Abstract
Summary: Many Proteobacteria use the chaperone/usher pathway to assemble proteinaceous filaments on the bacterial surface. These filaments can curl into fimbrial or nonfimbrial surface structures (e.g., a capsule or spore coat). This article reviews the phylogeny of operons belonging to the chaperone/usher assembly class to explore the utility of establishing a scheme for subdividing them into clades of phylogenetically related gene clusters. Based on usher amino acid sequence comparisons, our analysis shows that the chaperone/usher assembly class is subdivided into six major phylogenetic clades, which we have termed α-, β-, γ-, κ-, π-, and σ-fimbriae. Members of each clade share related operon structures and encode fimbrial subunits with similar protein domains. The proposed classification system offers a simple and convenient method for assigning newly discovered chaperone/usher systems to one of the six major phylogenetic groups.
INTRODUCTION
Proteinaceous nonflagellar filaments were found on the surface of Escherichia coli cells by electron microscopy in 1950 (139), and the term fimbriae was introduced in 1955 in a study demonstrating that these surface structures mediate the adherence of E. coli to eukaryotic cells (i.e., erythrocytes) (82). The term pili is an alternate designation for nonflagellar bacterial filaments, which was introduced in 1959 and is used by many investigators today (41). However, as the oldest valid published name for filamentous surface appendages functioning in adherence, the term fimbriae takes priority (248) and is therefore the one that will be used in this review article.
Since their first description, fimbriae have been identified in a large number of bacterial species. Over the years, the abundance of fimbriae among bacteria has motivated numerous efforts to develop classification schemes. Early attempts to group fimbriae based on morphology and functional characteristics (42, 81, 248) did not reflect the phylogenetic relatedness revealed subsequently by sequence analysis of the corresponding biosynthetic gene clusters and are thus mainly of historical interest. However, some fragments of these early nomenclatures have survived, including the term “type 1 fimbriae” for rigid filaments mediating mannose-sensitive hemagglutination (81) and the term “type IV fimbriae” (originally described as group 4) for a class of polar appendages mediating twitching motility (248). A morphological classification of fimbriae based on their diameters has been proposed more recently (252), but the large range of diameters reported for one and the same fimbrial type raises the question of whether the use of this dimension is a reliable parameter for classification. Furthermore, the sole use of arbitrary criteria (i.e., morphology and functional characteristics) for classifying fimbriae makes it uninformative to know their placement within the classification scheme.
Fimbriae can be distinguished serologically by bacterial agglutination (107), a method used extensively with E. coli to assign fimbrial (F) antigen numbers (246) or coli surface (CS) antigen numbers (102) to fimbrial structures. Although serology is well suited to generate lists of distinct appendages, the method does not lend itself to developing hierarchical classification schemes since it does not readily identify features shared by larger groups of phylogenetically related surface structures. Furthermore, morphological, functional, and serological classification schemes can be applied only to fimbriae expressed in vitro, an important shortcoming given the large amount of fimbrial gene sequences identified since the dawn of the genomic era. This problem is confounded by the observations for Salmonella enterica and E. coli that many of the fimbrial operons identified by whole-genome sequencing are poorly expressed under standard laboratory growth conditions (141, 203).
Classification systems that are based on the genealogy of fimbrial structures are superior in that a phylogenetic relationship has more predictive potential than does an arbitrary classification. Studies on the biochemistry and genetics of fimbrial biosynthesis have given rise to a nomenclature that distinguishes fimbriae based on their assembly mechanism. The major assembly classes present in gram-negative bacteria include those for conjugative fertility (F) fimbriae (F pili) (87, 192, 333), type IV fimbriae (e.g., toxin-coregulated pilus) (6, 134, 216, 240, 319), fimbriae assembled by the extracellular nucleation/precipitation pathway (curli) (126), and fimbriae assembled by the chaperone/usher-dependent pathway (162, 261, 290, 327, 344). This nomenclature has gained popularity because it defines classes of fimbriae in terms of ancestry and evolutionary descent. Members of an assembly class can be readily identified by sequence homology of their fimbrial biosynthesis genes, thus making this classification applicable to putative fimbrial gene clusters identified by whole-genome sequencing.
The chaperone/usher-dependent pathway is the fimbrial assembly class that contains the largest number of representatives by far. Names of fimbriae belonging to the chaperone/usher assembly class are a source of confusion, as they are derived from various serological, functional, and morphological schemes that mask the evolutionary descent of the respective biosynthesis gene clusters from a common ancestor. The establishment of a system to subdivide members of the chaperone/usher-dependent assembly class into clades of phylogenetically related gene clusters would promote clarity, universality, and stability in a theoretical context that is relevant to microbiology. The goal of this article is to explore the utility of such an approach.
CHARACTERISTICS OF THE CHAPERONE/USHER ASSEMBLY PATHWAY
Genes involved in the biosynthesis of fimbriae belonging to the chaperone/usher assembly class are almost invariably clustered into operons. These operons encode, at minimum, three different proteins: a major structural fimbrial subunit, a chaperone, and an usher protein. Fimbrial operons often contain additional genes encoding structural proteins (i.e., minor fimbrial subunits), assembly proteins (i.e., chaperones), or regulatory proteins. This article focuses on an analysis of genes encoding assembly and structural proteins to define groups on the basis of common ancestry.
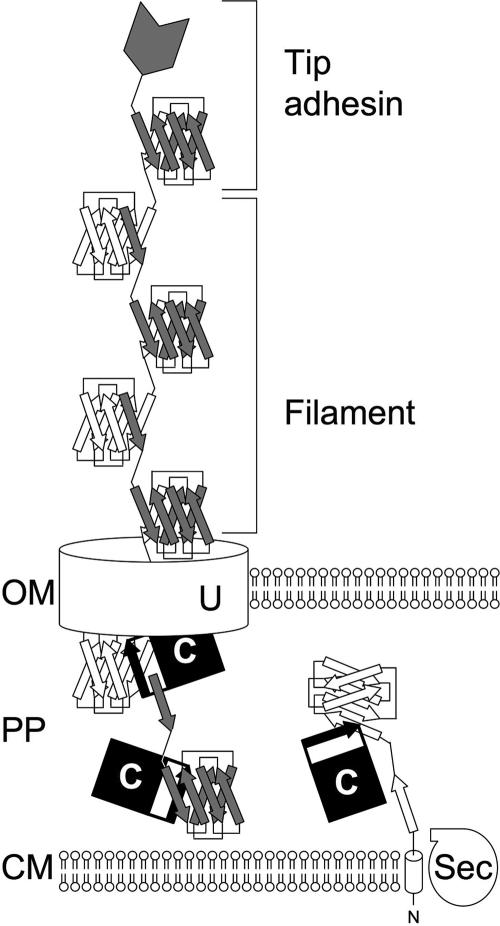
All assembly and structural proteins contain typical N-terminal signal sequences that allow their transport across the cytoplasmic membrane via the Sec general secretory pathway (210, 267). Upon the transport of fimbrial subunits into the periplasm and cleavage of the signal peptide, the formation of disulfide bonds may be catalyzed by periplasmic disulfide isomerases (e.g., DsbA and SrgA) (24, 36, 155). In the periplasm, fimbrial subunits interact with their cognate chaperones to ensure proper folding. Fimbrial subunits have immunoglobulin (Ig)-like folds, which lack the final antiparallel β-strand (G), creating a deep hydrophobic cleft. Chaperones bind fimbrial subunits by inserting their G1 donor β-strands into this hydrophobic cleft in a parallel orientation, thus creating a nonclassical Ig-like fold (Fig. 1) (59, 137, 264, 289). In the absence of the chaperone, fimbrial subunits form periplasmic aggregates that are rapidly degraded by the protease DegP (161, 251, 306).
FIG. 1.
Schematic drawing of fimbrial assembly by the chaperone/usher pathway. The N-terminal signal peptide of fimbrial subunits is cleaved during their transport across the cytoplasmic membrane (CM) by the general secretory pathway (Sec). In the periplasm (PP), the chaperone (C) completes the Ig-like fold of fimbrial subunits with its donor β-strand (black arrow). Interaction of the chaperone/fimbrial subunit complexes with the usher protein (U) facilitates the replacement of the chaperone donor β-strand with the N-terminal β-strand of a second subunit, thereby joining subunits into a filament that is transported across the outer membrane (OM). Filaments may curl into helical structures with two subunits per turn (thin, flexible fibrillae), as shown in this schematic drawing. Alternatively, subunits may assemble helices with 3 to 3.4 subunits per turn (thick, rigid fimbriae) or coil up into nonfimbrial, capsule-like structures on the cell surface. In some cases, fimbrial filaments carry a tip adhesin at their distal end, which mediates attachment.
