1803 | jk1823 | 30.7 | 21 | 60.3 | Siderophore biosynthesis genes and iron ABC-type transport systems; IS3515 at 5′ junction and IS3517 at 3′ junction | 427 |
Genomic island JKGI-4 is a defective composite transposon named Tn3596 (Table 3).
The chromosome of C. jeikeium K411 contains 24 different ISs (92 copies in all) of four different groups, the IS256, IS3, IS30, and IS110 families. Of these, 68 ISs are complete, suggesting that they can function in transposition, whereas 24 are partially deleted or disrupted by other mobile elements. Some ISs are integral parts of novel transposons that may play important roles in antibiotic resistance, iron homeostasis, or manganese uptake (Table 3; Fig. 9). Tn3598 is a composite transposon based on IS1249 and including the tetA-tetB gene pair known to mediate tetracycline resistance in corynebacteria (428). The TetAB proteins are representative of a new group of tetracycline resistance determinants that use ATP rather than the proton gradient as an energy source (73). Tn3598 also contains the uppP2 gene, encoding an undecaprenyl pyrophosphate phosphatase that may confer bacitracin resistance upon overexpression. Tn3599 carries the cmx gene, coding for a membrane protein of the major facilitator superfamily that confers chloramphenicol resistance (428). Furthermore, the 2.5-kb transposon Tn3595 encodes a protein of the glyoxylase/bleomycin resistance protein family.
Tn3596, at 33.6 kb, is the largest composite transposon in C. jeikeium. It encodes a nonribosomal peptide synthase and several other proteins apparently involved in siderophore biosynthesis, as well as a potential iron ABC transport system (427). Both transposase genes were disrupted by subsequent transposition events, suggesting that Tn3596 has lost the capability for autonomous transposition.
The composite transposons Tn3597a and Tn3597b differ only in the inversion of one copy of the IS. These transposons contain mntH genes that encode proteins of the Nramp (natural resistance-associated macrophage protein) family involved in manganese uptake. Tn3597a and Tn3597b also include utp genes encoding Nudix (nucleoside diphosphatases linked to some other moiety x) proteins, functioning in the hydrolysis of UTP and 5-methyl-UTP. The clustering of Nramp and Nudix genes suggested a common functional context of these genes in the overall defense against reactive oxygen species. Nramp proteins may help to protect the cell against reactive oxygen species by providing manganese for detoxification enzymes (205), whereas the Nudix hydrolases may act as surveillance system by hydrolyzing toxic or deleterious compounds possibly arising through DNA damage (30, 427, 488).
Extrachromosomal DNA Elements
The genetic characteristics of plasmids pEC2 and pEC3 of C. efficiens and pKW4 of C. jeikeium are summarized in Table 5. The replicases of pEC2, pEC3, and pKW4 have a number of features resembling those of the C. diphtheriae plasmid pNG2, including the RepA replication protein, indicating that they replicate by the rolling-circle mechanism (423-426, 429, 430, 496). Plasmids of this family have been found only in corynebacteria, except for pAP2 from Arcanobacterium pyogenes (212), but may, like pNG2, have a broad host range (212, 351, 390).
TABLE 5.
General features of corynebacterial plasmids sequenced in the course of genome projects
| Corynebacterium | Plasmid name | Plasmid size (bp) | % G+C content | No. of genes | Coding density (%) | Plasmid familya | Replication modec | Prominent features | Reference sequence | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| C. efficiens YS-314 | pEC2 | 23,743 | 54.4 | 15 | 73 | pNG2 | RCR | Cryptic | NC_004319 | 316 |
| pEC3 | 48,672 | 56.4 | 41 | 73 | pNG2 | RCR | mrr-like restriction system, genes involved in metal transport and homeostasis | NC_004320 | 316 | |
| C. jeikeium K411 | pKW4 | 14,323 | 53.8 | 16 | 71 | pNG2b | RCR | bls gene cluster and aucA gene for bacteriocin synthesis | NC_003080 | 427 |
The pEC2 plasmid of C. efficiens YS-314 is apparently a natural hybrid of two pNG2-like plasmids, since it contains two typical repA genes (CEP005 and CEP013). The fusion of replicons in corynebacteria is often mediated by ISs, as in the multiresistance mosaic plasmid pTP10 of Corynebacterium striatum M82B (428). pEC3 is also littered with ISs, particularly at the junctions of distinct functional segments. pEC3 encodes an mrr-like restriction system, two metallo-regulatory transcriptional repressors of the ArsR family, and possible copper resistance cation-transporting ATPases. pEC3 may therefore play a role in metal resistance and metal homeostasis (54).
The C. jeikeium K411 low-G+C plasmid pKW4 (427) is the prototype of a plasmid family implicated in the synthesis of a narrow-spectrum bacteriocin-like substance (BLS) (221, 222). The BLS is most likely encoded by the bls gene cluster (222). pKW4 also has a gene, aucA, encoding an additional BLS-like compound resembling aureocin A53 from Staphylococcus aureus (312). The latter BLS is active against staphylococci, including clinically significant S. aureus strains (311). Interestingly, the bacterial flora of the axillary region of the human skin is dominated by either Staphylococcus species or a dense population of corynebacteria (309, 369).
A recent sequencing study of 17 plasmids in the size range from 7.6 to 14.9 kb among 62 C. jeikeium isolates from different clinical settings placed all these plasmids in the pNG2 family (425). Other C. jeikeium plasmids have been documented, but no detailed genetic characterization was carried out (221-223, 344, 478).
The sequenced C. glutamicum strain (ATCC 13032) lacks plasmids, but systematic screening of C. glutamicum isolates has detected 24 different plasmids ranging in size from 2.4 to 95 kb (for reviews, see references 423 and 429). Most of the plasmids are cryptic, but four harbor resistance determinants against chloramphenicol, tetracycline, streptomycin, spectinomycin, and sulfonamides. HGT by plasmids is apparently the main source of antimicrobial resistance determinants in this species (423). Likewise, plasmid screening studies of C. diphtheriae revealed a number of plasmids, most of which were similar to pNG2, which harbors the erythromycin resistance gene erm(X) (385, 386, 424).
Corynebacterial Genomes and Biological Lifestyle
Adherence to pharyngeal epithelial cells by C. diphtheriae.
C. diphtheriae usually localizes in the upper respiratory tracts of humans and can cause local infections by ulcerating the mucosa and inducing the formation of inflammatory pseudomembrane (168). In addition, the release of the major virulence factor, diphtheria toxin, results in systemic intoxications, destroying the parenchyma of heart, liver, kidneys, and adrenal glands. Adherence to the host tissues plays a crucial role in extracellular mucosal pathogenesis and in the establishment of C. diphtheriae infections (286, 303). The minor pilins SpaB and SpaC of the SpaA-type pilus, but not the SpaA pilus fiber itself, act as adhesins to pharyngeal epithelial cells (284). The SpaA-type pilus is encoded on a pathogenicity island of C. diphtheriae NCTC 13129 that is characterized by local anomalies in the nucleotide composition (60, 435). The minor pilins are components of both the SpaA-type pilus and the cell wall of C. diphtheriae, thereby probably allowing both secure distant contact of piliated bacteria and intimate attachment onto pharyngeal epithelial cells (284). The tight intimate contact facilitates the delivery of diphtheria toxin to the host cell, creating a safe niche for the adherent bacteria after damaging the host cell through the inhibition of protein synthesis. Fimbria-mediated adhesion of C. diphtheriae might be enhanced by neuraminidase and trans-sialidase activities, unmasking receptors on the host cell surface (60, 286).
Adaptation to amino acid production by C. glutamicum and C. efficiens.
A comparative genomics study suggested that the common ancestor of corynebacteria already possessed almost all of the genes relevant for amino acid production. Thus, the evolutionary events of gene duplication, gene loss, and HGT must have been responsible for the functional differentiation in amino acid biosynthesis capabilities of environmental and pathogenic corynebacteria (317). In particular, differences between amino acid biosynthesis pathways of C. efficiens and C. glutamicum were detected by phylogenetic analysis, using amino acid biosynthesis-related genes. This set of genes included trpB (encoding the β chain of tryptophan synthase), ilvD (dihydroxy-acid dehydratase), aroQ (3-dehydroquinate dehydratase), glnA (glutamine synthetase I), and ocd (ornithine cyclodeaminase). Evaluation of the topologies of phylogenetic trees for the paralogous genes provided insights into the different evolutionary events that occurred in corynebacteria (317). The results suggest that duplication of trpB took place in the common ancestor of the corynebacteria and that gene loss was responsible for the single copy of this gene in C. glutamicum. Likewise, the additional ilvD gene of C. efficiens was acquired by ancient gene duplication rather than by HGT.
Adaptation to elevated temperatures by C. efficiens.
The 10% higher G+C content of C. efficiens compared with C. glutamicum may be due to the absence of a mutT gene, which affects the frequency of AT-to-GC transversions (97, 307). The 10 most frequently used codons in C. efficiens all have G or C in the third position, whereas none of 10 rarely used codons contains G or C in the third position. Furthermore, a comparison between orthologous proteins of C. efficiens and C. glutamicum revealed a tremendous bias in amino acid substitutions in C. efficiens, particularly towards arginine and glycine (317). The analysis of amino acid substitutions in 13 pairs of orthologous glutamate or lysine biosynthetic enzymes in C. glutamicum and C. efficiens revealed that three substitutions, lysine to arginine, serine to alanine, and serine to threonine, are important for thermostability (316). Thus, C. efficiens seems to have acquired thermostability through the accumulation of specific amino acid substitutions, which, in part, correlate with increased G+C content (316).
Adaptation to the lipophilic lifestyle by C. jeikeium.
The strict requirement of C. jeikeium for exogenous fatty acids (135) originates from the lack of a type I fatty acid synthase gene (423, 429). C. jeikeium possesses several fad genes encoding a complete β-oxidation pathway, which is absent from the three other available corynebacterial genomes. Furthermore, a fadH gene, encoding 2,4-dienoyl-CoA reductase, is apparently involved in the utilization of unsaturated fatty acids whose double bond extends from an even-numbered carbon atom (423, 429). C. jeikeium can utilize very few sugars as carbon and energy sources and so may depend largely on exogenous fatty acids (200). Fatty acids also provide building blocks for the synthesis of corynomycolic acids, which are major constituents of the cell envelopes of most corynebacterial species, including C. jeikeium (200).
The lipophilic phenotype of C. jeikeium may be related to its preference for the axillary, inguinal, and perineal areas of human skin (98), which are moist areas characterized by hydrolipid films composed of triglycerides, fatty acids, ceramides, cholesterol, and cholesterol esters (409, 427).
It is likely that some of the predicted virulence factors of C. jeikeium act to release additional exogenous fatty acids by damaging the plasma membrane of host cells. Such factors include a putative alkaline ceramidase with homology to a cholesterol esterase enzyme from Pseudomonas aeruginosa (314). This enzyme apparently catalyzes the hydrolysis of long-chain fatty acid esters from cholesterol and may thus be involved in fatty acid metabolism (315). Other virulence factors include an acid phosphatase with similarity to the AcpA protein of Francisella tularensis (364).
GENOMICS OF LEIFSONIA
General Features
The genus Leifsonia was created by Evtushenko et al. (120) to accommodate gram-positive, non-spore-forming, irregular rod- or filament-shaped, motile, mesophilic, and catalase-positive bacteria containing dl-2,4-diaminobutyric acid in their peptidoglycan layer. Currently, this genus includes seven species that have been isolated from different ecological niches, such as plants (L. poae, L. xyli subsp. xyli, and L. xyli subsp. cynodontis), soil (L. naganoensis and L. shinshuensis), distilled water (e.g., L. aquatica), and an Antarctic pond (L. rubra and L. aurea) (208, 361, 413). Along with other plant pathogens such as the gammaproteobacterium Xylella fastidiosa, L. xyli subsp. xyli belongs to a unique group of xylem-limited and fastidious bacterial pathogens and is the causative agent of the ratoon stunting disease, the main sugarcane disease worldwide (106, 347).
The genome of L. xyli subsp. xyli CTCB07 has a single circular chromosome of 2.5 Mb (Table 1), of which 70.6% represents protein-coding capacity (300). Consistent with this is the presence of pseudogenes in a number that is larger and more frequent than in any other completely sequenced plant-associated bacterium. Although pseudogenes are not as numerous as in M. leprae (84) (see above), they occur more frequently in L. xyli subsp. xyli CTCB07 than in other actinobacterial genome. Moreover, 3.5% of the genome is occupied by IS elements, of five distinct IS families, and transposases. A large number of presumed HGT-derived elements are also found in another plant pathogen, X. fastidiosa (334).
Extrachromosomal DNA Elements in Leifsonia
The only known Leifsonia plasmid, pCXC100 (51 kb), has been found in many strains of L. xyli subsp. cynodontis (269, 292, 431). Information on pCXC100 is limited: the RepA-encoding protein is homologous to that of the plasmids in the mycobacterial pLR7 family, and a region associated with plasmid stability comprises parA, which is necessary for plasmid resolution, and a gene encoding a hypothetical protein (269).
DNA Regions Acquired by HGT in Leifsonia
The Leifsonia genome contains four likely horizontally acquired DNA regions with deviant G+C contents, codon biases, and dinucleotide signatures. These encompass candidate pathogenicity genes such as pectinase-encoding genes flanked by transposable elements or polygalacturonases, which in other plant pathogens such as Xanthomonas can degrade pectin in plant cell walls, resulting in the maceration of host tissues (91). One 50-kb region, containing a high number of transposase-encoding genes, harbors a celA gene coding for a cellulase similar to CelA of Clavibacter spp., where the gene is a plasmid-borne virulence factor (202). This large DNA region also harbors a gene coding for a member of the delta fatty acid desaturase family. This protein could redirect the carotenoid biosynthetic pathway identified in this strain (12) to the biosynthesis of the plant hormone abscissic acid (ABA) (21, 220). ABA is a growth inhibitor of plant tissues, and the production and secretion of ABA may thus contribute to the stunting seen in plants infected with L. xyli subsp. xyli.
Prophage-Like Elements in Leifsonia
There are two prophage-like regions in the Leifsonia genome (300). The 33-kb LxxGII prophage appears to be inserted within a gene encoding a type IV secretion ATPase and is flanked by a 15-bp direct repeat. The 37-kb LxxGI4 prophage, which is bracketed by two 30-bp direct repeats, is integrated into a glycine tRNA gene. Notably, LxxGI4 carries a homolog of the plasmid-located pat-1 gene of C. michiganensis subsp. michiganensis, which plays a decisive causal role in plant wilting (115). Furthermore, the pat-1 gene is flanked at the 3′ end by a repetitive sequence whose presence appears to exacerbate the virulent phenotype (115). L. xyli subsp. xyli CTCB07 has a second pat-1 homolog, but only the copy present in LxxGI4 seems to be functional. However, this pat-1 copy lacks the repetitive sequence at the 3′ end, perhaps explaining why wilting is provoked only in very specific circumstances, i.e., in particular sugarcane varieties and under severe drought conditions.
L. xyli subsp. xyli Genome and Biological Lifestyle
Like other plant pathogens, L. xyli subsp. xyli CTCB07 has the potential to encode products (superoxide dismutase, catalase, iron dependent peroxidase, and alkyl hydroperoxide reductase) that would enable it to resist the reactive oxygen species synthesized as part of the host defense mechanisms. The genome also contains an gene encoding arginase, which converts arginine to urea and ornithine. Arginase may inhibit the production of the antimicrobial nitric oxide by plants (150). Interestingly, CTCB07 encodes a multidrug efflux pump similar to the AlbF system that allows self-protection of Xanthomonas albilineans against the albicidin toxin, presumably aiding survival in the complex ecosystem inhabited by the two organisms.
L. xyli subsp. xyli CTCB07 can use a range of sugars, including arabinose, fructose, galactose, glycerol, glucose, lactose, maltose, maltotriose, ribose, trehalose, and xylose, and it has a PTS for the transport of fructose and mannitol. Its limited ecological range is also obvious from the loss of genes involved in basic metabolism and synthesis of amino acids, including genes involved in the synthesis of cysteine and methionine.
GENOMICS OF THE MYCELIAL ACTINOBACTERIA: STREPTOMYCES, FRANKIA, AND THERMOBIFIDA
General Features
The many mycelial genera of actinobacteria include some of the most complex of known bacteria (295). Unfortunately, beyond taxonomic and phylogenetic analysis, most of these have received very little attention from academic scientists, and there are many conspicuous gaps in genome information. Just three mycelial genera, Streptomyces, Thermobifida, and Frankia, are represented in the genome databases, and here we consider these together, because genomic comparisons have revealed that their similar growth modes are congruent with their genomic relatedness, as if the mycelial growth habit had a common origin. Of the three genera, Streptomyces has received particular attention, for three main reasons. (i) Streptomycetes are extremely abundant and important in soil, where they are major agents in the cycling of carbon trapped in insoluble organic debris, especially from plants and fungi. They achieve this by the production of many and diverse hydrolytic exoenzymes. (ii) One strain, Streptomyces coelicolor A3(2), is a major model organism, particularly for its developmental complexity (see, e.g., reference 64). (iii) Streptomycetes are the most abundant natural source of antibiotics and other bioactive secondary metabolites and are therefore of great interest for medicine and industry (186). The Streptomyces species whose genome sequences are currently published or available online represent a fairly wide phylogenetic spread: S. coelicolor (the model streptomycete) and Streptomyces ambofaciens (studied mainly for its remarkable genetic instability) are closely related members of one of the two largest clusters, and S. avermitilis (the industrial producer of avermectin) and S. scabies (the agent of potato scab) are less closely related members of the other. The two clusters shared their last common ancestor probably about 220,000,000 years ago (A. C. Ward, personal communication). No genomes have been sequenced from a smaller, more remotely related cluster that includes the genetically studied producer of oxytetracycline, Streptomyces rimosus (340).
The genus Frankia has a special significance as the nitrogen-fixing partner in a symbiosis with certain nonleguminous plants, most notably of the genera Alnus, Casuarina, and Elaeaginus, permitting these plants to grow well in nitrogen-poor soils. The three main host range groups are phylogenetically close to each other (their last common ancestor existed about 120,000,000 years ago), and representatives of all three have been subjected to genome sequencing (319).
Thermobifida is moderately thermophilic, growing optimally at 55°C and acting as a major degrader of plant cell walls in heated organic materials such as compost heaps, rotting hay, manure piles, or mushroom growth medium. It is a source of heat-stable extracellular enzymes such as cellulases. Its spores can be allergenic and cause a condition called farmer's lung.
The literature on Streptomyces genomes is particularly extensive, whereas the Frankia and Thermobifida fusca genomes have been fully published only very recently, so this part of this review is inevitably biased towards Streptomyces, with an emphasis on the model organism S. coelicolor A3(2).
Architecture of Mycelial Actinobacterial Genomes
Streptomyces chromosomes, at around 8 to 10 Mb, are very large; for example, the chromosomes of the well-known free-living unicellular bacteria E. coli K-12 and Bacillus subtilis 168 are about half as large, both in DNA content and in gene numbers. Indeed, the S. coelicolor chromosome was the first case in which a bacterium was shown to carry more genes (i.e., 7,825) than the simple eukaryote Saccharomyces cerevisiae (which contains 6,203 recognized genes) (26). The situation is different for Frankia genomes: three have been sequenced, representing each of the three major host range types, revealing surprisingly disparate sizes (9.04, 7.5, and 5.5 Mb) (319). Large genomes are not always associated with mycelial growth; for example the T. fusca chromosome is only 3.64 Mb (281). Both Streptomyces and Frankia have particularly high GC contents (around 72 to 73%), while the T. fusca genome contains 67.5% GC (Table 6).
TABLE 6.
