Abstract
Summary: While reductionism has greatly advanced microbiology in the past 400 years, assembly of smaller pieces just could not explain the whole! Modern microbiologists are learning “system thinking” and “holism.” Such an approach is changing our understanding of microbial physiology and our ability to diagnose/treat microbial infections. This review uses oral microbial communities as a focal point to describe this new trend. With the common name “dental plaque,” oral microbial communities are some of the most complex microbial floras in the human body, consisting of more than 700 different bacterial species. For a very long time, oral microbiologists endeavored to use reductionism to identify the key genes or key pathogens responsible for oral microbial pathogenesis. The limitations of reductionism forced scientists to begin adopting new strategies using emerging concepts such as interspecies interaction, microbial community, biofilms, polymicrobial disease, etc. These new research directions indicate that the whole is much more than the simple sum of its parts, since the interactions between different parts resulted in many new physiological functions which cannot be observed with individual components. This review describes some of these interesting interspecies-interaction scenarios.
INTRODUCTION
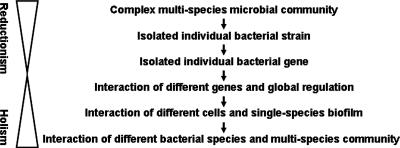
Like many other biological sciences, the study of microbiology has encompassed phases of “reductionism” and “holism.” For a long time, microbiologists took the reduction approaches to study individually isolated bacterial colonies from complex microbial communities and to analyze individually isolated bacterial genes from complex microbial genomes, hoping to understand the whole by examining smaller and smaller components (Fig. 1). While reductionism has greatly advanced microbiology in the past 400 years, assembly of smaller pieces just could not explain the whole! Recognizing that the whole is more than the simple sum of its parts, modern microbiologists are learning “system thinking” and “holism.” From global gene regulation to metagenomics to biofilms, microbiology is experiencing a new trend that emphasizes interactions of different elements within a community. Such an approach is changing our understanding of microbial physiology and our ability to diagnose/treat microbial infections. This review uses oral microbial communities as a focal point to describe this new trend and discuss its possible impact on future studies in microbiology.
FIG. 1.
Evolution of microbiological studies. Initial reductionism included zooming in from complex multispecies microbial communities to isolated genes from individual species. This process was followed by an expansion of scope based on the knowledge acquired through reductionism to achieve a holistic view of the interactions of different bacterial species and the multispecies communities in which they reside.
With the common name “dental plaque,” oral microbial communities are some of the most complex microbial floras in the human body, consisting of more than 700 different bacterial species (1, 155, 157). Extensive clinical studies have indicated that the oral microbial flora is responsible for two major human diseases: dental caries (tooth decay) and periodontitis (gum disease) (28, 119, 128, 148, 188). For a very long time, oral microbiologists endeavored to use reductionism to identify the key pathogens responsible for oral microbial pathogenesis (43, 67, 184). The limitations of reductionism forced scientists to begin adopting new strategies using emerging concepts such as interspecies interaction, microbial community, biofilms, polymicrobial disease, etc. These new research directions made oral microbiologists truly appreciate the fact that the whole is much more than the simple sum of its parts, since the interactions between different components resulted in many new physiological functions which cannot be observed with individual components. These interesting interspecies-interaction scenarios are the focus of this review.
ORAL MICROBIAL COMMUNITY
Oral Bacteria and Dental Plaque
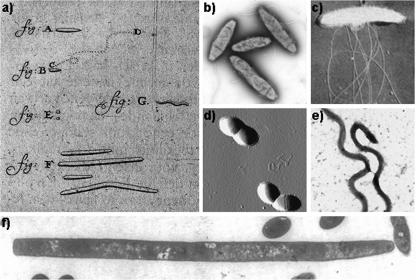
It is generally recognized that the advent of microbiological investigations began with the initial observations of bacteria by van Leeuwenhoek (1632-1723) using his primitive microscopes (57). In his notebook, he wrote “I didn't clean my teeth for three days and then took the material that had lodged in small amounts on the gums above my front teeth… . I found a few living animalcules.” The microbes sketched in his notebook are now known as some of the most abundant bacteria in the oral cavity, including cocci, fusiform bacteria, and spirochetes (Fig. 2). These fascinating observations at the birth of microbiology signaled the complexity of the oral microbial community.
FIG. 2.
Oral bacteria observed by Anton van Leeuwenhoek and their contemporary equivalents. (a) The original drawings by van Leeuwenhoek. The bacterium designated A was later suggested to be a rod-shaped motile bacterium such as Campylobacter rectus (b) (photo courtesy: National Research Council Canada); the highly motile species shown in his original drawing as B and moving quickly from C to D was likely Selenomonas sputigena (c) (electron micrograph, modified from reference 123 with permission) (187), whereas E was proposed to be oral cocci (d) (atomic force microscopy image). van Leeuwenhoek's original drawing G was disputed as being either a Spirillum sp. (not shown) or an oral spirochete such as Treponema denticola (e) (electron micrograph), and the long fusiform species designated F were proposed to be Leptotrichia (Leptotrix) buccalis (f) (electron micrograph, modified from reference 117 with permission).
W. D. Miller was the next important individual who greatly advanced oral microbiology. Practicing as a dentist, he spent his evenings and weekends at Robert Koch's laboratory for the purpose of identifying the “germs” that were responsible for tooth decay. In 1890, he published a book titled Microorganisms of the Human Mouth (138). In this book, he proposed a “chemicoparasitic” theory which suggested that dental plaque was made of microorganisms. In susceptible hosts who frequently consumed fermentable carbohydrates, microorganisms within dental plaque would convert these carbohydrates into acids, which would lead to the demineralization of teeth.
Building on Miller's chemicoparasitic theory, extensive clinical evidence was later obtained, indicating the etiological role of dental plaque in human oral diseases. Based on the locations relative to the gum line, dental plaque is generally divided into supragingival plaque and subgingival plaque (148). The supragingival plaque is known to be dominated by gram-positive streptococci, which are capable of building up microbial communities on smooth tooth surfaces. These communities, when fed the right metabolic substrate, are responsible for dental caries (tooth decay) (19). The subgingival plaque is known to be dominated by gram-negative anaerobic bacteria, which establish their community within periodontal pockets and are responsible for periodontitis (gum diseases) (149, 211). Removing dental plaque, as initially recommended by W. D. Miller, has been the cornerstone of the treatment and prevention of dental diseases.
Microbial Composition of Dental Plaque
W. D. Miller tried very hard to study the microbial composition of dental plaque but without much success, largely due to the limited bacterial isolation and culture techniques available in the 19th century. However, great progress has been made since then in understanding the microbial profiles within dental plaques. From the initial isolation of Streptococcus mutans by J. K. Clarke in 1924 to the latest large-scale 16S rRNA/DNA-based oral bacterial studies (1, 22, 157), over 700 bacterial species (gram-positive bacteria, gram-negative bacteria, and archaea) have been identified from the human oral cavity, making the oral microbial community one of the most complex microbial floras in the human body.
Based on our current understanding, supragingival plaque is dominated by gram-positive bacteria, including Streptococcus sanguinis, S. mutans, Streptococcus mitis, Streptococcus salivarius, and lactobacilli, while the subgingival plaque is made up primarily by gram-negative anaerobic bacteria, such as Aggregatibacter (Actinobacillus) actinomycetemcomitans, Tannerella forsythia, Campylobacter spp., Capnocytophoga spp., Eikenella corrodens, Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, and oral spirochetes such as Treponema denticola (1, 140, 157). In both cases, the microbial communities on teeth and gingival tissues can accumulate high concentrations of bacterial metabolites (e.g., fatty acid end products, ammonia, hydrogen peroxide, oxidants, and carbon dioxide) in their local environments, which influence the bacterial species within the microbial community, as well as the host (14).
Well-Organized Multispecies Microbial Communities within Dental Plaque
By the “reductionism” approach, oral microbiologists were initially hoping to understand dental plaque and its behavior through defining its microbial composition, i.e., making the connection between the behavior of specific oral bacterial species within dental plaque and the behavior of intact dental plaque. However, such hopes quickly evaporated, since from a structural and functional point of view, dental plaque is not merely the simple sum of its bacterial components. Instead, a sophisticated microbial community is established, which results in novel functions that are essential for biofilm architecture and microbial physiology (127, 128).
From a structural point of view, dental plaque shows a high degree of organization. During the process of dental plaque formation, some oral bacteria are early colonizers that express biochemical components allowing them to effectively adhere to targeted tissues (teeth or periodontal tissue). The later colonizers often contain components that enable them to adhere to the early colonizers, often bringing metabolic or other competitive advantages. Within an established dental plaque, specific bacterial species are often found located adjacent to each other or mixed together to form unique structures that often confer adherence or growth advantages. Previous comprehensive reviews by P. E. Kolenbrander et al. should be consulted for assessment of these important properties (95, 97).
From a microbial physiology aspect, oral microbial communities are classical examples of biofilms. Just as initially proposed by Costerton et al., the behaviors displayed by oral microbial organisms grown in liquid culture are very different from those of the same organisms grown on a solid surface or within a community such as dental plaque (24, 25). Many studies have indicated that bacteria present in biofilms have properties that are uniquely dependent upon such structures. This is of significant medical interest, since a number of studies have documented the increased resistance of oral bacteria within dental plaque to antimicrobial agents relative to that in planktonic growth (50, 60, 135). Additional confirmation of these differences has been provided by numerous investigations examining gene expression and protein synthesis. This has revealed that some properties of oral bacteria grown within biofilms are distinct from those of comparable planktonic cultures (7, 11).
Because of the multispecies nature of dental plaque, the oral microbial community is one of the best biofilm models for studying interspecies interactions (95). It is reasonable to assume that the interactions between the oral microbial residents may influence the properties of the whole community. In this regard, oral microbial communities (biofilms) may represent a micro example of the “Gaia” hypothesis (122); i.e., biofilms are analogous to the planet Earth, where the properties of the latter as a whole are determined by the interactions of all of the residents as well as interactions of the populations with the inanimate supporting structures.
Community-Based Microbial Pathogenesis
The successful isolation and characterization of microbial species from dental plaque naturally prompted oral microbiologists to connect specific bacterial species to certain diseases, according to Koch's postulates. For example, as a result of such strategies, the acid-producing oral bacterium S. mutans present in supragingival plaque was selected for its positive correlation with dental caries (71, 120). However, additional scientific data have suggested that such a simple correlation may be an oversimplification. Unlike many known medical pathogens that are “foreign invaders with specific virulence factors,” the oral “pathogens” such as S. mutans are part of the normal flora (1). While they express certain pathogenic factors (such as acid production in this case), a dynamic balance of both synergistic and antagonistic interactions with its neighboring bacteria plays an essential role in determining whether these pathogenic factors cause damage or not (92, 127). In other words, in the case of complex biofilms, it is not merely the presence of a single organism in a complex community which determines the properties of a biofilm, but it is the interactions between the biofilm residents which is crucial. As an example, in the presence of nearby base-producing bacteria, S. mutans in dental plaque may not be as pathogenic to the host. Thus, for dental caries, it is now generally recognized that this disease results not solely because of the presence of S. mutans or any single organism in dental plaque. Rather, it is the result of the interaction of multiple acid-producing organisms such as S. mutans with other biofilm residents (92, 127, 128). Such a community- and microbial ecology-based pathogenic theory serves as a new concept for understanding the relationship between dental plaque and the host in health or disease, as well as suggesting new strategies for disease treatment and prevention.
Multispecies communities like dental plaque can also produce polymicrobial infections in which microorganisms interact in a synergistic fashion, leading to pathogenesis. Periodontal diseases represent one of the best-documented polymicrobial infections. P. gingivalis, Treponema denticola, and Tannerella forsythia are strongly implicated clinically as a pathogenic consortium in the etiology of adult periodontitis (48, 189). It is anticipated that a better understanding of the mechanisms involved in periodontitis may provide opportunities to more critically evaluate the role of different virulence factors involved in these mixed infections. This may provide useful general information for investigators in the development of novel diagnostic, preventive, and treatment strategies against polymicrobial infections.
INTERACTIONS BETWEEN RESIDENTS IN DENTAL PLAQUE
The oral cavity contains complex, multispecies microbial communities. This suggests that the residents in this community should display extensive interactions while forming biofilm structures, carrying out physiological functions, and inducing microbial pathogenesis. This review focuses on these interesting interactions, which include (i) competition between bacteria for nutrients, (ii) synergistic interactions which may stimulate the growth or survival of one or more residents, (iii) production of an antagonist by one resident which inhibits the growth of another, (iv) neutralization of a virulence factor produced by one organism by another resident, and (v) interference in the growth-dependent signaling mechanisms of one organism by another. In a micro-Gaia community, these interactions could be envisaged as forms of “war and peace” among the bacterial residents of a biofilm. The implications of such effects are discussed below, especially in terms of lessons learned from the study of oral biofilms.