Interaction of the periplasmic chaperone/subunit complexes with the usher, an integral outer membrane protein, facilitates the release of fimbrial subunits and their secretion through the usher channel, with the most distal proteins being secreted first (26, 50, 113, 185, 195, 233, 234, 292, 328). The N terminus of subunits harbors a conserved β-strand motif similar to the G1 donor β-strand motif of chaperones. The assembly of subunits is thought to proceed through a donor strand exchange reaction in which the G1 donor β-strand of the chaperone, inserted with a parallel directionality into the subunit's hydrophobic cleft, is replaced by the N-terminal β-strand of a second subunit in the more energetically favorable antiparallel orientation, thereby completing a classical Ig-like fold and joining subunits into a fiber (Fig. 1) (59, 264, 289). For some operons belonging to the chaperone/usher assembly class, these fibers coil up into nonfimbrial, capsule-like structures on the cell surface. However, in most cases, chaperone/usher-type transport systems assemble their major subunits into fimbriae that can be visualized by electron microscopy. Depending on their orientation in the fimbrial filament, major subunits may assemble thin flexible fibrillae (with two subunits per helical turn) or thick rigid fimbriae (with 3 to 3.4 subunits per helical turn). Minor subunits may be incorporated into the fimbrial filament in the form of a tip adhesin, a tip fibrillum, or an adaptor protein joining these structures (12, 46, 154, 185). Fimbrial chaperone/usher transport systems are specific with regard to ushers interfacing with chaperone/subunit complexes from their own systems (278, 292). Even in examples where an usher can be complemented by a very close homologue from another system, the presence of both intact systems results in their operating in parental pairs with respect to chaperone/subunit recognition (176).
Tip adhesins are minor fimbrial subunits that fold their C-terminal and N-terminal amino acids into two separate domains. The C-terminal domain has the Ig-like fold of fimbrial subunits, which contains a cleft due to the absence of the final β-strand. Insertion of the N-terminal β-strand of a fimbrial subunit into this cleft completes the Ig-like fold and connects the tip adhesin to the fimbrial filament (Fig. 1). The N-terminal domain contains the receptor-binding site and is connected to the C-terminal domain with a short amino acid linker, thereby placing the receptor binding surfaces on the tip of the fimbrial filament (47, 75, 127, 142).
TOWARDS A PHYLOGENETIC NOMENCLATURE
Use of Sequence Comparison To Develop Classification Schemes
The chaperone/usher class contains one group, designated the alternate chaperone/usher family, whose members share significant sequence homology to each other but not to other members of this assembly class (286). Operons that do not belong to the alternate chaperone/usher family are sometimes classified as members of a classical chaperone/usher family. For each family, the assembly mechanism involves the export of fimbrial subunits through the action of a periplasmic chaperone and an outer membrane usher protein. Although sequence similarities between the classical and alternate chaperone/usher families are limited, their usher proteins share conserved domains (PFAM00577 and/or COG3188) identified through the Conserved Domain Database component of the National Center for Biotechnology Information (NCBI) Entrez query retrieval system (211, 212). PFAM and COG designations refer to conserved domain entries in the Protein Family (PFAM) and Clusters of Orthologous Groups (COG) databases, which represent motifs and recurrent patterns in protein sequences (94, 324, 325).
Phylogenetic groups of fimbriae can be defined based on sequence comparisons of structural proteins or assembly proteins. A classification scheme based on sequence comparisons of 21 major fimbrial subunits of E. coli and S. enterica distinguishes the alternate chaperone/usher family (defined as class 5) from the classical chaperone/usher family (classes 1, 2, and 3) and other assembly classes, including type IV fimbriae (class 4) and curli (class 6) (204). Phylogenetic trees constructed by sequence comparison of major subunits belonging to the alternate chaperone/usher family demonstrate their common ancestry (8, 102). One group within the classical chaperone/usher family, defined as class 2 (204), contains subunits that assemble into nonfimbrial surface structures, and their N-terminal donor β-strands share common features (369). Sequence comparison of fimbrial subunits is superior to classification schemes based on serology, morphology, or function, as it defines groups by their descent from a common ancestor. However, a shortcoming of this approach is that an operon may contain several subunits that group to distant branches of a phylogenetic tree, which poses a problem for using this method as the sole criterion for classifying putative fimbrial operons since the identity of a major subunit may not be apparent from the nucleotide sequence.
Comparison of assembly proteins belonging to the classical chaperone/usher family reinforces the idea that fimbriae can be clustered into groups with common phylogenies. Computer modeling of the three-dimensional structure of 16 chaperones distinguishes two subfamilies based on the length of an amino acid sequence that corresponds to a loop that connects the F1 and G1 β-strands in the E. coli PapD crystal structure (371). A subsequent study on 26 chaperones defined proteins containing a relatively long F1-G1 loop as the FGL chaperone subfamily (e.g., MyfB, PsaB, Caf1M, CS3-1, AggD, AfaB, SefB, and CssC) and proteins with a short F1-G1 loop as the FGS chaperone subfamily (143). Interestingly, the FGL chaperone subfamily assembles thin fibrillae or nonfimbrial surface structures, while the FGS chaperone subfamily assembles fimbrial filaments (143, 371). Sequence comparison of 31 chaperones by the neighbor-joining method suggests that the classical chaperone/usher family can be divided into several clades, each including members that apparently share a common ancestor that is not shared by another protein outside of the clade (35). This phylogenetic tree suggests that members of the FGL chaperone subfamily (i.e., MyfB, PsaB, Caf1M, CS3-1, AggD, AfaB, NfaE, SefB, and CssC) share a common ancestor. However, this analysis also shows that the FGS chaperone subfamily cannot be defined by a single node or branch on the phylogenetic tree, suggesting that further subdivision is needed to explicitly categorize the respective fimbriae into clades on the basis of common ancestry. Sequence comparison of chaperones may not be ideally suited for developing such a subdivision because some fimbrial operons encode more than one chaperone, thus raising the question as to which protein should be used to assign the respective operon to a phylogenetic group.
The Fimbrial Usher Protein Family
Comparison of usher sequences has been used to derive phylogenetic trees of members of the chaperone/usher assembly class (8, 367). This approach has the advantage that the resulting definition of phylogenetic groups is unambiguous, because all fimbrial operons belonging to the chaperone/usher assembly class contain only a single usher gene, while multiple genes encoding subunits or chaperones may be present.
The fimbrial usher protein (FUP) family is a large and rapidly growing group of proteins that are distributed among genera of the phyla Proteobacteria, Cyanobacteria, and Deinococcus-Thermus (367). To make sense of the diversity within this large protein family, it is helpful to split this group into clades, thus creating a nonoverlapping hierarchical arrangement.
An initial comparison of 58 members of the FUP family distinguished 10 clusters on the basis of common ancestry (367). A revised phylogenetic tree of the FUP family constructed by comparing 189 proteins containing the usher domains PFAM00577 and/or COG3188 is shown in Fig. 2 (a list of the 189 usher proteins, NCBI Entrez accession numbers for protein sequences used in the usher analysis, and GenBank or RefSeq DNA accession numbers from which operon pictures in Fig. 3 to 11 were derived are provided in Table S1 in the supplemental material). Ushers were gathered for analysis by performing a multitude of BLAST searches with various usher amino acid sequences. The 189 ushers analyzed comprise previously reported fimbriae with sufficiently sequenced DNA to visualize the operons to which they belong and those ushers originating from conceptual translations from completed genome sequences. Subunits and chaperones not found in the local DNA surrounding an usher were not included in this analysis.
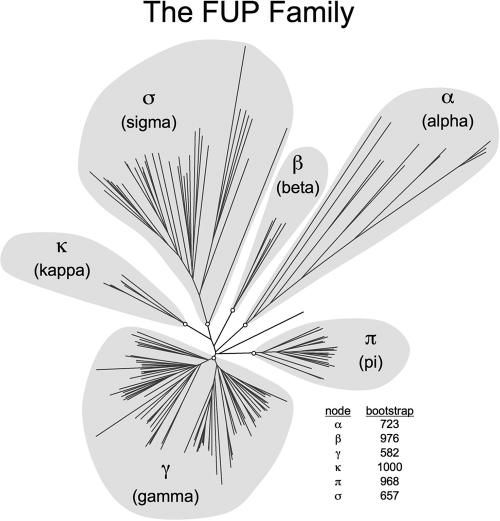
FIG. 2.
Phylogenetic tree of the FUP family. The graph shows an unrooted phenogram generated using usher amino acid sequences listed in Table S1 in the supplemental material. Ushers are grouped into six fimbrial clades (highlighted in gray) termed α, β, γ, κ, π, and σ. Bootstrap values of nodes defining these clades (indicated by open circles at the base of each fimbrial clade) are shown. The phylogenetic tree and bootstrap values were generated as follows. Usher amino acid sequences were aligned using ClustalW (MacVector 7.2.3) with a BLOSUM 30 matrix for pairwise alignment and a BLOSUM series matrix for multiple alignment, both with default settings. The phylogeny inference package software (PHYLIP 3.65) developed by Felsenstein (93) was used to perform the remaining analyses. Bootstrapping of the aligned usher sequences was performed in triplicate using the bootstrap algorithm of BOOTSTRAP to determine the number of instances, out of 1,000 sets analyzed (using random seeds 123, 345, and 567), where the members of each clade grouped completely behind the displayed node. Protein distance matrices were generated with PROTDIST using a Jones-Taylor-Thornton matrix with default settings for the unrooted tree or set to analyze 1,000 data sets for bootstrapping. Neighbor joining was performed with NEIGHBOR with default settings or set to analyze 1,000 data sets using the random seeds 345, 567, and 789, respectively, for the above-mentioned bootstrapping runs. Consensus trees (not shown) to interpret the bootstrap data were generated using the majority-rule-extended algorithm of CONSENSE. The unrooted phenogram displayed was generated using DRAWTREE without tree improvement iteration.
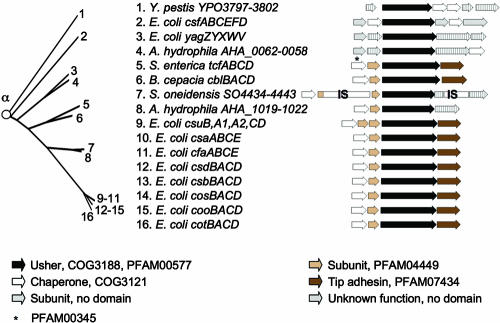
FIG. 3.