State of genomic sequencing of streptomycetes
| Microorganism | Accession no. | Main reference | Comments |
|---|---|---|---|
| S. coelicolor [M145 derivative of A3(2)] | AL645882 (http://streptomyces.org.uk) | (26) | 8.66 Mb, 72.1% GC, TIRs 21.7 kb, 7,826 genes, 146 (2%) with TTA; plasmids SCP1 (linear, 365 kb) and SCP2 (circular, 31 kb) |
| S. avermitilis ATCC 31267 | BA000030 (http://avermitilis.ls.kitasato-u.ac.jp) | (194) | 9.02 Mb, 70.7% GC, TIRs 174 bp, 7,575 genes, 260 (3%) with TTA; plasmid SAP1 (linear, 94 kb) |
| S. venezuelae ATCC 10595 | Diversa Corp., unpublished; M. J. Bibb, personal communication | Sequence completed but not publicly available | |
| S. scabies ATCC49173 | http://www.sanger.ac.uk/Projects/S_scabies/ | R. Loria and S. D. Bentley, personal communication | 10.1 Mb, TIRs 18.5 kb |
| S. peucetius ATCC 27952 | (331) | 8.7 Mb; sequence completed but not publicly available | |
| S. ambofaciens ATCC 23877 | AM238663 (left end), AM238664 (right end) | (76) | TIRs ca. 198 kb; complete sequence of chromosome ends (1,544 kb and 1,367 kb), partial sequence of central region |
| S. griseus IFO13350 | Y. Ohnishi and S. Horinouchi, personal communication | Sequence completed in 2006, to be published |
Another remarkable feature of Streptomyces chromosomes is their linearity (274), a feature that had not been apparent from the extensive genetic linkage mapping of S. coelicolor (for reviews, see references (185 and 464). Although linearity has been demonstrated for the chromosomes of several different mycelial actinobacteria, including species of Saccharopolyspora, Actinoplanes, Micromonospora, and Nocardia (61, 69, 362), the genomes of Frankia and T. fusca are circular, so linearity is not closely associated with mycelial growth. Indeed, the recently published chromosome sequence of the actinobacterial species Rhodococcus sp. strain RHA1, which is not mycelial, is both very large (9.7 Mb) and linear, with telomeres (i.e., chromosome ends) like those of streptomycetes (289). Linear chromosomes have also been reported for a few other bacteria, in which the details of the structures of telomeres, and hence of replication, are different (464). The replication of linear Streptomyces chromosomes and plasmids (70) is initiated from a fairly centrally located replication origin rich in DnaA box sequences and proceeds bidirectionally towards the telomeres (396). The telomeres themselves are replicated by a mechanism that includes priming from a terminal protein covalently bound to the 5′ ends (70). The approximately 250 to 320 nucleotides at these ends possess a characteristic and complex secondary structure. Linearity of the Streptomyces chromosome is thought to have originated by single-crossover recombination between an initially circular chromosome and a linear plasmid (67, 69, 464). There are several examples of exchange of ends between chromosomes and linear plasmids, to give hybrid molecules with different right and left ends. Most commonly, though, both ends of linear plasmids or chromosomes are the same, giving recognizable terminal inverted repeats (TIRs) called TIR-L (at the conventional left end) and TIR-R (at the other end). TIR lengths among available sequenced Streptomyces chromosomes cover a 1,000-fold range: 174 bp for S. avermitilis, 18,488 bp for S. scabies, 21,653 bp for S. coelicolor M145, and approximately 198 kb for S. ambofaciens. Even within one strain, the lengths of the chromosomal TIRs can change through unequal recombination between ends (e.g., in S. ambofaciens [76]) or recombination within a TIR, such as might occur between different copies of a transposable element. An example of this in S. coelicolor resulted in the loss of about 1 Mb from one of the TIRs present in the parental A3(2) strain during the derivation of the plasmid-free strain M145 that was used as the source of DNA for genome sequencing (Fig. 10) (470). This can complicate the interpretation of experiments involving sequences present in the 1-Mb sequence: for example, a sequence chosen to be close to, but not within, TIR-R in the sequenced M145 genome was used as a hybridization probe specific for the right end of the chromosome in fluorescence in situ hybridization experiments (490), but the sequence has subsequently turned out also to be present at the left-hand end of the particular strain used for this cytological work (J1508) (C. W. Chen, personal communication). The conclusion of Yang and Losick (490), that the two chromosome ends are located close to each other at the growth stages studied, is therefore questionable.
FIG. 10.
Loss of a large segment of duplicated DNA from S. coelicolor A3(2). Strain M145, used for genome sequencing, is a prototrophic, plasmid-free derivative obtained from the wild-type strain A3(2). After sequencing, it was discovered that A3(2) has much longer chromosomal TIRs than M145, and it was deduced that the reduction in length was probably because of homologous recombination between copies of ORF SCO0020. Black areas, conserved central unique region; hatched areas, region duplicated in A3(2) but not M145; white areas, extent of duplication in M145; dotted lines, genes omitted for clarity.
Comparative Genomics of Mycelial Actinobacterial Genomes
Whole-genome synteny plots show that there has been remarkable conservation of the overall position and orientation of common genes in the chromosomes of S. coelicolor and S. avermitilis. The two chromosomes differ at the overall genome scale by four inversion events, each arranged fairly symmetrically around the centrally placed replication origin (194). Recent near-complete sequence analysis of the genome of S. ambofaciens, which is phylogenetically close to S. coelicolor, has suggested that two of these inversions took place before the divergence of S. coelicolor and S. ambofaciens, while the other two are assumed to be more recent events (Fig. 11). However, the fact that the inversion points correspond to species-specific DNA makes unambiguous interpretation of such data difficult and could mean that rearrangements are less efficiently counterselected at these points than elsewhere (75).
FIG. 11.
Inversions in the lineage of Streptomyces and Thermobifida fusca chromosomes. Dotted lines indicate the nonconserved ends of linear Streptomyces chromosomes (the T. fusca chromosome is circular). The origin of replication is indicated by a shaded oval. Connected arrows indicate inversion events deduced from overall synteny plots between chromosomes (65, 75, 194, 442). The diagram should be taken as an approximation of both the positions and frequencies of such events. There is only one clear example of a substantial inversion event that does not span the origin of replication (boldface arrows).
Ikeda et al. (194) found that about two-thirds (5,283) of the genes of S. coelicolor and S. avermitilis were conserved orthologs, as judged by reciprocal BLAST analysis. In a similar analysis, using slightly more stringent parameters, we found that 4,837 genes are orthologous between these two species (G. Chandra and K. F. Chater, unpublished data). Similar comparisons can be made with other emerging genome sequences, to more closely approach the “basic gene complement” of streptomycetes. Thus, when S. scabies is added into the equation, the number of genes conserved among the three species falls to 4,190, and a four-way analysis, taking advantage of privileged access to the unpublished sequence of S. venezuelae (M. J. Bibb, personal communication), reduces the number further to 3,566.
Normand et al. (319) obtained comparable results for the three sequenced Frankia genomes: of the 4,499 genes in the smallest of the three genomes, 2,810 were present in all three.
We also carried out wider reciprocal BLAST comparisons of Streptomyces genomes with the genomes of E. coli and B. subtilis, which shared 1,277 genes with each other in our analysis (Chandra and Chater, unpublished data). Of those 1,277 genes, 638 were also shared among the four sequenced streptomycetes (532 of the remainder were absent from all four streptomycetes, while the other 107 were absent from one to three of the four). Thus, only about 17% of the 3,566 genes common to four Streptomyces genomes are also present in E. coli and B. subtilis. Of the 1,277 genes common to E. coli and B. subtilis, 727 are present in T. fusca and 661 in at least one Frankia genome. Among these, 416 are present in all actinobacterial genomes (Chandra and Chater, unpublished data).
Multiply represented metabolic genes.
Probably reflecting the complex morphological and physiological differentiation of streptomycetes, their genomes contain several examples of apparent redundancy of metabolic genes. The most studied involves two nearly identical clusters (SCO5443 to -5440 and SCO7335 to -7332) that contain genes encoding functions involved in carbon storage transactions, with each cluster being specific for different hyphal cell type (51, 387), but others include gene sets for enzymes of the pentose phosphate pathway (SCO1935 to -1939 and SCO6657 to -6663) and multiple fabH-like genes for the first step in fatty acid biosynthesis (several of these are associated with secondary metabolism gene sets).
Genes unexpectedly missing from mycelial Actinobacteria.
The determinants of two of the three subunits of exonuclease V (the recB and recC genes) are notably absent from streptomycetes, Frankia, and T. fusca, though they are present in other actinobacteria such as mycobacteria.
The XerCD pathway for the resolution of circular chromosomes after replication is specifically absent from streptomycetes, in agreement with the linearity of their chromosomes. The widely conserved ftsA gene, which is involved in cell division, is generally absent from actinobacteria, as are minC and minE, which play an important part in determining the choice of division site in many unicellular bacteria.
Conservons and transposons.
Bentley et al. (26) noted the presence in S. coelicolor of 13 paralogous sets of four genes, which they called conservons, that encode membrane-bound complexes resembling eukaryotic G protein-coupled regulatory systems (234). In each set, the first gene, cvnA, encodes a sensor kinase-like ATPase, and the fourth, cvnD, encodes a guanine nucleotide-binding GTPase. The cvn9 conservon has a conditional role in determining the timing of development and antibiotic production. Most Streptomyces genomes have about a dozen conservons, while other mycelial or partially mycelial actinobacteria have several (T. fusca has five, Frankia Ccl3 contains three, and N. farcinica harbors seven such conservons), in contrast to most mycobacteria, which possess just a single conservon. Interestingly, four of the S. coelicolor conservons are next to cytochrome P450 genes (see below).
Genes encoding predicted transposases are abundant in Streptomyces chromosomes (78 in S. coelicolor and 99 in S. avermitilis). Nevertheless, only a few of these appear to possess all the necessary genetic elements supporting transposability. There is a strong bias of transposase genes towards the subtelomeric end regions. Since some linear plasmids (notably SCP1 [28]) are also rich in transposase genes, it is likely that some of these genes have come to move between replicons by wholesale exchange of large DNA segments (especially of ends) rather than by direct transposition. In Frankia genomes, there is remarkable variability in the abundance of such genes: considering the combined totals of integrase and transposase genes (269 for the 9.4-Mb genome, 46 for the 7.5-Mb one, and 187 for the smallest), Normand et al. (319) speculated that the two more recently derived strains had acquired their relatively expanded and contracted genomes as a result of the increased plasticity conferred by the activity of transposable elements and their provision of scattered regions of sequence identity.
DNA Regions in Mycelial Actinobacterial Genomes Acquired by HGT
Despite the gross synteny between the central regions of Streptomyces genomes, there are hundreds of insertion-deletion (indel) differences, mostly involving one or a few genes, between S. coelicolor and S. avermitilis, often making it difficult to recognize synteny at the level of small groups of genes (see references 65 and 75 for examples). Streptomycetes also have numerous larger islands of species-specific DNA. Thus, 4 islands of >80 kb were peculiar to S. ambofaciens, while 12 islands of 12 to 149 kb were specific for S. coelicolor, as was found through comparative analysis of the two species (75). Before other Streptomyces genome sequences became available, Bentley et al. (26) defined 14 islands of likely laterally acquired DNA in the S. coelicolor genome on the basis of gene content, atypical GC content, or location next to a tRNA determinant; half of these turned out to be shared with S. ambofaciens (75).
Further significant variation is provided by plasmid-chromosome exchanges such as those already referred to. It is therefore not surprising that pairwise synteny plots show that the genes in the “subtelomeric” “arms” of Streptomyces chromosomes are much less conserved between species than are those in the central regions, or “cores,” and that the cores contain most of the genes conserved with other actinobacteria (26, 76, 194). The boundaries between these regions are not well defined. Choulet et al. (76), considering S. ambofaciens, S. coelicolor, and S. avermitilis, showed that there is in fact a long gradient of indel differences between the core and arm regions. This is consistent with the interpretation by Chater and Chandra (65) of the way in which laterally acquired genes in the arms can be expected to migrate inwards towards the core over evolutionary time if they confer adaptive benefits that are subject to comparatively frequent selection. A corollary of this is that the conserved core region is longer when the more closely related S. coelicolor and S. ambofaciens genomes are compared than when either of those species is compared with S. avermitilis (76). It is interesting to note that the three differently sized Frankia genomes also show a strong tendency for species-specific genes to be located near the replication terminus, even though the Frankia genomes are circular (319).
Streptomyces Extrachromosomal Elements
Many plasmids from streptomycetes have been described (for a review, see reference 225). As already mentioned, linear plasmids, often very large, are widespread and diverse among streptomycetes (67), but circular plasmids are also found. Most Streptomyces plasmids are self-transmissible but phenotypically cryptic. Sequences of a range of them have been published. Focusing first on organisms whose entire genomes have been characterized, wild-type S. coelicolor A3(2) contains the 365-kb linear plasmid SCP1, which encodes the production of an antibiotic (see below) (25), and the 31-kb circular cryptic plasmid SCP2 (173). Both plasmids have been implicated in chromosome mobilization (185), and they can also interact with each other via a common transposable element (67). SCP1 also bears traces of the endwise integration of another linear plasmid within its length (25). SCP2 (in its SCP2* form) derivatives have been important in cloning, particularly as low-copy-number, high-capacity vectors for S. coelicolor (225). S. avermitilis also contains a 94-kb cryptic linear plasmid, SAP1 (194). Several small, high-copy-number cryptic plasmids of otherwise little-studied streptomycetes have been characterized, all replicating via a rolling-circle mechanism. Among these, pIJ101, from a strain of S. lividans, has found many uses in gene cloning and expression, and pSG5, from S. ghanaensis, has been used as the basis of a gene disruption system, because it exhibits temperature-sensitive replication (225).
Prophage-Like Elements in Streptomyces
There are no obvious prophages in the S. coelicolor A3(2) genome, though there are integrated plasmids of the SLP1/pSAM2 family, which can excise from their tRNA attachment sites and self-transfer as double-stranded DNA on contact with strains lacking the relevant plasmid (185, 345). One likely prophage can be found in the S. avermitilis genome (coordinates 6681496 to 6721444). Two putative integrase determinants are present in this element, a situation also found in some streptococcal phages. The prophage also has a homolog of the spd genes of Streptomyces plasmids, which make it possible for such plasmids, after transfer to a new host, to spread within a mycelium by transfer to adjacent hyphal compartments (see reference 185 for a review). A spd-like gene is also present in phiC31 (M. C. Smith, personal communication), which is by far the most studied of the substantial number of Streptomyces phages analyzed after their isolation from soil or release from lysogens (10, 279). The genomes of several of these have been sequenced, i.e., phiC31 (395), the phi31-homo-immune phage phiBT1 (NCBI source NC_004664), and VWB (444). All three sequenced phages belong to the Siphoviridae family and resemble Sfi21-like prophages in their modular organization (57). phiC31 has a remarkable system of prophage repression, with repressor targets in each of the multiple promoters spread through the early region, and these promoters have an unusual, and phage-specific, character (196, 480).
phiC31 has provided valuable systems for gene cloning and molecular analysis in streptomycetes (225), and its site-specific integration system is widely used in higher-organism genetics (102). Most Streptomyces phages have a host range confined to streptomycetes, with different extents of host specificity within the genus; phiC31 infects about half of the strains tested (279).
Mycelial Actinobacterial Genomes and Biological Lifestyle
Ecology.
Streptomycetes are highly successful and widely distributed soil organisms, playing important global roles in the recycling of organic matter held in relatively intractable macromolecular forms, such as the walls of plants and fungi. The mycelial growth habit probably assists in penetrating such materials. It is not surprising that Streptomyces genomes encode huge numbers of predicted secreted proteins; the ∼800 in S. coelicolor include 147 hydrolases, of which 7 are cellulases and 5 are chitinases (26). Although many of these are probably exported as unfolded proteins through the major secretory pathway, a remarkably high number (around 20% in S. coelicolor) were predicted to be exported in the folded state via the twin arginine translocation (Tat) pathway, the importance of which has been backed up by substantial experimental evidence for at least 28 of these proteins, using proteomics and a Tat-specific reporter system (475). Interestingly, very few of the 28 are expected to require metal or other cofactors, which was hitherto considered to be a typical feature of Tat-dependent proteins. It seems, therefore, that the Tat pathway is a general protein export system in Streptomyces.
In addition to the transport of proteins, streptomycetes need to transport many smaller molecules, both outwards in the case of antibiotics, siderophores, and other secondary metabolites and inwards in order to profit from the activities of their hydrolytic exoenzymes. Some 614 S. coelicolor gene products were annotated as being associated with transport, including many ABC transporter components (26).
The importance of the extracellular context of streptomycetes is also reflected in the possession by S. coelicolor of nearly 50 genes for different “extracytoplasmic function” sigma factors (26), as well as of those for 84 sensor kinases (and for 80 response regulators, 67 of which lie adjacent to sensor kinase-encoding genes and are therefore assumed to represent individual two-component systems) (192). Further sensory pathways presumably involve some of the (at least) 34 deduced serine-threonine protein kinases (26, 341). Similar numbers of such regulatory genes are found in other sequenced Streptomyces genomes.
Frankia and Thermobifida, though less generally abundant, are also free-living soil organisms, and they both have many transport systems and extracellular enzymes. Thus, T. fusca has 45 genes that are presumed to encode hydrolytic enzymes for oligo- or polysaccharides, including six cellulases. Many of the T. fusca exoenzymes are presumed to be transported by the Tat system (as yet, no such analysis has been published for Frankia) (281).
Several genes specific for the symbiotic, nitrogen-fixing alternative lifestyle are present in the genomes of Frankia species. Most of these genes are common to all three sequenced chromosomes, and they are located in scattered positions. This is in contrast to the plasmid location of symbiotic genes in rhizobial genomes (319).
Secondary metabolism.
Streptomycetes have been the most abundant source of clinically important antibiotics since the discovery of actinomycin D, streptothricin, and streptomycin in the 1940s by Waksman and coworkers (for a review, see reference 186). One major reason for sequencing their genomes was to uncover their coding capacity for secondary metabolites. This was rewarded by the discovery of 23 such gene sets in the S. coelicolor chromosome (26, 62) and 30 in that of S. avermitilis (194, 324). Many of these gene sets are present in one genome but not in the other. Indeed, the same position in different chromosomes can be occupied by different secondary metabolism clusters: for example, the pks1 cluster of S. avermitilis (SAV7356 to -7422) is replaced in S. ambofaciens by a different secondary metabolism cluster of 28 genes and in S. coelicolor by a 31-gene insertion (SCO0850 to -0880) (75).
The subtelomeric chromosome arms are comparatively rich in gene clusters for secondary metabolism, especially those that are species specific. More widely present genes for secondary metabolism, such as those for biosynthesis of pentalenolactone, some siderophores, and the odor compound geosmin, typically fall in syntenous locations within the central core region: (26, 194, 324). Certain linear plasmids also carry such clusters, providing a plausible route for their lateral transfer between chromosomes residing in different streptomycetes—it has already been pointed out that the ends of chromosomes and plasmids can undergo genetic exchange. In one such instance, the pPZG101 plasmid of Streptomyces rimosus appears to have picked up one end of its host's chromosome, including the genes for the biosynthesis of oxytetracycline (154, 330). Strikingly, the sequence of the 211-kb linear plasmid pSLA2-L of Streptomyces rochei carries five gene sets for secondary metabolism, making up about two-thirds of the plasmid (296), while SCP1, a 365-kb linear plasmid of S. coelicolor, carries the methylenomycin biosynthetic genes (25). In the case of SCP1, several independent and different kinds of integration into the chromosome have been demonstrated, some resulting in the insertion of SCP1 via its ends into the central region of the chromosome, but also one in which a recombinational exchange resulted in two large chromosome-SCP1 hybrid molecules with complementary structures (for a review see reference 67). Remarkably, a methylenomycin biosynthetic cluster almost identical to that found in SCP1 is present on pSV1, a circular plasmid of Streptomyces violaceusruber SANK 95570, which otherwise has very little sequence in common with SCP1 (489).
P450 cytochromes (CYPs).
Streptomyces genomes, and those of most other actinobacteria, encode unusually large numbers of CYPs (S. coelicolor, 18; S. avermitilis, 33; S. scabies, 25; S. peucetius, 19; M. tuberculosis, 20; M. smegmatis, 39; M. bovis, 18; M. vanbaalenii, 51; and N. farcinica, 26 [194, 249, 331; D. C. Lamb, personal communication]). The frequency of CYPs in the different Frankia genomes relates approximately exponentially to size, with the largest genome (9.04 Mb) having 47 annotated CYP genes, the next largest (7.5 Mb) 22, and the smallest (5.4 Mb) 13.
There is little overlap in the CYP contents of streptomycetes and mycobacteria. Even the one apparently conserved family, CYP125, is present in only some members of each genus (linkage of the CYP125 gene to genes for lipid degradation suggests a role in lipid breakdown). More than 200 Streptomyces CYPs are known at the time of this writing, from at least 46 species. There are no CYP genes in E. coli K-12 and the majority of gram-negative bacteria or in some actinobacterial genomes (T. whipplei and C. diphtheriae), while M. leprae has only one.
The main function of CYPs is the monooxygenation of various substrates, which include many naturally occurring antagonists and xenobiotics. About 10% of Streptomyces CYP genes are clustered with genes for ferredoxins, and a substantial number are associated with gene clusters for secondary metabolite production (whereas many of those of mycobacteria are probably connected to the biosynthesis of the complex lipids that these species produce). The 18 genes for cytosolic CYPs of S. coelicolor A3(2) have all been shown to be expressed in standard culture conditions, and all 18 have been expressed in E. coli (250). Control of CYP activity in S. coelicolor A3(2) involves temporal expression of ferredoxin reductases, with maximal activity being achieved via the correct interactions of P450, ferredoxin, and ferredoxin reductase, a previously unrecognized mechanism for regulating CYP function (262). Unusual CYP forms are associated with a specific group of repeated elements of streptomycetes, termed conservons (see above).
Development.