Nutrients as the Basis for Bacterial Interspecies Interactions within Biofilms
One important factor in determining the bacterial composition of a biofilm is clearly the availability of nutrients. Organisms that have adapted to particular environments such as the oral cavity have evolved metabolic pathways to efficiently utilize the available nutrients in each specific ecological niche. For the residents within oral biofilms, nutrients are available from the periodic intake of food, saliva, and nutrients provided by other organisms as well as polysaccharides present in dental plaque (8). Relevant to the pathogenesis of dental caries is the demonstration that S. mutans is able to metabolize sucrose more efficiently than other common oral bacteria. This may be an important factor in its role in becoming a resident in caries-associated biofilms (71). With many human diets being composed of significant proportions of sucrose, especially in developed countries, S. mutans may have a competitive advantage relative to other oral bacteria. In addition, S. mutans' rapid conversion of sucrose to lactic acid also provides S. mutans with an additional mechanism for competing with many organisms due to its relatively strong aciduric properties (41, 63). S. mutans' ability to convert sucrose to adherent glucan molecules also promotes the attachment of these organisms to teeth (59). Bacteria lacking this property may be nonspecifically trapped by these glucans into a developing plaque.
Porphyromonas gingivalis, one of the major etiological agents of periodontal disease, often coexists with other periodontopathic bacteria, such as Prevotella intermedia, F. nucleatum, Tannerella forsythia, and Treponema denticola (43, 139-141). In an early study, Grenier and Maryrand demonstrated cross-feeding between P. gingivalis and T. denticola (65). They observed that when grown in coculture, P. gingivalis can promote its own growth by metabolizing the succinate produced by T. denticola. In addition, isobutyric acid excreted by P. gingivalis can stimulate the growth of T. denticola (64). In a more recent study, Yoneda et al. demonstrated that cell extracts from both F. nucleatum and T. forsythia can also stimulate the growth of P. gingivalis, although the growth-promoting factors have not yet been identified (213).
Another important nutrient source for oral bacteria are the proteins present in human saliva (12, 176, 205) and gingival crevicular fluid (66). These can be produced by oral host tissues as well as by bacteria. Those organisms that are highly proteolytic will have a competitive advantage for growth under conditions where salivary proteins are a limiting source of nutrients. Since this may be the case for bacteria growing in subgingival dental plaque (below the gum line), it is suggested that one of reasons why the gram-negative anaerobic organism P. gingivalis can readily colonize this region and is associated with periodontitis (inflammation or destruction of gum tissue) is that it is a highly proteolytic organism (132, 161). Other, relatively nonproteolytic organisms that can colocalize with P. gingivalis in biofilms may also benefit from the potent hydrolytic activity of these organisms (31). Thus, it should be expected that organisms which share such symbiotic relationships would be found in close proximity within biofilms.
Synergistic or mutualistic interactions between two organisms in biofilms that appear to be dependent upon saliva have been demonstrated for Actinomyces naeslundii and Streptococcus oralis (150). Either organism alone is a poor colonizer of saliva-coated surfaces, but together they form extensive biofilms on these same surfaces. This appears to result from the combined metabolic activities of the two organisms in metabolizing salivary components.
General Metabolic Products Which Influence Biofilm Resident Interactions
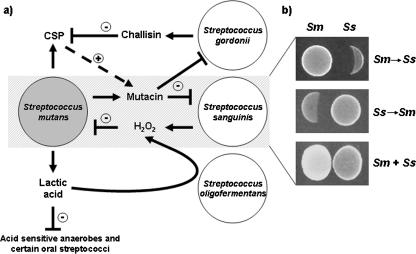
It is also likely that the metabolic products (secondary metabolites) of one organism have effects on other organisms within the same biofilms. For example, dental plaque which contains relatively high proportions of S. mutans generally yields low levels of S. sanguinis strains (121). The former organisms can metabolize sugars to lactic acid, which is the strongest acid product of most bacteria. Since S. mutans strains are generally more aciduric than members of the S. sanguinis family, the production of lactic acid favors the growth of S. mutans relative to that of other oral streptococci (Fig. 3). On the other hand, members of the S. sanguinis group are producers of hydrogen peroxide, a nonspecific antimicrobial agent that has an antagonistic effect on other coresidents, such as S. mutans, which does not express effective systems for metabolizing this toxic product (15, 73, 101). In addition, many anaerobic bacteria, including those which are associated with human periodontitis, are also sensitive to the oxidant (80). Therefore, relatively high proportions of S. sanguinis are generally found in dental plaque with lower levels of S. mutans or periodontopathic organisms such as P. gingivalis (75). Furthermore, the presence of members of the S. sanguinis group in dental plaque is generally correlated with low levels of periodontopathic organisms and thus is conducive to good oral health (75).
FIG. 3.
Illustration of interspecies interactions between Streptococcus mutans and selected oral streptococci that have been studied on a molecular level. (a) Interactions of S. mutans with S. gordonii, S. oligofermentans, and S. sanguinis. Depicted is the production of lactic acid, mutacin, and CSP, which also enhances mutacin production by S. mutans. Lactic acid has been found to inhibit proliferation of a variety of acid-sensitive anaerobe species, but it was recently discovered to serve as a substrate for S. oligofermentans for H2O2 production. This compound is also generated by S. sanguinis and was previously shown to inhibit growth of S. mutans. S. sanguinis and S. gordonii are sensitive to the mutacins produced by S. mutans. In contrast to S. sanguinis, which can inhibit S. mutans by its ability to produce H2O2, S. gordonii utilizes challisin to reduce mutacin production by reducing the levels of the stimulating factor CSP. The dashed arrow in combination with + indicates stimulation, solid arrows symbolize production, and ⊣ in combination with − designates inhibition. The shaded area indicates the timing-sensitive interactions between S. mutans and S. sanguinis illustrated in panel b. (b) Importance of timing for reciprocal inhibition of S. mutans (Sm) and S. sanguinis (Ss). Given a 1-day growth advantage, S. mutans inhibits S. sanguinis via its mutacin production (upper panel), whereas S. sanguinis controls establishment of S. mutans by its ability to generate H2O2 when given the same 1-day advantage (middle panel). Without the benefit to either of establishing itself earlier, both species coexist without any apparent mutual inhibition (lower panel). For further details, see the text.
In addition to antagonistic effects, metabolic products of one organism may promote the growth of other organisms. The lactic acid produced by S. mutans can be readily metabolized by members of the Veillonella family (137). It has also been observed that Veillonella species are present in early developing dental plaque (151). More recently, it has been observed that S. oligofermentans can also metabolize the lactic acid produced by S. mutans (201) (Fig. 3). S. oligofermentans was first isolated from the dental plaque of caries-free human subjects, and its abundance within dental plaque was found to be negatively correlated with that of S. mutans. This noncariogenic oral streptococcus was also shown to inhibit the growth of S. mutans. The mechanism of this inhibitory effect is quite unique. S. oligofermentans, using the enzymatic activity of lactate oxidase, converts lactic acid, a normally growth-inhibitory by-product secreted by S. mutans, into hydrogen peroxide, a substance which is highly toxic to the latter organism (201).
Cooperative metabolic interactions are also suggested by the observation that certain bacterial species can modify the local microenvironment, making it more suitable for the growth of other species. For example, it has been observed that F. nucleatum and P. intermedia can grow at a wide pH range of 5.0 to 7.0, while P. gingivalis is susceptible to pH levels of below 6.5. A recent study by Takahashi demonstrated that by using glutamic and aspartic acids, the two major amino acids from crevicular fluid and saliva, as fermentation substrates, F. nucleatum and P. intermedia can generate ammonia as well as organic acids (195). This contributes to a more neutral pH in dental plaque and prevents severe drops in pH even in the presence of lactic acid bacteria and fermentable carbohydrates. Thus, acid-sensitive species such as P. gingivalis are protected against acid attack (195).
Other metabolic by-products which could influence the interactions of bacteria within biofilms are the gases involved in metabolism. Prime examples of this influence are the metabolism of oxygen and carbon dioxide. Highly anaerobic organisms, such as those involved in periodontitis, proliferate in the absence of high concentrations of oxygen. Therefore, organisms which are able to metabolize oxygen would favor the growth of nearby anaerobic organisms. Other than maintaining an optimum pH in the microenvironment for P. gingivalis, F. nucleatum also contributes to generating a reducing and capnophilic microenvironment that is necessary for the growth of P. gingivalis (39). Although both F. nucleatum and P. gingivalis are strictly anaerobic bacteria, their susceptibilities to oxygen are quite distinct. P. gingivalis is very sensitive to oxygen, while F. nucleatum can tolerate oxygen levels of as high as 20%. Interestingly, when cocultivated with F. nucleatum in chemostat culture, P. gingivalis was able to grow even in gaseous environments containing 20% oxygen. This is likely due to the active NADH oxidase/peroxidase activity of F. nucleatum which is able to metabolize both molecular oxygen and hydrogen peroxide, creating an optimum reducing microenvironment for P. gingivalis. At the same time, F. nucleatum can also satisfy the requirement for carbon dioxide for growth by P. gingivalis (39).
Bacteriocins and Their Role in Interspecies Interaction within Oral Biofilms
Bacteria are also able to generate products which may exert either specific or nonspecific effects on other bacteria. Prime examples of specific effects are those involving the expression of bacteriocins by some bacteria. Bacteriocins are proteinaceous toxins that have been found in all major lineages of bacteria. Unlike traditional antibiotics, they often have a narrow killing spectrum and inhibit the growth of related organisms (16, 23, 133, 168, 193). As this relates to dental plaque, S. mutans strains have been shown to produce a number of distinct bacteriocins, also termed mutacins (20, 163, 164). Of all the oral bacteria, streptococci have shown the greatest capacity for bacteriocin production (147). Indeed, many oral streptococcal species have been shown to produce a variety of peptide bacteriocins. S. mutans is able to produce at least five different bacteriocins, mutacins I to V. Mutacins I, II, and III belong to the lanthionine-containing lantibiotics with a relatively broad antimicrobial spectrum, while mutacins IV and V are unmodified nonlantibiotics, with mutacin IV specifically active against members of the mitis group of oral streptococci (20, 163, 164). A more recent genome search of S. mutans strain UA159 revealed at least nine other putative mutacin-encoding genes, suggesting a large repertoire or “arsenal” that can be used against its competitors (70). Numerous studies have suggested that the mutacin activity of S. mutans could be related to its prevalence and successful establishment in the dental biofilm (101, 147). Some of these bacteriocins are also able to inhibit the growth of S. sanguinis strains, and these inhibitors may be responsible, in part, for the presence of relatively low levels of S. sanguinis strains in plaque containing high proportions of S. mutans (101). Thus, the expression of bacteriocins by some biofilm residents may determine which other organisms are coresidents in these structures.
Bacteriocins may also affect interspecies interactions by acting as analogues of signaling molecules. For example, the lantibiotic bacteriocins produced by Streptococcus pyogenes and Streptococcus salivarius are structurally similar and can interact with the two-component signaling systems of each other (204). Both organisms can colonize mucosal surfaces and may inhibit the growth of the other via antagonizing growth-dependent signaling. In this manner, the commensal S. salivarius strains may prevent biofilm formation by virulent S. pyogenes strains.
It has also been suggested that antimicrobial agents other than bacteriocins can have effects on the signal transduction systems of some bacteria (77). Such agents include antibiotics initially characterized as growth inhibitors. They may have previously unrecognized signaling activity at sublethal concentrations. If this is true, such interactions could have effects on the expression of virulence as well as biofilm-dependent genes. These provocative possibilities should be amenable to investigations with current biofilm model systems.
Many oral bacteria have been shown to use bacteriocin-like compounds to compete with other species. Using overlay and agar diffusion methods, Teanpaisan et al. demonstrated the production of, and sensitivity to, bacteriocin-like activity among 44 strains of black-pigmented anaerobes, including P. gingivalis, P. intermedia, and Prevotella nigrescens, isolated from periodontal sites (198). Subsequently, the same group isolated and identified the genetic determinant for nigrescin, a 41-kDa novel bacteriocin which is produced by P. nigrescens and displays a bactericidal effect against P. gingivalis, P. intermedia, T. forsythia, and Actinomyces spp. (84).