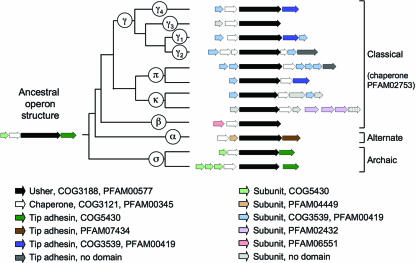
Phylogenetic relationship of operons belonging to the α-fimbriae. The branch of the FUP tree representing α-fimbriae is shown on the left. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. IS, homology to insertion sequence (IS) element.
FIG. 11.
Phylogenetic tree of fimbrial chaperones. An unrooted phenogram illustrating the phylogenetic relatedness of the chaperones associated with the ushers listed in Table S1 in the supplemental material is shown. Chaperone amino acid sequences were aligned and analyzed in the same fashion as that described in the legend of Fig. 2. Although PapJ has been shown to act in some capacity as a periplasmic chaperone (326), it and its homologues (PixJ, SfpJ, and PrfJ) were not included in this analysis, as they do not possess a fimbrial chaperone domain, nor do they share sequence similarity with other fimbrial proteins. The six fimbrial clades (α, β, γ, κ, π, and σ) defined by sequence comparison of usher proteins (Fig. 2) are indicated in the phenogram using gray highlighting. Arrows and circled branches indicate operons containing multiple chaperone genes and are inserted to the right and below the phenogram. The positions of chaperone genes in fimbrial operons (open arrows) in the phenogram are indicated by lowercase letters or by black arrows.
The phylogenetic analysis of 189 usher proteins suggests a classification into clades, which is similar but not identical to that proposed based on analysis of 58 usher proteins (367). To avoid confusion with this previous division into clusters 1 to 10 (367) we propose a nomenclature using Greek letters to refer to individual clades. This approach also distinguishes the FUP classification system from previous nomenclatures using Roman or Arabic numerals to define fimbriae of groups 1 to 6 (248), classes 1 to 6 (204), or types 1 to 6 (81).
Using a node-based definition, the FUP family can be divided into six clades, designated α-, β-, γ-, κ-, π-, and σ-fimbriae, each stemming from a common ancestor represented by a node in the phylogenetic tree (Fig. 2). The γ-fimbrial clade is further subdivided into four clades, termed γ1-, γ2-, γ3-, and γ4-fimbriae. The α-, κ-, π-, and σ-fimbrial clade names were assigned arbitrarily to recall a particular characteristic of the clade or a prominent member as follows: α-fimbriae, alternate chaperone/usher family; κ-fimbriae, K88 (F4) fimbriae; π-fimbriae, pyelonephritis-associated fimbriae (P fimbriae); and σ-fimbriae, spore coat protein U from Myxococcus xanthus. The β- and γ-fimbriae were assigned names alphabetically.
This subdivision of the FUP family largely confirms the subdivisions proposed initially (367), but former FUP clusters 4 and 5 now form a single clade (γ3-fimbriae). The subdivision of the chaperone/usher class into six FUP clades confirms that the alternate chaperone/usher family (α-fimbriae) (8) contains operons that stem from a common ancestor (Fig. 2). The FGL chaperone subfamily forms a monophyletic group (γ3-fimbriae) within the classical chaperone/usher family (143). However, the FGS chaperone subfamily is composed of several clades (β-, γ1-, γ2-, γ4-, κ-, and π-fimbriae) that are not more closely related to each other than to the FGL chaperone subfamily (γ3-fimbriae). The analysis also reveals the existence of a major FUP clade (σ-fimbriae) that was represented only by two usher proteins in a previous analysis (367) and whose members share limited or no sequence homology to members of the alternate chaperone/usher family (α-fimbriae) or the classical chaperone/usher family (β-, γ-, κ-, and π-fimbriae). An analysis of each FUP clade presented below shows that this classification scheme defines groups whose members share common characteristics, thus illustrating the utility of the proposed phylogenetic nomenclature.
THE ALTERNATE CHAPERONE/USHER FAMILY
The α-Fimbriae: Class 5 Fimbriae
The α-fimbrial clade includes the alternate chaperone/usher family (286), a group also known as class 5 fimbriae (204); the colonization factor antigen I (CFA/I)-like group (102); or FUP cluster 8 (367). Phylogenetic trees constructed by comparing fimbrial subunits, chaperones, and ushers demonstrate the common ancestry of members of this family (8, 102). With only 15 representatives from genera belonging to the Gammaproteobacteria and one representative from the Betaproteobacteria, the α-fimbriae are a relatively small but highly divergent group within the FUP family (Fig. 3). Previous studies (8, 102) listed ushers of Burkholderia cepacia (CblC) (282, 332), E. coli (CfaC, CooC, CosC, CotC, CsaC, CsbC, CsdC, and CsuC) (7, 8, 97, 99, 100, 125, 164), S. enterica serotype Typhi (TcfC) (72, 95), and Yersinia pestis (YPO3798) (254) as being members of the alternate chaperone/usher family. The α-fimbrial clade defined in this study includes additional usher proteins, one of which, CsfC (CS5 fimbriae), had not previously been classified as a member of the alternate chaperone/usher family. The α-fimbrial clade thus includes, but is not identical to, the group previously defined as class 5 fimbriae or CFA/I-like fimbriae (8, 102, 204). New members of the α-fimbriae include AHA_0060 and AHA_1021 from Aeromonas hydrophila (300), CsfC and YagX from E. coli (33, 85, 266), and SO_4439 from Shewanella oneidensis (66, 130). Many of the fimbriae belonging to operons of the α-fimbrial clade have been characterized morphologically.
A well-studied member of the α-fimbriae is the coo operon, encoding CS1 fimbriae of human enterotoxigenic E. coli (286). CS1 fimbriae are rigid, 6- to 7-nm fimbrial filaments that do not contain a visible tip fibrillum (102, 194). The fimbrial shaft is formed by the major subunit CooA and carries the CooD tip adhesin at its distal end, both of which are stabilized in the periplasm prior to surface assembly by the chaperone CooB (259, 285, 347). Other fimbriae of human enterotoxigenic E. coli that belong to the α-fimbrial clade include CFA/I (CfaC) (125), CS2 (CotC) (100), CS4 (CsaC) (7), CS5 (CsfC) (60, 85), CS14 (CsuC) (8), CS17 (CsbC) (8), CS19 (CsdC) (8), and PFC071 (CosC) (8). Similar to CS1 fimbriae, most of these organelles comprise a rigid fimbrial shaft of polymerized major subunits (6 to 7 nm in diameter) and an apparently tip-localized minor subunit, which mediates adherence (90, 102, 120, 220, 286). A notable exception is the csf operon, which encodes CS5 fimbriae composed of 2-nm flexible fibrillae (83-85). The only representative of the α-fimbriae that is present in a member of the Betaproteobacteria, the cable pilus (cable fimbriae) of B. cepacia, exhibits a morphological appearance that has been described as giant (2 to 4 μm in length) intertwined fibers (281). Expression of the cbl operon in E. coli results in an elaboration of fimbriae that are morphologically similar to other fimbrial structures belonging to the α-fimbrial clade (282).
The CFA/I, CS1, CS2, CS4, CS5, CS14, CS17, CS19, and PCF071 antigens of human enterotoxigenic E. coli isolates are carried by plasmids (8, 60, 89, 92, 97, 98, 120, 219, 270, 308, 358). The presence of these fimbrial antigens in human isolates is thought to reflect their selective binding to human tissue (102). Adhesion to small intestinal enterocytes or colonic epithelial carcinoma cells implicates CFA/I, CS1, CS2, CS4, and CS5 in mediating the colonization of the human intestine (67, 169, 170, 179, 180, 346). Volunteer studies provide direct evidence of a contribution of CFA/I fimbriae to the colonization of the human intestine with enterotoxigenic E. coli (91). The fact that all fimbrial operons of the α-fimbriae that are present in E. coli encode human-specific colonization factors is somewhat surprising, since it is not apparent why variants with tip adhesins that would bind receptors present in other vertebrate host species were not selected during diversification of this group. Interestingly, another member of the α-fimbrial clade, TcfC, is present in strictly human-adapted pathogens, including S. enterica serotype Typhi and S. enterica serotype Paratyphi A (95, 217, 336). However, fimbriae encoded by the chromosomally located tcf operon have not been functionally characterized.
Operons encoding CFA/I (cfa), CS1 (coo), CS2 (cot), CS4 (csf), CS17 (csb), CS19 (csd), PCF071 (cos), cable fimbriae (cbl), and serotype Typhi colonization factor (tcf) share the same organization, in which a gene encoding a chaperone is followed by genes encoding a major subunit, an usher, and a tip adhesin (7, 8, 100, 125, 336) (Fig. 3). The operon encoding CS14 (csu) has the same operon structure but contains two major subunit genes arranged in tandem (8, 101). Of 189 usher proteins analyzed, only CsuC (CS14 fimbriae) exhibited an E value for the PFAM00577 and COG3188 protein families that was above the cutoff of 0.01 used in this study (E values of 8.6 for PFAM00577 and 0.013 for COG3188) (212). The major fimbrial subunits and tip adhesins of most α-fimbriae contain the conserved protein domains PFAM04449 and PFAM07434, respectively. These protein domains are not found in fimbrial subunits of any other fimbrial clade. The more divergent members of the α-fimbrial clade are located in operons with different gene orders that encode fimbrial subunits with no significant homology to the PFAM04449 or PFAM07434 domains (Fig. 3), which may explain why these operons were not previously classified as being members of the alternate chaperone/usher family. Some chaperones of the α-fimbriae exhibited homology to the protein family COG3121, while only one (TcfA) had homology to the protein family PFAM00345, and no homology to PFAM02753 was detected. CooB and CsfB did not exhibit homology to any of the chaperone protein families, but experimental data show that these proteins function as chaperones for the biosynthesis of CS1 and CS5 fimbriae, respectively (84, 347). Similarly, no COG3121, PFAM00345, or PFAM02753 domains were detected in AHA_1019, CblB, CosB, and YPO3800, and the putative functions of these proteins were deduced from their sequence homologies to known chaperone proteins.