Streptomycetes are the only complex actinobacteria whose development has been studied in detail. Many of the genes involved in the formation and sporulation of their aerial hyphae have been identified, and they were recently subjected to a comparative genomic analysis (65). Here we summarize the main take-home messages. The gene numbers for S. coelicolor developmental genes have been included here to help future cross-referencing.
It is postulated that at the end of the main growth phase, morphological differentiation to give rise to a sporulating aerial mycelium is the least favorable of various physiological options and that the inability to achieve each of these options is signaled by the production of an extracellular signal, under the influence of a so-called “bld” gene. Mutations in such bld genes prevent signal emission and hence prevent differentiation unless the signal is provided from an outside source, such as the wild type or a different bld mutant growing close by. Thus, the decision to form an aerial mycelium appears to be the endpoint of a series of checkpoints. At least one bld gene, bldB (SCO5723), seems to operate outside of this checkpoint cascade. Most of the known bld genes are represented only in the genomes of mycelial genera of actinomycetes, with bldB, bldH (adpA) (SCO2792), bldK (SCO5112 to -5116), and bldM (SCO4768) being confined to Streptomyces and bldN (SCO3323) also being found in Frankia, while bldC (SCO4091), bldD (SCO1489), and bldG (SCO3549) are present in all three genera.
When this series of signals confirms that aerial growth is necessary, several different types of structural proteins are produced, which assemble at the interface of the air with the medium or mycelial surface and permit aerial growth by forming a surface layer on the emerging aerial branches (118, 143, 476). These surface proteins include (i) somewhat species-specific small peptides, such as SapB, which are processed from a larger gene product (in S. coelicolor that of ramS, one of five genes in the ram cluster SCO6681 to -6685); (ii) the chaplins, a large family of closely related amphipathic proteins (SCO1674 to -1675, -1800, -2699, -2705, -2716 to -2717, and -7257); and (iii) the rodlins (SCO2718 to -2719), which are needed for the formation of the rodlet structures observed on the surfaces of spores and aerial hyphae of some species. It appears that SapB-like proteins and rodlins are peculiar to streptomycetes (although rodlins are absent from S. avermitilis), while chaplins are found in all three mycelial species. None of the surface proteins have been found outside of mycelial actinobacteria.
Once the aerial hyphae emerge, further developmental genes, mostly called whi genes, control the processes of sporulation septation and spore maturation. One section of this regulatory network involves whiG (SCO5621), which specifies a homolog of the fliA-specified sigma factor needed for motility in many motile single-celled bacteria. The WhiG sigma factor directly activates the whiH (SCO5819) and whiI (SCO6029) sporulation regulatory genes. These three genes, all of which are needed for sporulation septation, are absent from all actinomycetes except streptomycetes, with the single exception that a whiG-like gene is present in Leishmania xyli, the only motile actinobacterial species that has been subjected to genome sequencing.
A parallel section of the Whi network preceding sporulation septation involves whiA (SCO1950), homologs of which are universal among (and confined to) gram-positive bacteria, and whiB (SCO3034), homologs of which are confined to (and universal among) actinobacteria. No role for whiA-like genes has been identified in other bacteria, despite their wide occurrence. The whiB ortholog of mycobacteria (e.g., whmD of M. smegmatis) is important for cell division, and whiB of S. coelicolor can complement a whmD mutant of M. smegmatis, implying that the mode of action of the orthologs has been retained over evolutionary time even though their developmental contexts differ (352). It seems that in Streptomyces whiA and whiB have become unimportant for growth and have instead been sequestered for development. This may have been made possible by the change from a predominantly single-celled growth habit, in which increased biomass and cell division/separation are inextricably linked, to an obligatory mycelial growth habit, in which cell division (and especially that leading to cell separation) is largely suppressed.
For the maturation of prespore compartments into normal spores, at least two further regulatory genes are needed. One of these, sigF (SCO4035), encodes a sigma factor of a gram-positive-specific subclass, whose best-known members are specified by B. subtilis, and include the SigB stress-responsive sigma factor and two forespore-specific sigma factors (SigF and SigG). There are eight other members of this family in S. coelicolor, and one of them, encoded by sigN (SCO4034, immediately downstream of sigF), appears to be crucial for the differentiation of a specific aerial hyphal compartment immediately below the sporulating tip compartment (103). Another, SigB (SCO0600), is involved in stress responses (258). In B. subtilis, most members of this family are regulated by anti-sigma/anti-anti-sigma cascades, the components of which are encoded by genes immediately adjacent to the sigma factor determinant (68). In streptomycetes, on the other hand, most of the genes that encode this sigma factor type are separated from genes encoding predicted antagonists. Possibly, this may indicate some degree of promiscuity in the antagonistic interactions in streptomycetes, in contrast to a comparatively high specificity in low-GC gram-positive bacteria.
The second known late sporulation regulatory gene is whiD (SCO4767). WhiD is a Wbl protein, a paralog of WhiB (298). WhiD orthologs are widespread among actinobacteria. In M. tuberculosis, the WhiD ortholog WhiB3 has a role in pathogenicity, and some evidence suggests a direct interaction with the principal sigma factor (410). It is therefore interesting that three of the wbl genes of S. coelicolor are located within two genes of sigma factor determinants, and one, wblP of the S. coelicolor plasmid SCP1, encodes a Wbl-sigma fusion protein (25). WhiD contains a redox-sensitive iron-sulfur cluster coordinated by four cysteine residues that are highly conserved among Wbl proteins and are needed for their biological function (4, 204, 352). Remarkably, two members of the family have been recently shown to have protein disulfide reductase activity (4, 138). Three other Wbl proteins are widely conserved among actinobacteria. WblC (SCO5190) activates multiple resistance to antibiotics and other inhibitors in streptomycetes and mycobacteria and so possibly does the same in all organisms that possess it (304). WblA has very recently been found to have effects on secondary metabolism and to be needed for a very early stage of sporulation in streptomycetes (K. Fowler, B. Gust, K. Findlay, N. Bird, and K. F. Chater, unpublished data; S. H. Kang, J. Huang, H. N. Lee, Y. A. Hur, S. N. Cohen, and E. S. Kim, personal communication). The role of WblE (SCO5240) is unclear, since a wblE mutant of S. coelicolor exhibits no obvious phenotype (182); however, a mutant of the corynebacterial ortholog showed increased sensitivity to heat and oxidative stresses (227).
The various stages of aerial growth and development involve temporally and spatially specific modulation of peptidoglycan synthesis and metabolism. Evidence is growing that this involves another family of paralogs, the most studied of which are ssgA (SCO3926) and ssgB (SCO1541) (318). With the exception of ssgB, which is also found in T. fusca and Frankia, most of the paralogs are found only in streptomycetes. In S. coelicolor, mutations in any of the paralogs affect different stages of aerial mycelium development (318). Other Actinobacteria-specific genes having significant effects on development have also been described but are less extensively characterized. Several of these appear to be widespread among nondifferentiating actinobacteria, including samR (SCO2935), crgA (SCO3845), and paralogous members of the bldB (SCO5723) and whiJ (SCO5443) families (65).
Based on the distribution of these developmental genes among actinobacterial genomes, Chater and Chandra (65) proposed a scheme for the evolution of development in actinobacteria, which is presented in a modified form in Fig. 12.
FIG. 12.
Sequential acquisition of developmental genes during the evolution of mycelial actinobacteria. The actinobacterial lineage, based on the occurrence of orthologs of S. coelicolor developmental genes, is shown in the shaded area. (Modified from reference 65 with permission of the publisher.)
Specialized use of the rare UUA leucine codon.
Correlating with the high GC content of most actinobacterial DNA, codon usage is highly skewed towards codon alternatives ending in G or C. In the case of leucine, there are six alternative codons, one of which, UUA, contains neither G nor C. Examination of the available Streptomyces genome sequences suggests that only one tRNA can translate UUA codons. Remarkably, strains with mutations or deletion of bldA, the determinant of the UUA-reading tRNA from S. coelicolor, grow vigorously, indicating either that no TTA-containing genes are essential for growth or that some other tRNA can in fact translate UUA codons (253). The former turns out to be the case, since bldA mutants are impaired in the expression of TTA-containing genes (265). Although they do not impede growth, bldA mutations of S. coelicolor do have phenotypic consequences: the colonies fail to form aerial mycelium on most laboratory media, and they also fail to make several different antibiotics. These processes are typically stationary-phase associated, and indeed the bldA tRNA shows increased abundance in stationary phase, whereas other tRNAs are more abundant during rapid growth (266, 437). From such observations, the idea grew that bldA has a regulatory role in relation to the biology of the stationary phase (reviewed in references 66 and 264).
Of the 7,825 annotated genes in the S. coelicolor chromosome, 145 contain TTA codons. Only 42 of these genes give reciprocal BLAST hits with genes in S. avermitilis. Thus, the large majority of TTA-containing genes are likely to have been acquired by lateral gene transfer, such that genes with TTA codon are found 10 times more often among putatively laterally transferred genes than among conserved genes. In support of this, 31 were located within islands thought to have been acquired by HGT (270). Of the 42 genes common to S. coelicolor and S. avermitilis, TTA codons are present in both orthologs in 12 cases (270). Just 5 of these 12 genes are also conserved, with their TTA codons, in S. scabies and S. venezuelae (Chandra and Chater, unpublished data). Chater and Chandra (65) have suggested that such conserved TTA-containing genes, and a putative regulatory role of bldA, are an ancient feature of streptomycetes, possibly having been present at the earliest stages of their evolution. This notion is supported by the finding that bldA mutants of phylogenetically distant streptomycetes have similar pleiotropic phenotypes. One of the conserved TTA-containing genes, adpA, was shown to be the principal route through which bldA exerts its influence on morphological development (313, 418), whereas the effects of bldA mutation on secondary metabolism seem often to be mediated by pathway-specific (and therefore somewhat organism-specific) TTA-containing regulatory genes (63). Li et al. (270) systematically inactivated 12 of the TTA-containing genes of S. coelicolor conserved in S. avermitilis as well as 9 of the nonconserved ones, but they could find no clear phenotypic effects. They suggested that such genes were likely to be of particular adaptive value and phenotypically significant, mainly in specific ecological situations that are not mimicked by normal laboratory conditions.
Chater and Chandra (65) found that this specialized use of TTA codons did not seem to be shared by other actinobacteria sequenced so far, since TTA-containing genes were generally much more frequent in those genomes and included certain genes considered to be required for growth-associated processes. Since that review, Frankia genome sequences have become available, and the comparatively close relatedness of Frankia to streptomycetes, and the similarly high GC content, makes it interesting to consider the possible significance of TTA codons in these genomes. TTA-containing genes are somewhat more frequent in the two Frankia spp analyzed than in any of four streptomycetes, and none of them corresponds to any of the TTA-containing genes conserved among the four Streptomyces genomes (Chandra and Chater, unpublished data). The positions of TTA codons within Streptomyces genes show a particularly strong bias towards the early parts of the genes in streptomycetes compared with other actinobacteria, a position that may be optimal for the modulation of translation (65, 133, 265). Figure 13 shows that this effect is also more marked in Streptomyces than in Frankia, again suggesting that the specialized role of TTA codons in streptomycetes is not shared with Frankia.
FIG. 13.
Relative intragenic positions of TTA codons in streptomycetes, Frankia, and T. fusca. Blue, S. coelicolor (Sco); pink, S. avermitilis (Sav); brown, S. scabies (Ssc); lilac, S. venezuelae (Sven); dark green, Frankia alni (Fal); light green, Frankia sp. strain Ccl3 (Fra); light brown, T. fusca (Tfu); red, E. coli (Eco); pale pink, M. tuberculosis (Mtb). TTA codons show a more pronounced bias in distribution towards the start of the gene in Streptomyces genomes.
COMPARATIVE GENOMICS OF ACTINOBACTERIA
Synteny of Actinobacterial Genomes
Due to the profound diversity and the taxonomic distance between the various Actinobacteria, protein rather than DNA alignments have to be employed in order to provide a meaningful global view of the existing relationships. Alignments of very closely related genomes usually show a continuous diagonal line starting from one corner of the xy plot figure up to the opposite corner, indicating a high degree of synteny of the genes that are conserved. Nonconserved genes, due to either insertions or deletions, contribute spaces rather than signals to these plots. Departures from the simple diagonal plot may reflect the occurrence of genomic rearrangements such as inversions, transpositions, duplications, or large-scale deletions that have occurred since the last common ancestor of the two strains being compared. The most obvious such departure is the formation of an X-shaped plot, reflecting inversions that include the replication origin (50). This latter can be seen from genome comparison of strains T. whipplei TW08/27 and T. whipplei Twist. Although the close relatedness of these two strains is obvious from their continuous regions of alignment, there is a large region in the alignment going in the opposite direction, from the upper left corner to the lower right corner following an X-shaped distribution (Fig. 4).
Using whole-genome alignments at the protein level (Fig. 8; see Fig. S3 in the supplemental material), the first stages in evolutionary divergence can be seen even between two separately maintained descendants of a single isolate of C. glutamicum ATCC 13032, which show a small interruption in an otherwise near-perfect alignment (Fig. 8; see Fig. S3a in the supplemental material). Interspecific comparisons of corynebacterial genomes also show strong synteny, but the diagonal plot is somewhat distorted by the accumulated indel differences scattered through the genome (see Fig. S3b in the supplemental material). These give rise to a larger number of points off the main diagonal, at least some of which are caused by hits between paralogs rather than orthologs. Alignments between two independently isolated strains of M. tuberculosis also show high synteny, though a modest number of off-diagonal points again reflect indel differences (see Fig. S3c in the supplemental material). The equally high level of synteny observed between M. bovis and M. tuberculosis strains (see Fig. S3d in the supplemental material) may suggest that M. bovis is an ecotype of M. tuberculosis (50, 188) rather than a separate species. In contrast, despite high sequence identity at the level of individual genes, the genome alignment between M. avium subsp. paratuberculosis and M. bovis reveals many inversion events separating the organisms and therefore a greater evolutionary distance (see Fig. S3e in the supplemental material). Whole-genome alignments of Mycobacterium sp. strain MC, a Mycobacterium strain isolated from the environment, indicate that it is more related to M. bovis and M. tuberculosis than to M. avium (data not shown). Alignments of any of these mycobacterial genomes with that of M. leprae show much less synteny, in line with their different GC contents (M. leprae, 57.8%; M. tuberculosis, 65.6%) and lower conservation at the level of individual gene products (see Fig. S3f in the supplemental material). This diversity may reflect the fact that M. leprae is not subject to many of the selective forces that impinge on free-living organisms.
High levels of synteny and sequence conservation can still be observed in some cases when genome sequences of different families of the order Actinomycetales are aligned. The human pathogen N. farcinica shows closer relatedness to Mycobacterium (see Fig. S3g in the supplemental material) and Corynebacterium than to other Actinomycetales. Genome-wide comparison of a Frankia sp. with T. fusca XY produced defined alignments (see Fig. S3h to i in the supplemental material), displaying an X-shaped distribution that encompasses large genome regions. Comparisons of circular chromosomes with linear ones are complicated by the different conventions for presenting the two kinds of genomes: the numbering of genes in circular genomes conventionally starts from the replication origin, whereas in linear genomes the sequence, and hence the gene numbering, runs from end to end, with the replication origin being approximately central. Thus, if a circular genome and a linear genome were otherwise perfectly syntenous, the resulting synteny plot would be shaped as a V (or an inverted V). Any inversions including the origin would introduce a rhomboid form to the plot. This is seen when Streptomyces genomes are compared with those of any of the other available actinobacterial genomes (see Fig. S3h to i in the supplemental material), though comparisons between Streptomyces species are still expected to generate diagonal/X-shaped plots (194). All intergeneric comparisons reveal multiple short regions of synteny interrupted by inversions including the replication origin, with very few examples of inversions that do not include the origin (which would be seen as additional X-shaped crossover points on the main diagonals). The synteny is generally somewhat obscured by the much-increased number of points that are located off the diagonal, which are mostly explained not by the different arrangement of orthologs, but rather by the greater number of “hits” between paralogs (which become more likely both with increasing numbers of paralogs and with greater sequence divergence between the orthologous components of genomes).
Actinobacterial Core Genome Sequences: Phylogenomics
A core can be defined as the set of all genes shared as orthologs by all members of an evolutionarily coherent group. They are relatively unlikely to have experienced HGT, which make them suitable targets as molecular markers to infer phylogeny. The larger the phylogenetic distance between organisms, the lower the number of core genes that can be compared; e.g., a minimal set of 60 core genes was obtained when comparing 100 genomes of Bacteria, Archaea, and Eukarya. For actinobacteria, the clustering method MCL, based on BLAST results, provides 123 proteins which can be useful for reconstructing actinobacterial phylogeny (Table 7). Although different analytical procedures may give different numbers of core proteins, the use of core sets derived by different procedures in phylogenomic analysis can be expected to yield comparable results. The large phylogenetic distances across actinobacteria are reflected in the relatively low number of orthologs found using the MCL clustering method. The phylogenetic trees obtained with of each of these genes are not all consistent, so a consensus tree (Fig. 14) was built using SplitsTree software (191). As expected, genome sequences belonging to different strains of the same species showed no distance, sharing the same topology (though the T. whipplei TW08/27 and T. whipplei Twist branches do not fully overlap, despite the fact that the two strains share >99% nucleotide identity [357]). On the other hand, different species in the same genera should not overlap, since their protein sequences record the history of an older speciation process. Based on this criterion, M. bovis and M. tuberculosis might both be considered to be members of the M. tuberculosis complex.
TABLE 7.