Bacteriocin or bacteriocin-like activities have also been reported and well documented for other oral bacteria, such as P. intermedia (194), Capnocytophaga ochracea (145), A. actinomycetemcomitans (72), Haemophilus influenzae (116), F. nucleatum (56), and E. corrodens (2). However, their biochemical properties and genetic determinants have not yet been well characterized. Recently, the genome sequence of T. denticola further revealed at least three potential bacteriocin secretion systems, suggesting possible bacteriocin production by this species as well (178).
Some recent molecular and genetic studies have revealed that bacteriocin production is a well-regulated event which is affected by both environmental conditions, such as cell density, nutritional availability, and pH, and genetic factors (100, 101, 136, 162, 203). These regulatory systems ensure that bacteriocins are produced at the right time and place so that they can be effectively used for “war and peace” activities within an oral microbial community.
Microbial community stability can be achieved only when a natural balance is established among different species within the same biological niche, and this balance is often the result of the constant “war and peace” activities experienced by all the members of the biocommunity. The production of, and sensitivity to, certain bacteriocin or bacteriocin-like activities among oral bacteria could enable bacteria to select their neighbors, promote the establishment of a community with specific bacterial species, and play an important role in the ecological balance of the oral ecosystem.
Other Biofilm Properties Influenced by Bacterial Interactions
Several recent studies have further suggested that interactions between biofilm bacteria could influence their relative antimicrobial susceptibilities (85, 86). These results demonstrated that in vitro biofilms containing both S. mutans and Veillonella parvula were more resistant to the antimicrobial agent chlorhexidine than were the monospecies biofilms of either. Although the molecular basis for such increased resistance was not determined, it was suggested that the clustering of these two organisms within biofilms may be responsible for this property.
Another strategy involving interactions affecting the antimicrobial susceptibility of a biofilm resident is exemplified by that involving S. mutans and S. gordonii. These two commensal oral streptococci are normally found in inverse proportions in dental plaque (121). It has recently been demonstrated that S. gordonii can antagonize several of the quorum sensing-dependent mechanisms of S. mutans by inactivating the competence-stimulating peptide (CSP) of the latter organism (209) (Fig. 3). In addition, it was shown recently that the resistance of S. mutans to a variety of antimicrobial agents was diminished by inactivating the CSP (130). Thus, the presence of S. gordonii in biofilms may antagonize the colocalization of S. mutans in these structures by increasing the sensitivity of the latter organism to endogenous antimicrobial agents such as histatins, a group of antimicrobial peptides often found in saliva. While such a mechanism for antagonism has not yet been directly demonstrated in vivo, it does indicate another possible outcome of biofilm resident interactions.
Synergism between two or more different oral bacterial species has been reported to enhance virulence in many animal models. When coinfected with P. gingivalis and T. forsythia in a murine abscess model, animals develop larger lesions and higher mortality rates than after single-species infection (213). Synergistic effects have also been reported between P. gingivalis and F. nucleatum (44, 49), P. gingivalis and T. denticola (90), and P. gingivalis and A. actinomycetemcomitans (17) in murine abscess models. Although cooperative metabolic interactions might not be able to fully explain the enhanced virulence of multispecies infections compared to monoinfections, they could play an important role in obtaining increased growth of pathogens, thus promoting the initiation and acceleration of the infection process.
Bacterial Interactions Influence the Localization of Residents within Biofilms
The demonstration of a multitude of different possible interactions between biofilm residents suggests that the net effect of these interactions may influence the localization or colocalization of certain organisms within the biofilm structure. For example, using fluorescently stained cells, it was observed that P. gingivalis colonized in vitro biofilms of S. gordonii especially in regions where the latter organisms accumulated (103). This observation is consistent with the known interactions between these two organisms as described above. In addition, such a correlation between in vitro studies and in vivo systems was obtained with Veillonella species and streptococci (151).
A number of direct visualization studies have further indicated that some organisms are clustered within biofilms and are not randomly distributed (96, 98). This might occur as a result of colocalization of the “pioneer” organisms of biofilms by means of some of the positive interactions described above. In addition, initial negative effects of one organism on another may lead to relative sequestering of these organisms from one another. Therefore, one would predict that in dental plaque, S. mutans and S. sanguinis strains would be observed in distinct clusters. However, this has not yet been directly demonstrated within plaque in vivo.
Bacterial Coaggregation Influences Localization within Biofilms
The clustering of organisms within biofilms can most readily be rationalized for those organisms whose interactions are mediated primarily by coaggregation. Beginning with the pioneering studies of Gibbons and coworkers (58), it was demonstrated that a number of oral bacteria specifically aggregated with each other but not with other organisms. For example, strains of P. gingivalis were shown to aggregate with several oral streptococci (185). It was also demonstrated that P. gingivalis could colonize biofilms composed of these oral streptococci both in vitro and in vivo. This suggested that this might be an important mechanism whereby P. gingivalis could become incorporated into dental plaque that was composed primarily of the initial streptococcal colonizers. Subsequently, many other coaggregating partners have been identified among oral bacteria, as previously described (94).
P. gingivalis-oral streptococcal coaggregations augment the ability of each organism to colonize into biofilms. However, several recent investigations have demonstrated that some organisms which do not form biofilms by themselves, at least in vitro, do so in the presence of other potential partners. The oral treponeme T. denticola does not appear to form biofilms on most inert surfaces (208), in contrast to P. gingivalis, which does so readily. However, in the presence of the latter organism, the treponeme is incorporated into the biofilm. This model appears to mimic the situation in subgingival dental plaque, where T. denticola is found more external relative to P. gingivalis (81). Similarly, the periodontopathogen T. forsythia is a weak colonizer of inert surfaces but will become incorporated into biofilms in the presence of F. nucleatum (179). These synergistic effects appear to be relevant to biofilm formation in vivo, since both pairs of organisms are frequently found associated in the same plaque samples (189).
Interactions Mediated by Signaling Molecules Influence Biofilm Formation
Several approaches have demonstrated that signal transduction mechanisms may play a role in monospecies biofilm formation (33, 34). Therefore, interference by other antecedent biofilm residents that can metabolize or transport the signaling molecules would antagonize biofilm formation by the producing organisms. No such interactions have yet to be demonstrated for oral biofilm residents. Nevertheless, due to the apparent ubiquity of the autoinducer 2 (AI-2) synthase gene, luxS, in many different organisms (5), including oral bacteria (54), the quorum-sensing regulatory molecule AI-2 has been suggested as a potential signaling molecule between heterogeneous bacteria within biofilms. A number of investigations have shown the ability of an organism to complement luxS-mediated defects in other species residing in oral cavity (214), suggesting the interspecies nature of the AI-2 signaling molecule. Recently, the AI-2 signaling molecule has been shown to be required for mixed biofilm formation in vitro (134). In this model system, the presence of AI-2 has been demonstrated to be required for biofilm formation by P. gingivalis-S. gordonii pairs. Therefore, luxS mutants of both strains are not able to form mixed biofilms, while a mutation in either strain still allows for such a property. As indicated above, such interactions may be important in the incorporation of P. gingivalis into early biofilms dominated by gram-positive organisms. In addition, it was demonstrated that AI-2 also mediates mutualistic biofilm formation by S. oralis and A. naeslundii strains (167). That study further indicated that there was an optimal concentration of AI-2, above or below which biofilm formation was suppressed. Thus, in at least two cases AI-2 appears to mediate interactions which could occur within oral biofilms.
In a recent study, Merritt et al. found that deletion of luxS in S. mutans abolished the production of the bacteriocin, mutacin I, at high cell density (136). Through microarray analysis, a putative transcription repressor annotated as Smu1274 in the Los Alamos National Laboratory Oral Pathogens Sequence Database was identified, which was strongly induced in the luxS mutant. Characterization of Smu1274, which was referred to as irvA, suggested that it may act as an inducible repressor to suppress mutacin I gene expression. A luxS irvA double mutant regained the ability to produce mutacin I, whereas a constitutive irvA-producing strain was impaired in mutacin I production. These findings suggest a role for quorum sensing in bacteriocin production, revealing a novel regulatory pathway for S. mutans for conducting “war and peace” activities in an oral microbial community.
Many of the oral streptococci, including S. mutans, S. gordonii, and S. sanguinis, express signal transduction systems mediated by the CSP. These molecules appear to be highly species specific, and the CSP of one organism is not likely to interfere with the activity of another distinct CSP molecule. Nevertheless, it has been demonstrated recently that some strains of S. gordonii are able to inhibit CSP-dependent properties of S. mutans (209). This appears to be mediated by a protease, challisin, expressed by S. gordonii. Challisin inactivates the CSP produced by S. mutans (Fig. 3). This raises the possibility that other highly proteolytic dental plaque residents might also interfere with the signal transduction mechanisms of the cariogenic streptococcus as well as other bacteria utilizing peptide signaling molecules. This has been demonstrated for P. gingivalis, which is able to interfere with competence-dependent transformation of S. mutans (B.-Y. Wang, personal communication).
Signaling between the dental plaque residents S. gordonii and Veillonella atypica has also recently been described using in vitro biofilm systems (47). Although the identity of the signaling molecule involved has not yet been demonstrated, these studies documented the importance of colocalization in signaling and the key role of localized concentrations of a signaling molecule.
Genetic Exchange between Biofilm Residents
Another interaction which could have major consequences for the physiology of biofilms as well as evolutionary consequences is genetic exchange between biofilm residents. Such interactions have been speculated upon for a number of years now, and evidence for such exchanges has been gradually emerging (35, 107). The accumulation of data cataloging the genome sequences of multiple bacteria makes clear the ubiquity of horizontal gene transfer in many ecological niches. It is likely that the environment within biofilms, including dental plaque, is conducive for genetic exchange because of the close proximity of the residents.
Genome sequencing of some oral bacteria has also demonstrated the presence of “pathogenicity islands” on the chromosomes of organisms such as P. gingivalis, with G+C ratios distinct from the remainder of the chromosome (26). This suggests the transfer of such specific genetic sequences from other organisms into P. gingivalis. Very recently, using an in vitro assay, Tribble et al. demonstrated that P. gingivalis is able to transfer chromosomal DNA between different strains (202). Although such exchanges have not yet been directly demonstrated in dental plaque, the close proximity of bacteria in these biofilms makes such interactions highly plausible. The exchange of antibiotic resistance markers between oral bacteria and transient bacteria destined for nonoral sites could also have significant medical relevance.
Potential mechanisms mediating genetic exchange in biofilms could include conjugation, transformation, and transduction. Conjugation involving oral streptococci was demonstrated over 30 years ago, although it was not directly shown in biofilms (109). Likewise, transformation of plasmids between oral streptococci involving planktonic cultures has been documented (107). More recently, it has been shown with in vitro biofilms that a shuttle plasmid present in T. denticola could be transformed into S. gordonii. Therefore, it is reasonable to speculate that chromosomal fragments could be exchanged between naturally transformable oral streptococci residing in biofilms. This has been further suggested by the demonstration that the lysis of cells within in vitro biofilms of S. mutans releases DNA fragments which can transform nearby intact cells (111). Although transduction has been demonstrated in individual oral bacteria (212), genetic exchange between different species mediated by bacteriophage has not yet been observed. Because of these results with individual oral biofilm bacteria, it is likely that genetic exchange can occur in dental plaque.
One of the more fascinating examples regarding genetic exchange in the oral bacterial community is the recent study reported by Kreth et al. (100). In that report, it was observed that S. mutans CSP induced coordinated expression of competence and mutacin production genes. In mixed cultures of S. mutans and S. gordonii harboring a shuttle plasmid, plasmid DNA transfer from S. gordonii to S. mutans in a CSP- and mutacin IV-dependent manner was observed. Further analysis demonstrated increased DNA release from S. gordonii upon addition of the partially purified mutacin IV extract. On the basis of these findings, it was proposed that S. mutans, which resides in a multispecies oral biofilm, may utilize competence-induced bacteriocin production to acquire transforming DNA from other species living in the same ecological niche. This hypothesis is consistent with the well-documented observation that great genomic diversity exists among different S. mutans strains. This diversity may have resulted from extensive horizontal gene transfer.
Modulation of Virulence Factors by Interspecies Interactions in Biofilms
As indicated above, bacterial interactions can affect the growth of individual organisms or groups of related organisms. In addition, such interactions may have specific effects in terms of the virulence properties of biofilm residents which could influence the overall pathogenicity of such structures. In this regard, organisms in dental plaque which are capable of neutralizing the acidic end products of S. mutans would tend to reduce the cariogenicity of these biofilms. As indicated above, this could result from the metabolism of lactic acid or from the production of alkaline end products. The latter could involve the production of ammonia or expression of urease by some plaque organisms (142). Since saliva normally contains significant levels of urea, the enzyme urease could convert this substrate to ammonia.