THE CLASSICAL CHAPERONE/USHER FAMILY
The β-Fimbriae
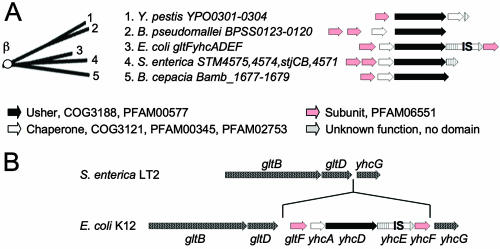
The β-fimbrial clade was previously defined as FUP cluster 7 (367), which contained a single representative, YhcD of E. coli K-12 (33). Additional members of the β-fimbriae identified since that initial report include Bamb_1679 of B. cepacia, BPSS0120 of Burkholderia pseudomallei, StjB of S. enterica serotype Typhimurium, and YPO0302 of Y. pestis (62, 136, 218, 254) (Fig. 4). Unfortunately, all putative fimbrial operons encoding usher proteins of the β-fimbriae were identified by whole-genome sequencing and remain uncharacterized.
FIG. 4.
Phylogenetic relationship of operons belonging to the β-fimbriae. (A) The branch of the FUP tree representing β-fimbriae is shown on the left. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. IS, homology to IS element. (B) Comparison of the gltB-yhcG intergenic regions in E. coli strain K-12 and S. enterica serotype Typhimurium strain LT2. Genes are indicated by arrows.
No structural or functional information is available for this small but distinct group of putative fimbrial operons, and none contain genes with homology to fimbrial subunits present in other fimbrial clades. However, in each operon, the genes encoding a chaperone and an usher are preceded by at least one open reading frame that encodes a protein with a domain of unknown function (PFAM06551) (Fig. 4A). The E. coli gltF yhcADEF operon encodes two proteins with a PFAM06551 domain, GltF and YhcF. GltF was initially proposed to be a regulator of glutamate synthase (GltBD) in E. coli (52, 53), but this hypothesis was refuted by subsequent studies (115, 118). Comparisons of the corresponding DNA regions of E. coli K-12 and S. enterica serotype Typhimurium (Fig. 4B) illustrate that the putative gltF yhcADEF fimbrial operon is in fact an insertion downstream of the glutamate synthase genes (gltBD). Given the presence of genes encoding PFAM06551-containing proteins in all operons belonging to the β-fimbrial clade, it is likely that the PFAM06551 protein domain defines fimbrial subunits characteristic of β-fimbriae. Operons of the β-fimbriae do not contain genes resembling typical tip adhesins. The absence of tip adhesins is characteristic of operons that assemble thin fibrillae or nonfimbrial surface structures (see sections below on γ3-fimbriae and κ-fimbriae). The apparent lack of genes resembling tip adhesins thus suggests that operons of the β-fimbrial clade encode nonfimbrial or fibrillar structures.
The γ-Fimbriae
The γ1-fimbriae: type 1 fimbria-like operons.
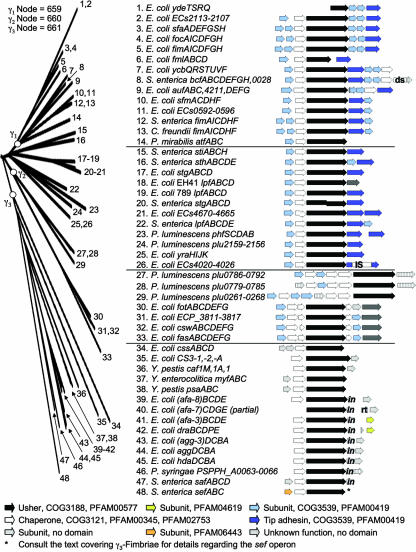
The γ1-fimbrial clade corresponds to FUP cluster 1 (367) and consists of ushers present in several bacterial species, all belonging to a single family within the Gammaproteobacteria (the Enterobacteriaceae). Representatives are present in Citrobacter freundii (FimD) (133), E. coli (AufC, ECs0594, ECs2110, FimD, FmlC, FocD, SfaF, SfmD, YcbS, and YdeT) (17, 33, 45, 56, 73, 128, 177, 202, 203, 271, 274, 297, 340, 352), Proteus mirabilis (AtfC) (213, 214), and S. enterica serotype Typhimurium (BcfC and FimD) (218, 337). A number of usher proteins are closely related to the γ1-fimbrial clade but are not connected by a node with a good bootstrapping value (Fig. 5). These usher proteins will be discussed together with the γ1-fimbriae in the following paragraphs and include ECs4022, ECs4667, LpfC[789], LpfC[EH41], StgC, and YraJ of E. coli (33, 77, 128, 146, 203, 208, 334, 335); PhfD and plu2157 of Photorhabdus luminescens (79); LpfC, SthB, and StiC of S. enterica serotype Typhimurium (28, 218); and StgC of S. enterica serotype Typhi (72, 336). Many of the fimbriae encoded by operons belonging to, or closely related to, the γ1-fimbrial clade are well studied, with type 1 fimbriae being perhaps the best-known representatives.
FIG. 5.
Phylogenetic relationship of operons belonging to the γ1-, γ2-, and γ3-fimbriae. The branch of the FUP tree representing γ1-, γ2-, and γ3-fimbriae is shown on the left. The bootstrap value for the node defining each subclade is displayed at the top and was generated in the analysis performed for Fig. 2. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. IS, homology to IS element; in, homology to invasin domain PFAM05775; rt, homology to reverse transcriptase; ds, homology to DsbA.
Type 1 fimbriae are defined as rigid fimbriae that are 7 to 10 nm in diameter and mediate mannose-sensitive hemagglutination (80). This designation can be confusing, since naturally occurring mutations resulting in a single-amino-acid substitution in the S. enterica FimH tip adhesin can result in a loss of mannose-sensitive hemagglutination (175). Furthermore, gene clusters encoding type 1 fimbriae of S. enterica and E. coli are both referred to as fim operons, although these are not orthologous gene clusters. The S. enterica serotype Typhimurium fimAICDHF operon has an orthologue, termed sfmACDHF, which is present at the corresponding location in the E. coli K-12 chromosome. Comparison of usher sequences illustrates the close relationship between S. enterica FimD and E. coli SfmD (Fig. 5). However, the sfmACDHF operon is cryptic, and type 1 fimbriae of E. coli are encoded by a paralogous operon, fimBEAICDFGH, which is located at a separate region of the chromosome (33, 218). Nonetheless, the E. coli sfmACDHF and fimBEAICDFGH operons share enough sequence similarities for both to be included in the γ1-fimbrial clade.
E. coli type 1 fimbriae bind to mannosylated glycoproteins in the bladder and are a virulence factor that is important for cystitis caused by uropathogenic E. coli (10, 229, 309, 310). They are composed of a rigid helical fimbrial shaft that ends in a short tip fibrillum (163). The helical shaft has a diameter of 7 nm and consists of 3.4 copies of the FimA major subunit per turn (42, 122). The short tip fibrillum is composed of the FimF and FimG minor fimbrial subunits and carries the FimH tip adhesin at its distal end (122, 163, 279, 291).
The γ1-fimbriae also contain the ushers of S fimbriae (SfaF) (181) and type 1C fimbriae (FocD) (271, 340) from uropathogenic E. coli. The presence of S fimbriae and type 1C fimbriae in E. coli is associated with urinary tract infection (160, 168, 257, 258, 302). Fimbriae assembled by SfaF and FocD are morphologically similar to type 1 fimbriae but exhibit different binding activities. S fimbriae attach to sialic acid-containing receptors and mediate neuraminidase-sensitive hemagglutination (181, 255), while type 1C fimbriae bind GalNAc(β1-4)β-galactosidase residues and do not mediate hemagglutination (172, 178, 340). Uropathogenic P. mirabilis isolates also contain a member of the γ1-fimbrial clade, the ambient-temperature fimbria (atf) operon; however, the encoded adhesin is not required for cystitis in a mouse model (213, 214, 373).
Gene clusters that are closely related to the γ1-fimbriae include the lpf operons of S. enterica (28), enterohemorrhagic E. coli (77, 334, 335), enteropathogenic E. coli (232), and extraintestinal pathogenic E. coli (146). In S. enterica serotype Typhimurium, the lpf operon has been implicated in intestinal colonization in a mouse model (29, 351).
The core structure of operons belonging to the γ1-fimbriae is composed of a major subunit gene followed by genes encoding a chaperone and an usher and a minor subunit encoding a tip adhesin. Additional minor subunits may be present at different positions in the operon (Fig. 5). Subunits of γ1-fimbriae contain the PFAM00419 and COG3539 fimbrial conserved domains, which are also found in subunits of γ2-fimbriae, γ4-fimbriae, π-fimbriae, and some of the κ-fimbriae.