Protein-coding genes in the hypothetical minimal actinobacterial cells
| ORF designationa | Prominent features |
|---|---|
| BL0053 | Mrp; COG family, ATPases involved in chromosome partitioning; PFAM_ID, DUF59; PFAM_ID, fer4_NifH |
| BL0065 | Involved in peptide bond synthesis; alters affinity of the ribosome for aminoacyl-tRNA |
| BL0067 | Composed of two chains; the small (or glutamine) chain promotes the hydrolysis of glutamine to ammonia, which is used by the large (or ammonia) chain to synthesize carbamoyl phosphate |
| BL0068 | CarB |
| BL0070 | Gmp kinase; COG family, guanylate kinase; PFAM_ID, Guanylate_kin |
| BL0097 | Catalyzes the NADP-dependent rearrangement and reduction of 1-deoxy-d-xylulose-5-phosphate (DXP) to 2-C-methyl-d-erythritol 4-phosphate |
| BL0100 | Involved in DNA repair and recF pathway recombination |
| BL0118 | GTPase; similar structure to tubulin; forms ring-shaped polymers at the site of cell division; other proteins such as FtsA, ZipA, and ZapA interact with and regulate FtsZ function |
| BL0193 | COG family, Mg-dependent DNase; PFAM_ID, TatD_DNase |
| BL0291 | Pantetheine-phosphate adenylyltransferase; PpaT; dephospho-CoA pyrophosphorylase; CoaD; COG family, phosphopantetheine adenylyltransferase; PFAM_ID, Cytidylyltransf |
| BL0295 | RNase III; RncS; COG family, double-stranded-RNA-specific RNase; PFAM_ID, Ribonuclease_3; PFAM_ID, dsrm |
| BL0305 | RpsP; COG family, ribosomal protein S16; PFAM_ID, Ribosomal_S16 |
| BL0327 | M1g-methyltransferase; tRNA [GM37] methyltransferase; TrmD; COG family, tRNA-(guanine-N1)-methyltransferase; PFAM_ID, tRNA_m1G_MT |
| BL0328 | COG family, N6-adenine-specific methylase |
| BL0358 | AtpG; COG family, F0F1-type ATP synthase gamma subunit; PFAM_ID, ATP-synt |
| BL0395 | Catalyzes formation of valyl-tRNA(Val) from valine and tRNA(Val) |
| BL0402 | Allows formation of correctly charged Asn-tRNA(Asn) or Gln-tRNA(Gln) through the transamidation of misacylated Asp-tRNA(Asn) or Glu-tRNA(Gln) in organisms which lack either or both of asparaginyl-tRNA or glutaminyl-tRNA synthetases |
| BL0413 | RplI; COG family, ribosomal protein L9; PFAM_ID, Ribosomal_L9 |
| BL0431 | DnaB; COG family, replicative DNA helicase; PFAM_ID, DnaB |
| BL0499 | COG family, recombinational DNA repair protein; PFAM_ID, RecR; PFAM_ID, Toprim |
| BL0500 | Plays a role in assembling DnaB onto the primer template and interacts with the core polymerase |
| BL0519 | Hsp-70 cofactor; GrpE; COG family, molecular chaperone GrpE (heat shock protein); PFAM_ID, GrpE |
| BL0549 | Catalyzes formation of N6-(1,2,-dicarboxyethyl)-AMP from l-aspartate, IMP, and GTP in AMP biosynthesis |
| BL0637 | COG family, recombinational DNA repair ATPase |
| BL0640 | DnaA; COG family, ATPase involved in DNA replication initiation; PFAM_ID, AAA; PFAM_ID, bac_DNAA |
| BL0648 | ParB; COG family, predicted transcriptional regulators; PFAM_ID, ParBc |
| BL0703 | UvrC; COG family, nuclease subunit of the excinuclease complex; PFAM_ID, Exci_endo_N; PFAM_ID, UVR; PFAM_ID, HHH |
| BL0706 | COG family, uncharacterized Bcr; PFAM_ID, DUF199 |
| BL0707 | Catalyzes formation of 3-phospho-d-glyceroyl phosphate from 3-phospho-d-glycerate |
| BL0726 | COG family, uncharacterized Acr; PFAM_ID, DUF28 |
| BL0727 | Cleaves the cruciform structure in supercoiled DNA by nicking strands with the same polarity at sites symmetrically opposed at the junction in the homologous arms |
| BL0728 | COG family, Holliday junction resolvasome DNA-binding subunit; PFAM_ID, RuvA; PFAM_ID, RuvA_II |
| BL0729 | COG family, Holliday junction resolvasome helicase subunit; PFAM_ID, AAA |
| BL0735 | Involved in de novo purine biosynthesis |
| BL0848 | Binds to the ribosome on the universally conserved alpha-sarcin loop |
| BL0849 | RpsT; COG family, ribosomal protein S20; PFAM_ID, Ribosomal_S20p |
| BL0859 | Bex; essential protein in Escherichia coli that is involved in many cellular processes; GTPase; binds cell membrane through apparent C-terminal domain |
| BL0861 | COG family, predicted metal-dependent hydrolase; PFAM_ID, UPF0054 |
| BL0870 | COG family, iron-regulated ABC transporter ATPase subunit SufC; PFAM_ID, ABC_tran |
| BL0878 | Catalyzes formation of chorismate from 5-O-(1-carboxyvinyl)-3-phosphoshikimate in aromatic amino acid biosynthesis |
| BL0881 | COG family, predicted endonuclease involved in recombination (possible Holliday junction resolvase in mycoplasmas and B. subtilis) |
| BL0926 | COG family, predicted GTPase |
| BL0943 | Binds and unfolds substrates as part of the ClpXP protease |
| BL0947 | Tig; RopA; peptidyl-prolyl cis/trans-isomerase; promotes folding of newly synthesized proteins; binds ribosomal 50S subunit; forms a homodimer |
| BL0960 | Contains glutamine-hydrolyzing domain and glutamine amidotransferase; GMP-binding domain; functions to produce GMP from XMP in the IMP pathway |
| BL0964 | N-Acetylglucosamine-1-phosphate uridyltransferase; Tms protein; GlmU; COG family, N-acetylglucosamine-1-phosphate uridyltransferase (contains nucleotidyltransferase and I-patch acetyltransferase domains); PFAM_ID, NTP_transferase |
| BL0990 | UvrABC repair system catalyzes recognition and processing of DNA lesions; beta-hairpin of the UvrB subunit is inserted between the strands, where it probes for the presence of a lesion |
| BL0992 | Binds mRNA to the ribosomes and plays a role in delivering transfer mRNA to stalled ribosomes |
| BL1032 | Transfers an adenyl group from ATP to NaMN to form nicotinic acid adenine dinucleotide (NaAD), which is then converted to the ubiquitous compound NAD by NAD synthetase; essential enzyme in bacteria |
| BL1043 | Recombination protein N; COG family, ATPases involved in DNA repair |
| BL1051 | Tyrosine-tRNA ligase; TyrRS; TyrS; COG family, tyrosyl-tRNA synthetase; PFAM_ID, tRNA-synt_1b; PFAM_ID, S4 |
| BL1066 | Phenylalanine-tRNA ligase beta chain; PheRS; PheT; COG family, emap domain |
| BL1067 | Catalyzes a two-step reaction, first charging a phenylalanine molecule by linking its carboxyl group to the alpha-phosphate of ATP, followed by transfer of the aminoacyl-adenylate to its tRNA; forms a heterotetramer of alpha(2)beta(2); binds two magnesiusm |
| BL1099 | One of the primary rRNA-binding proteins; binds directly to 16S rRNA, where it nucleates assembly of the head domain of the 30S subunit; is located at the subunit interface close to the decoding center |
| BL1100 | Important for translational accuracy; interacts with and stabilizes bases of the 16S rRNA that are involved in tRNA selection in the A site and with the mRNA backbone; located at the interface of the 30S and 50S subunits, it traverses the body of the 30S subunit |
| BL1108 | PurL |
| BL1121 | Glutamine phosphoribosylpyrophosphate amidotransferase; PurF; COG family, glutamine phosphoribosylpyrophosphate amidotransferase; PFAM_ID, GATase_2; PFAM_ID, Pribosyltran |
| BL1122 | Catalyzes formation of 1-(5-phosphoribosyl)-5-aminoimidazole from 2-(formamido)-N1- (5-phosphoribosyl)acetamidine and ATP in purine biosynthesis |
| BL1123 | GarS; glycinamide ribonucleotide synthetase; phosphoribosylglycinamide synthetase; PurD; COG family, phosphoribosylamine-glycine ligase; PFAM_ID, GARS_N; PFAM_ID, GARS_B; PFAM_ID, GARS; PFAM_ID, GARS_C |
| BL1130 | Air carboxylase; AirC; PurE; COG family, phosphoribosylcarboxyaminoimidazole (ncair) mutase; PFAM_ID, AIRC |
| BL1180 | Binds to ssrA RNA and is required for its successful binding to ribosomes |
| BL1182 | FtsX; COG family, cell division protein; PFAM_ID, DUF214 |
| BL1187 | MrsA; COG family, phosphomannomutase; PFAM_ID, PGM_PMM_I; PFAM_ID, PGM_PMM_II; PFAM_ID, PGM_PMM_III; PFAM_ID, PGM_PMM |
| BL1192 | COG family, predicted hydrolase of the metallo-beta-lactamase superfamily; PFAM_ID, lactamase_B; PFAM_ID, UPF0036 |
| BL1204 | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates |
| BL1275 | HK; ThrB; COG family, homoserine kinase; PFAM_ID, GHMP_kinases |
| BL1282 | RplU; COG family, ribosomal protein L21; PFAM_ID, Ribosomal_L21p |
| BL1283 | Function of this ribosomal subunit is unknown |
| BL1284 | Obg; COG family, predicted GTPase; PFAM_ID, GTP1_OBG |
| BL1288 | NusG; COG family, transcription antiterminator; PFAM_ID, NusG |
| BL1312 | COG family, predicted transcriptional regulator; consists of a Zn-ribbon and ATP-cone domains; PFAM_ID, DUF193 |
| BL1316 | COG family, predicted S-adenosylmethionine-dependent methyltransferase involved in cell envelope biogenesis; PFAM_ID, Methyltransf_5 |
| BL1320 | Catalyzes binding of UDP-N-acetylmuramoyl(pentapeptide) to bactoprenol to produce lipid I in peptidoglycan synthesis |
| BL1321 | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase; d-glutamic acid-adding enzyme; MurD; COG family, UDP-N-acetylmuramoylalanine-d-glutamate ligase; PFAM_ID, Mur_ligase; PFAM_ID, Mur_ligase_C |
| BL1323 | UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide)pyrophosphoryl-undecaprenol N-acetylglucosamine transferase; involved in cell wall formation; inner membrane associated; last step of peptidoglycan synthesis |
| BL1324 | UDP-N-acetylmuramoyl-l-alanine synthetase; MurC; COG family, UDP-N-acetylmuramate-alanine ligase; PFAM_ID, Mur_ligase; PFAM_ID, Mur_ligase; PFAM_ID, Mur_ligase_C |
| BL1366 | InfC; COG family, translation initiation factor If3; PFAM_ID, IF3 |
| BL1366a | RpmI; COG family, ribosomal protein L35; PFAM_ID, Ribosomal_L35p |
| BL1367 | Protein binds directly to 23S rRNA and is necessary for the in vitro assembly process of the 50S ribosomal subunit |
| BL1415 | Catalyzes hydrolysis of ATP in the presence of single-stranded DNA, ATP-dependent uptake of single-stranded DNA by duplex DNA, and ATP-dependent hybridization of homologous single-stranded DNAs |
| BL1438 | dUTPase; dUTP pyrophosphatase; DutP; COG family, dUTPase; PFAM_ID, dUTPase |
| BL1450 | Leucine-tRNA ligase; LeuRS; charges leucine by linking carboxyl group to alpha-phosphate of ATP and then transfers aminoacyl-adenylate to its tRNA |
| BL1503 | Essential for binding of S1 to the small ribosomal subunit |
| BL1504 | EF-Ts; functions during elongation stage of protein translation; forms a dimer; associates with EF-Tu-GDP complex and promotes exchange of GDP to GTP, resulting in regeneration of the active form of EF-Tu |
| BL1505 | UK; UMP kinase; UMP kinase; PyrH; COG family, uridylate kinase; PFAM_ID, aakinase |
| BL1506 | Ribosome-releasing factor; Frr; COG family, ribosome-recycling factor; PFAM_ID, RRF |
| BL1507 | CDP-diglyceride synthetase; CDP-diglyceride pyrophosphorylase; CDP-diacylglycerol synthase; CdsA; COG family, CDP-diglyceride synthetase; PFAM_ID, Cytidylyltrans |
| BL1545a | RpsO; COG family, ribosomal protein S15p/S13e; PFAM_ID, Ribosomal_S15 |
| BL1546 | Polynucleotide phosphorylase; Pnpase; COG family, polyribonucleotide nucleotidyltransferase (polynucleotide phosphorylase); PFAM_ID, RNase_PH; PFAM_ID, RNase_PH; PFAM_ID, KH-domain; PFAM_ID, S1 |
| BL1549 | Binds the two ribosomal protein L7/L12 dimers and anchors them to the large ribosomal subunit |
| BL1550 | RplL; COG family, ribosomal protein L7/L12; PFAM_ID, Ribosomal_L12 |
| BL1558 | GroES; COG family, cochaperonin GroES (Hsp10); PFAM_ID, cpn10 |
| BL1571 | L13 is associated with the antitermination complex |
| BL1572 | RpsI; COG family, ribosomal protein S9; PFAM_ID, Ribosomal_S9 |
| BL1577 | Involved in the binding of tRNA to the ribosomes |
| BL1578 | RplC; COG family, ribosomal protein L3; PFAM_ID, Ribosomal_L3 |
| BL1579 | Subunit important during the early stages of 50S assembly, initially binding near the 5′ end of the 23S rRNA |
| BL1580 | Binds signal recognition particle and trigger factor at the peptide exit area on the 50S ribosome |
| BL1581 | One of the primary rRNA-binding proteins; required for association of the 30S and 50S subunits to form the 70S ribosome, for tRNA binding, and for peptide bond formation |
| BL1582 | RpsS; COG family, ribosomal protein S19; PFAM_ID, Ribosomal_S19 |
| BL1583 | RplV; COG family, ribosomal protein L22; PFAM_ID, Ribosomal_L22 |
| BL1584 | Forms a complex with S10 and S14; binds the lower part of the 30S subunit head and the mRNA in the complete ribosome to position it for translation |
| BL1586 | Located in the peptidyl transferase center and may be involved in peptidyl transferase activity |
| BL1588 | RpmC; COG family, ribosomal protein L29; PFAM_ID, Ribosomal_L29 |
| BL1589 | RpsQ; COG family, ribosomal protein S17; PFAM_ID, Ribosomal_S17 |
| BL1590 | RplN; COG family, ribosomal protein L14; PFAM_ID, Ribosomal_L14 |
| BL1591 | RplX; COG family, ribosomal protein L24; PFAM_ID, Ribosomal_L24 |
| BL1592 | Mediates attachment of the 5S RNA into the large ribosomal subunit, where it forms part of the central protuberance |
| BL1594 | Binds directly to 16S rRNA central domain, where it helps coordinate assembly of the platform of the 30S subunit |
| BL1595 | RplF; COG family, ribosomal protein L6; PFAM_ID, Ribosomal_L6 |
| BL1596 | RplR; COG family, ribosomal protein L18; PFAM_ID, Ribosomal_L18p |
| BL1597 | Located at the back of the 30S subunit body, where it stabilizes the conformation of the head with respect to the body; contacts S4 and S8; with S4 and S12 plays a role in translational accuracy |
| BL1599 | Associates with 23S rRNA and is involved in 50S ribosome assembly |
| BL1600 | With SecE and SecG forms a protein conduction channel in the inner membrane |
| BL1604 | Located at the top of the head of the 30S subunit, it contacts several helices of the 16S rRNA |
| BL1604.1 | Located on the platform of the 30S subunit, it bridges several disparate RNA helices of the 16S rRNA; forms part of the Shine-Dalgarno cleft in the 70S ribosome |
| BL1607 | RplQ; COG family, ribosomal protein L17; PFAM_ID, Ribosomal_L17 |
| BL1615 | Binds to RNA polymerase and RNA; stimulates rho-independent termination by promoting hairpin formation; also interacts with N protein and prevents hairpin formation, thereby preventing termination |
| BL1616 | Protects formylmethionyl-tRNA from spontaneous hydrolysis and promotes its binding to the 30S ribosomal subunits during initiation of protein synthesis; also involved in the hydrolysis of GTP during the formation of the 70S ribosomal complex |
| BL1618 | tRNA pseudouridine 55 synthase; Psi55 synthase; pseudouridylate synthase; uracil hydrolyase; TruB; COG family, pseudouridine synthase; PFAM_ID, TruB_N |
| BL1619 | Fad pyrophosphorylase; Fad synthetase; RibF; COG family, FAD synthase; PFAM_ID, FAD_Synth |
| BL1777 | Catalyzes formation of isoleucyl-tRNA(Ile) from isoleucine and tRNA(Ile) |
| BL0642 | RpmH; COG family, ribosomal protein L34; PFAM_ID, Ribosomal_L34 |
The ORF designations refer to those of B. longum biotype longum NCC2705 (384).
FIG. 14.
Phylogenomic tree of the Actinobacteria phylum. The tree is based on the 123 protein sequences representing the minimal core gene sequences of Actinobacteria described in Table 7.
The longest branch in both the supertree and the 16S rRNA gene tree includes Symbiobacterium thermophilum and Rubrobacter xylanophilus; of these two, S. thermophilum is still taxonomically unclassified, though it was initially considered to be actinobacterial based on GC content and 16S rRNA gene sequence (440).
These observations provide a starting point for the reclassification of Actinobacteridae using whole-genome information, which is already considerable and is set to increase rapidly.
IMPACT OF ACTINOBACTERIAL GENOMICS ON TAXONOMY
It has become obvious that the genome sequence of only a single strain does not depict the overall diversity of a species. For example, E. coli genomes available in 2002 displayed up to 29% sequence divergence (81). On the basis of such findings, Lan and Reves (251) introduced the term of species genome, which includes all the genes present in the characterized strains of a species. The species genome consists of a core of genes present in most of the strains and a supplementary set of genes, which are usually strain specific. Obligate intracellular parasites such as Tropheryma, Buchnera, or Chlamydophila are highly adapted to the physiologically stable environments of their host cells and tend to contain very few supplementary genes (113), while an extremely variable supplementary gene set is typically found in free-living bacteria that are highly adaptable to different environmental conditions (79).
The specific level of genome conservation between bacterial taxa can be used as a measure of evolutionary divergence (81, 243). The ranks of genus and family are the least well defined, at least when a genomic similarity measure is considered. The broadest distribution of genome conservation score is observed within the genus rank. The similarity between species belonging to the same genus can also vary considerably (e.g., M. tuberculosis and M. bovis are 96% similar, whereas Mycoplasma gallisepticum and Mycoplasma penetrans are only 16% similar).
New Approaches to Investigate Taxonomic Relationships in Actinobacteria Based on Whole-Genome Sequences
The DNA-DNA reassociation method has been a cornerstone of modern bacterial taxonomy. However, besides problems associated with reproducibility of this methodology, DNA reassociation values do not represent actual sequence identity or gene content differences, since DNA heteroduplexes will form only between strands that show at least 80% sequence complementarity (377). Another, universally accepted method used in bacterial taxonomy is comparing 16S rRNA gene sequences. The limitation of this approach is that there are no threshold values for 16S rRNA gene identity for species recognition, while a number of cases are known where two taxa display high 16S rRNA gene sequence identity but low DNA-DNA binding values (128, 405). Furthermore, high levels of intragenomic heterogeneity between multiple 16S rRNA operons in a given genome, as well as HGT of the 16S rRNA gene between bacteria, have been documented (388, 440). Finally, considerable differences in the mutation rates between different lineages have also been observed for 16S rRNA gene sequence, thus leading to artifacts in the phylogenetic tree construction (481).
Since Darwin's theory of evolution, taxonomists have wanted to obtain a hierarchical classification that reflects the evolutionary relationships between individual species, reconstructing the history of life while also enhancing our understanding of current life in the context of the evolutionary forces that have helped to shape it (114). For the Actinobacteria, earlier phylogenies using morphological and chemotaxonomic characteristics were found to be unreliable (119), and these were superseded by trees based on 16S rRNA sequences (406). In these trees, actinobacterial species form compact groups, and it has been difficult to solve the precise branching order or interrelationships among different constituent subgroups (406). Thus, other alternative molecular markers such as recA, tuf, atpD, rpoB, sod, gyrB, or molecular chaperone-encoding genes such as groEL, dnaK, grpE, clpP, and hsp65 have been proposed to infer phylogeny in Actinobacteria taxa, such as bifidobacteria (210, 448-461) or mycobacteria (32, 49, 149, 217, 226, 497). However, molecular phylogenies based on a single gene often lead to apparently conflicting results (374).
With the availability of complete genome sequences it has become possible to reconstruct phylogenies on the basis of a much larger data set per species, allowing a more reliable and representative inference of the tree of life. Several approaches to build trees from complete genomes have been introduced (43, 104, 125, 160, 191, 348, 397, 433, 483, 491). Genome-based trees can be divided into (i) alignment-free genome trees based on statistical properties of the complete genome, (ii) gene content trees based on the presence and absence of genes, (iii) genome trees based on average sequence similarity, (iv) genome trees based on chromosomal gene order, and (v) phylogenomic trees based either on the collection of phylogenetic trees derived from shared gene families or on a concatenated alignment of those families (399).
Another strategy used to understand the interrelationship between bacteria is to analyze conserved inserts or deletions, i.e., indels, in widely distributed proteins that are characteristic of the different groups of bacteria (for reviews, see references 164 and 165). Several indels, which constitute distinctive molecular markers, have been identified for phyla such as Proteobacteria, Chlamidiae, Cyanobacteria, and “Deinococcus-Thermus” (157, 163, 166). The taxonomic resolution of some of the techniques mentioned above, in particular with respect to the Actinobacteria group, may vary (Fig. 15). However, it should be kept in mind that more large-scale studies and a larger number of actinobacterial genomes are required to corroborate such taxonomic inferences. Recently, three conserved indels, which correspond to a deletion of 2 aa in cytochrome c oxidase subunit 1 (Cox1), a 4-aa insert (except in the case of T. whipplei, which contains a 3-aa insert in the same position) in CTP synthetase, and a 5-aa insert (with the exceptions of T. fusca, P. acnes, and Rubrobacterium xylanophilus) in glutamyl-tRNA synthetase (GluRS), were found to be distinctive characteristics of Actinobacteria (136).
FIG. 15.
Taxonomic resolution of genomic approaches for assessing taxonomic relationships based on whole-genome sequences. These data are based on the results presented in the references cited in the text.
Furthermore, a large insert of 79 to 100 bp in actinobacterial 23S rRNA is absent from all other groups of bacteria (136, 375). Based on the distribution of all of the above-mentioned signatures, it can be inferred that these indels were introduced in a common ancestor of Actinobacteria and thus represent molecular signatures that are very helpful in circumscribing the phylum Actinobacteria while useful for the placement of deep-branching species within this group (136). A recent comprehensive analysis of four actinobacterial genomes, i.e., M. leprae TN (84), L. xyli subsp. xyli strain CTCB07 (300), B. longum biotype longum NCC2705 (384), and T. fusca YX (281), identified 233 proteins that are unique in this cluster of actinobacterial genomes and do not have homologs in any other currently available bacterial genome (137). (We note that the inclusion in this set of four strains of M. leprae has resulted in the exclusion of a significant number of genes found in all free-living actinobacteria.) Most of these proteins are of unknown function, except for WhiB, which has roles in development in Streptomyces and cell division in Mycobacterium smegmatis (400), and the transcriptional regulator of mercuric compounds MerR (360). Genes related to whiB have also been found in some actinophages, which may have acquired them from Actinobacteria (336). Among these 233 Actinobacteria-specific proteins, the genes encoding six proteins are absent from the B. longum biotype longum genome. Of these six genes, one (encoding a homolog of the hypothetical protein ML1781) was also absent in any of the sequenced corynebacterial genomes. The latter finding suggests that this protein was introduced in an actinobacterial antecedent after the divergence of Bifidobacterium and subsequently lost in a common ancestor of Corynebacterium (137). These proteins provide useful indications that bifidobacteria most likely constitute one of the earliest branching lineages within Actinobacteria. Another group of 11 Actinobacteria-specific proteins are specific mainly for the Corynebacterium-Mycobacterium-Nocardia subgroups, Streptomyces, Thermobifida, and Frankia but are not specified by species belonging to Bifidobacterium or Micrococcineae (L. xyli, T. whipplei, Arthrobacter, and Brevibacterium). Similarly, 13 proteins were found to be encoded by members of the CMN subgroup but were not found in other bacteria (137) (Fig. 16). Finally, 14 orthologous proteins are specified by both Mycobacterium and Nocardia species but are not encoded by any other microorganism, including Corynebacterium, suggesting a recent origin of the Mycobacterium and Nocardia subgroup (137) (Fig. 16).