The expression of virulence factors by bacteria is, in general, tightly regulated. In some instances, such regulation is dependent upon quorum-sensing mechanisms (153, 206). Such mechanisms have been shown either to be dependent upon specific regulators such as those involved in streptococcal and Pseudomonas aeruginosa quorum sensing or to involve regulators such as AI-2, which are expressed in a variety of organisms (18, 136, 153, 206). Therefore, biofilm organisms which could reduce the relative concentrations of the regulatory molecules could also have profound effects on the pathogenicity of individual biofilms. For example, bacteriocin production by S. mutans has been shown to be dependent upon the relative levels of its quorum-sensitive regulator CSP (206). Bacteriocins can be considered to be virulence factors because they can modulate the growth of related noncariogenic streptococci (101), and therefore, reducing the concentration of the S. mutans CSP in biofilms could lead to a less cariogenic biofilm. Since noncariogenic S. gordonii strains can inactivate the S. mutans CSP, the presence of the former organism in biofilms could modulate the virulence of these biofilms. In addition, it is likely that CSP modulates other virulence properties of S. mutans (27, 115).
One of the virulence properties of the periodontopathogen A. actinomycetemcomitans is the elaboration of a leukotoxin (191). The expression of this protein has also been shown to be dependent upon AI-2 levels (52). Therefore, coresident bacteria which can reduce the levels of this signaling molecule could influence the expression of this toxin. Although oral bacteria which can inactivate AI-2 have not yet been identified, many biofilm residents likely contain active AI-2 binding systems (54). Therefore, the competition of biofilm organisms for localized concentrations of AI-2 could influence the virulence of specific biofilms.
Influence of Host Factors on Oral Microbes
As a micro-Gaia system, the viability of biofilms is dependent not only on interactions between the resident organisms but also on interactions of these constituents with the nonmicrobial environment. One obvious influence on biofilms in humans is the provision of potential nutrients by the host. In the oral cavity, the continuous production of saliva by the salivary glands provides a potential source of nutrients for some oral biofilm bacteria. These oral fluids contain proteins, glycoproteins, peptides, and minerals (calcium and iron) which can stimulate the growth of oral biofilm residents (8). Gingival crevicular fluid is another endogenous nutrient source. It contains host proteins such as albumin, glycoproteins, and heme-containing molecules. This source of nutrients greatly influences the microbes that reside in the subgingival crevice (66). In addition, some of the glycoproteins in the oral fluids can become incorporated into the tooth pellicle and may serve as receptors for the adhesins of pioneer plaque-forming bacteria (108). Thus, these molecules can play a major role in determining which organisms are able to colonize specific sites in the oral cavity.
Bacterial accumulation at the gingival margin of teeth as a result of poor oral hygiene can also lead to inflammation in the surrounding tissue, provoking serum leakage into the oral cavity. Red blood cells containing hemoglobin can serve as an important source of hemin for some oral bacteria, such as P. gingivalis (9, 181). Such leakage can be an important factor in the conversion of dental plaque dominated by gram-positive organisms into one containing large proportions of anaerobic gram-negative bacteria such as P. gingivalis, which are associated with periodontitis.
Host tissues surrounding the oral cavity may also secrete antimicrobial agents such as histatins (87) and defensins (30). A number of oral bacteria, including S. mutans, have been shown to be susceptible to the histatins produced by salivary glands (160). However, some bacteria, including T. denticola, have demonstrated resistance to specific defensins (10). Since bacteria residing in biofilms are generally more resistant to antimicrobial agents than their planktonic counterparts, it is not yet clear if these endogenous antimicrobial agents play a significant role in modulating the properties of oral biofilms.
Host cells lining the oral cavity may also secrete immune modulators that could influence the properties of dental plaque (32). Salivary secretions contain immunoglobulin A antibodies produced from the secretory glands lining the oral cavity. Some of these may be directed against antigens expressed by oral biofilm residents. However, it is not yet clear if such antibodies can penetrate the plaque matrix to interfere with bacterial properties. Nevertheless, the induction of such antibodies as a result of passive or active immunization could interfere with tooth colonization by potential oral pathogens present in saliva and serve as a rational basis for the development of anticaries or antiperiodontitis vaccines (93).
COMMUNITY-BASED ASSAYS: NEW TECHNIQUES FOR STUDYING ORAL BIOFILM
Since the interactions between different components create many new physiological functions that cannot be observed with individual components, it is important to develop community-based assays that allow researchers to identify the microbial composition, multispecies architecture, and associated physiology. However, for a long time, our understanding of the microbial world has been hampered by the intrinsic limitation of the conventional culture-dependent methods. According to Staley and Konopka, fewer than 1% of the organisms are able to grow under laboratory conditions (192), suggesting that our views of the complexity and genetic diversity of microbial communities based on cultivation strategies are severely biased. Fortunately, a number of DNA-based assays, such as 16S rRNA gene sequencing, checkerboard, or genomic approaches have helped to overcome this limitation, allowing us to obtain a clearer picture of microbial communities in terms of their structural complexity and genetic diversity.
16S rRNA Gene Sequencing Approaches
Woese and coworkers discovered almost three decades ago that the conservation as well as the variation within the sequences of 16S rRNA-encoding genes allows for the construction of evolutionary trees for bacteria to the species level (53, 210). This finding in combination with the development of PCR methods in 1985 opened the door for culture-independent analyses and classification of previously unknown members of many different microbial communities, including the resident flora of oral biofilms (170, 196).
The first application of this cloning- and sequencing-based method in the oral microbial field revealed an astonishing diversity of largely uncultivated oral spirochetes present in a single patient (21). The expansion of this approach to all bacterial species present in subgingival biofilms collected from a broader patient pool with a diverse range of periodontal conditions culminated in the discovery of more than 300 different phylotypes (102, 157). Some of these newly recognized species represented potential novel periodontal pathogens, since they were detected predominantly in diseased sites. Additionally, a study focusing on spirochetes revealed the presence of almost 60 different species of oral treponemes, most of which have yet to be cultured (38). In later studies the inventory of oral biofilms was extended to species growing on different oral surfaces and in saliva (1, 88, 172, 173). An even greater species diversity became apparent when patients with distinct conditions such as human immunodeficiency virus infection (156) or suffering from noma, a potentially fatal oral gangrene, were added (158). 16S rRNA profiling was recently applied to assess the role of oral bacteria in ventilator-associated pneumonia, which affects more than a quarter of patients on respiratory support (3). Interestingly, a study focusing on the normal flora of healthy individuals was not undertaken until only 2 years ago despite its importance in actually determining disease (1). Compiling all of the data currently available, the bacterial variety found colonizing the human oral cavity has now reached more than 700 different species (155), many of which are specific to a particular oral surface.
In addition to the studies described above, which created a vast inventory of oral species, more focused approaches were designed for the quantitative assessment of individual species that would allow the identification of candidate (novel) pathogens as well as beneficial species (105). The same group performed a thorough analysis of the species distribution during a longitudinal study of periodontally healthy subjects as well as patients with either improving or worsening periodontal health. The microbial communities colonizing patients with no significant change in their periodontal status, especially the healthy ones, were found to be very stable, whereas disease progression was accompanied by the largest shifts in species composition (106). Previously suspected (novel) health- and disease-associated species were confirmed through this study.
PCR-Based High-Throughput Approaches
The past decade has witnessed a virtual explosion in the development of high-throughput tools for microbial community analysis. Some of these methods directly examine nucleic acids isolated from samples of microbial communities, such as microarrays (186), while others are PCR based, such as denaturing gradient gel electrophoresis (DGGE) (146) or temperature gradient gel electrophoresis (215), terminal restriction fragment length polymorphism (T-RFLP) (118, 200), automated ribosomal intergenic spacer analysis (13), and denaturing high-performance liquid chromatography (DHPLC) (4, 40). Most of these approaches were originally developed for analyzing environmental microbial communities and have been used to study human microbial ecology, including oral microbial analysis.
DGGE.
DGGE is a PCR-based approach for microbial community analysis, where marker genes (genes with both conserved and variable regions, such as 16S rRNA genes) are PCR amplified from DNA pools isolated from specific microbial communities. PCR product mixtures with different sequences are applied to DGGE, where DNAs with different sequence compositions (GC content) will denature at different denaturant concentrations and resolve into distinct bands on the gel, resulting in a pattern of bands (51), with each band theoretically representing a different bacterial population present in the community. The resulting profile reflects the structure of the community, including its complexity and the relative abundance of each detected species. If needed, bands of interest can be excised from the gel, cloned, and sequenced to identify the specific bacterial species.
DGGE has been widely used to analyze environmental microbial communities (42, 83, 146) and human microbial ecology, such as the microbial flora in the gastrointestinal (61, 180, 216) and vaginal (36, 37, 166) tracts. It is a useful tool for assessing the diversity of the microbiota associated with humans and can monitor the shift in microbial ecology under certain pathogenic conditions (216). It also facilitates the identification of pathogens by comparing the community structures in samples taken from patients and healthy individuals (166).
DGGE has also proven to be a useful tool for analyzing the microbiota in the human oral cavity (112, 113, 165, 217). It was used to conduct surveys of microbial diversity in human saliva (112, 165) and to study oral bacterial community structure associated with severe dental caries (114), endodontic infections (124, 182, 183), and periodontitis (55, 217), as well as to evaluate the oral microbial changes after prophylactic treatments (113). Due to its broad-range nature, this method facilitates the detection of previously unknown fastidious or uncultivable microbial species (177), and it is less labor-intensive than the traditional cloning methods. Like many other molecular approaches, DGGE has limitations, such as the difficulty in attaining reproducible results and quantitative analysis of the community. Nevertheless, it is a useful tool in studying and analyzing the human oral microbiota.
T-RFLP.
T-RFLP is an alternative molecular tool that has been extensively used for the assessment of environmental microbial communities and quick comparison of the complexity and genetic diversity of different ecosystems (171, 200). Like DGGE, T-RFLP is also a PCR-based approach, in which PCR amplification of certain marker genes, such as 16S rRNA genes, using fluorescently labeled primers results in PCR product mixtures with fluorescent tags. Since marker genes amplified from different bacterial species might have variable sequences with different numbers of endonuclease restriction sites at different positions, the hydrolysis of the PCR fragments with certain endonucleases will produce a band pattern, or “fingerprint,” characteristic of the microbial community studied. Since T-RF lengths can be predicted from known 16S rRNA or other marker gene sequences, it is relatively easy to create a database, and the results of each T-RFLP run can be analyzed by matching and comparing peaks to an existing database, thus facilitating the interpretation of the results.
Since its first application in analyzing microbial diversity in 1997 (118), T-RFLP has become one of the most useful tools for studying environmental microbial communities and human microbial ecology (110, 131, 171, 175, 199, 200). The past decade has also witnessed the application of T-RFLP in the assessment of the oral microbial flora, including characterizing structural profiles of microbial communities in human saliva (175), subgingiva (174), and infected root canals (78), as well as monitoring the changes in the subgingival bacterial flora after clinical treatment (174). However, because of its requirements for powerful computational hardware, novel software, and large databases, this approach has yet to reach its full potential.
DHPLC.
DHPLC is a fairly new technology for microbial community analysis. Like DGGE, it was originally used to detect DNA sequence variation such as point mutations (79), and it was first adapted by Domann et al. for analyzing the microbial profiles in urinary tract specimens from the recipients of renal transplants (40). Since then, this technique has been used to study genetic diversity and monitor population changes in microbial communities (4, 61, 62).
DHPLC is a PCR-dependent method, like DGGE, in which marker genes (such as 16S rRNA genes) are amplified by PCR, and the separation of the PCR products using DHPLC is based on the elution of heat-induced, partially denatured double-stranded DNA from the cartridge matrix of the system. DNA is first attached to the beads in the matrix by interaction with triethylammonium acetate, an ion-pairing reagent which binds more tightly to the double-stranded DNA molecules. At certain temperatures, DNAs with differing sequences experience different degrees of melting, thus affecting their ability to be retained on the matrix. When washed with gradient buffers, PCR products with different GC contents (thus having variable melting domains) display different retention times. DNA with differing sequences will elute and pass through a UV detector in a time-dependent manner, resulting in a unique “fingerprint” for each microbial community analyzed. DHPLC has demonstrated high sensitivity and reproducibility in detecting 16S rRNA gene variants in the PCR mixture, as reflected by the number and the intensity of peaks recorded in the samples. It also allows for the identification of species of interest in the sample by isolating and sequencing DNA collected from the corresponding eluted fraction.