The γ2-fimbriae.
The γ2-fimbrial clade represents FUP cluster 2 (367) and contains usher proteins assembling the CS12 fimbriae (CswD) (322), the CS18 fimbriae (FotD) (138), and the F6 fimbrial antigen (987P fimbriae) (FasD) (152, 295) of E. coli. Uncharacterized members include ECP_3814 of E. coli (135) and plu0268, plu0784, and plu0791 of P. luminescens (79) (Fig. 5).
The best-studied member of the γ2-fimbriae is the fas operon, which encodes the F6 fimbrial antigen (987P fimbriae) (295). The fas operon is commonly carried by plasmids in porcine isolates of enterotoxigenic E. coli (296). F6 fimbriae are rigid filaments that are 7 nm in diameter (153). Attachment is mediated by a tip adhesin, FasG, which is linked by a minor subunit, FasF, to the rigid fimbrial shaft composed of the major subunit FasA (49, 173). Assembly of F6 fimbriae involves three chaperones, FasB, FasC, and FasE. The FasB chaperone interacts with the major fimbrial subunit FasA but not with FasG. Conversely, FasC interacts only with FasG but not FasA, suggesting that chaperones are subunit specific (86). F6 fimbriae bind to receptors found in the brush border of piglet intestinal cells (57, 58, 71, 171), which have recently been identified as histone H1 proteins (372).
CS12 and CS18 are fimbriae that are present in human isolates of enterotoxigenic E. coli (132, 138). CS18 fimbriae are rigid filaments that are 6 to 8 nm in diameter (345). The morphologies of F6 fimbriae and CS18 fimbriae suggest that a type 1 fimbria-like appearance may be a shared feature of adhesins belonging to the γ2-fimbrial clade.
A distinguishing characteristic of operons belonging to the γ2-fimbriae is the presence of three chaperone genes (Fig. 5). While all three chaperones of the plu0261-268, plu0779-0785, and plu0786-0792 operons each contain three chaperone domains (COG3121, PFAM00345, and PFAM02753), the chaperones of the csw, ECP_3811 to ECP_3817, fas, and fot operons are more varied. The first chaperone in the operon (CswB, ECP_3812, FasB, and FotB) contains three chaperone domains (COG3121, PFAM00345, and PFAM02753), while the third chaperone (CswE, ECP_3815, FasE, and FotE) contains only two (COG3121 and PFAM00345). The second chaperone contains a COG3121 (ECP_3813 [FasC]) or PFAM00345 domain (FasC) or no domain at all (CswC and FotC), in which case the putative function of these proteins was assigned based on sequence homology to the known chaperone FasC (86). As with γ1-fimbriae, most of the fimbrial subunits of γ2-fimbriae contain the conserved domains PFAM00419 and COG3539. However, unlike γ1-fimbriae, the PFAM00419 and COG3539 domains are not present in the C-terminal half of tip adhesins.
The γ3-fimbriae: the FGL chaperone subfamily.
Representatives of the γ3-fimbrial clade include AfaC-3, AfaC-7, AfaC-8, AggC, Agg3C, CS3-2, CssD, DraC, and HdaC of E. coli (32, 39, 76, 105, 158, 187, 189, 190, 238, 239, 293, 361, 363, 366); PSPPH_A0064 of Pseudomonas syringae (159); SefC of S. enterica serotype Enteritidis (61); SefC of S. enterica serotype Paratyphi A (217); SafC of S. enterica serotype Typhimurium (95, 218); MyfC of Yersinia enterocolitica (150); and Caf1A and PsaC of Y. pestis (199, 254, 370) (Fig. 5). The operons assigned to the γ3-fimbriae based on sequence homology of their usher sequences are identical to those belonging to the FGL chaperone subfamily, defined by structural similarities of their chaperones (143). The γ3-fimbriae include two groups previously defined based on sequence comparisons of usher proteins, FUP clusters 4 and 5 (367). Furthermore, one group of fimbriae previously defined by sequence similarities of fimbrial major subunits (i.e., class 2) (204) consists of representatives that all belong to the γ3-fimbrial clade.
The fact that the γ3-fimbrial clade is readily distinguishable from other subfamilies on the basis of shared features in both their assembly proteins (i.e., chaperones and ushers) (143, 367, 371) and their structural proteins (i.e., major subunits) (204, 369) may reflect the distinct morphology of the assembled surface structures. Some members of the γ3-fimbriae assemble their major subunits into nonfimbrial structures on the cell surface (e.g., afa, nfa, css, and caf operons) (4, 65, 124, 131, 183, 205, 361, 369). These nonfimbrial structures are thought to be composed of fibrillae that are too thin to be resolved by electron microscopy. Other members of the γ3-fimbrial clade assemble very thin (2 to 3 nm in diameter), flexible, fibrillar organelles that can be visualized by electron microscopy (e.g., dra, agg, psa, myf, cs3, and sef operons) (61, 150, 194, 200, 230, 236, 293, 353).
Generally, organelles belonging to the γ3-fimbriae are composed of only one or two structural subunits and do not contain specialized subunits with homology to tip adhesins. The dra, daa, nfa, and afa operons encode adhesins of the Dr family, which mediate agglutination of erythrocytes carrying the Dr blood group antigen by binding to decay-accelerating factor (also known as CD55) and carcinoembryonic antigen-related proteins (9, 31, 140, 182, 188, 237-239, 260, 262, 342). The afa-3 operon encodes a nonfimbrial structure in which the AfaE major subunit assembles into fibers that are capped by the AfaD minor subunit (65). Studies of the close homologue DraE suggest that major subunit assembly can also proceed in the absence of the minor subunit cap (368). The AfaD minor subunit is an invasin that mediates bacterial entry into HeLa cervical carcinoma cells (104, 116, 165) via an interaction with an α5β1 integrin (121). However, binding to the decay-accelerating factor or carcinoembryonic antigen-related receptor of Dr family adhesins is mediated by the AfaE major subunit (31, 103, 182, 342, 343). The distinction between major subunits involved in adhesion and minor subunits involved in invasion may not be so rigid for all Dr family adhesins, as the major subunit DraE has been implicated in facilitating cell invasion (68). Members of the Dr family are commonly present in diffusely adhering E. coli isolates associated with diarrhea or urinary tract infections (299).
The γ3-fimbrial clade contains two adhesins that are associated with human isolates of enterotoxigenic E. coli, CS3 and CS6. The CS3 antigen mediates attachment to human colonic epithelial cancer cells (231) and forms thin, flexible fibrillae with a diameter of 2 nm on the bacterial surface (194). The css operon encodes the CS6 antigen of human enterotoxigenic E. coli, which is present on the cell surface but has a morphology that is beyond the limits of resolution by electron microscopy (205, 361). The css operon contains two major subunit genes, and the encoded proteins (CssA and CssB) are present at a 3:1 ratio on the bacterial surface (363, 366). The assembly of this surface structure results in the binding of E. coli to human enterocytes (132).
In Y. pestis, the caf operon encodes the fraction 1 (F1) capsule-like antigen, which confers antiphagocytic properties (78). The major subunit Caf1 assembles into fibers that form a large gel-like capsule on the bacterial surface (18, 369).
Operons of the γ3-fimbriae share an arrangement of genes encoding a major subunit, a chaperone, an usher, and a minor subunit. Some operons lack a subunit gene at the beginning of the operon, and the major subunit is encoded by a gene located downstream of the usher gene. Unlike other members of the γ-fimbriae, fimbrial subunits of the γ3-fimbriae do not contain the conserved domains PFAM00419 and COG3539. The S. enterica sef operon shown in Fig. 5 is from the genome of serotype Paratyphi A and does not contain a minor subunit. However, sef operons present in other S. enterica serotypes (e.g., serotype Enteritidis) carry a minor subunit gene (sefD) (61) encoding a protein containing a PFAM05775 domain. The PFAM05775 domain is also found in all invasins (i.e., minor subunits) of the Dr family of adhesins.
The γ4-fimbriae: Acu, Hif, and Mrk.
The γ4-fimbrial clade corresponds to FUP family 10 (367) and contains usher proteins of bacterial genera representing two classes, the Betaproteobacteria and the Gammaproteobacteria. The majority of operons belonging to the γ4-fimbriae were identified by whole-genome sequencing, and only a few members, the acu, hif, and mrk operons, are well characterized (Fig. 6). Representatives of the γ4-fimbrial clade are present in Acinetobacter spp. (ACIAD0121, ACIAD0389, and AcuC) (23, 110), Bordetella avium (FimC) (311), Bordetella pertussis (FhaA or FimC) (201, 253), B. cepacia (Bamb_2831 and Bamb_4024) (62), B. pseudomallei (BPSS0093) (136), Burkholderia xenovorans (Bxe_B0728, Bxe_B2970, and Bxe_C1151), (54), Chromobacterium violaceum (CV_1296) (70), E. coli (ECs0022, ECs0143, ECs1278, ECs2915, HtrE, UTI89_C4905, and YehB) (33, 56, 128, 203, 268), Haemophilus spp. (GhfC, HafC, HifC[86-0295], and HifC[AM30]) (43, 117, 269, 341, 349), Klebsiella pneumoniae (MrkC) (5), P. luminescens (plu0505) (79), Pseudomonas aeruginosa (CupA3 and CupB3) (278, 317, 339), Pseudomonas fluorescens (PFL_1464) (256), Ralstonia eutropha (H16_B2090) (263), Ralstonia metallidurans (Rmet_0574, Rmet_1665, Rmet_4252, and Rmet_4959), S. enterica serotype Typhi (StaC) (72, 336), S. enterica serotype Typhimurium (StbC and StcC) (218), Xylella fastidiosa (XF0081) (305), and Y. pestis (YPO1709, YPO1920, and YPO3879) (254).