FIG. 16.
Diagram representing the distribution patterns of various Actinobacteria-specific proteins. The numbers of signature proteins are indicated.
In addition to providing novel molecular markers, as alternatives to the 16S rRNA gene, that are distinctive for the entire Actinobacteria taxon, major subgroups within this phylum (e.g., Micrococcineae, CMN subgroup, Streptosporangineae, and Streptomycineae) can be distinguished (137). Due to their specificity for Actinobacteria or specific groups within this phylum, genes encoding group-specific proteins provide a very useful source of sequences for specific bacterial tracing (e.g., against human pathogens such as M. tuberculosis, M. leprae, and N. farcinica). Furthermore, the distribution pattern of these Actinobacteria-specific proteins supplies valuable information concerning the relative branching order and interrelationships between various subgroups within the Actinobacteria phylum. Recently, a tentative model of the branching order of Actinobacteria subgroups that would reflect the evolutionary stages was proposed (137). This analysis indicated that Rubrobacterales constitutes one of the deepest branches within the phylum Actinobacteria, followed by the emergence of Bifidobacteriales and Micrococcinae. Genera belonging to the suborders Streptomycineae, Streptosporangineae, Frankiniae, and Corynebacterineae (CMN subgroup) represent late-branching groups (137) (Fig. 16).
Actinobacterial Taxonomy Based on Multilocus Approach
Although sequence comparisons of conserved macromolecules have become a routine practice in prokaryotic taxonomy, several studies have indicated that single-gene trees may not adequately reflect phylogenetic relationships, because of possible HGT, variable mutation rates, and variable rates of recombination. Trees based on large combined alignments of conserved orthologous proteins, i.e., the supertree, are highly robust. Based on the consensus view introduced by Philippe and Douady (342), only proteins encoded by the so-called hard-core genes, i.e., those not subject to detectable HGT, and by the soft-core genes, which includes genes that are not commonly susceptible to HGT, should be used to build a supertree. These two different categories of genes allow phylogenetic inferences at distant, short, and intermediate evolutionary scales. Furthermore, other criteria must be taken into account in the selection of these genes, i.e., there must be universal distribution among genomes, the genes must be unique within a given genome, and the individual gene sequences must be long enough to contain sufficient information but short enough to allow sequencing in a convenient manner (493). The multigene approach fulfils the recent recommendations of the ad hoc committee for the reevaluation of the definition of the bacterial species (405). The concatenation of four gene fragments encompassing the 16S rRNA gene, hsp65, rpoB, and sod have been used to create a supertree which infers phylogeny among members of the Mycobacterium genus (112). The concatenation of these genes significantly increased the resolution and robustness of the tree. In fact, species such as Mycobacterium fortuitum and M. avium are poorly resolved using a single-gene-based tree, i.e., 16S rRNA gene-based tree, whereas these taxa are well separated by a supertree approach (112). Recently, another example of an actinobacterial supertree was proposed (448). The concatenation of fragments from seven conserved genes, i.e., clpC, dnaB, dnaG, dnaJ1, purF, rpoC, and xfp, generated a robust and highly discriminatory supertree that has been used to infer phylogeny in the genus Bifidobacterium. This phylogenetic investigation revealed that the bifidobacterial strains isolated from an insect hindgut, i.e., B. asteroides and Bifidobacterium coryneforme, constitute the closest relatives of the ancestor of the genus Bifidobacterium (448).
CONCLUSIONS
Access to large sets of genome sequences of Actinobacteria has significantly improved our understanding of the evolutionary path followed by this group of bacteria. Moreover, genomic and comparative genomic analyses have revealed key gene regions in Actinobacteria that are worthy of continued investigation for their potential roles in pathogenesis or bioprocessing. As more genome sequences become available, we can expect a more thorough understanding of phylogeny, further expansion of our insights into behavior and developmental processes, and better delineation of the genetic traits that distinguish the high-G+C gram-positive bacteria. In this context the availability of novel high-throughput sequencing capabilities will enable scientists to embark on environmental genomic (metagenomic) studies in which extensive but random DNA surveys of a particular environment are executed (372). Such analyses will help to form a clear picture of actinobacterial diversity.
Genomics has been shown to be a powerful means to study microbial pathogenicity (i.e., pathogenomics), to identify the genetic determinants involved in the health-promoting effects of probiotic bacteria (i.e., probiogenomics), and to characterize the molecular pathways for the production of biomolecules in industrial processes.
The ongoing proliferation of whole-genome sequences is a stepping stone for systems biology (2), which aims to study the integrated network constituted by the complete repertoire of genes (genome), the population of transcripts (transcriptome), the population of proteins (proteome), the population of metabolites (metabolome), and fluxes of an organism or cell, in relation to intrinsic and environmental stimuli (2, 34, 184). It is too early to speculate on how such a systems biological approach will affect our understanding of genome functionality in Actinobacteria. However, functional genome analysis promises to reveal genetic networks that orchestrate complex microbial responses to a variety of conditions that are critical to growth, metabolic activity, survival, and communication and signaling.
A final issue is represented by the future impact of the availability of a large number of complete genome sequences on bacterial taxonomy and the prokaryotic species concept. It is clear that novel insights into the current classification scheme will arise as the increasing number of available genome sequences will allow the bacterial taxonomists to evaluate the potential of genome-based taxonomical approaches, i.e., gene content, gene order, comparative sequence analysis of conserved macromolecules, presence/absence analysis, and nucleotide composition (79), to deduce relationships between taxa. It can be expected that the availability of a large number of completely sequenced genomes will facilitate the development of universal genome sequence analysis schemes, which will allow the adoption of a more natural species concept.
Supplementary Material
Acknowledgments
This work was financially supported by the Italian Award for Outstanding Young Researcher scheme “Incentivazione alla mobilità di studiosi stranieri e italiani residente all'estero” 2005-2009 and a Marie Curie Reintegration Grant (MERG-CT-2005-03080) to M.V., by an IRCSET Embark postdoctoral fellowship to C.C., and by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork.
We thank D. C. Lamb for information on P450 cytochromes and M. C. Smith for comments on Streptomyces phages. We thank all the students and coworkers who have contributed so much data and enthusiasm over the years. The help of Elena Foroni, Angela Ribbera, and Francesca Turroni is most gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Abou-Zeid, C., T. Garbe, R. Lathigra, H. G. Wiker, M. Harboe, G. A. Rook, and D. B. Young. 1991. Genetic and immunological analysis of Mycobacterium tuberculosis fibronectin-binding proteins. Infect. Immun. 59:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aderem, A. 2005. Systems biology: its practice and challenges. Cell 121:511-513. [DOI] [PubMed] [Google Scholar]
- 3.Adindla, S., and L. Guruprasad. 2003. Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. J. Biosci. 28:169-179. [DOI] [PubMed] [Google Scholar]
- 4.Alam, M. S., S. K. Garg, and P. Agrawal. 2007. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol. Microbiol. 63:1414-1431. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Martin, P., A. B. Florez, and B. Mayo. 2006. Screening for plasmids among human bifidobacteria species: sequencing and analysis of pBC1 from Bifidobacterium catenulatum L48. Plasmid 57:165-174. [DOI] [PubMed] [Google Scholar]
- 6.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson, J. O., and S. G. Andersson. 2001. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol. Biol Evol. 18:829-839. [DOI] [PubMed] [Google Scholar]
- 8.Andersson, S. G. 2000. The genomics gamble. Nat. Genet. 26:134-135. [DOI] [PubMed] [Google Scholar]
- 9.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 10.Anne, J., W. Wohlleben, H. J. Burkardt, R. Springer, and A. Puhler. 1984. Morphological and molecular characterization of several actinophages isolated from soil which lyse Streptomyces cattleya or S. venezuelae. J. Gen. Microbiol. 130:2639-2649. [DOI] [PubMed] [Google Scholar]
- 11.Aoyama, Y., T. Horiuchi, O. Gotoh, M. Noshiro, and Y. Yoshida. 1998. CYP51-like gene of Mycobacterium tuberculosis actually encodes a P450 similar to eukaryotic CYP51. J. Biochem. (Tokyo) 124:694-696. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51:629-659. [DOI] [PubMed] [Google Scholar]
- 13.Arruda, S., G. Bomfim, R. Knights, T. Huima-Byron, and L. W. Riley. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454-1457. [DOI] [PubMed] [Google Scholar]
- 14.Ashkenazi, H., Z. Malik, Y. Harth, and Y. Nitzan. 2003. Eradication of Propionibacterium acnes by its endogenic porphyrins after illumination with high intensity blue light. FEMS Immunol. Med. Microbiol. 35:17-24. [DOI] [PubMed] [Google Scholar]
- 15.Atlas, R. 1997. Principles of microbiology. WCB McGrill-Hill, New York, NY.
- 16.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 17.Bannantine, J. P., J. K. Hansen, M. L. Paustian, A. Amonsin, L. L. Li, J. R. Stabel, and V. Kapur. 2004. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barksdale, L. 1970. Corynebacterium diphtheriae and its relatives. Bacteriol. Rev. 34:378-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 20.Barreiro, C., E. Gonzalez-Lavado, S. Brand, A. Tauch, and J. F. Martin. 2005. Heat shock proteome analysis of wild-type Corynebacterium glutamicum ATCC 13032 and a spontaneous mutant lacking GroEL1, a dispensable chaperone. J. Bacteriol. 187:884-889. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bartley, G. E., and P. A. Scolnik. 1995. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becq, J., M. C. Gutierrez, V. Rosas-Magallanes, J. Rauzier, B. Gicquel, O. Neyrolles, and P. Deschavanne. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol., in press. [DOI] [PubMed]
- 23.Beggs, M. L., J. T. Crawford, and K. D. Eisenach. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 177:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 25.Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser, and K. F. Chater. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol. Microbiol. 51:1615-1628. [DOI] [PubMed] [Google Scholar]
- 26.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 27.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, D. E. Harris, A. von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. G. Barrell, J. Parkhill, and D. A. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 28.Bentley, S. D., and J. Parkhill. 2004. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 38:771-792. [DOI] [PubMed] [Google Scholar]
- 29.Berdy, J. 2005. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 58:1-26. [DOI] [PubMed] [Google Scholar]
- 30.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 31.Bickle, T. A., and D. H. Kruger. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bork, P., and L. Serrano. 2005. Towards cellular systems in 4D. Cell 121:507-509. [DOI] [PubMed] [Google Scholar]
- 35.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 36.Bouhnik, Y., L. Raskine, G. Simoneau, E. Vicaut, C. Neut, B. Flourie, F. Brouns, and F. R. Bornet. 2004. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: a double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am. J. Clin. Nutr. 80:1658-1664. [DOI] [PubMed] [Google Scholar]
- 37.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan, M. J., G. Delogu, Y. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs, Jr. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 69:7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 40.Brinkman, F. S., J. L. Blanchard, A. Cherkasov, Y. Av-Gay, R. C. Brunham, R. C. Fernandez, B. B. Finlay, S. P. Otto, B. F. Ouellette, P. J. Keeling, A. M. Rose, R. E. Hancock, S. J. Jones, and H. Greberg. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown, J. M., M. M. McNeil, and E. P. Desmond. 1999. Nocardia, Rhodococcus, Gordona, Actinomadura, Streptomyces, and other actinomycetes of medical importance, p. 370-398. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 43.Brown, J. R., C. J. Douady, M. J. Italia, W. E. Marshall, and M. J. Stanhope. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281-285. [DOI] [PubMed] [Google Scholar]
- 44.Brown, K. L., G. J. Sarkis, C. Wadsworth, and G. F. Hatfull. 1997. Transcriptional silencing by the mycobacteriophage L5 repressor. EMBO J. 16:5914-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruggemann, H. 2005. Insights in the pathogenic potential of Propionibacterium acnes from its complete genome. Semin. Cutan. Med. Surg. 24:67-72. [DOI] [PubMed] [Google Scholar]
- 46.Bruggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Durre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671-673. [DOI] [PubMed] [Google Scholar]
- 47.Brune, I., K. Brinkrolf, J. Kalinowski, A. Puhler, and A. Tauch. 2005. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brune, I., H. Werner, A. T. Huser, J. Kalinowski, A. Puhler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana. 2001. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruton, C. J., K. A. Plaskitt, and K. F. Chater. 1995. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolor A3(2). Mol. Microbiol. 18:89-99. [DOI] [PubMed] [Google Scholar]
- 52.Bukovska, G., L. Klucar, C. Vlcek, J. Adamovic, J. Turna, and J. Timko. 2006. Complete nucleotide sequence and genome analysis of bacteriophage BFK20—a lytic phage of the industrial producer Brevibacterium flavum. Virology 348:57-71. [DOI] [PubMed] [Google Scholar]
- 53.Burkhart, C. N., and C. G. Burkhart. 2003. Microbiology's principle of biofilms as a major factor in the pathogenesis of acne vulgaris. Int. J. Dermatol. 42:925-927. [DOI] [PubMed] [Google Scholar]
- 54.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 55.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 56.Campbell, A. M. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Candela, M., B. Vitali, D. Matteuzzi, and P. Brigidi. 2004. Evaluation of the rrn operon copy number in Bifidobacterium using real-time PCR. Lett. Appl. Microbiol. 38:229-232. [DOI] [PubMed] [Google Scholar]
- 59.Cases, I., V. de Lorenzo, and C. A. Ouzounis. 2003. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 11:248-253. [DOI] [PubMed] [Google Scholar]
- 60.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaconas, G., and C. W. Chen. 2005. Linear chromosomes in Bacteria: no longer going round in circles, p. 525-539. In N. P. Higgins (ed.), The bacterial chromosomes. American Society for Microbiology, Washington, DC.
- 62.Challis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chater, K. F. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B 361:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 65.Chater, K. F., and G. Chandra. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651-672. [DOI] [PubMed] [Google Scholar]
- 66.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 67.Chater, K. F., and H. Kinashi. 2007. Streptomyces linear plasmids: their discovery, functions, interactions with other replicons, and evolutionary significance, p. 237-278. In F. Meinhardt and F. Klassen (ed.), Microbial linear plasmids. Microbiology monographs. Springer, New York, NY.
- 68.Chen, C. C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 69.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12:192-196. [DOI] [PubMed] [Google Scholar]
- 70.Chen, C. W. 2007. Streptomyces linear plasmids: replication and telomeres, p. 145-160. In F. Meinhardt and F. Klassen (ed.), Microbial linear plasmids. Microbiology monographs. Springer, New York, NY.
- 71.Chen, H. H., W. J. Li, S. K. Tang, R. M. Kroppenstedt, E. Stackebrandt, L. H. Xu, and C. L. Jiang. 2004. Corynebacterium halotolerans sp. nov., isolated from saline soil in the west of China. Int. J. Syst. Evol. Microbiol. 54:779-782. [DOI] [PubMed] [Google Scholar]
- 72.Chopin, M. C., A. Chopin, and E. Bidnenko. 2005. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8:473-479. [DOI] [PubMed] [Google Scholar]
- 73.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. Murthy, V. M. Katoch, and S. E. Hasnain. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 71:6338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choulet, F., B. Aigle, A. Gallois, S. Mangenot, C. Gerbaud, C. Truong, F. X. Francou, C. Fourrier, M. Guerineau, B. Decaris, V. Barbe, J. L. Pernodet, and P. Leblond. 2006. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 23:2361-2369. [DOI] [PubMed] [Google Scholar]
- 76.Choulet, F., A. Gallois, B. Aigle, S. Mangenot, C. Gerbaud, C. Truong, F. X. Francou, F. Borges, C. Fourrier, M. Guerineau, B. Decaris, V. Barbe, J. L. Pernodet, and P. Leblond. 2006. Intraspecific variability of the terminal inverted repeats of the linear chromosome of Streptomyces ambofaciens. J. Bacteriol. 188:6599-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Christensen, B. B., T. Atlung, and F. G. Hansen. 1999. DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J. Bacteriol. 181:2683-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coco, W. M., W. E. Levinson, M. J. Crist, H. J. Hektor, A. Darzins, P. T. Pienkos, C. H. Squires, and D. J. Monticello. 2001. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 19:354-359. [DOI] [PubMed] [Google Scholar]
- 79.Coenye, T., D. Gevers, Y. Van de Peer, P. Vandamme, and J. Swings. 2005. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 29:147-167. [DOI] [PubMed] [Google Scholar]
- 80.Cohan, F. M. 2001. Bacterial species and speciation. Syst. Biol. 50:513-524. [DOI] [PubMed] [Google Scholar]
- 81.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 82.Cole, S. T. 1999. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Lett. 452:7-10. [DOI] [PubMed] [Google Scholar]
- 83.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 84.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 85.Cole, S. T., P. Supply, and N. Honore. 2001. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72:449-461. [PubMed] [Google Scholar]
- 86.Collins, C. H. 2000. The bovine tubercle bacillus. Br. J. Biomed. Sci. 57:234-240. [PubMed] [Google Scholar]
- 87.Collins, C. H., J. M. Grange, and M. D. Yates. 1984. Mycobacteria in water. J. Appl. Bacteriol. 57:193-211. [DOI] [PubMed] [Google Scholar]
- 88.Collins, D. M., R. P. Kawakami, G. W. de Lisle, L. Pascopella, B. R. Bloom, and W. R. Jacobs, Jr. 1995. Mutation of the principal sigma factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 92:8036-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collins, M. D., and C. S. Cummins. 1986. Genus Corynebacterium. Williams & Wilkins, Baltimore, MD.
- 90.Collins, M. D., L. Hoyles, G. Foster, and E. Falsen. 2004. Corynebacterium caspium sp. nov., from a Caspian seal (Phoca caspica). Int. J. Syst. Evol. Microbiol. 54:925-928. [DOI] [PubMed] [Google Scholar]
- 91.Collmer, A., and N. T. Keen. 1986. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24:383-409. [Google Scholar]
- 92.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 72:2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Converse, S. E., and J. S. Cox. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 187:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Corfield, T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2:509-521. [DOI] [PubMed] [Google Scholar]
- 95.Corneau, N., E. Emond, and G. LaPointe. 2004. Molecular characterization of three plasmids from Bifidobacterium longum. Plasmid 51:87-100. [DOI] [PubMed] [Google Scholar]
- 96.Cove, J. H., K. T. Holland, and W. J. Cunliffe. 1983. Effects of oxygen concentration on biomass production, maximum specific growth rate and extracellular enzyme production by three species of cutaneous propionibacteria grown in continuous culture. J. Gen. Microbiol. 129:3327-3334. [DOI] [PubMed] [Google Scholar]
- 97.Cox, E. C., and C. Yanofsky. 1967. Altered base ratios in the DNA of an Escherichia coli mutator strain. Proc. Natl. Acad. Sci. USA 58:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coyle, M. B., and B. A. Lipsky. 1990. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin. Microbiol. Rev. 3:227-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Csukas, Z., B. Banizs, and F. Rozgonyi. 2004. Studies on the cytotoxic effects of Propionibacterium acnes strains isolated from cornea. Microb. Pathog. 36:171-174. [DOI] [PubMed] [Google Scholar]
- 100.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 102.Dafhnis-Calas, F., Z. Xu, S. Haines, S. K. Malla, M. C. Smith, and W. R. Brown. 2005. Iterative in vivo assembly of large and complex transgenes by combining the activities of phiC31 integrase and Cre recombinase. Nucleic Acids Res. 33:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dalton, K. A., A. Thibessard, K. Findlay, and G. H. Kelemen. SigN, an RNA polymerase sigma factor, controls aerial development and defines a novel compartment, the stem, during morphogenesis of Streptomyces coelicor. Mol. Microbiol., in press.