In the past decade, DHPLC has been successfully used for profiling microbial communities (4), analyzing human intestinal microbiota (61), and detecting clinically relevant microbial species (62). Recently, this approach has been successfully adapted in J. W. Costerton's laboratory at the University of Southern California for assessing the oral microbial flora (personal communication).
Ibis T5000, a high-throughput tool for analyzing microbial communities.
Recently, a new automated platform for pathogen identification and strain typing was developed and put on the market (45). The Ibis T5000 universal biosensor is a unique system for quick and accurate identification of different microorganisms in samples. This technology is based on the use of broad-range primers to amplify PCR products from multiple genomic regions of all the microbes within the sample, and the unambiguous nucleotide base composition signature obtained from mass spectrometry is used to identify the microbe(s) in the sample (45).
Checkerboard Approaches
Around the same time that the 16S rRNA-based identifications described above were first employed to study oral microorganisms, Socransky et al. described the checkerboard DNA-DNA hybridization method, which revolutionized epidemiological studies in a manner similar to that of whole-population 16S rRNA cloning and sequencing (190). The checkerboard approach enables simultaneous profiling of multiple species (up to 40) within the same plaque sample and the processing of a large number of plaque samples on the same blot in a semiquantitative manner. In the original version of this method, DNA preparations of whole plaque samples were fixed in lines across a nylon membrane with a slot blot apparatus. These experimental samples were then hybridized with labeled whole-genome probes of up to 40 different oral microorganisms that were placed on the blot at a 90° angle with respect to the original lanes containing the whole plaque samples. This hybridization pattern ensured that each plaque sample would be probed with each species-specific probe.
This large-scale high-throughput screening approach was applied to examine thousands of plaque samples collected from hundreds of patients. The initial work by Socransky and coworkers enabled the designation of distinct “complexes” that are indicative of periodontal disease or health status (190). Since then, a number of studies have taken advantage of this relatively sensitive and efficient method to determine species distribution profiles in a variety of different settings. Even though the majority of studies focused on a more detailed understanding of the biofilm composition contributing to periodontal diseases (29, 144, 152), extensive analysis of other oral surfaces was performed as well (125). The prevalence of certain species in health and disease was evaluated for a range of oral infections (caries, periodontal, and endodontic infections) and for certain systemic conditions (e.g., diabetes) (76) or lifestyle choices (e.g., smoking) (68). In addition to the analysis of bacterial prevalence in a specific oral community, the outcomes of therapeutic treatments were also compared (69).
The original “direct” checkerboard hybridization is limited to the identification of cultivable species, since whole-genome probes have to be used in order to ensure sensitivity and selectivity as well as enable semiquantitative analysis of the results. To overcome this limitation, PCR-based “reverse-capture” checkerboard hybridization was introduced (154). In this modification, the 16S rRNA of any species of interest (cultivable or noncultivable) is PCR amplified and spotted on the blots. Then, the 16S rRNA genes of the bacteria present in the sample are PCR amplified with labeled universal primers and serve as probes for the blots. The tradeoff for the ability to examine the distribution of any species of interest is the loss of sensitivity and quantitative assessment due to the amplification step in the “reverse-capture” version of checkerboard hybridization. Nevertheless, this modified checkerboard approach has been used in numerous large-scale studies ranging form the detailed examination of different species of oral streptococci (154) to species profiles in early childhood caries (6).
In order to increase the number of species that can be simultaneously identified in plaque samples during large-scale studies, the human oral microbe identification microarray is currently being developed. These microarrays can identify 200 different oral bacterial species and were recently shown to reveal significant differences in the species profiles of subjects with periodontal disease in comparison to healthy individuals (155).
Genomic and Metagenomic Approaches
The availability of high-throughput DNA sequencing technology together with the rapid expansion of bacterial genome data has now made it feasible to identify the primary bacterial residents in a specific ecological site. This has recently been accomplished for deep-sea samples (207) as well as for samples from beneath the earth's surface (159). Therefore, it is now feasible to identify essentially all of the residents in dental plaque as well as to compare those from healthy and diseased sites. Currently, such an approach is in progress, and the results should become available in the next few years. It is anticipated that such comparisons will assist in identifying potential pathogenic organisms which may not have been detected using currently available technologies such as direct culturing or 16S rRNA analysis. The latter technology has suggested that there may be oral pathogenic or pathogenicity-associated organisms which have not been detected up to now (104).
Genomic Expression Approaches
In addition to identifying pathogenic bacteria within a biofilm, it is also important to identify the genes from such organisms which help define the virulence of biofilms. Such information is crucial in determining how the expression of such genes is regulated and how this might be leveraged in novel therapeutic approaches. In addition, it may be that the relative level of a particular potential pathogen within a biofilm is not the rate-limiting factor for pathogenicity but that this may depend upon the relative expression of a particular set of genes. This, in turn, could be dependent upon the presence of other biofilm residents as described above. The identification of key virulence genes would allow for an examination of how they are regulated using in vitro and in vivo plaque model systems in combination with transcription analyses. One could then envision the development of limited microarrays containing key sets of virulence genes from a variety of different potential pathogens which could be used to analyze the complex interactions which likely occur within dental plaque under distinct environmental conditions.
Beyond these DNA-based techniques, advanced biotechnology has made possible additional culture-independent methods that are beyond the scope of this review. From monoclonal antibodies to diagnostic proteins/peptides to nanotechnology, many new community-based research tools are being developed and applied to oral microbial research. This will greatly help us to better understand the oral flora structure and genetic variation; allow for the identification of novel, yet-to-be cultivated commensal or oral pathogens; and facilitate the elucidation of the etiology of certain oral infectious diseases.
NEW APPROACHES FOR CONTROLLING COMMUNITY-BASED ORAL MICROBIAL PATHOGENESIS
Current dental therapy is focused primarily on removing whole dental plaque. Since we now know that dental plaque is made of large numbers of commensal bacteria together with a limited number of oral pathogens, such an approach may not be effective, since the “remove all and kill all” approach creates niches for pathogens to repopulate the oral cavity. With our better understanding of the oral microbial community, especially our understanding of the importance of the balance between oral pathogens and commensal residents, there is now interest in new approaches for selective inhibition of oral pathogens and modulation of the microbial composition of dental plaque to control community-based oral microbial pathogenesis. In the past several years, oral microbiology has become a leading area for developing these technologies, which should also be useful for other community-based microbial pathogenesis.
Inhibiting Adherence with Antagonists
An obvious approach to inhibiting the virulence of biofilms would be to prevent the incorporation of potentially pathogenic organisms into biofilms. A number of approaches have been pursued to accomplish this in terms of human dental caries. Since S. mutans strains are strongly associated with this disease, reducing the colonization of these organisms on the tooth surface should have a significant impact on the development of dental caries. Several mechanisms are likely involved in the attachment of these organisms to teeth, including nonspecific binding and physical trapping in the retentive surfaces of teeth, specific interactions with components of the tooth pellicle, and dietary sucrose-mediated glucan-enhanced attachment (95, 97). In this regard, a cell surface protein of S. mutans termed SpaP or Ag I/II has been identified as an adhesin which interacts with the tooth pellicle (82). Furthermore, a dodecapeptide analogue of the active binding site of SpaP which inhibits attachment of S. mutans to teeth both in vivo and in vitro has been synthesized (89). Therefore, such analogues are potential therapeutic agents which could be incorporated into toothpastes or mouth rinses to reduce the incidence of dental caries.
Since sucrose is an important factor in dental caries, the utilization of sucrose analogues may have potential value in reducing caries susceptibility (129). Likewise, the enzyme dextranase, which reduces the synthesis of adhesive glucan molecules from sucrose by S. mutans, has been incorporated into toothpastes and has been suggested to reduce the incidence of caries in children (91). However, at present, none of these approaches has been introduced as a general therapeutic approach for controlling cavity formation.
Passive Immunization
One general approach for controlling bacterial pathogens has been the attempt to develop specific vaccination strategies. In the case of dental caries, this possibility has been investigated for the past four decades (169). Despite promising results using experimental animal models, it appears unlikely that active immunization approaches will be further pursued in most developed countries because of economic and ethical concerns. As an alternative to active immunization, passive immunization strategies have been recently proposed (93). However, it is unclear at present whether or not these approaches will lead to a general strategy for immunizing susceptible children against dental caries in the future.
Replacement Therapy
One interesting strategy which has been proposed in regard to reducing the incidence of dental caries is that of the replacement of cariogenic S. mutans by competitor noncariogenic strains. This strategy, originally proposed and subsequently developed by J. Hillman and colleagues (74), involves the introduction of a noncariogenic S. mutans strain which produces a bacteriocin active against other S. mutans strains into the oral cavity to replace the naturally occurring cariogenic strains. Both in vitro and animal model assessments of this strategy suggest that it has utility in reducing tooth colonization by cariogenic S. mutans strains (74). This approach is currently awaiting evaluation for its efficacy in humans.
Regulating the Levels of Nonpathogenic Bacteria to Influence Virulence
Since the incorporation of some organisms into biofilms is dependent upon other antecedent biofilm residents, it may be possible to identify such dependencies for specific pathogens and target these antecedent organisms for elimination. Since the incorporation of several gram-negative anaerobes involved in periodontitis into biofilms appears to be partially dependent upon ubiquitous F. nucleatum strains (97), strategies which specifically prevent biofilm formation by the latter organism could then influence the development of periodontitis. Likewise, enhancing the colonization and growth of organisms which are known to antagonize the cariogenic potential of S. mutans (S. gordonii, S. salivarius, etc.) should have similar effects on dental caries development. However, it has not yet been demonstrated that this is feasible in humans.
Probiotic Approaches
The strategy developed by Hillman and coworkers to replace cariogenic S. mutans strains with a noncariogenic mutant also depends upon bacteriocin (mutacin 1140) production by the latter strain. This represents an example of a probiotic approach involving production of a bacteriocin effective against a virulent bacterium.
Another potential probiotic approach for reducing dental caries involves the utilization of oral streptococci which are able to metabolize arginine or urea to ammonia (126). Since such organisms naturally occur in dental plaque and therefore qualify as GRAS (generally regarded as safe) organisms, it may be possible to use these organisms in a probiotic approach for controlling dental caries. Previous investigations in animal models suggested that coinfection of rats with oral streptococci such as S. salivarius TOVE-R and S. mutans did reduce dental caries incidence relative to infection with the latter organisms alone (197). However, since multiple interactions occur between oral streptococci, as described above, it is not clear how this attenuation of caries development occurred.
Although there have been a few reports regarding the existence of bacteriocins which are active against gram-negative anaerobes (84, 143), none that are active against suspected periodontopathogens have yet been identified. If such bacteriocins with efficacy against these organisms can be identified, it may be possible to genetically engineer GRAS organisms to actively produce such bacteriocins for incorporation into dietary supplements as a means of controlling periodontitis.
Previously, it was demonstrated that the presence of organisms associated with periodontitis was markedly reduced when members of the sanguis group of streptococci were present (75). This antagonistic effect was likely due to the active production of hydrogen peroxide by the latter organisms. These properties are consistent with the observed inverse proportions of oral streptococci relative to gram-negative anaerobes found in dental plaque (99). Thus, the deliberate implantation of specific oral streptococci or the encouragement of their growth in dental plaque can be considered to be a probiotic approach for encouraging the shift from a pathogenic to a nonpathogenic biofilm.
Interference with Signaling Mechanisms
As indicated above, a number of laboratories have demonstrated that several pathogenic properties of S. mutans are regulated by quorum-sensing mechanisms involving CSP as the signaling molecule (27, 206). Therefore, the presence of organisms which interfere with such signaling may have a beneficial effect in terms of dental caries. As described above, S. gordonii was demonstrated to interfere with quorum sensing in S. mutans by inactivating the CSP of the latter organisms (209). Therefore, this may be one of the mechanisms employed by S. gordonii in interfering with the virulence of S. mutans in animal models. Thus, GRAS organisms such as S. gordonii may prove useful in probiotic approaches for controlling dental caries.
Also as indicated above, the AI-2 signaling molecule can modulate the virulence properties of several bacteria, including biofilm formation. However, the ubiquitous occurrence of quorum-sensing mechanisms mediated by such regulators in many organisms suggests that it may be difficult to utilize AI-2 antagonists or agonists to target specific pathogens. In addition, it is likely that local gradients of AI-2 occur within biofilms, and exogenous alteration of these levels would also likely alter the interactions between the resident bacteria. Therefore, it is difficult to predict what overall effects such alterations would have on biofilms. For these reasons, it would be more reasonable to exploit regulatory mechanisms which are specific for targeted pathogens, i.e., quorum sensing in S. mutans mediated by its specific CSP, to target pathogenic bacteria in biofilms.