FIG. 6.
Phylogenetic relationship of operons belonging to the γ4-fimbriae. The branch of the FUP tree representing γ4-fimbriae is shown on the left. The bootstrap value for the node defining the γ4-fimbriae is displayed at the top and was generated in the analysis performed for Fig. 2. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom.
The node connecting usher proteins BPSL1628 of B. pseudomallei, CupC3 of P. aeruginosa, EtfC of Edwardsiella ictaluri, EtfC of Edwardsiella tarda, PFL_3924 of P. fluorescens, and PSPPH_1201 and PSPPH_2011 of P. syringae (136, 159, 256, 278, 283, 284, 317, 339), as a sister group to the γ4-fimbrial clade, has a low bootstrapping value (37.3%), suggesting that there is insufficient information to decide where to place them among the γ-fimbriae (Fig. 2 and 6). One member, CupC, assembles structures exhibiting a fimbrial morphology (278), but the remaining members of this group remain uncharacterized.
The mrk operon of K. pneumoniae encodes thin (2- to 4-nm-wide) fimbriae (5), termed type 3 fimbriae based on their ability to mediate mannose-resistant agglutination of tannic acid-treated erythrocytes (80). The fimbrial shaft is composed of the major subunit MrkA, while the mrkD gene encodes a minor subunit that is located at the tip of the fimbrial filaments and mediates adhesion (106, 157, 191, 323). The hif operon of H. influenzae (341, 349) encodes fimbriae that agglutinate erythrocytes carrying the blood group AnWj antigen (64) and promotes colonization of the upper respiratory tract (108, 109, 184, 350). The 7-nm-wide fimbrial filaments are composed of a helical shaft containing three copies of the HifA major subunit per turn (228). The filaments carry a short tip fibrillum composed of the minor subunit HifD (312), which contains the minor subunit HifE located at its tip (221). AcuC, the usher protein assembling thin fimbriae (pili) of Acinetobacter spp., is closely related to MrkC and HifC (Fig. 6). The acuADCG operon encodes thin (2- to 3-nm) fimbriae that are packed into right-handed bundles (110). The fimABCD operon of B. pertussis (201, 355-357) encodes serotype 2 fimbriae, which have diameters of 5 to 6 nm and are composed of a helical shaft containing 2.5 copies of the major subunit per turn (313).
The core operon structure present in all members of the γ4-fimbrial clade consists of genes encoding a major subunit, a chaperone, and an usher (Fig. 6). In the majority of operons, a gene encoding a minor fimbrial subunit resembling the MrkD and HifE tip adhesins is present downstream of the usher gene. In some cases, additional minor subunits may be present, which assemble into a tip fibrillum (e.g., HifD) (314). A subgroup of operons carries a second chaperone gene downstream of the gene encoding the tip adhesin. Like γ1-fimbriae and γ2-fimbriae, operons belonging to the γ4-fimbriae encode subunits containing the conserved domains PFAM00419 and COG3539.
The κ-Fimbriae: Flexible Fibrillae
All usher proteins belonging to the κ-fimbrial clade are present in bacteria belonging to the family Enterobacteriaceae (Fig. 7). The κ-fimbriae (FUP cluster 6) (367) comprise usher proteins required for the assembly of the F4 fimbrial antigen (K88 fimbriae) (FaeD) (20, 22, 224, 245, 315, 316, 338), the F5 fimbrial antigen (K99 fimbriae) (FanD) (1, 20, 149, 151, 247, 275-277, 303), and the F18 fimbrial antigen (F107 fimbriae) (FedB) (147, 148, 307) found in porcine and bovine isolates of enterotoxigenic E. coli (51, 362). Furthermore, this group includes adherence factor/rabbit 1 (AF/R1) fimbriae (AfrB) (48), an uncharacterized operon of E. coli (CshB), the locus for diffuse adherence (LdaD) of atypical enteropathogenic E. coli (294), plasmid-encoded fimbriae (PefC) of S. enterica serotype Typhimurium (96, 218), and an intestinal colonization factor of rabbit enteropathogenic E. coli (RalD) (2).
FIG. 7.
Phylogenetic relationship of operons belonging to the κ-fimbriae. The branch of the FUP tree representing κ-fimbriae is shown on the left. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom.
In addition to sequence homology of their usher proteins, members of the κ-fimbrial clade share morphological characteristics; that is, each operon encodes thin (2- to 5-nm-diameter) flexible fibrillae (2, 96, 123, 272) that can be readily distinguished morphologically from the thicker, rigid fimbriae assembled by operons of the γ- or π-fimbrial clades. The only exception is the ldaCDEFGHI operon, which encodes subunits that are assembled into a nonfimbrial structure on the bacterial surface (294). In the case of F18 fibrillae, the major subunit FedA is assembled into a helix containing two subunits per turn (123). Minor fimbrial subunits have been implicated in the adherence of F18 fimbriae and AF/R1 fimbriae (48, 148, 307). However, in the case of F4 and F5 fimbriae, minor subunits are dispensable for adherence (22, 303, 304), and the major fimbrial subunits (FaeG and FanC) have been implicated in determining the binding activity of their respective adhesins (21, 156, 250).
Many members of the κ-fimbriae that are present in E. coli or S. enterica are carried by conjugative plasmids (2, 3, 40, 96, 242, 301, 359, 360). Conjugative plasmids with similarity to the well-characterized fertility (F) plasmid are found in a considerable fraction of E. coli isolates (38). An F-like OriT is also present on the virulence plasmids of S. enterica serotype Typhimurium, serotype Enteritidis, and serotype Gallinarum (249). Comparative sequencing of genes (traD, traY, and finO) involved in the DNA transfer of E. coli F-like plasmids reveals close homology to the corresponding genes on the virulence plasmid of S. enterica serotype Typhimurium, which carries the pef operon (37). The average pairwise differences between F-plasmid genes (traD, traY, and finO) present in E. coli and S. enterica isolates are similar to those observed between different isolates within each species. These data suggest that the genera Escherichia and Salmonella share an F-like plasmid pool (37). The plasmids contained within the shared F-like plasmid pool are mosaic in structure, with different regions acquired from different sources (38). For example, the DNA regions upstream and downstream of the pef operon are conserved between the F-like virulence plasmid of S. enterica serotype Typhimurium and the S. enterica serotype Gallinarum virulence plasmid. However, the virulence plasmid of S. enterica serotype Gallinarum carries a different fimbrial operon at the corresponding position. Interestingly, sequence analysis shows that the fimbrial proteins encoded by the S. enterica serotype Gallinarum virulence plasmid share high sequence identity to proteins of the E. coli F4 fimbrial antigen (76% identity to FaeH and 70% identity to FaeI) (280). Collectively, these data suggest that E. coli and S. enterica have exchanged fimbrial operons belonging to the κ-fimbrial clade by interspecies conjugative transfer of F-like plasmids.
Members of the κ-fimbriae share similarities in their operon structures and lack a gene encoding a tip adhesin (Fig. 7). Operons belonging to the κ-fimbrial clade also share a common core structure composed of genes encoding a major subunit, an usher, and a chaperone (Fig. 7). Some operons encode subunits that contain the conserved domains PFAM00419 and COG3539 present in subunits of π-fimbriae and γ-fimbriae. However, fimbrial subunits encoded by the csh, fae, lda, and ral operons contain the PFAM02432 domain, which is present only in members of the κ-fimbriae (Fig. 7).
The π-Fimbriae: P-Fimbria-Like Operons
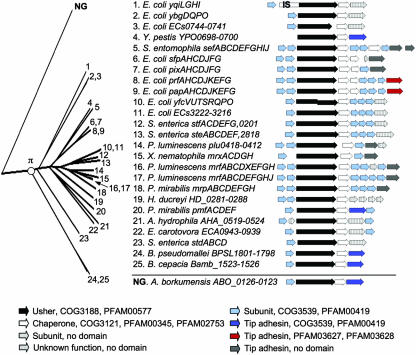
Usher proteins of the π-fimbrial clade (FUP cluster 3) (367) are found in bacteria belonging to the Betaproteobacteria and the Gammaproteobacteria. Members of the π-fimbriae are present in A. hydrophila (AHA_0521) (300), B. cepacia (Bamb_1524) (62), B. pseudomallei (BPSL1800) (136), Erwinia carotovora (ECA0942) (30), E. coli (PapC, PrfC, PixC, SfpC, YbgQ, YfcUT, YqiG, ECs0743, and ECs3221) (33, 44, 56, 74, 128, 203, 206, 226, 235, 321), Haemophilus ducreyi (HD_0283), P. luminescens (MrfC[K122], MrfC[TTO1], and plu0416) (79, 223), P. mirabilis (MrpC and PmfC) (13, 16, 215, 243), S. enterica serotype Paratyphi A (SteB) (217), S. enterica serotype Typhimurium (StdB and StfC) (88, 218, 225), Serratia entomophila (SefC) (144, 145), Xenorhabdus nematophila (MrxC) (129), and Y. pestis (YPO0698) (254) (Fig. 8). While most members of the π-fimbriae remain poorly characterized, the pap and mrp operons are well studied. The pap operon in particular has served as a paradigm to elucidate some of the mechanisms by which the chaperone/usher pathway assembles fimbrial organelles.