- 104.Daubin, V., M. Gouy, and G. Perriere. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Daubin, V., and G. Perriere. 2003. G+C3 structuring along the genome: a common feature in prokaryotes. Mol. Biol. Evol. 20:471-483. [DOI] [PubMed] [Google Scholar]
- 106.Davis, M. J., A. G. Gillaspie, R. W. Harris, and R. H. Lawson. 1980. Ratoon stunting disease of sugarcane: isolation of the casual bacterium. Science 210:1365-1367. [DOI] [PubMed] [Google Scholar]
- 107.DeBoy, R. T., E. F. Mongodin, J. B. Emerson, and K. E. Nelson. 2006. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 188:2364-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Degnan, B. A., and G. T. Macfarlane. 1993. Transport and metabolism of glucose and arabinose in Bifidobacterium breve. Arch. Microbiol. 160:144-151. [DOI] [PubMed] [Google Scholar]
- 109.Delogu, G., and M. J. Brennan. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 69:5606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Mot, R., I. Nagy, A. De Schrijver, P. Pattanapipitpaisal, G. Schoofs, and J. Vanderleyden. 1997. Structural analysis of the 6 kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcus shuttle vectors. Microbiology 143:3137-3147. [DOI] [PubMed] [Google Scholar]
- 112.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293-302. [DOI] [PubMed] [Google Scholar]
- 113.Dobrindt, U., and J. Hacker. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4:550-557. [DOI] [PubMed] [Google Scholar]
- 114.Dobzhansky, T. 1973. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teacher 35:125-129. [Google Scholar]
- 115.Dreier, J., D. Meletzus, and R. Eichenlaub. 1997. Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 10:195-206. [DOI] [PubMed] [Google Scholar]
- 116.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM10140(T) and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 117.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elliot, M. A., and N. J. Talbot. 2004. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 7:594-601. [DOI] [PubMed] [Google Scholar]
- 119.Embley, T. M., and E. Stackebrandt. 1994. The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 48:257-289. [DOI] [PubMed] [Google Scholar]
- 120.Evtushenko, L. I., L. V. Dorofeeva, S. A. Subbotin, J. R. Cole, and J. M. Tiedje. 2000. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of ‘Corynebacterium aquaticum’ Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984) gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 50:371-380. [DOI] [PubMed] [Google Scholar]
- 121.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernandez-Garayzabal, J. F., A. I. Vela, R. Egido, R. A. Hutson, M. P. Lanzarot, M. Fernandez-Garcia, and M. D. Collins. 2004. Corynebacterium ciconiae sp. nov., isolated from the trachea of black storks (Ciconia nigra). Int. J. Syst. Evol. Microbiol. 54:2191-2195. [DOI] [PubMed] [Google Scholar]
- 124.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Forde, A., and G. F. Fitzgerald. 2003. Molecular organization of exopolysaccharide (EPS) encoding genes on the lactococcal bacteriophage adsorption blocking plasmid, pCI658. Plasmid 49:130-142. [DOI] [PubMed] [Google Scholar]
- 128.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 129.Frank, A. C., and J. R. Lobry. 1999. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238:65-77. [DOI] [PubMed] [Google Scholar]
- 130.Fredricks, D. N., and D. A. Relman. 2001. Localization of Tropheryma whippelii rRNA in tissues from patients with Whipple's disease. J. Infect. Dis. 183:1229-1237. [DOI] [PubMed] [Google Scholar]
- 131.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 132.Fudou, R., Y. Jojima, A. Seto, K. Yamada, E. Kimura, T. Nakamatsu, A. Hiraishi, and S. Yamanaka. 2002. Corynebacterium efficiens sp. nov., a glutamic-acid-producing species from soil and vegetables. Int. J. Syst. Evol. Microbiol. 52:1127-1131. [DOI] [PubMed] [Google Scholar]
- 133.Fuglsang, A. 2005. Intragenic position of UUA codons in streptomycetes. Microbiology 151:3150-3152. [DOI] [PubMed] [Google Scholar]
- 134.Funke, G., P. A. Lawson, and M. D. Collins. 1995. Heterogeneity within human-derived Centers for Disease Control and Prevention (CDC) coryneform group ANF-1-like bacteria and description of Corynebacterium auris sp. nov. Int. J. Syst. Bacteriol. 45:735-739. [DOI] [PubMed] [Google Scholar]
- 135.Funke, G., A. von Graevenitz, J. E. Clarridge III, and K. A. Bernard. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao, B., and R. S. Gupta. 2005. Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 55:2401-2412. [DOI] [PubMed] [Google Scholar]
- 137.Gao, B., R. Paramanathan, and R. S. Gupta. 2006. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Leeuwenhoek 90:69-91. [DOI] [PubMed] [Google Scholar]
- 138.Garg, S. K., M. S. Alam, K. V. Kishan, and P. Agrawal. 2007. Expression and characterization of alpha-(1,4)-glucan branching enzyme Rv1326c of Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 51:198-208. [DOI] [PubMed] [Google Scholar]
- 139.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gase, K., J. J. Ferretti, C. Primeaux, and W. M. McShan. 1999. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 67:4725-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188:1526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gavigan, J. A., J. A. Ainsa, E. Perez, I. Otal, and C. Martin. 1997. Isolation by genetic labeling of a new mycobacterial plasmid, pJAZ38, from Mycobacterium fortuitum. J. Bacteriol. 179:4115-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gebbink, M. F., D. Claessen, B. Bouma, L. Dijkhuizen, and H. A. Wosten. 2005. Amyloids—a functional coat for microorganisms. Nat. Rev. Microbiol. 3:333-341. [DOI] [PubMed] [Google Scholar]
- 144.George, K. M., D. Chatterjee, G. Gunawardana, D. Welty, J. Hayman, R. Lee, and P. L. Small. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854-857. [DOI] [PubMed] [Google Scholar]
- 145.Ghadiali, A. H., M. Strother, S. A. Naser, E. J. Manning, and S. Sreevatsan. 2004. Mycobacterium avium subsp. paratuberculosis strains isolated from Crohn's disease patients and animal species exhibit similar polymorphic locus patterns. J. Clin. Microbiol. 42:5345-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 147.Gil, R., F. J. Silva, J. Pereto, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68:518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 150.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98:13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gogarten, J. P., W. F. Doolittle, and J. G. Lawrence. 2002. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19:2226-2238. [DOI] [PubMed] [Google Scholar]
- 152.Goodfellow, M., and S. T. Williams. 1983. Ecology of actinomycetes. Annu. Rev. Microbiol. 37:189-216. [DOI] [PubMed] [Google Scholar]
- 153.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 154.Gravius, B., D. Glocker, J. Pigac, K. Pandza, D. Hranueli, and J. Cullum. 1994. The 387 kb linear plasmid pPZG101 of Streptomyces rimosus and its interactions with the chromosome. Microbiology 140:2271-2277. [DOI] [PubMed] [Google Scholar]
- 155.Gribbon, E. M., W. J. Cunliffe, and K. T. Holland. 1993. Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 139:1745-1751. [DOI] [PubMed] [Google Scholar]
- 156.Gribbon, E. M., J. G. Shoesmith, W. J. Cunliffe, and K. T. Holland. 1994. The microaerophily and photosensitivity of Propionibacterium acnes. J. Appl. Bacteriol. 77:583-590. [DOI] [PubMed] [Google Scholar]
- 157.Griffiths, E., and R. S. Gupta. 2004. Distinctive protein signatures provide molecular markers and evidence for the monophyletic nature of the Deinococcus-Thermus phylum. J. Bacteriol. 186:3097-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Griffiths, E., and R. S. Gupta. 2006. Lateral transfers of serine hydroxymethyltransferase (glyA) and UDP-N-acetylglucosamine enolpyruvyl transferase (murA) genes from free-living Actinobacteria to the parasitic chlamydiae. J. Mol. Evol 63:283-296. [DOI] [PubMed] [Google Scholar]
- 159.Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grishin, N. V., Y. I. Wolf, and E. V. Koonin. 2000. From complete genomes to measures of substitution rate variability within and between proteins. Genome Res. 10:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 162.Guglielmetti, S., M. Karp, D. Mora, I. Tamagnini, and C. Parini. 2007. Molecular characterization of Bifidobacterium longum biovar longum NAL8 plasmids and construction of a novel replicon screening system. Appl. Microbiol. Biotechnol. 74:1053-1061. [DOI] [PubMed] [Google Scholar]
- 163.Gupta, R. S. 2004. The phylogeny and signature sequences characteristics of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit. Rev. Microbiol. 30:123-143. [DOI] [PubMed] [Google Scholar]
- 164.Gupta, R. S. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367-402. [DOI] [PubMed] [Google Scholar]
- 165.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gupta, R. S., M. Pereira, C. Chandrasekera, and V. Johari. 2003. Molecular signatures in protein sequences that are characteristic of cyanobacteria and plastid homologues. Int. J. Syst. Evol. Microbiol. 53:1833-1842. [DOI] [PubMed] [Google Scholar]
- 167.Gutierrez, M. C., S. Brisse, R. Brosch, M. Fabre, B. Omais, M. Marmiesse, P. Supply, and V. Vincent. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 1:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hadfield, T. L., P. McEvoy, Y. Polotsky, V. A. Tzinserling, and A. A. Yakovlev. 2000. The pathology of diphtheria. J. Infect. Dis. 181(Suppl. 1):S116-S120. [DOI] [PubMed] [Google Scholar]
- 169.Haile, Y., G. Bjune, and H. G. Wiker. 2002. Expression of the mceA, esat-6 and hspX genes in Mycobacterium tuberculosis and their responses to aerobic conditions and to restricted oxygen supply. Microbiology 148:3881-3886. [DOI] [PubMed] [Google Scholar]
- 170.Hansmeier, N., F. W. Bartels, R. Ros, D. Anselmetti, A. Tauch, A. Puhler, and J. Kalinowski. 2004. Classification of hyper-variable Corynebacterium glutamicum surface-layer proteins by sequence analyses and atomic force microscopy. J. Biotechnol. 112:177-193. [DOI] [PubMed] [Google Scholar]
- 171.Harris, J. E., and A. M. Lammerding. 2001. Crohn's disease and Mycobacterium avium subsp. paratuberculosis: current issues. J. Food Prot. 64:2103-2110. [DOI] [PubMed] [Google Scholar]
- 172.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395-405. [DOI] [PubMed] [Google Scholar]
- 173.Haug, I., A. Weissenborn, D. Brolle, S. Bentley, T. Kieser, and J. Altenbuchner. 2003. Streptomyces coelicolor A3(2) plasmid SCP2*: deductions from the complete sequence. Microbiology 149:505-513. [DOI] [PubMed] [Google Scholar]
- 174.Hayman, J. 1991. Postulated epidemiology of Mycobacterium ulcerans infection. Int. J. Epidemiol. 20:1093-1098. [DOI] [PubMed] [Google Scholar]
- 175.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hinz, S. W., M. I. Pastink, L. A. van den Broek, J. P. Vincken, and A. G. Voragen. 2005. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 71:5501-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hirasawa, K., Y. Ishii, M. Kobayashi, K. Koizumi, and K. Maruhashi. 2001. Improvement of desulfurization activity in Rhodococcus erythropolis KA2-5-1 by genetic engineering. Biosci. Biotechnol. Biochem. 65:239-246. [DOI] [PubMed] [Google Scholar]
- 178.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Holland, K. T., O. Aldana, R. A. Bojar, W. J. Cunliffe, E. A. Eady, D. B. Holland, E. Ingham, C. McGeown, A. Till, and C. Walters. 1998. Propionibacterium acnes and acne. Dermatology 196:67-68. [DOI] [PubMed] [Google Scholar]
- 181.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181(Suppl. 1):S156-S167. [DOI] [PubMed] [Google Scholar]
- 182.Homerova, D., J. Sevcikova, and J. Kormanec. 2003. Characterization of the Streptomyces coelicolor A3(2) wblE gene, encoding a homologue of the sporulation transcription factor. Folia Microbiol. (Praha) 48:489-495. [DOI] [PubMed] [Google Scholar]
- 183.Hong, S. H., T. Y. Kim, and S. Y. Lee. 2004. Phylogenetic analysis based on genome-scale metabolic pathway reaction content. Appl. Microbiol. Biotechnol. 65:203-210. [DOI] [PubMed] [Google Scholar]
- 184.Hood, L., J. R. Heath, M. E. Phelps, and B. Lin. 2004. Systems biology and new technologies enable predictive and preventative medicine. Science 306:640-643. [DOI] [PubMed] [Google Scholar]
- 185.Hopwood, D. A. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40:1-23. [DOI] [PubMed] [Google Scholar]
- 186.Hopwood, D. A. 2007. Streptomyces in nature and medicine. Oxford University Press, New York, NY.
- 187.Hoskins, L. C., M. Agustines, W. B. McKee, E. T. Boulding, M. Kriaris, and G. Niedermeyer. 1985. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J. Clin. Investig. 75:944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Huard, R. C., M. Fabre, P. de Haas, L. C. Lazzarini, D. van Soolingen, D. Cousins, and J. L. Ho. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J. Bacteriol. 188:4271-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hubble, J. P., T. Cao, J. A. Kjelstrom, W. C. Koller, and B. L. Beaman. 1995. Nocardia species as an etiologic agent in Parkinson's disease: serological testing in a case-control study. J. Clin. Microbiol. 33:2768-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hummerjohann, J., S. Laudenbach, J. Retey, T. Leisinger, and M. A. Kertesz. 2000. The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol. 182:2055-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huson, D. H., and M. Steel. 2004. Phylogenetic trees based on gene content. Bioinformatics 20:2044-2049. [DOI] [PubMed] [Google Scholar]
- 192.Hutchings, M., P. A. Hoskisson, G. Chandra, and M. J. Buttner. 2004. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicor A3(2). Microbiology 150:2795-2806. [DOI] [PubMed] [Google Scholar]
- 193.Huynh, T. T., A. D. Walling, M. A. Miller, T. K. Leung, Y. Leclerc, and L. Dragtakis. 1995. Propionibacterium acnes endocarditis. Can. J. Cardiol. 11:785-787. [PubMed] [Google Scholar]
- 194.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 195.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 196.Ingham, C. J., H. J. Crombie, C. J. Bruton, K. F. Chater, N. M. Hartley, G. J. Murphy, and M. C. Smith. 1993. Multiple novel promoters from the early region in the Streptomyces temperate phage phi C31 are activated during lytic development. Mol. Microbiol. 9:1267-1274. [DOI] [PubMed] [Google Scholar]
- 197.Ingham, E. 1999. The immunology of Propionibacterium acnes and acne. Curr. Opin. Infect. Dis. 12:191-197. [DOI] [PubMed] [Google Scholar]
- 198.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ivanov, D., C. Emonet, F. Foata, M. Affolter, M. Delley, M. Fisseha, S. Blum-Sperisen, S. Kochhar, and F. Arigoni. 2006. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281:17246-17252. [DOI] [PubMed] [Google Scholar]
- 200.Jackman, P. J. H., D. G. Pitcher, S. Pelczynska, and P. Borman. 1987. Classification of corynebacteria associated with endocarditis (group JK) as Corynebacterium jeikeium sp. nov. Syst. Appl. Microbiol. 9:83-90. [Google Scholar]
- 201.Jacobson, R. R., and J. L. Krahenbuhl. 1999. Leprosy. Lancet 353:655-660. [DOI] [PubMed] [Google Scholar]
- 202.Jahr, H., J. Dreier, D. Meletzus, R. Bahro, and R. Eichenlaub. 2000. The endo-beta-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant-Microbe Interact. 13:703-714. [DOI] [PubMed] [Google Scholar]
- 203.Jakab, E., R. Zbinden, J. Gubler, C. Ruef, A. von Graevenitz, and M. Krause. 1996. Severe infections caused by Propionibacterium acnes: an underestimated pathogen in late postoperative infections. Yale J. Biol. Med. 69:477-482. [PMC free article] [PubMed] [Google Scholar]
- 204.Jakimowicz, P., M. R. Cheesman, W. R. Bishai, K. F. Chater, A. J. Thomson, and M. J. Buttner. 2005. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J. Biol. Chem. 280:8309-8315. [DOI] [PubMed] [Google Scholar]
- 205.Jakubovics, N. S., and H. F. Jenkinson. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709-1718. [DOI] [PubMed] [Google Scholar]
- 206.Jang, K. H., P. J. Chambers, and M. L. Britz. 1996. Analysis of nucleotide methylation in DNA from Corynebacterium glutamicum and related species. FEMS Microbiol. Lett. 136:309-315. [DOI] [PubMed] [Google Scholar]
- 207.Jansen, R., J. D. Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 208.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jappe, U., E. Ingham, J. Henwood, and K. T. Holland. 2002. Propionibacterium acnes and inflammation in acne; P. acnes has T-cell mitogenic activity. Br. J. Dermatol. 146:202-209. [DOI] [PubMed] [Google Scholar]
- 210.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 211.Jore, J. P., N. van Luijk, R. G. Luiten, M. J. van der Werf, and P. H. Pouwels. 2001. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67:499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jost, B. H., A. C. Field, H. T. Trinh, J. G. Songer, and S. J. Billington. 2003. Tylosin resistance in Arcanobacterium pyogenes is encoded by an erm X determinant. Antimicrob. Agents Chemother. 47:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kalinowski, J. 2005. The genomes of amino acid-producing bacteria. CRC Press, Boca Raton, FL.