Targeted Antimicrobial Therapy via a Novel STAMP Technology
Eckert et al. (46) reasoned that, with the exception of a limited number of pathogens, the majority of microorganisms within the indigenous oral microflora are benign or beneficial. Currently available antimicrobials exhibit broad killing spectra with regard to bacterial genus and species. Indiscriminate killing of microbes by these conventional antimicrobials disrupts the entire ecological balance of the indigenous microbial flora and may result in negative clinical consequences. These investigators developed a novel technology for a new class of antimicrobials called specifically targeted antimicrobial peptides (STAMPs). A STAMP is a fusion peptide with two moieties: a killing moiety made of a nonspecific antimicrobial peptide and a targeting moiety made of a species-specific binding peptide. The targeting moiety provides specific binding to a selected pathogen and facilitates the targeted delivery of the attached antimicrobial peptide. In one of their recently published papers (46), they explored the possibility of utilizing a pheromone produced by S. mutans, namely, CSP, as a STAMP targeting domain to mediate S. mutans-specific delivery of an antimicrobial peptide domain. They discovered that STAMPs constructed with peptides derived from CSP were potent against S. mutans grown in liquid or biofilm states but did not affect other oral streptococci tested. Further studies showed that an 8-amino-acid region within the CSP sequence was sufficient for targeted delivery of the antimicrobial peptide domain to S. mutans. The STAMPs were capable of eliminating S. mutans from multispecies biofilms without affecting closely related noncariogenic oral streptococci, indicating the potential of these molecules to be developed into “probiotic” antibiotics which could selectively eliminate pathogens while preserving the protective benefits of a healthy normal flora. This proof-of-principle demonstration utilizing S. mutans suggests that it may be possible to develop other STAMPs which are specifically targeted to other biofilm pathogens.
CONCLUSIONS
Although this review is focused on oral microbial communities, it is now clear that many of the specific bacteria which are of medical and environmental concern reside in similar multispecies biofilm structures. Since most of these are present in heterogeneous biofilms, of which oral biofilms appear to be among the most complex, it is likely that these micro-Gaia communities exhibit properties which are dependent upon how the resident organisms interact. Information from the study of both oral and nonoral biofilms over the past decade has suggested a wide variety of such interactions. Interactions involving potential human pathogens as well as environmentally significant organisms are therefore of prime importance in this regard. Our ability to both identify the resident organisms in biofilms and decipher the interactions involving key components of these biofilms has rapidly increased within the past decade. Nevertheless, it is recognized that our understanding of the interactions which occur between biofilm residents is still in its infancy. Continued expansion of such information in the near future will be dependent on the development of new technologies designed to simultaneously identify multiple properties within biofilms. Such information could then serve as a basis for exogenously modulating the interactions between biofilm constituents, resulting in novel approaches for controlling biofilm activities.
Acknowledgments
We acknowledge researchers in our laboratories who have greatly contributed to the some of the information cited in this review. In addition, we specifically acknowledge J. W. Costerton for his assistance in the preparation of this review.
Some of the cited studies from our laboratories were supported by NIH grants DE03258 (H.K.), DE09821 (H.K.), GM54666 (W.S.), and MD01831 (M.A. and W.S.) and by a UC discovery grant.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apolonio, A. C. M., M. A. R. Carvalho, R. N. R. Ribas, L. G. Sousa-Gaia, K. V. Santos, M. A. Lana, J. R. Nicoli, and L. M. Farias. 2007. Production of antagonistic substance by Eikenella corrodens isolated from the oral cavity of human beings with and without periodontal disease. J. Appl. Microbiol. 103:245-251. [DOI] [PubMed] [Google Scholar]
- 3.Bahrani-Mougeot, F. K., B. J. Paster, S. Coleman, S. Barbuto, M. T. Brennan, J. Noll, T. Kennedy, P. C. Fox, and P. B. Lockhart. 2007. Molecular analysis of oral and respiratory bacterial species associated with ventilator-associated pneumonia. J. Clin. Microbiol. 45:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Profiling and monitoring of microbial populations by denaturing high-performance liquid chromatography. J. Microbiol. Methods 61:399-412. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 6.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, C., I. Allan, S. K. Ford, M. Wilson, and R. McNab. 2004. Biofilm-specific surface properties and protein expression in oral Streptococcus sanguis. Arch. Oral Biol. 49:295-304. [DOI] [PubMed] [Google Scholar]
- 8.Bowden, G. H. W. 1997. Nutritional influences on biofilm development. Adv. Dent. Res. 11:81-99. [DOI] [PubMed] [Google Scholar]
- 9.Bramanti, T. E., and S. C. Holt. 1991. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J. Bacteriol. 173:7330-7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brissette, C. A., and S. A. Lukehart. 2007. Mechanisms of decreased susceptibility to beta-defensins by Treponema denticola. Infect. Immun. 75:2307-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burne, R. A., Y. M. Chen, and J. E. C. Penders. 1997. Analysis of gene expression in Streptococcus mutans in biofilms in vitro. Adv. Dent. Res. 11:100-109. [DOI] [PubMed] [Google Scholar]
- 12.Byers, H. L., E. Tarelli, K. A. Homer, and D. Beighton. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469-479. [DOI] [PubMed] [Google Scholar]
- 13.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlsson, J. 1997. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 11:75-80. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson, J., and M. B. Edlund. 1987. Pyruvate oxidase in Streptococcus sanguis under various growth conditions. Oral Microbiol. Immunol. 2:10-14. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633-684. [DOI] [PubMed] [Google Scholar]
- 17.Chen, P. B., L. B., Davern, J. Katz, J. H. Eldrdge, and S. M. Michalek. 1996. Host response induced by co-infection with Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in a murine model. Oral Microbiol. Immunol. 11:274-281. [DOI] [PubMed] [Google Scholar]
- 18.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 19.Chhour, K. L., M. A. Nadkarni, R. Byun, F. E. Martin, N. A. Jacques, and N. Hunter. 2005. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 43:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chikindas, M. L., J. Novak, A. J. Driessen, W. N. Konings, K. M. Schilling, and P. W. Caufield. 1995. Mutacin II, a bactericidal antibiotic from Streptococcus mutans. Antimicrob. Agents Chemother. 39:2656-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke, J. K. 1924. On the bacterial factor in the etiology of dental caries. Br. J. Exp. Pathol. 5:141-147. [Google Scholar]
- 23.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 24.Costerton, J. W., G. G. Geesey, and G. K. Cheng. 1978. How bacteria stick. Sci. Am. 238:86-95. [DOI] [PubMed] [Google Scholar]
- 25.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, K. R. Keber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 26.Curtis, M. A., S. A. Hawley, and J. Aduse-Opoku. 1999. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J. Periodont. Res. 34:400-405. [DOI] [PubMed] [Google Scholar]
- 27.Cvitkovitch, D. G., Y. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 112:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlen, G. 1993. Role of suspected periodontopathogens in microbiological monitoring of periodontitis. Adv. Dent. Res. 7:163-174. [DOI] [PubMed] [Google Scholar]
- 29.Dahlen, G., and A. Leonhardt. 2006. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol. Immunol. 21:6-11. [DOI] [PubMed] [Google Scholar]
- 30.Dale, B. A., and S. Krisanaprakornkit. 2001. Defensin antimicrobial peptides in the oral cavity. J. Oral Pathol. Med. 30:321-327. [DOI] [PubMed] [Google Scholar]
- 31.Darenfed, H., D. Grenier, and D. Mayrand. 1999. Acquisition of plasmin activity by Fusobacterium nucleatum subsp. nucleatum and potential contribution to tissue destruction during periodontitis. Infect. Immun. 67:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darveau, R. P. 2000. Oral innate host defense responses: interactions with microbial communities and their role in the development of disease, p. 169-218. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Science Press, Wymondham, United Kingdom.
- 33.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costeron, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 35.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 36.Devillard, E., J. Burton, J. Hannond, D. Lam, and G. Reid. 2005. Complexity of vaginal microflora as analyzed by PCR denaturing gradient gel electrophoresis in a patient with recurrent bacterial vaginosis. Infect. Dis. Obstet. Gynecol. 13:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devillard, E., J. Burton, J. Hannond, D. Lam, and G. Reid. 2004. Novel insight into the vaginal microflora in postmenopausal women under hormone replacement therapy as analyzed by PCR denaturing gradient gel electrophoresis. Euro. J. Obstet. Gynaecol. Reprod. Biol. 117:76-81. [DOI] [PubMed] [Google Scholar]
- 38.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 39.Diaz, P. I., P. S. Zilm, and A. H. Rogers. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbondioxide-depleted environments. Microbiology 148:467-472. [DOI] [PubMed] [Google Scholar]
- 40.Domann, E., G. Hong, C. Imirzalioglu, S. Turschner, J. Kuhle, C. Watzel, T. Hain, H. Hossain, and T. Chakraborty. 2003. Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J. Clin. Microbiol. 41:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dorana, A., S. Kneist, and J. Verran. 2004. Ecology control: in vitro inhibition of anaerobic bacterial by oral streptococci. Microb. Ecol. Health. Dis. 16:23-27. [Google Scholar]
- 42.Duineveld, B. M., A. S. Rosado, J. D. van Elsas, and J. A. van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 44.Ebersole, J. L., F. Feuille, L. Kesavalu, and S. C. Holt. 1997. Host modulation of tissue destruction caused by periodontopathogens. Effects on a mixed microbial infection composed of Porphyromonas gingivalis and Fusobacterium nucleatum. Microb. Pathog. 23:23-32. [DOI] [PubMed] [Google Scholar]
- 45.Ecker, D. J., J. J. Drader, J. Gutierrez, A. Gutierrez, J. C. Hannis, A. Schink, R. Sampath, L. B. Blyn, M. W. Eshoo, T. A. Hall, M. Tobarmosquera, Y. Jiang, K. A. Sannes-Lowery, L. L. Cummins, B. Libby, D. J. Walcott, C. Massire, R. Ranken, S. Manalili, C. Ivy, R. Melton, H. Levene, V. Harpin, F. Li, N. White, M. Pear, J. A. Ecker, V. Samant, D. Knize, D. Robbins, K. Rudnick, F. Hajjar, and S. A. Hofstadler. 2006. The Ibis T5000 universal biosensor: an automated platform for pathogen identification and strain typing. J. Assoc. Lab. Auto. 11:341-351. [Google Scholar]
- 46.Eckert, R., J. He, D. K. Yarbrough, F. Qi, M. H. Anderson, and W. Shi. 2006. Targeted killing of Streptococcus mutans by a pheromone-guided “Smart” antimicrobial peptide. Antimicrob. Agents Chemother. 50:3651-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng, Z., and A. Weinberg. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontology 2000 40:50-76. [DOI] [PubMed] [Google Scholar]
- 49.Feuille, F., I. L. Ebersole, L. Kesavalu, M. J. Steffen, and S. C. Holt. 1996. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum. Infect. Immun. 64:2094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fine, D. H., D. Furgang, and M. L. Barnett. 2001. Comparative antimicrobial activities of antiseptic mouthrinses against isogenic planktonic and biofilm forms of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 28:697-700. [DOI] [PubMed] [Google Scholar]
- 51.Fischer, S. G., and L. S. Lerman. 1983. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc. Natl. Acad. Sci. USA 80:1579-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209:457-463. [DOI] [PubMed] [Google Scholar]
- 54.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum-sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujimoto, C., H. Maeda, S. Kokeguchi, S. Takashiba, F. Nishimura, H. Arai, K. Fukui, and Y. Murayama. 2003. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 38:440-445. [DOI] [PubMed] [Google Scholar]
- 56.Gaetti-Jardim, E., Jr., and M. J. Avila-Campos. 1999. Bacteriocin-like activity of oral Fusobacterium nucleatum isolated from human and non-human primates. Rev. Microbiol. 30:342-346. [Google Scholar]
- 57.Gest, H. 2004. The discovery of microorganisms by Robert Hooke and Antoni van Leeuwenhoek. Fellows of the royal society. Notes Res. R. Soc. London 58:187-201. [DOI] [PubMed] [Google Scholar]