FIG. 8.
Phylogenetic relationship of operons belonging to the π-fimbriae. The branch of the FUP tree representing π-fimbriae is shown on the left. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. NG, not grouped but related to π-fimbriae; IS, homology to IS element.
The pap operon encodes pyelonephritis-associated (P) fimbriae that allow uropathogenic E. coli to bind Galα(1-4)Gal moieties present in glycolipids expressed by kidney epithelial cells (34, 310, 312, 318, 320, 321). P fimbriae are composed of a rigid helical fimbrial shaft that ends in a flexible tip fibrillum (46, 185). The helical shaft has a diameter of 8 nm and is composed of 3.3 copies of the PapA major subunit per turn (46, 227). It is thought to be connected to the outer membrane by the minor subunit PapH (12). The tip fibrillum composed of the minor subunit PapE is 2 nm wide and carries the tip adhesin PapG in a single copy at its distal end (185, 207, 273). The tip adhesin PapG is joined to the tip fibrillum by the PapF minor subunit, which serves as an adaptor protein (154, 185). Another minor subunit encoded by the pap operon, PapK, serves as an adaptor protein to link the tip fibrillum to the helical fimbrial shaft (154). While PapD is the periplasmic chaperone for the P-fimbrial subunits (25, 186, 306, 365), PapJ plays an auxiliary role by ensuring the proper integration of PapA into the fimbrial shaft (326). E. coli fimbrial operons that are closely related to pap include prf (P-related fimbriae) (226), pix (Pix fimbriae) (206), and sfp (Sfp fimbriae) (44). While P-related fimbriae and Pix fimbriae are morphologically similar to P fimbriae, Sfp fimbriae are only 3 to 5 nm in diameter (44).
Mannose-resistant/Proteus-like (MR/P) fimbriae encoded by the mrp operon of P. mirabilis contribute to the colonization of the urinary tract and the development of pyelonephritis in a mouse model (14, 16). MR/P fimbriae decorate the bacterial surface as rigid fimbrial filaments with diameters of 6 to 7 nm, which are composed of the major subunit MrpA (15, 243). The tip adhesin, MrpH, mediates the binding of MR/P fimbriae to erythrocytes (196) and is linked to the fimbrial shaft by the minor subunit MrpG (198). The minor subunit MrpB terminates fimbrial assembly and is thought to be a functional homologue of PapH (197).
Genes encoding a major fimbrial subunit, an usher, and a chaperone form the core structure of operons belonging to the π-fimbrial clade (Fig. 8). This gene order is shared with the κ-fimbriae but is distinct from core structures found in all other FUP clades in which the chaperone normally precedes the usher. Each operon contains a gene located downstream of the chaperone gene and encodes either a protein with homology to tip adhesins (e.g., PapG and MrpH) or a similarly sized protein of unknown function. A large subgroup within the π-fimbriae contains an additional three minor subunit genes that are located between the genes encoding the chaperone and the tip adhesin. Major subunits, minor subunits, and some of the tip adhesins contain the conserved domains PFAM00419 and COG3539, which are also present in subunits of the κ-fimbriae and γ-fimbriae. Distinct protein domains have been described for the N-terminal (PFAM03627) and C-terminal (PFAM03628) portions of the tip adhesins of P fimbriae and P-related fimbriae (Fig. 8).
An usher protein from Alcanivorax borkumensis (ABO_0125) (298), a member of the order Oceanospirillales, forms a sister group of the π-fimbriae (Fig. 8). However, the corresponding node is supported by a low bootstrapping value (35.9%), suggesting that not enough information is available at this time to decide whether the operon is related more closely to the π- or the κ-fimbrial clade based upon the usher sequence alone. Nevertheless, the presence of a gene resembling a tip adhesin (ABO_0123) at the end of the operon suggests that it is most closely related to the π-fimbrial clade.
ARCHAIC CHAPERONE/USHER SYSTEMS
The σ-Fimbriae
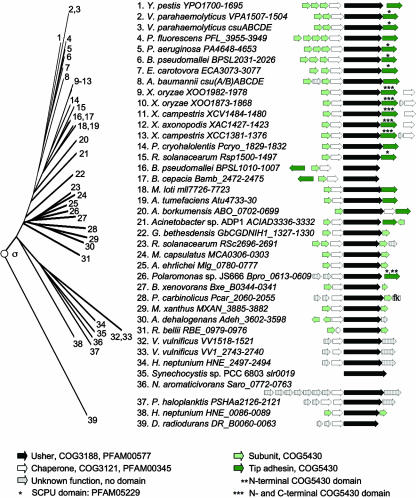
The σ-fimbrial clade contains usher proteins that contain a PFAM00577 domain but that share limited sequence similarity to usher proteins of the alternate chaperone/usher family (α-fimbriae) or the classical chaperone/usher family (clades β, γ, κ, and π). The group was initially proposed as FUP cluster 9, which contained only two usher proteins, Orf1 (PA4652) of Pseudomonas aeruginosa and CsuD of Vibrio parahaemolyticus, both members of the Gammaproteobacteria (367). Our analysis expands the σ-fimbriae considerably to include representatives of the phyla Proteobacteria, Cyanobacteria, and Deinococcus-Thermus, making it the most widely distributed FUP cluster (Fig. 9). This wide phylogenetic distribution suggests that the σ-fimbrial clade represents an archaic group within the chaperone/usher assembly class.
FIG. 9.
Phylogenetic relationship of operons belonging to the σ-fimbriae. The branch of the FUP tree representing σ-fimbriae is shown on the left. Numbers at the end of each branch of the phylogenetic tree correspond to the numbers given for each operon in the center. The gene order for each operon is shown on the right (arrows). The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. fk, homology to flagellum hook-associated protein; SCPU, spore coat protein U.
Within the Gammaproteobacteria, representatives of the σ-fimbriae are present in A. borkumensis (ABO_0701), Acinetobacter spp. (ACIAD3334), Acinetobacter baumannii (CsuD), E. carotovora (ECA3076), Methylococcus capsulatus (MCA0304), Alkalilimnicola ehrlichei (Mlg_0778), Pseudoalteromonas haloplanktis (PSHAa2122), P. aeruginosa (PA4652), P. fluorescens (PFL_3950), Psychrobacter cryohalolentis (Pcryo_1831), Vibrio vulnificus (VV1_2741 and VV1520), Vibrio parahaemolyticus (CsuD and VPA1504), Xanthomonas axonopodis (XAC1425), Xanthomonas campestris (XCC1379 and XCV1482), Xanthomonas oryzae (XOO1871[MAFF311018] and XOO1980[KACC10331]), and Y. pestis (YPO1696) (19, 23, 30, 55, 69, 174, 193, 209, 222, 244, 254, 256, 298, 317, 329, 331, 348). Members of the σ-fimbrial clade are present in several Betaproteobacteria including B. cepacia (Bamb_2475), B. pseudomallei (BPSL1007 and BPSL2027), B. xenovorans (Bxe_B0342), Polaromonas spp. (Bpro_0610), and Ralstonia solanacearum (RSp1498 and RSc2693) (54, 62, 63, 136, 287). Within the Alphaproteobacteria, σ-fimbriae are present in Agrobacterium tumefaciens (Atu4731), Granulibacter bethesdensis (GbCGDNIH1_1329), Hyphomonas neptunium (HNE_0088 and HNE_2495), Mesorhizobium loti (mll7724), Novosphingobium aromaticivorans (Saro_0764), and Rickettsia bellii (RBE_0977) (11, 114, 119, 166, 241, 364). Members of the σ-fimbrial clade are present in three Deltaproteobacteria, including Anaeromyxobacter dehalogenans (Adeh_3599), Pelobacter carbinolicus (Pcar_2057), and Myxococcus xanthus (MXAN_3883) (111, 112, 288). Finally, usher proteins of the σ-fimbriae are found one each in the phyla Deinococcus-Thermus (DR_B0063 of Deinococcus radiodurans) (354) and Cyanobacteria (Slr0019 of Synechocystis spp.) (167). Although the phylum Deinococcus-Thermus is classified as being gram positive, D. radiodurans has a complex cell wall profile that includes an outer membrane-like structure (330), which may explain the requirement for a specialized secretion system of the chaperone/usher type to transport proteins to the cell surface.
Given the wide phylogenetic distribution of operons belonging to the σ-fimbriae, surprisingly little is known about the morphology or function of the encoded surface structures. The little that we know about this clade is derived from studies of A. baumannii and M. xanthus, which implicate σ-fimbriae in biofilm formation and spore coat formation, respectively.
Analysis of biofilm-deficient transposon mutants of A. baumannii demonstrates that the csu fimbrial operon is required for biofilm formation and for the elaboration of fimbriae on the bacterial surface (331). These data suggest that at least some of the members of the σ-fimbriae assemble their subunits into fimbrial filaments. The open reading frames MXAN_3885 to MXAN_3882 in the M. xanthus genome encode a member of the σ-fimbrial clade (111). The first gene in this operon encodes protein U, a spore coat protein produced at the late stage of development of M. xanthus (112). These data suggest that some members of the σ-fimbrial clade may assemble their subunits into nonfimbrial structures on the bacterial surface. However, much work remains to be done to generate basic insights into the properties and characteristics of the archaic σ-fimbrial clade.