- 214.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 215.Kalinowski, J., J. Cremer, B. Bachmann, L. Eggeling, H. Sahm, and A. Puhler. 1991. Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol. Microbiol. 5:1197-1204. [DOI] [PubMed] [Google Scholar]
- 216.Kalkus, J., R. Menne, M. Reh, and H. G. Schlegel. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology 144:1271-1279. [DOI] [PubMed] [Google Scholar]
- 217.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Katayama, T., A. Sakuma, T. Kimura, Y. Makimura, J. Hiratake, K. Sakata, T. Yamanoi, H. Kumagai, and K. Yamamoto. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-l-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kato-Maeda, M., P. J. Bifani, B. N. Kreiswirth, and P. M. Small. 2001. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J. Clin. Investig. 107:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kende, H., and J. Zeevaart. 1997. The five “classical” plant hormones. Plant Cell 9:1197-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kerry-Williams, S. M., and W. C. Noble. 1984. Plasmid-associated bacteriocin production in a JK-type coryneform bacterium. FEMS Microbiol. Lett. 25:179-182. [Google Scholar]
- 222.Kerry-Williams, S. M., and W. C. Noble. 1986. Plasmids in group JK coryneform bacteria isolated in a single hospital. J. Hyg. (London) 97:255-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Khabbaz, R. F., J. B. Kaper, M. R. Moody, S. C. Schimpff, and J. H. Tenney. 1986. Molecular epidemiology of group JK Corynebacterium on a cancer ward: lack of evidence for patient-to-patient transmission. J. Infect. Dis. 154:95-99. [DOI] [PubMed] [Google Scholar]
- 224.Kiatpapan, P., Y. Hashimoto, H. Nakamura, Y. Z. Piao, H. Ono, M. Yamashita, and Y. Murooka. 2000. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 66:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 226.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, G. T. Chae, E. C. Kim, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kim, T. H., J. S. Park, H. J. Kim, Y. Kim, P. Kim, and H. S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 228.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Production of l-glutamic acid by various microorgonisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 229.Kirby, C., A. Waring, T. J. Griffin, J. O. Falkinham III, N. D. Grindley, and K. M. Derbyshire. 2002. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol. Microbiol. 43:173-186. [DOI] [PubMed] [Google Scholar]
- 230.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Klasson, L., and S. G. Andersson. 2004. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 12:37-43. [DOI] [PubMed] [Google Scholar]
- 232.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 233.Kolsto, A. B. 1997. Dynamic bacterial genome organization. Mol. Microbiol. 24:241-248. [DOI] [PubMed] [Google Scholar]
- 234.Komatsu, M., Y. Kuwahara, A. Hiroishi, K. Hosono, T. Beppu, and K. Ueda. 2003. Cloning of the conserved regulatory operon by its aerial mycelium-inducing activity in an amfR mutant of Streptomyces griseus. Gene 306:79-89. [DOI] [PubMed] [Google Scholar]
- 235.Konstantinidis, K. T., and J. M. Tiedje. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 101:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Korbel, J. O., B. Snel, M. A. Huynen, and P. Bork. 2002. SHOT: a web server for the construction of genome phylogenies. Trends Genet. 18:158-162. [DOI] [PubMed] [Google Scholar]
- 237.Koreck, A., A. Pivarcsi, A. Dobozy, and L. Kemeny. 2003. The role of innate immunity in the pathogenesis of acne. Dermatology 206:96-105. [DOI] [PubMed] [Google Scholar]
- 238.Kostichka, K., L. Tao, M. Bramucci, J. F. Tomb, V. Nagarajan, and Q. Cheng. 2003. A small cryptic plasmid from Rhodococcus erythropolis: characterization and utility for gene expression. Appl. Microbiol. Biotechnol. 62:61-68. [DOI] [PubMed] [Google Scholar]
- 239.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555-558. [DOI] [PubMed] [Google Scholar]
- 241.Kulakov, L. A., M. J. Larkin, and A. N. Kulakova. 1997. Cryptic plasmid pKA22 isolated from the naphthalene degrading derivative of Rhodococcus rhodochrous NCIMB13064. Plasmid 38:61-69. [DOI] [PubMed] [Google Scholar]
- 242.Kumar, A., A. Chandolia, U. Chaudhry, V. Brahmachari, and M. Bose. 2005. Comparison of mammalian cell entry operons of mycobacteria: in silico analysis and expression profiling. FEMS Immunol. Med. Microbiol. 43:185-195. [DOI] [PubMed] [Google Scholar]
- 243.Kunin, V., and C. A. Ouzounis. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13:1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kunisawa, T. 1995. Identification and chromosomal distribution of DNA sequence segments conserved since divergence of Escherichia coli and Bacillus subtilis. J. Mol. Evol. 40:585-593. [DOI] [PubMed] [Google Scholar]
- 245.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 247.Kunz, C., S. Rudloff, W. Baier, N. Klein, and S. Strobel. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20:699-722. [DOI] [PubMed] [Google Scholar]
- 248.Labo, M., L. Gusberti, E. D. Rossi, P. Speziale, and G. Riccardi. 1998. Determination of a 15437 bp nucleotide sequence around the inhA gene of Mycobacterium avium and similarity analysis of the products of putative ORFs. Microbiology 144:807-814. [DOI] [PubMed] [Google Scholar]
- 249.Lamb, D. C., H. Ikeda, D. R. Nelson, J. Ishikawa, T. Skaug, C. Jackson, S. Omura, M. R. Waterman, and S. L. Kelly. 2003. Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem. Biophys. Res. Commun. 307:610-619. [DOI] [PubMed] [Google Scholar]
- 250.Lamb, D. C., T. Skaug, H. L. Song, C. J. Jackson, L. M. Podust, M. R. Waterman, D. B. Kell, D. E. Kelly, and S. L. Kelly. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277:24000-24005. [DOI] [PubMed] [Google Scholar]
- 251.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 252.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 253.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 254.Leahy, S. C., D. G. Higgins, G. F. Fitzgerald, and D. van Sinderen. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303-1315. [DOI] [PubMed] [Google Scholar]
- 255.Lechevalier, H. A., and M. P. Lechevalier. 1967. Biology of actinomycetes. Annu. Rev. Microbiol. 21:71-100. [DOI] [PubMed] [Google Scholar]
- 256.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 257.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183:2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lee, E. J., N. Karoonuthaisiri, H. S. Kim, J. H. Park, C. J. Cha, C. M. Kao, and J. H. Roe. 2005. A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57:1252-1264. [DOI] [PubMed] [Google Scholar]
- 259.Lee, H. W., Y. S. Park, J. S. Jung, and W. S. Shin. 2002. Chitosan oligosaccharides, dp 2-8, have prebiotic effect on the Bifidobacterium bifidium and Lactobacillus sp. Anaerobe 8:319-324. [DOI] [PubMed] [Google Scholar]
- 260.Lee, J. H., and D. J. O'Sullivan. 2006. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 72:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lei, L., M. R. Waterman, A. J. Fulco, S. L. Kelly, and D. C. Lamb. 2004. Availability of specific reductases controls the temporal activity of the cytochrome P450 complement of Streptomyces coelicolor A3(2). Proc. Natl. Acad. Sci. USA 101:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lerat, E., and H. Ochman. 2004. Psi-Phi: exploring the outer limits of bacterial pseudogenes. Genome Res. 14:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Leskiw, B. K., M. J. Bibb, and K. F. Chater. 1991. The use of a rare codon specifically during development? Mol. Microbiol. 5:2861-2867. [DOI] [PubMed] [Google Scholar]
- 265.Leskiw, B. K., E. J. Lawlor, J. M. Fernandez-Abalos, and K. F. Chater. 1991. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA 88:2461-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Leskiw, B. K., R. Mah, E. J. Lawlor, and K. F. Chater. 1993. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J. Bacteriol. 175:1995-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Leuchtenberger, W., K. Huthmacher, and K. Drauz. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69:1-8. [DOI] [PubMed] [Google Scholar]
- 268.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Li, T. Y., P. Yin, Y. Zhou, Y. Zhang, Y. Y. Zhang, and T. A. Chen. 2004. Characterization of the replicon of a 51-kb native plasmid from the gram-positive bacterium Leifsonia xyli subsp. cynodontis. FEMS Microbiol. Lett. 236:33-39. [DOI] [PubMed] [Google Scholar]
- 270.Li, W., J. Wu, W. Tao, C. Zhao, Y. Wang, X. He, G. Chandra, X. Zhou, Z. Deng, K. F. Chater, and M. Tao. 2007. A genetic and bioinformatic analysis of Streptomyces coelicor genes containing TTA codons, possible targets for regulation by developmentally significant tRNA. FEMS Microbiol. Lett. 266:20-28. [DOI] [PubMed] [Google Scholar]
- 271.Li, Y., E. Miltner, M. Wu, M. Petrofsky, and L. E. Bermudez. 2005. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell. Microbiol. 7:539-548. [DOI] [PubMed] [Google Scholar]
- 272.Liebl, W. 2005. Corynebacterium taxonomy. CRC Press, Boca Raton, FL.
- 273.Lievin, V., I. Peiffer, S. Hudault, F. Rochat, D. Brassart, J. R. Neeser, and A. L. Servin. 2000. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lin, Y. S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 275.Liu, M., R. Deora, S. R. Doulatov, M. Gingery, F. A. Eiserling, A. Preston, D. J. Maskell, R. W. Simons, P. A. Cotter, J. Parkhill, and J. F. Miller. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091-2094. [DOI] [PubMed] [Google Scholar]
- 276.Liu, M., F. H. van Enckevort, and R. J. Siezen. 2005. Genome update: lactic acid bacteria genome sequencing is booming. Microbiology 151:3811-3814. [DOI] [PubMed] [Google Scholar]
- 277.Liu, Y. T., C. M. Su, C. H. Lee, M. J. Sui, Y. H. Chang, W. P. Lin, W. T. Wu, and C. Y. Chen. 2000. Cloning and characterization of the replicon of the Nocardia italica plasmid, pNI100. Plasmid 43:223-229. [DOI] [PubMed] [Google Scholar]
- 278.Lobry, J. R., and J. M. Louarn. 2003. Polarisation of prokaryotic chromosomes. Curr. Opin. Microbiol. 6:101-108. [DOI] [PubMed] [Google Scholar]
- 279.Lomovskaya, N. D., K. F. Chater, and N. M. Mkrtumian. 1980. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol. Rev. 44:206-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Loria, R., J. Kers, and M. Joshi. 2006. Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 44:469-487. [DOI] [PubMed] [Google Scholar]
- 281.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. Dibartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Maiwald, M., H. J. Ditton, A. von Herbay, F. A. Rainey, and E. Stackebrandt. 1996. Reassessment of the phylogenetic position of the bacterium associated with Whipple's disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int. J. Syst. Bacteriol. 46:1078-1082. [DOI] [PubMed] [Google Scholar]
- 283.Maiwald, M., F. Schuhmacher, H. J. Ditton, and A. von Herbay. 1998. Environmental occurrence of the Whipple's disease bacterium (Tropheryma whippelii). Appl. Environ. Microbiol. 64:760-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64:111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Mattarelli, P., B. Biavati, A. Alessandrini, F. Crociani, and V. Scardovi. 1994. Characterization of the plasmid pVS809 from Bifidobacterium globosum. New Microbiol. 17:327-331. [PubMed] [Google Scholar]
- 286.Mattos-Guaraldi, A. L., L. C. Duarte Formiga, and G. A. Pereira. 2000. Cell surface components and adhesion in Corynebacterium diphtheriae. Microbes Infect. 2:1507-1512. [DOI] [PubMed] [Google Scholar]
- 287.Maze, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.McLean, M. J., K. H. Wolfe, and K. M. Devine. 1998. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol 47:691-696. [DOI] [PubMed] [Google Scholar]
- 289.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 291.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 292.Metzler, M. C., Y. P. Zhang, and T. A. Chen. 1992. Transformation of the gram-positive bacterium Clavibacter xyli subsp. cynodontis by electroporation with plasmids from the IncP incompatibility group. J. Bacteriol. 174:4500-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 294.Miskin, J. E., A. M. Farrell, W. J. Cunliffe, and K. T. Holland. 1997. Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by gehA. Microbiology 143:1745-1755. [DOI] [PubMed] [Google Scholar]
- 295.Miyadoh, S. 1997. Atlas of actinomycetes. Society for Actinomycetes Japan, Asakura Publishing Co., Tokyo, Japan.
- 296.Mochizuki, S., K. Hiratsu, M. Suwa, T. Ishii, F. Sugino, K. Yamada, and H. Kinashi. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 297.Mojica, F. J., C. Diez-Villasenor, E. Soria, and G. Juez. 2000. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36:244-246. [DOI] [PubMed] [Google Scholar]
- 298.Molle, V., W. J. Palframan, K. C. Findlay, and M. J. Buttner. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:1286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Monot, M., N. Honore, T. Garnier, R. Araoz, J. Y. Coppee, C. Lacroix, S. Sow, J. S. Spencer, R. W. Truman, D. L. Williams, R. Gelber, M. Virmond, B. Flageul, S. N. Cho, B. Ji, A. Paniz-Mondolfi, J. Convit, S. Young, P. E. Fine, V. Rasolofo, P. J. Brennan, and S. T. Cole. 2005. On the origin of leprosy. Science 308:1040-1042. [DOI] [PubMed] [Google Scholar]
- 300.Monteiro-Vitorello, C. B., L. E. Camargo, M. A. Van Sluys, J. P. Kitajima, D. Truffi, A. M. do Amaral, R. Harakava, J. C. de Oliveira, D. Wood, M. C. de Oliveira, C. Miyaki, M. A. Takita, A. C. da Silva, L. R. Furlan, D. M. Carraro, G. Camarotte, N. F. Almeida, Jr., H. Carrer, L. L. Coutinho, H. A. El-Dorry, M. I. Ferro, P. R. Gagliardi, E. Giglioti, M. H. Goldman, G. H. Goldman, E. T. Kimura, E. S. Ferro, E. E. Kuramae, E. G. Lemos, M. V. Lemos, S. M. Mauro, M. A. Machado, C. L. Marino, C. F. Menck, L. R. Nunes, R. C. Oliveira, G. G. Pereira, W. Siqueira, A. A. de Souza, S. M. Tsai, A. S. Zanca, A. J. Simpson, S. M. Brumbley, and J. C. Setubal. 2004. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant-Microbe Interact. 17:827-836. [DOI] [PubMed] [Google Scholar]
- 301.Montes, L. F., and W. H. Wilborn. 1970. Fine structure of Corynebacterium acnes. J. Investig. Dermatol. 54:338-345. [DOI] [PubMed] [Google Scholar]
- 302.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 303.Moreira Lde, O., A. F. Andrade, M. D. Vale, S. M. Souza, R. Hirata, Jr., L. M. Asad, N. R. Asad, L. H. Monteiro-Leal, J. O. Previato, and A. L. Mattos-Guaraldi. 2003. Effects of iron limitation on adherence and cell surface carbohydrates of Corynebacterium diphtheriae strains. Appl. Environ. Microbiol. 69:5907-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Morris, R. P., L. Nguyen, J. Gatfield, K. Visconti, K. Nguyen, D. Schnappinger, S. Ehrt, Y. Liu, L. Heifets, J. Pieters, G. Schoolnik, and C. J. Thompson. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 102:12200-12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Mostowy, S., J. Inwald, S. Gordon, C. Martin, R. Warren, K. Kremer, D. Cousins, and M. A. Behr. 2005. Revisiting the evolution of Mycobacterium bovis. J. Bacteriol. 187:6386-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Nakamura, Y., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 (supplement). DNA Res. 9:135-148. [DOI] [PubMed] [Google Scholar]
- 307.Nakamura, Y., Y. Nishio, K. Ikeo, and T. Gojobori. 2003. The genome stability in Corynebacterium species due to lack of the recombinational repair system. Gene 317:149-155. [DOI] [PubMed] [Google Scholar]
- 308.Nakano, T., M. Sugawara, and H. Kawakami. 2001. Sialic acid in human milk: composition and functions. Acta Paediatr. Taiwan 42:11-17. [PubMed] [Google Scholar]
- 308a.Nakashima, N., and T. Tamura. 2004. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 70:5557-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Natsch, A., H. Gfeller, P. Gygax, J. Schmid, and G. Acuna. 2003. A specific bacterial aminoacylase cleaves odorant precursors secreted in the human axilla. J. Biol. Chem. 278:5718-5727. [DOI] [PubMed] [Google Scholar]
- 310.Nesbit, C. E., M. E. Levin, M. K. Donnelly-Wu, and G. F. Hatfull. 1995. Transcriptional regulation of repressor synthesis in mycobacteriophage L5. Mol. Microbiol. 17:1045-1056. [DOI] [PubMed] [Google Scholar]
- 311.Netz, D. J., C. Bastos Mdo, and H. G. Sahl. 2002. Mode of action of the antimicrobial peptide aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 68:5274-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Netz, D. J., R. Pohl, A. G. Beck-Sickinger, T. Selmer, A. J. Pierik, C. Bastos Mdo, and H. G. Sahl. 2002. Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319:745-756. [DOI] [PubMed] [Google Scholar]
- 313.Nguyen, K. T., J. Tenor, H. Stettler, L. T. Nguyen, L. D. Nguyen, and C. J. Thompson. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Nieuwenhuizen, W. F., S. van Leeuwen, R. W. Jack, M. R. Egmond, and F. Gotz. 2003. Molecular cloning and characterization of the alkaline ceramidase from Pseudomonas aeruginosa PA01. Protein Expr. Purif. 30:94-104. [DOI] [PubMed] [Google Scholar]
- 315.Nishimura, M., and M. Sugiyama. 1994. Cloning and sequence analysis of a Streptomyces cholesterol esterase gene. Appl. Microbiol. Biotechnol. 41:419-424. [DOI] [PubMed] [Google Scholar]
- 316.Nishio, Y., Y. Nakamura, Y. Kawarabayasi, Y. Usuda, E. Kimura, S. Sugimoto, K. Matsui, A. Yamagishi, H. Kikuchi, K. Ikeo, and T. Gojobori. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 13:1572-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Nishio, Y., Y. Nakamura, Y. Usuda, S. Sugimoto, K. Matsui, Y. Kawarabayasi, H. Kikuchi, T. Gojobori, and K. Ikeo. 2004. Evolutionary process of amino acid biosynthesis in Corynebacterium at the whole genome level. Mol. Biol. Evol. 21:1683-1691. [DOI] [PubMed] [Google Scholar]
- 318.Noens, E. E., V. Mersinias, B. A. Traag, C. P. Smith, H. K. Koerten, and G. P. van Wezel. 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58:929-944. [DOI] [PubMed] [Google Scholar]
- 319.Normand, P., P. Lapierre, L. S. Tisa, J. P. Gogarten, N. Alloisio, E. Bagnarol, C. A. Bassi, A. M. Berry, D. M. Bickhart, N. Choisne, A. Couloux, B. Cournoyer, S. Cruveiller, V. Daubin, N. Demange, M. P. Francino, E. Goltsman, Y. Huang, O. R. Kopp, L. Labarre, A. Lapidus, C. Lavire, J. Marechal, M. Martinez, J. E. Mastronunzio, B. C. Mullin, J. Niemann, P. Pujic, T. Rawnsley, Z. Rouy, C. Schenowitz, A. Sellstedt, F. Tavares, J. P. Tomkins, D. Vallenet, C. Valverde, L. G. Wall, Y. Wang, C. Medigue, and D. R. Benson. 2007. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 17:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ochman, H., and L. M. Davalos. 2006. The nature and dynamics of bacterial genomes. Science 311:1730-1733. [DOI] [PubMed] [Google Scholar]
- 321.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 322.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 323.Ohnishi, J., S. Mitsuhashi, M. Hayashi, S. Ando, H. Yokoi, K. Ochiai, and M. Ikeda. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58:217-223. [DOI] [PubMed] [Google Scholar]
- 324.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Onishi, N., and T. Tanaka. 1997. Purification and characterization of galactooligosaccharide-producing beta-galactosidase from Sirobasidium magnum. Lett. Appl. Microbiol. 24:82-86. [Google Scholar]
- 326.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2004. Analysis of genes that encode DtxR-like transcriptional regulators in pathogenic and saprophytic corynebacterial species. Infect. Immun. 72:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.O'Riordan, K., and G. F. Fitzgerald. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174:285-294. [DOI] [PubMed] [Google Scholar]
- 328.Otsuka, Y., Y. Kawamura, T. Koyama, H. Iihara, K. Ohkusu, and T. Ezaki. 2005. Corynebacterium resistens sp. nov., a new multidrug-resistant coryneform bacterium isolated from human infections. J. Clin. Microbiol. 43:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 330.Pandza, S., G. Biukovic, A. Paravic, A. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 331.Parajuli, N., D. B. Basnet, H. Chan Lee, J. K. Sohng, and K. Liou. 2004. Genome analyses of Streptomyces peucetius ATCC 27952 for the identification and comparison of cytochrome P450 complement with other Streptomyces. Arch. Biochem. Biophys. 425:233-241. [DOI] [PubMed] [Google Scholar]
- 332.Park, M. S., K. H. Lee, and G. E. Ji. 1997. Isolation and characterization of two plasmids from Bifidobacterium longum. Lett. Appl. Microbiol. 25:5-7. [DOI] [PubMed] [Google Scholar]
- 333.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145:585-592. [DOI] [PubMed] [Google Scholar]
- 334.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 335.Pascual, C., P. A. Lawson, J. A. Farrow, M. N. Gimenez, and M. D. Collins. 1995. Phylogenetic analysis of the genus Corynebacterium based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:724-728. [DOI] [PubMed] [Google Scholar]
- 336.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 337.Peng, X., K. Brugger, B. Shen, L. Chen, Q. She, and R. A. Garrett. 2003. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 185:2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Perez, P. F., Y. Minnaard, E. A. Disalvo, and G. L. De Antoni. 1998. Surface properties of bifidobacterial strains of human origin. Appl. Environ. Microbiol. 64:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pethe, K., D. L. Swenson, S. Alonso, J. Anderson, C. Wang, and D. G. Russell. 2004. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. USA 101:13642-13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Petkovic, H., J. Cullum, D. Hranueli, I. S. Hunter, N. Peric-Concha, J. Pigac, A. Thamchaipenet, D. Vujaklija, and P. F. Long. 2006. Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol. Mol. Biol. Rev. 70:704-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Petrickova, K., and M. Petricek. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology 149:1609-1621. [DOI] [PubMed] [Google Scholar]
- 342.Philippe, H., and C. J. Douady. 2003. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 6:498-505. [DOI] [PubMed] [Google Scholar]
- 343.Picardeau, M., and V. Vincent. 1998. Mycobacterial linear plasmids have an invertron-like structure related to other linear replicons in actinomycetes. Microbiology 144:1981-1988. [DOI] [PubMed] [Google Scholar]
- 344.Pitcher, D., A. Johnson, F. Allerberger, N. Woodford, and R. George. 1990. An investigation of nosocomial infection with Corynebacterium jeikeium in surgical patients using a ribosomal RNA gene probe. Eur. J. Clin. Microbiol. Infect. Dis. 9:643-648. [DOI] [PubMed] [Google Scholar]
- 345.Possoz, C., C. Ribard, J. Gagnat, J. L. Pernodet, and M. Guerineau. 2001. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 42:159-166. [DOI] [PubMed] [Google Scholar]