- 58.Gibbons, R. J., and M. Nygaard. 1970. Interbacterial aggregation of plaque bacteria. Arch. Oral Biol. 15:1397-1400. [DOI] [PubMed] [Google Scholar]
- 59.Gibbons, R. J., and M. Nygaard. 1968. Synthesis of insoluble dextran and its significance in the formation of gelatinous deposits by plaque-forming streptococci. Arch. Oral Biol. 13:1249-1262. [DOI] [PubMed] [Google Scholar]
- 60.Gilbert, P. J. D., and I. Foley. 1997. Biofilm susceptibility to antimicrobials. Adv. Dent. Res. 11:160-167. [DOI] [PubMed] [Google Scholar]
- 61.Goldenberg, O., S. Herrmann, G. Marjoram, M. Noyer-Weidner, G. Hong, S. Bereswill, and U. B. Gobel. 2007. Molecular monitoring of the intestinal flora by denaturing high performance liquid chromatography. J. Microbiol. Methods 68:94-105. [DOI] [PubMed] [Google Scholar]
- 62.Goldenberg, O., S. Herrmann, T. Adam, G. Marjoram, G. Hong, U. B. Gobel, and B. Graf. 2005. Use of denaturing high-performance liquid chromatography for rapid detection and identification of seven Candida species. J. Clin. Microbiol. 43:5912-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grenier, D. 1996. Antagonistic effect of oral bacteria towards Treponema denticola. J. Clin. Periodontol. 34:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grenier, D. 1992. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect. Immun. 60:5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grenier, D., and D. Maryrand. 1986. Nutritional relationships between oral bacteria. Infect. Immun. 53:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffiths, G. S. 2003. Formation, collection and significance of gingival crevice fluid. Periodontology 2000. 31:32-42. [DOI] [PubMed] [Google Scholar]
- 67.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontology 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 68.Haffajee, A. D., and S. S. Socransky. 2001. Relationship of cigarette smoking to the subgingival microbiota. J. Clin. Periodontol. 28:377-388. [DOI] [PubMed] [Google Scholar]
- 69.Haffajee, A. D. M. A. C., A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 70.Hale, J. D. F., Y. T. Ting, R. W. Jack, J. R. Tagg, and N. C. K. Heng. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71:7613-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammond, B. F., S. E. Lillard, and R. H. Stevens. 1987. A bacteriocin of Actinobacillus actinomycetemcomitans. Infect. Immun. 55:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higuchi, M. 1984. The effect of oxygen on the growth and mannitol fermentation of Streptococcus mutans. J. Gen. Microbiol. 130:1819-1826. [DOI] [PubMed] [Google Scholar]
- 74.Hillman, J. D. 2002. Genetically modified Streptococcus mutans for prevention of dental caries. Antoine Leewenhoek 82:361-366. [PubMed] [Google Scholar]
- 75.Hillman, J. D., S. S. Socransky, and M. Shivers. 1985. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch. Oral Biol. 30:791-795. [DOI] [PubMed] [Google Scholar]
- 76.Hintao, J., R. Teanpaisan, V. Chongsuvivatwong, C. Ratarasan, and G. Dahlen. 2007. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol. Immunol. 22:175-181. [DOI] [PubMed] [Google Scholar]
- 77.Hoffman, L., D. D. D'Argenio, M. Bader, and S. Miller. 2007. Microbial recognition of antibiotics: ecological, physiological, and therapeutic implications. Microbe 2:175-182. [Google Scholar]
- 78.Hommez, G., R. Verhelst, G. Claeys, and R. De Moor. 2004. Investigation of the effect of the coronal restoration quality on the composition of the root canal microflora in teeth with apical periodontitis by means of T-RFLP analysis. Int. Endod. J. 37:819-827. [DOI] [PubMed] [Google Scholar]
- 79.Hoogendoorn, B., N. Norton, G. Kirov, N. Williams, M. Hamshere, G. Spurlock, J. Austin, M. Stephens, P. Buckland, M. Owen, and M. O'Donovan. 2000. Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum. Genet. 107:488-493. [DOI] [PubMed] [Google Scholar]
- 80.Ihalin, R., V. Loimaranta, M. Lenander-Lumikari, and J. Tenovuo. 2001. The sensitivity of Porphyromonas gingivalis and Fusobacterium nucleatum to different (pseudo)halide-peroxidase combinations compared with mutans streptococci. J. Med. Microbiol. 50:42-48. [DOI] [PubMed] [Google Scholar]
- 81.Ishihara, K., A. Nabuchi, R. Ito, K. Miyachi, H. K. Kuramitsu, and K. Okuda. 2004. Correlation between detection rates of periodontopathic bacterial DNA in carotid coronary stenotic artery plaque and in dental plaque samples. J. Clin. Microbiol. 42:1313-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 83.Jensen, S., L. Ovreas, F. L. Daae, and V. Torsvik. 1998. Diversity in methane enrichments from agricultural soil revealed by DGGE separation of PCR amplified 16S rDNA fragments. FEMS Microbiol. Ecol. 26:17-26. [Google Scholar]
- 84.Kaewsrichan, J., C. W. I. Douglas, J. Nissen-Meyer, G. Fimland, and R. Teanpaisan. 2004. Characterization of a bacteriocin produced by Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 39:451-458. [DOI] [PubMed] [Google Scholar]
- 85.Kara, D., S. B. I. Luppens, and J. M. Cate. 2006. Differences between single- and dual-species biofilms of Streptococcus mutans and Veillonella parvula in growth, acidogenicity and susceptibility to chlorhexidine. Eur. J. Oral. Sci. 114:58-63. [DOI] [PubMed] [Google Scholar]
- 86.Kara, D., S. B. I. Luppens, J. van Marle, R. Ozok, and J. M. ten Cate. 2007. Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine. FEMS Microbiol. Lett. 271:90-97. [DOI] [PubMed] [Google Scholar]
- 87.Kavanagh, K., and S. Dowd. 2004. Histatins: antimicrobial peptides with therapeutic potential. J. Pharm. Pharmacol. 56:285. [DOI] [PubMed] [Google Scholar]
- 88.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly, C. G., J. S. Younson, B. Y. Hikmat, S. M. Todryk, M. Czisch, P. I. Harris, I. R. Flindall, C. Newby, A. I. Mallet, J. K. Ma, and T. Lehner. 1999. A synthetic peptide adhesion epitope as a novel antimicrobial agent. Nat. Biotechnol. 17:42-47. [DOI] [PubMed] [Google Scholar]
- 90.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 1998. Virulence of polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol. Immunol. 13:373-377. [DOI] [PubMed] [Google Scholar]
- 91.Keyes, P. H., M. A. Hicks, B. M. Goldman, R. M. McCabe, and, and R. J. Fitzgerald. 1971. Dispersion of dextranous bacterial plaques on human teeth with dextranase. J. Am. Dent. Assoc. 82:136-141. [DOI] [PubMed] [Google Scholar]
- 92.Kleinberg, I. 2002. A mixed-bacterial ecological approach to understanding the role of oral bacteria in dental caries causation: an alternate to Streptococcus mutans and the specific-plaque hypothesis. Crit. Rev. Oral Biol. Med. 13:108-125. [DOI] [PubMed] [Google Scholar]
- 93.Koga, T., T. Oho, Y. Shimzaki, and Y. Nakano. 2002. Immunization against dental caries. Vaccine 20:2027-2044. [DOI] [PubMed] [Google Scholar]
- 94.Kolenbrander, P. E. 1988. Intergeneric coaggregation among human oral baceria and ecology of dental plaque. Annu. Rev. Microbiol. 42:627-686. [DOI] [PubMed] [Google Scholar]
- 95.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 96.Kolenbrander, P. E., P. G. Egland, P. I. Diaz, and J. R. J. Palmer. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13:11-15. [DOI] [PubMed] [Google Scholar]
- 97.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kolenbrander, P. E., R. N. Andersen, K. Kazmerzak, R. Wu, and R. J. Palmer. 1999. Spatial organization of oral bacteria in biofilms. Methods Enzymol. 310:322-332. [DOI] [PubMed] [Google Scholar]
- 99.Koll-Klais, P., R. Mandar, E. Leibur, and M. Mikelsaar. 2005. Oral microbial ecology in chronic periodontitis and periodontal health. Microb. Ecol. Health Dis. 17:146-155. [DOI] [PubMed] [Google Scholar]
- 100.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57:392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuboniwa, M., G. D. Tribble, C. E. Lamer, A. O. Killic, L. Tao, M. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60:121-139. [DOI] [PubMed] [Google Scholar]
- 104.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 105.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar, P. S., E. J. Leys, J. M. Bryk, F. J. Martinez, M. L. Moeschberger, and A. L. Griffen. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuramitsu, H. K., and V. Trapa. 1984. Genetic exchange between oral streptococci during mixed growth. J. Gen. Microbiol. 130:2497-2500. [DOI] [PubMed] [Google Scholar]
- 108.Lamont, R. J., and H. F. Jenkinson. 2000. Adhesins as an ecological determinant in the oral cavity, p. 131-168. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bactrial ecology: the molecular basis. Horizon Science Press, Wymondham, United Kingdom.
- 109.LeBlanc, D. J., R. J. Hawley, L. N. Lee, and E. St. Martin, Jr. 1978. “Conjugal”transfer of plasmid DNA among oral streptococci. Proc. Natl. Acad. Sci. USA 75:3484-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li, F., A. Meredith, and J. W. Lampe. 2007. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J. Microbiol. Methods 68:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li, Y.-H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li, Y., C. Ku, J. Xu, D. Saxena, and P. Caufields. 2005. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J. Dent. Res. 84:559-564. [DOI] [PubMed] [Google Scholar]
- 113.Li, Y., D. Saxena, V. Barnes, H. M. Trivedi, and T. Xu. 2006. Polymerase chain reaction-based denaturing gradient gel electrophoresis in the evaluation of oral microbiota. Oral Microbiol. Immunol. 21:333-339. [DOI] [PubMed] [Google Scholar]
- 114.Li, Y., Y. Ge, D. Saxena, and P. W. Caufield. 2007. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J. Clin. Microbiol. 45:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.LiPuma, J. J., H. Richman, and T. L. Stull. 1990. Haemocin, the bacteriocin produced by Haemophilus influenzae: species distribution and role in colonization. Infect. Immun. 58:1600-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Listgarten, M. A., and C.-H. Lai. 1975. Unusual cell wall ultrastructure of Leptotrichia buccalis. J. Bacteriol. 123:747-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu, W., T. Marsh, H. Cheng, and L. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loe, H., E. Theilade, and S. Jensen. 1965. Experimental gingivitis in man. J. Periodontol. 36:177-187. [DOI] [PubMed] [Google Scholar]
- 120.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Loesche, W. J., J. Rowan, L. H. Straffon, and P. J. Loos. 1975. Association of Streptococcus mutans with human dental decay. Infect. Immun. 11:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lovelock, J. E. 1965. A physical basis for life detection experiments. Nature 207:568. [DOI] [PubMed] [Google Scholar]
- 123.Macdonald, J. B., E. M. Madlener, and S. S. Socransky. 1959. Observations on Spirillum sputigenum and its relationship to Selenomonas species with special reference to flagellation. J. Bacteriol. 77:559-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Machado de Oliveira, J. C., J. F. Siqueira, I. N. Rocas, J. C. Baumgartner, T. Xia, R. S. Peixoto, and A. S. Rosado. 2007. Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol. Immunol. 22:14-18. [DOI] [PubMed] [Google Scholar]
- 125.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 126.Marquis, R. E., R. A. Burne, D. T. Parsonsons, and A. E. Casiano-Colon. 1993. Arginine deiminase and alkaline generation in plaque, p. 309-317. In W. H. Bowen and L. A. Tabak (ed.), Cariology for the nineties. University of Rochester Press, Rochester, NY.
- 127.Marsh, P. D. 2005. Dental plaque: biological significance of a biofilm and community life-style. J. Clin. Periodontol. 32:7-15. [DOI] [PubMed] [Google Scholar]
- 128.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 129.Matsukubo, T., and I. Takazoe. 2006. Sucrose substitutes and their role in caries prevention. Int. Dent. J. 56:119-130. [DOI] [PubMed] [Google Scholar]
- 130.Matsumoto-Nakano, M., and H. K. Kuramitsu. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 188:8095-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Matsumoto, M., M. Sakamoto, H. Hayashi, and Y. Benno. 2005. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colonic microbiota. J. Microbiol. Methods 61:305-319. [DOI] [PubMed] [Google Scholar]
- 132.Mayrand, D., C. Mouton, and D. Grenier. 1991. Chemical and biological properties of cell-surface components of oral blackpigmented Bacteroides species, p. 99-115. In S. Hamada, S. C. Holt, and J. R. McGhee (ed.), Periodontal disease: pathogens and host immune responses. Quintessence Publishing Co., Tokyo, Japan.
- 133.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 134.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mercuri, L. G. 2006. Microbial biofilms: a potential source for alloplastic device failure. J. Oral Maxillofac. Surg. 64:1303-1309. [DOI] [PubMed] [Google Scholar]
- 136.Merritt, J., J. Kreth, W. Shi, and F. Qi. 2005. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory output. Mol. Microbiol. 57:960-969. [DOI] [PubMed] [Google Scholar]
- 137.Mikx, F. H., and J. S. van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20:407-410. [DOI] [PubMed] [Google Scholar]
- 138.Miller, W. D. 1890. The micro-organisms of the human mouth. Graphische Anstalt Schuler AG, Biel, Switzerland.
- 139.Moore, W., E. L. V. Holdeman, E. P. Cato, R. M. Smibert, J. A. Burmeister, K. G. Palcanis, and R. R. Ranney. 1982. Bacteriology of experimental gingivitis in young adult humans. Infect. Immun. 38:651-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moore, W., E. L. V. Holdeman, E. P. Cato, R. M. Smibert, J. A. Burmeister, K. G. Palcanis, and R. R. Ranney. 1983. Bacteriology of moderate (chronic) periodontitis in mature adult human. Infect. Immun. 42:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moore, W., E. L. V. Holdeman, E. P. Cato, R. M. Smibert, J. A. Burmeister, K. G. Palcanis, and R. R. Ranney. 1985. Comparative bacteriology of juvenile periodontitis. Infect. Immun. 48:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morou-Bermudez, E., and R. A. Burne. 1999. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect. Immun. 67:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mota-Meira, M., G. Lapointe, C. Lacroix, and M. C. Lavoie. 2000. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 44:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nacimento, C. M., J. P. Watanabe, and Y. I. Ito. 2006. DNA checkerboard method for bacterial pathogen identification in oral diseases. Int. J. Morphol. 24:619-624. [Google Scholar]
- 145.Nakamura, Y., Y. Shibata, R. Shimura, and S. Fujimura. 1992. Isolation and properties of the Capnocytophaga ochracea bacteriocin. Oral Microbiol. Immunol. 7:96-99. [DOI] [PubMed] [Google Scholar]
- 146.Nakatsu, C. H. 2007. Soil microbial community analysis using denaturing gradient gel electrophoresis. Soil Sci. Soc. Am. J. 71:562-571. [Google Scholar]
- 147.Nes, I. F., D. B. Diep, and H. Holo. 2007. Bacteriocin diversity in streptococcus and enterococcus. J. Bacteriol. 189:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontology 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- 149.Osterberg, S. K. A., S. Z. Sudo, and L. E. A. Folke. 1976. Microbial succession in subgingival plaque of man. J. Periodontal Res. 11:243-255. [DOI] [PubMed] [Google Scholar]
- 150.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Palmer, R. J., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188:4117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Papapanoul, P. N., V. Baelum, W. M. Luan, X. Chen, O. Fejerskov, and G. Dahlen. 1997. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J. Periodontol. 68:651-666. [DOI] [PubMed] [Google Scholar]
- 153.Passador, L., K. D. Tucker, K. R. Guertin, M. P. Journet, A. S. Kende, and B. H. Iglewski. 1996. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 178:5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 155.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 156.Paster, B. J., M. K. Russell, T. Alpagot, A. M. Lee, S. K. Boches, J. L. Galvin, and F. E. Dewhirst. 2002. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann. Periodontol. 7:8-16. [DOI] [PubMed] [Google Scholar]
- 157.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Paster, B. J., W. A. Falkler, Jr., C. O. Enwonwu, E. O. Idigbe, K. O. Savage, V. A. Levanos, M. A. Tamer, R. L. Ericson, C. N. Lau, and F. E. Dewhirst. 2002. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J. Clin. Microbiol. 40:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pedersen, K. 2001. Diversity and activity of microorganisms in deep igneous rock aquifers of the Fennoscandian Shield, p. 97-139. In J. K. Fredrickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss Inc., New York, NY.
- 160.Piludu, M., M. S. Lantini, M. Cossu, M. Piras, F. G. Oppenheim, E. J. Helmerhorst, W. Siqueira, and A. R. Hand. 2006. Salivary histatins in humandeep posterior lingual glands (of von Ebner). Arch. Oral Biol. 51:967-973. [DOI] [PubMed] [Google Scholar]
- 161.Potempa, J., N. Pavloff, and J. Travis. 1995. Porphyromonas gingivalis: a protein/gene accounting audit. Trends Microbiol. 3:430-434. [DOI] [PubMed] [Google Scholar]
- 162.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH gene diminishes mutacin production and competence development, alters sucrose dependent biofilm formation, and reduces stress tolerance in Streptococcus mutans. Infect. Immun. 72:4895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produce both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analysis of mutacin III biosynthesis genes. Appl. Environ Microbiol. 65:3880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rasiah, L. W., S. Anderson, and C. Sissons. 2005. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch. Oral Biol. 50:779-787. [DOI] [PubMed] [Google Scholar]
- 166.Reid, G., J. Burton, J. A. Hammond, and A. Bruce. 2004. A PCR-DGGE method identified urogenital pathogenic bacteria in vaginal smears of patients with bacterial vaginosis. J. Med. Food 7:223-228. [DOI] [PubMed] [Google Scholar]
- 167.Rickard, A. H., R. J. Palmer, D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 168.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 169.Russell, M. N., N. K. Childers, S. M. Michalek, D. J. Smith, and M. A. Taubman. 2004. A caries vaccine? The state of the science of immunization against dental caries. Caries Res. 38:230-235. [DOI] [PubMed] [Google Scholar]
- 170.Saiki, R., S. Scharf, F. Faloona, K. Mullis, G. Horn, H. Erlich, and N. Arnheim. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 171.Sakamoto, M., H. Hayashi, and Y. Benno. 2003. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol. Immunol. 47:133-142. [DOI] [PubMed] [Google Scholar]
- 172.Sakamoto, M., M. Umeda, and Y. Benno. 2005. Molecular analysis of human oral microbiota. J. Periodontal Res. 40:277-285. [DOI] [PubMed] [Google Scholar]
- 173.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 174.Sakamoto, M., Y. Huang, M. Ohnishi, M. Umeda, I. Ishikawa, and Y. Benno. 2004. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J. Med. Microbiol. 53:563-571. [DOI] [PubMed] [Google Scholar]
- 175.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2003. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J. Med. Microbiol. 52:79-89. [DOI] [PubMed] [Google Scholar]
- 176.Scannapieco, F. A. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 5:203-248. [DOI] [PubMed] [Google Scholar]
- 177.Schabereiter-Gurtner, C., S. Maca, S. Rolleke, K. Nigl, J. Lukas, A. Hirschl, W. Lubitz, and T. Barisani-Asenbauer. 2001. 16S rDNA-based identification of bacteria from conjunctival swabs by PCR and DGGE fingerprinting. Investig. Ophthalmol. Vis. Sci. 42:1164-1171. [PubMed] [Google Scholar]
- 178.Seshadri, R., G. S. A. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E. Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, I. T. Paulsen, and, and R. Haselkorn. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA. 101:5646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sharma, A., S. Inagaki, W. Sigurdson, and H. K. Kuramitsu. 2005. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20:39-42. [DOI] [PubMed] [Google Scholar]
- 180.Shen, J., B. Zhang, G. Wei, X. Pang, H. Wei, M. Li, Y. Zhang, and L. Zhao. 2006. Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 72:5232-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shizukuishi, S., K. Tazaki, E. Inoshita, K. Kataoka, T. Hanioka, and A. Amano. 1995. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol. Lett. 131:313-317. [DOI] [PubMed] [Google Scholar]
- 182.Siqueira, J. F., I. N. Rocas, and A. S. Rosado. 2004. Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol. Immunol. 19:363-370. [DOI] [PubMed] [Google Scholar]
- 183.Siqueira, J. J. F., I. N. Rocas, and A. S. Rosado. 2005. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of endodontic infections. J. Endodontol. 31:775-782. [DOI] [PubMed] [Google Scholar]
- 184.Slots, J., and M. A. Listgarten. 1988. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J. Clin. Periodontol. 15:15. [DOI] [PubMed] [Google Scholar]
- 185.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Smit, P., and J. Heniger. 1975. Antoni van Leeuwenhoek (1632-1723) and the discovery of bacteria. Antonie Leeuwenhoek 41:217-228. [PubMed] [Google Scholar]
- 188.Socransky, S. S. 1979. Criteria for the infectious agents in dental caries and periodontal disease. J. Clin. Periodontol. 6:16-21. [DOI] [PubMed] [Google Scholar]
- 189.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 190.Socransky, S. S., C. Smith, L. Martin, B. J. Paster, F. E. Dewhirst, and A. E. Levin. 1994. “Checkerboard” DNA-DNA hybridization. BioTechniques 17:788-792. [PubMed] [Google Scholar]
- 191.Spitznagel, J., Jr., E. Kraig, and D. Kolodrubetz. 1991. Regulation of leukotoxin in leukotoxic and nonleukotoxic strains of Actinobacillus actinomycetemcomitans. Infect. Immun. 59:1394-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 193.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Takada, K., M. Hirasawa, and T. Ikeda. 1991. Isolation and purification of bacteriocin from Prevotella intermedia (Bacteroides intermedius). J. Clin. Periodontol. 62:439-444. [DOI] [PubMed] [Google Scholar]
- 195.Takahashi, N. 2003. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol. Immunol. 18:109-113. [DOI] [PubMed] [Google Scholar]
- 196.Tanner, A., M. F. Maiden, B. J. Paster, and F. E. Dewhirst. 1994. The impact of 16S ribosomal RNA-based phylogeny on the taxonomy of oral bacteria. Periodontology 2000 5:26-51. [DOI] [PubMed] [Google Scholar]
- 197.Tanzer, J. M., A. B. Kurasz, and J. Clive. 1985. Inhibition of ecological emergence of mutans streptococci naturally transmitted between rats and consequent caries inhibition by Streptococcus salivarius TOVE-R infection. Infect. Immun. 49:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Teanpaisan, R., A. M. Baxter, and C. W. Douglas. 1998. Production and sensitivity of bacteriocin-like activity among Porphyromonas gingivalis, Prevotella intermedia and Pr. nigrescens strains isolated from periodontal sites. J. Med. Microbiol. 47:585-589. [DOI] [PubMed] [Google Scholar]
- 199.Thies, F. L., W. Konig, and B. Konig. 2007. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J. Med. Microbiol. 56:755-761. [DOI] [PubMed] [Google Scholar]
- 200.Thies, J. E. 2007. Soil microbial community analysis using terminal restriction fragment length polymorphisms. Soil Sci. Soc. Am. J. 71:579-591. [Google Scholar]
- 201.Tong, H., W. Chen, J. Merritt, F. Qi, W. Shi, and X. Dong. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63:872-880. [DOI] [PubMed] [Google Scholar]
- 202.Tribble, G. D., G. J. Lamont, A. Progulske-Fox, and R. J. Lamont. 2007. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J. Bacteriol. 189:6382-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tsang, P., J. Merritt, T. Nguyen, W. Shi, and F. Qi. 2005. Identification of genes associated with mutacin I production in Streptococcus mutans using random insertional mutagenesis. Microbiology 151:3947-3955. [DOI] [PubMed] [Google Scholar]
- 204.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.van der Hoeven, J. S., C. W. A. Van der Kieboom, and P. J. M. Camp. 1990. Utilization of mucin by oral Streptococcus species. Antonie Leeuwenhoek 57:165-172. [DOI] [PubMed] [Google Scholar]
- 206.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 208.Vesey, P. M., and H. K. Kuramitsu. 2004. Genetic analysis of Treponema denticola ATCC 35405 biofilm formation. Microbiology 150:2401-2407. [DOI] [PubMed] [Google Scholar]
- 209.Wang, B.-Y., and H. K. Kuramitsu. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722-732. [DOI] [PubMed] [Google Scholar]
- 212.Yeung, M. K. 1999. Molecular and genetic analysis of Actinomyces species. Oral Biol. Med. 10:120-138. [DOI] [PubMed] [Google Scholar]
- 213.Yoneda, M., T. Hirofuju, and H. Anan. 2001. Mixed infection of Porphyromonas gingivalis and Bacteriodes forsythus in a murine abscess model. J. Periodontal Res. 36:237-243. [DOI] [PubMed] [Google Scholar]
- 214.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yoshino, K., K. Nishigaki, and Y. Husimi. 1991. Temperature sweep gel electrophoresis: a simple method to detect point mutations. Nucleic Acids Res. 19:3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang, M., B. Liu, Y. Zhang, H. Wei, Y. Lei, and L. Zhao. 2007. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J. Clin. Microbiol. 45:496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zijnge, V., H. J. M. Harmsen, J. W. Kleinfelder, M. E. van der Rest, J. E. Degener, and G. W. Welling. 2003. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol. Immunol. 18:59-65. [DOI] [PubMed] [Google Scholar]