- 346.Provost, F., M. V. Blanc, B. L. Beaman, and P. Boiron. 1996. Occurrence of plasmids in pathogenic strains of Nocardia. J. Med. Microbiol. 45:344-348. [DOI] [PubMed] [Google Scholar]
- 347.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 348.Qi, J., B. Wang, and B. I. Hao. 2004. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J. Mol. Evol. 58:1-11. [DOI] [PubMed] [Google Scholar]
- 349.Qin, M., H. Taniguchi, and Y. Mizuguchi. 1994. Analysis of the replication region of a mycobacterial plasmid, pMSC262. J. Bacteriol. 176:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Qin, M. H., M. V. Madiraju, and M. Rajagopalan. 1999. Characterization of the functional replication origin of Mycobacterium tuberculosis. Gene 233:121-130. [DOI] [PubMed] [Google Scholar]
- 351.Radford, A. J., and A. L. Hodgson. 1991. Construction and characterization of a Mycobacterium-Escherichia coli shuttle vector. Plasmid 25:149-153. [DOI] [PubMed] [Google Scholar]
- 352.Raghunand, T. R., and W. R. Bishai. 2006. Mapping essential domains of Mycobacterium smegmatis WhmD: insights into WhiB structure and function. J. Bacteriol. 188:6966-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 354.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital-Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 355.Raoult, D., B. La Scola, P. Lecocq, H. Lepidi, and P. E. Fournier. 2001. Culture and immunological detection of Tropheryma whippelii from the duodenum of a patient with Whipple disease. JAMA 285:1039-1043. [DOI] [PubMed] [Google Scholar]
- 356.Raoult, D., H. Lepidi, and J. R. Harle. 2001. Tropheryma whipplei circulating in blood monocytes. N. Engl. J. Med. 345:548. [DOI] [PubMed] [Google Scholar]
- 357.Raoult, D., H. Ogata, S. Audic, C. Robert, K. Suhre, M. Drancourt, and J. M. Claverie. 2003. Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res. 13:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Rastogi, N., E. Legrand, and C. Sola. 2001. The mycobacteria: an introduction to nomenclature and pathogenesis. Rev. Sci. Tech. 20:21-54. [DOI] [PubMed] [Google Scholar]
- 359.Ratti, G., A. Covacci, and R. Rappuoli. 1997. A tRNA(2Arg) gene of Corynebacterium diphtheriae is the chromosomal integration site for toxinogenic bacteriophages. Mol. Microbiol. 25:1179-1181. [DOI] [PubMed] [Google Scholar]
- 360.Ravel, J., J. DiRuggiero, F. T. Robb, and R. T. Hill. 2000. Cloning and sequence analysis of the mercury resistance operon of Streptomyces sp. strain CHR28 reveals a novel putative second regulatory gene. J. Bacteriol. 182:2345-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Reddy, G. S., J. S. Prakash, R. Srinivas, G. I. Matsumoto, and S. Shivaji. 2003. Leifsonia rubra sp. nov. and Leifsonia aurea sp. nov., psychrophiles from a pond in Antarctica. Int. J. Syst. Evol. Microbiol. 53:977-984. [DOI] [PubMed] [Google Scholar]
- 362.Redenbach, M., J. Scheel, and U. Schmidt. 2000. Chromosome topology and genome size of selected actinomycetes species. Antonie Leeuwenhoek 78:227-235. [DOI] [PubMed] [Google Scholar]
- 363.Rehberger, T. G., and B. A. Glatz. 1990. Characterization of Propionibacterium plasmids. Appl. Environ. Microbiol. 56:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Reilly, T. J., G. S. Baron, F. E. Nano, and M. S. Kuhlenschmidt. 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 271:10973-10983. [DOI] [PubMed] [Google Scholar]
- 365.Ren, Q., and I. T. Paulsen. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Renesto, P., N. Crapoulet, H. Ogata, B. La Scola, G. Vestris, J. M. Claverie, and D. Raoult. 2003. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362:447-449. [DOI] [PubMed] [Google Scholar]
- 367.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, R. G. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 277:21598-21603. [DOI] [PubMed] [Google Scholar]
- 368.Riegel, P., R. Creti, R. Mattei, A. Nieri, and C. von Hunolstein. 2006. Isolation of Corynebacterium tuscaniae sp. nov. from blood cultures of a patient with endocarditis. J. Clin. Microbiol. 44:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 370.Roberfroid, M. B. 2001. Prebiotics: preferential substrates for specific germs? Am. J. Clin. Nutr. 73:406S-409S. [DOI] [PubMed] [Google Scholar]
- 371.Rocha, E. P. 2003. DNA repeats lead to the accelerated loss of gene order in bacteria. Trends Genet. 19:600-603. [DOI] [PubMed] [Google Scholar]
- 372.Rodriguez-Valera, F. 2004. Environmental genomics, the big picture? FEMS Microbiol. Lett. 231:153-158. [DOI] [PubMed] [Google Scholar]
- 373.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Rokas, A., B. L. Williams, N. King, and S. B. Carroll. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798-804. [DOI] [PubMed] [Google Scholar]
- 375.Roller, C., W. Ludwig, and K. H. Schleifer. 1992. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J. Gen. Microbiol. 138:1167-1175. [DOI] [PubMed] [Google Scholar]
- 376.Rosas-Magallanes, V., P. Deschavanne, L. Quintana-Murci, R. Brosch, B. Gicquel, and O. Neyrolles. 2006. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol. Biol. Evol 23:1129-1135. [DOI] [PubMed] [Google Scholar]
- 377.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 378.Rossi, M., P. Brigidi, A. Gonzalez Vara y Rodriguez, and D. Matteuzzi. 1996. Characterization of the plasmid pMB1 from Bifidobacterium longum and its use for shuttle vector construction. Res. Microbiol. 147:133-143. [DOI] [PubMed] [Google Scholar]
- 379.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2005. Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Salzberg, S. L., A. J. Salzberg, A. R. Kerlavage, and J. F. Tomb. 1998. Skewed oligomers and origins of replication. Gene 217:57-67. [DOI] [PubMed] [Google Scholar]
- 382.Schafer, A., J. Kalinowski, and A. Puhler. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl. Environ. Microbiol. 60:756-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Schafer, A., A. Schwarzer, J. Kalinowski, and A. Puhler. 1994. Cloning and characterization of a DNA region encoding a stress-sensitive restriction system from Corynebacterium glutamicum ATCC 13032 and analysis of its role in intergeneric conjugation with Escherichia coli. J. Bacteriol. 176:7309-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Schiller, J., N. Groman, and M. Coyle. 1980. Plasmids in Corynebacterium diphtheriae and diphtheroids mediating erythromycin resistance. Antimicrob. Agents Chemother. 18:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Schiller, J., M. Strom, N. Groman, and M. Coyle. 1983. Relationship between pNG2, an Emr plasmid in Corynebacterium diphtheriae, and plasmids in aerobic skin coryneforms. Antimicrob. Agents Chemother. 24:892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Schneider, D., C. J. Bruton, and K. F. Chater. 2000. Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:543-553. [DOI] [PubMed] [Google Scholar]
- 388.Schouls, L. M., C. S. Schot, and J. A. Jacobs. 2003. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J. Bacteriol. 185:7241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Schrempf, H. 2001. Recognition and degradation of chitin by streptomycetes. Antonie Leeuwenhoek 79:285-289. [DOI] [PubMed] [Google Scholar]
- 390.Serwold-Davis, T. M., N. Groman, and M. Rabin. 1987. Transformation of Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium glutamicum, and Escherichia coli with the C. diphtheriae plasmid pNG2. Proc. Natl. Acad. Sci. USA 84:4964-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 392.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 393.Shigemori, H., H. Komaki, K. Yazawa, Y. Mikami, A. Nemoto, Y. Tanaka, T. Sasaki, Y. In, T. Ishida, and J. Kobayashi. 1998. Brasilicardin A. A novel tricyclic metabolite with potent immunosuppressive activity from actinomycete Nocardia brasiliensis. J. Org. Chem. 63:6900-6904. [DOI] [PubMed] [Google Scholar]
- 394.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 395.Smith, M. C., R. N. Burns, S. E. Wilson, and M. A. Gregory. 1999. The complete genome sequence of the Streptomyces temperate phage straight phiC31: evolutionary relationships to other viruses. Nucleic Acids Res. 27:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Smulczyk-Krawczyszyn, A., D. Jakimowicz, B. Ruban-Osmialowska, A. Zawilak-Pawlik, J. Majka, K. Chater, and J. Zakrzewska-Czerwinska. 2006. Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: from in silico to in vivo studies. J. Bacteriol. 188:6184-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 398.Snel, B., P. Bork, and M. A. Huynen. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12:17-25. [DOI] [PubMed] [Google Scholar]
- 399.Snel, B., M. A. Huynen, and B. E. Dutilh. 2005. Genome trees and the nature of genome evolution. Annu. Rev. Microbiol. 59:191-209. [DOI] [PubMed] [Google Scholar]
- 400.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 401.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Stach, J. E., and A. T. Bull. 2005. Estimating and comparing the diversity of marine actinobacteria. Antonie Leeuwenhoek 87:3-9. [DOI] [PubMed] [Google Scholar]
- 404.Stackebrandt, S. P. 2000. The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY.
- 405.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 406.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Pauker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 47:1134-1139. [DOI] [PubMed] [Google Scholar]
- 407.Stanton, C., R. P. Ross, G. F. Fitzgerald, and D. Van Sinderen. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 16:198-203. [DOI] [PubMed] [Google Scholar]
- 408.Steiner, B., S. Romero-Steiner, D. Cruce, and R. George. 1997. Cloning and sequencing of the hyaluronate lyase gene from Propionibacterium acnes. Can. J. Microbiol. 43:315-321. [DOI] [PubMed] [Google Scholar]
- 409.Stewart, M. E. 1992. Sebaceous gland lipids. Semin. Dermatol. 11:100-105. [PubMed] [Google Scholar]
- 410.Steyn, A. J., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Stinear, T. P., A. Mve-Obiang, P. L. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 413.Suzuki, K. I., M. Suzuki, J. Sasaki, Y. H. Park, and K. K. Komagata. 1999. Leifsonia gen. nov., a genus for 2,4-diaminobutyric acid-containing actinomycetes to accommodate “Corynebacterium aquaticum” Leifson 1962 and Clavibacter xyli subsp. cynodontis Davis et al. 1984. J. Gen. Appl. Microbiol. 45:253-262. [DOI] [PubMed] [Google Scholar]
- 414.Suzuki, N., N. Okai, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2006. High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol. 72:3750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Suzuki, N., S. Okayama, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2005. Large-scale engineering of the Corynebacterium glutamicum genome. Appl. Environ. Microbiol. 71:3369-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Suzuki, N., Y. Tsuge, M. Inui, and H. Yukawa. 2005. Cre/loxP-mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 67:225-233. [DOI] [PubMed] [Google Scholar]
- 417.Swierczynski, A., and H. Ton-That. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 188:6318-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, M. J. Bibb, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475-486. [DOI] [PubMed] [Google Scholar]
- 419.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 420.Tamas, I., L. M. Klasson, J. P. Sandstrom, and S. G. Andersson. 2001. Mutualists and parasites: how to paint yourself into a (metabolic) corner. FEBS Lett. 498:135-139. [DOI] [PubMed] [Google Scholar]
- 421.Tanaka, K., K. Samura, and Y. Kano. 2005. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69:422-425. [DOI] [PubMed] [Google Scholar]
- 422.Tanaka, Y., H. Komaki, K. Yazawa, Y. Mikami, A. Nemoto, T. Tojyo, K. Kadowaki, H. Shigemori, and J. Kobayashi. 1997. Brasilinolide A, a new macrolide antibiotic produced by Nocardia brasiliensis: producing strain, isolation and biological activity. J. Antibiot. (Tokyo) 50:1036-1041. [DOI] [PubMed] [Google Scholar]
- 423.Tauch, A. 2005. Native plasmids of amino acid-producing corynebacteria. CRC Press, Boca Raton, FL.
- 424.Tauch, A., N. Bischoff, I. Brune, and J. Kalinowski. 2003. Insights into the genetic organization of the Corynebacterium diphtheriae erythromycin resistance plasmid pNG2 deduced from its complete nucleotide sequence. Plasmid 49:63-74. [DOI] [PubMed] [Google Scholar]
- 425.Tauch, A., N. Bischoff, A. Puhler, and J. Kalinowski. 2004. Comparative genomics identified two conserved DNA modules in a corynebacterial plasmid family present in clinical isolates of the opportunistic human pathogen Corynebacterium jeikeium. Plasmid 52:102-118. [DOI] [PubMed] [Google Scholar]
- 426.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48:117-129. [DOI] [PubMed] [Google Scholar]
- 427.Tauch, A., O. Kaiser, T. Hain, A. Goesmann, B. Weisshaar, A. Albersmeier, T. Bekel, N. Bischoff, I. Brune, T. Chakraborty, J. Kalinowski, F. Meyer, O. Rupp, S. Schneiker, P. Viehoever, and A. Puhler. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 187:4671-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Tauch, A., S. Krieft, J. Kalinowski, and A. Puhler. 2000. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol. Gen. Genet. 263:1-11. [DOI] [PubMed] [Google Scholar]
- 429.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2003. Plasmids in Corynebacterium glutamicum and their molecular classification by comparative genomics. J. Biotechnol. 104:27-40. [DOI] [PubMed] [Google Scholar]
- 430.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2000. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid 44:285-291. [DOI] [PubMed] [Google Scholar]
- 431.Taylor, J., R. S. Stearman, and B. B. Uratani. 1993. Development of a native plasmid as a cloning vector in Clavibacter xyli subsp. cynodontis. Plasmid 29:241-244. [DOI] [PubMed] [Google Scholar]
- 432.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 433.Tekaia, F., A. Lazcano, and B. Dujon. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9:550-557. [PMC free article] [PubMed] [Google Scholar]
- 434.Titball, R. W. 1998. Bacterial phospholipases. Symp. Ser. Soc. Appl. Microbiol. 27:127S-137S. [PubMed] [Google Scholar]
- 435.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 436.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 437.Trepanier, N. K., G. D. Nguyen, P. J. Leedell, and B. K. Leskiw. 1997. Use of polymerase chain reaction to identify a leucyl tRNA in Streptomyces coelicolor. Gene 193:59-63. [DOI] [PubMed] [Google Scholar]
- 438.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Udaka, S. 1960. Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J. Bacteriol. 79:754-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Valway, S. E., M. P. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. S. Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 442.Vandepoele, K., Y. Saeys, C. Simillion, J. Raes, and Y. Van De Peer. 2002. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 12:1792-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.van der Werf, T. S., W. T. van der Graaf, J. W. Tappero, and K. Asiedu. 1999. Mycobacterium ulcerans infection. Lancet 354:1013-1018. [DOI] [PubMed] [Google Scholar]
- 444.Van Dessel, W., L. Van Mellaert, H. Liesegang, C. Raasch, S. De Keersmaeker, N. Geukens, E. Lammertyn, W. Streit, and J. Anne. 2005. Complete genomic nucleotide sequence and analysis of the temperate bacteriophage VWB. Virology 331:325-337. [DOI] [PubMed] [Google Scholar]
- 445.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.van Pittius, N. C. G., S. L. Sampson, H. Lee, Y. Kim, P. D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Vaughan, E. E., H. G. Heilig, K. Ben-Amor, and W. M. de Vos. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 448.Ventura, M., C. Canchaya, A. Del Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783-2792. [DOI] [PubMed] [Google Scholar]
- 449.Ventura, M., C. Canchaya, G. F. Fitzgerald, R. S. Gupta, and D. van Sinderen. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Leeuwenhoek 91:351-372. [DOI] [PubMed] [Google Scholar]
- 450.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Ventura, M., C. Canchaya, Z. Zhang, V. Bernini, G. F. Fitzgerald, and D. van Sinderen. 2006. How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30:734-759. [DOI] [PubMed] [Google Scholar]
- 453.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 455.Ventura, M., J. H. Lee, C. Canchaya, R. Zink, S. Leahy, J. A. Moreno-Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, D. J. O'Sullivan, and D. van Sinderen. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 71:8692-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 459.Ventura, M., Z. Zhang, M. Cronin, C. Canchaya, J. G. Kenny, G. F. Fitzgerald, and D. van Sinderen. 2005. The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003. J. Bacteriol. 187:8411-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Vertes, A. A., Y. Asai, M. Inui, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1994. Transposon mutagenesis of coryneform bacteria. Mol. Gen. Genet. 245:397-405. [DOI] [PubMed] [Google Scholar]
- 463.Vertes, A. A., M. Inui, and H. Yukawa. 2005. Manipulating corynebacteria, from individual genes to chromosomes. Appl. Environ. Microbiol. 71:7633-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 465.Wang, B., J. Brand-Miller, P. McVeagh, and P. Petocz. 2001. Concentration and distribution of sialic acid in human milk and infant formulas. Am. J. Clin. Nutr. 74:510-515. [DOI] [PubMed] [Google Scholar]
- 466.Wang, F. S., T. S. Whittam, and R. K. Selander. 1997. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J. Bacteriol. 179:6551-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Ward, R. E., M. Ninonuevo, D. A. Mills, C. B. Lebrilla, and J. B. German. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 72:4497-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Wayne, L. G., D. J. Brenner, R. R Colwell, et al. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 469.Wayne, L. G., and L. G. Hayes. 1998. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:127-132. [DOI] [PubMed] [Google Scholar]
- 470.Weaver, D., N. Karoonuthaisiri, H. H. Tsai, C. H. Huang, M. L. Ho, S. Gai, K. G. Patel, J. Huang, S. N. Cohen, D. A. Hopwood, C. W. Chen, and C. M. Kao. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535-1550. [DOI] [PubMed] [Google Scholar]
- 471.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 472.Webster, G. F., J. J. Leyden, R. A. Musson, and S. D. Douglas. 1985. Susceptibility of Propionibacterium acnes to killing and degradation by human neutrophils and monocytes in vitro. Infect. Immun. 49:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Westerhof, W., G. N. Relyveld, M. M. Kingswijk, P. de Man, and H. E. Menke. 2004. Propionibacterium acnes and the pathogenesis of progressive macular hypomelanosis. Arch. Dermatol. 140:210-214. [DOI] [PubMed] [Google Scholar]
- 474.Whale, G. A., I. C. Sutcliffe, A. R. Morrisson, E. L. Pretswell, and N. Emmison. 2004. Purification and characterisation of lipoglycan macroamphiphiles from Propionibacterium acnes. Antonie Leeuwenhoek 86:77-85. [DOI] [PubMed] [Google Scholar]
- 475.Widdick, D. A., K. Dilks, G. Chandra, A. Bottrill, M. Naldrett, M. Pohlschroder, and T. Palmer. 2006. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 103:17927-17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Willey, J. M., A. Willems, S. Kodani, and J. R. Nodwell. 2006. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 59:731-742. [DOI] [PubMed] [Google Scholar]
- 477.Williams, L. E., C. Detter, K. Barry, A. Lapidus, and A. O. Summers. 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72:4899-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Williams, S. M., and W. C. Noble. 1988. Plasmids in coryneform bacteria of human origin. J. Appl. Bacteriol. 64:475-482. [DOI] [PubMed] [Google Scholar]
- 479.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 480.Wilson, S. E., C. J. Ingham, I. S. Hunter, and M. C. Smith. 1995. Control of lytic development in the Streptomyces temperate phage phi C31. Mol. Microbiol. 16:131-143. [DOI] [PubMed] [Google Scholar]
- 481.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Wolf, Y. I., I. B. Rogozin, N. V. Grishin, R. L. Tatusov, and E. V. Koonin. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BCM Evol. Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Wolf, Y. I., I. B. Rogozin, A. S. Kondrashov, and E. V. Koonin. 2001. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 11:356-372. [DOI] [PubMed] [Google Scholar]
- 485.Wolinsky, E., and T. K. Rynearson. 1968. Mycobacteria in soil and their relation to disease-associated strains. Am. Rev. Respir. Dis. 97:1032-1037. [DOI] [PubMed] [Google Scholar]
- 486.Wren, B. W. 2000. Microbial genome analysis: insights into virulence, host adaptation and evolution. Nat. Rev. Genet. 1:30-39. [DOI] [PubMed] [Google Scholar]
- 487.Wu, G., L. Nie, and W. Zhang. 2006. Predicted highly expressed genes in Nocardia farcinica and the implication for its primary metabolism and nocardial virulence. Antonie Leeuwenhoek 89:135-146. [DOI] [PubMed] [Google Scholar]
- 488.Xu, W., J. Shen, C. A. Dunn, and M. J. Bessman. 2003. A new subfamily of the Nudix hydrolase superfamily active on 5-methyl-UTP (ribo-TTP) and UTP. J. Biol. Chem. 278:37492-37496. [DOI] [PubMed] [Google Scholar]
- 489.Yamasaki, M., Y. Ikuto, A. Ohira, K. Chater, and H. Kinashi. 2003. Limited regions of homology between linear and circular plasmids encoding methylenomycin biosynthesis in two independently isolated streptomycetes. Microbiology 149:1351-1356. [DOI] [PubMed] [Google Scholar]
- 490.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Yang, S., R. F. Doolittle, and P. E. Bourne. 2005. Phylogeny determined by protein domain content. Proc. Natl. Acad. Sci. USA 102:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]
- 493.Zeigler, D. R. 2003. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 53:1893-1900. [DOI] [PubMed] [Google Scholar]
- 494.Zhang, R., and C. T. Zhang. 2005. Genomic islands in the Corynebacterium efficiens genome. Appl. Environ. Microbiol. 71:3126-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Zhang, R., and C. T. Zhang. 2004. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics 20:612-622. [DOI] [PubMed] [Google Scholar]
- 496.Zhang, Y., J. Praszkier, A. Hodgson, and A. J. Pittard. 1994. Molecular analysis and characterization of a broad-host-range plasmid, pEP2. J. Bacteriol. 176:5718-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Zolg, J. W., and S. Philippi-Schulz. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Zumarraga, M., F. Bigi, A. Alito, M. I. Romano, and A. Cataldi. 1999. A 12.7 kb fragment of the Mycobacterium tuberculosis genome is not present in Mycobacterium bovis. Microbiology 145:893-897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.