The core operon structure of σ-fimbriae consists of genes encoding a major subunit, a chaperone, an usher, and a tip adhesin (Fig. 9). Most subunits of σ-fimbriae have significant homology to COG5430, a conserved protein domain that is characteristic of archaic chaperone usher systems. Chaperones of the σ-fimbriae contain either a COG3121 domain only or a COG3121 domain and a PFAM00345 domain.
EVOLUTION OF FIMBRIAL OPERONS
Comparative analysis shows that fimbrial operons represent hypervariable DNA regions in S. enterica genomes, indicative of their frequent horizontal transfer between different organisms (lateral gene transfer) (27, 265, 336). This variability is still greater between species, as illustrated by the fact that of the 12 chaperone/usher-type fimbrial operons present in S. enterica serotype Typhimurium, only fim and stc are well conserved in the closely related organism E. coli (33, 218). Their limited phylogenetic distributions and frequent lateral transfers between species make fimbrial operons not suited for inferring relationships between bacteria. Instead, the utility of delineating the genealogy of fimbrial operons lies in its value for predicting their evolutionary relationship and the morphology of the encoded surface structures. For instance, operons of the γ3- and κ-fimbrial clades encode exclusively fibrillae and nonfimbrial structures, while operons belonging to the α-, γ1-, γ2-, γ4-, and π-fimbrial clades exclusively assemble fimbrial structures.
It is informative to know the placement of members of the chaperone/usher assembly class within the usher-based classification scheme to make sense of the diversity in their operon structures. The proposed classification scheme divides the classical chaperone/usher family into clades of fimbrial operons with related gene orders (Fig. 3 to 9). In most cases, the divergence of operon structures within each clade can be explained by proposing a single duplication, acquisition, or deletion event, which illustrates that the suggested evolutionary scenario is highly parsimonious. Thus, the gene order within fimbrial operons concurs in supporting the division of the chaperone/usher assembly class into clades based on the sequence homology of usher proteins.
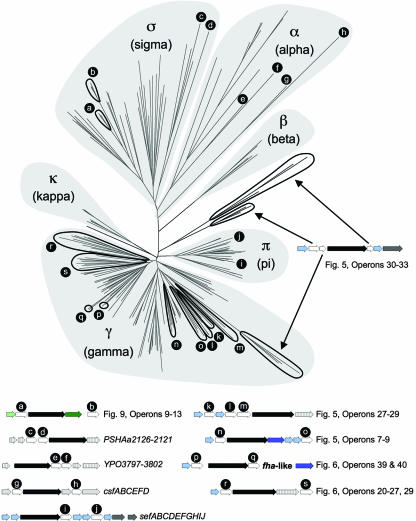
When the most common operon structures within each clade are placed at the end of each of the corresponding branches on the FUP tree, an evolutionary scenario explaining the divergence of operon structures from a common precursor can be derived (Fig. 10). The exact relationship between major clusters in the FUP tree (Fig. 2) is currently not clear, since the nodes connecting the α-, β-, γ-, κ-, π-, and σ-fimbrial clades are supported by low bootstrapping values. However, the operon structures of κ- and π-fimbriae are related to each other, as both share a core structure composed of genes encoding a major subunit, an usher, and a chaperone. Furthermore, a close relationship of the γ-fimbriae to the κ- and π-fimbriae is indicated by the presence of a PFAM00419 domain exclusively in subunits of operons belonging to these three clades (Fig. 10). These data suggest that members of the γ-, κ-, and π-fimbrial clades form a monophyletic group, which will be referred to as the γκπ cluster from here on. The resulting subdivision of the chaperone/usher assembly class into four major groups is supported by the presence of distinct protein domains in their respective fimbrial subunits, including PFAM00419 in the γκπ cluster, PFAM06551 in the β-fimbriae, PFAM04449 in the α-fimbriae, and COG5430 in the σ-fimbriae. The γκπ cluster and the β-fimbriae together comprise the previously defined classical chaperone/usher family. The low bootstrapping values of nodes connecting members of the classical chaperone/usher family to the α-fimbriae or the σ-fimbriae made it impossible to infer whether the classical chaperone/usher family represents a monophyletic group (Fig. 2). However, a monophyletic origin of this group is supported by the finding that the chaperones encoded by operons of the classical chaperone/usher family all share a protein domain (PFAM02753) that is absent from chaperones of the α-fimbriae and the σ-fimbriae (Fig. 10).
FIG. 10.
Evolution of fimbrial operons of the chaperone/usher assembly class from a hypothetical ancestor. The predicted functions for each gene product and the sequence homologies to protein families are indicated in the legend at the bottom. For a detailed discussion, see the text.
The α-fimbrial operons are composed of genes encoding a chaperone, a major subunit, an usher, and a tip adhesin. The α-fimbriae are the only group in which the first two genes in the operon comprise a chaperone followed by a subunit, and this gene order is not found in the most divergent members of this group (e.g., the csf operon, encoding CS5 fimbriae of E. coli) (Fig. 3). Based on these observations, it is likely that the operon structure characteristic of most α-fimbriae evolved after this group had already diverged from the other lineages. In contrast, a common core structure can be found in at least some members of the remaining three groups (i.e., β-fimbriae, the γκπ cluster, and σ-fimbriae). The γκπ cluster and σ-fimbriae contain many operons in which a major subunit gene is followed by genes encoding a chaperone, an usher, and a tip adhesin. This operon structure likely gave rise to that found in β-fimbriae, in which a gene encoding a tip adhesin is absent (Fig. 10). These considerations suggest that a major subunit gene followed by genes encoding a chaperone, an usher, and a tip adhesin may represent the ancestral operon structure of the chaperone/usher assembly class. A divergence from an ancestral gene cluster was not apparent from previous comparative studies on fimbrial operons.
Only two representatives of the chaperone/usher assembly class (both members of the σ-fimbriae) are present in bacteria outside the phylum Proteobacteria, one in the phylum Cyanobacteria (Synechocystis sp.) and one in the phylum Deinococcus-Thermus (D. radiodurans). Thus, the chaperone/usher assembly class is largely restricted to the phylum Proteobacteria (367). Within this phylum, fimbrial operons belonging to the σ-fimbrial clade are distributed most widely, with representatives being present in the Alpha-, Beta-, Gamma-, and Deltaproteobacteria. In contrast, representatives of the α-, β-, γ-, κ-, and π-fimbriae are restricted to two classes, the Beta- and Gammaproteobacteria. This distribution suggests that σ-fimbriae may be phylogenetically older than the other fimbrial clades, thus making COG5430 a likely candidate for a protein domain present in fimbrial subunits of a hypothetical ancestor of the chaperone/usher assembly class (Fig. 10).
Fimbrial operons contain conserved subunit protein domains characteristic of either the α-fimbriae (PFAM04449), the β-fimbriae (PFAM06551), the γκπ cluster (PFAM00419), or the σ-fimbriae (COG5430) but never a mixture thereof, suggesting that the diversity in operon structures found within each clade never involved lateral exchange of subunit genes among these four major groups. Major fimbrial subunits and the C-terminal domains of tip adhesins both have an incomplete Ig-like fold containing a cleft in which a donor β-strand is inserted to incorporate the subunit into a fimbrial filament (Fig. 1). It is thus not surprising that the conserved protein domain, which is present in the major fimbrial subunit, is also commonly present in the C-terminal domain of the corresponding tip adhesin (Fig. 10). The N-terminal domain of tip adhesins contains the receptor-binding site and also has an Ig-like fold (47). With respect to the α-, β-, γ-, κ-, and π-fimbriae, the N-terminal domain of tip adhesins never contains the conserved protein domain present in its C terminus. Interestingly, some operons belonging to the σ-fimbriae, the most archaic group within the chaperone/usher assembly class, encode tip adhesins that contain COG5430 domains in both the C-terminal and the N-terminal domains of their tip adhesin (Fig. 9). This observation suggests that tip adhesins arose from a gene fusion event involving two fimbrial subunit genes.
A phylogenetic tree constructed by comparing chaperone proteins is shown in Fig. 11. The phylogenetic analysis of chaperone proteins suggests a division into six groups that are in most cases identical to the α-, β-, γ-, κ-, π-, and σ-fimbriae. The only exception is the classification of CsfB and CsfF (the chaperones of CS5 fimbriae) as a sister group to the α- and σ-fimbriae (Fig. 11). In the FUP classification, CsfC (the usher of CS5 fimbriae) is a member of the α-fimbriae (Fig. 3). Overall, an analysis of chaperone proteins provides compelling support for the proposed FUP nomenclature. Although sequence comparisons of chaperone proteins yielded similar results, an analysis of the FUP family is better suited for deriving a classification scheme, because operons encoding more than one chaperone complicate the analysis. A closer examination of fimbrial operons that contain more than one chaperone shows that in most cases, chaperones encoded by the same operon form sister groups on the chaperone tree (Fig. 11). These data suggest that the evolution of the respective fimbrial operons involved a duplication of their chaperone gene. One notable exception is a subgroup of γ2-fimbrial operons, including csw, ECP_3811 to ECP_3817, fas, and fot (Fig. 3). These operons encode three chaperones each, only one of which groups with other chaperones of the γ2-fimbriae. The remaining two chaperones form a distinct branch on the chaperone tree that does not belong to any group defined in the FUP tree (Fig. 11). A possible interpretation of this result is that the respective γ2 operons evolved by acquiring (and subsequently duplicating) a chaperone gene from a donor that does not resemble any of the currently sequenced members of the chaperone/usher assembly class. This represents the only example detected in our analysis in which an operon structure may have been altered by horizontal gene transfer.
CONCLUSIONS
Assembly proteins (i.e., chaperones and ushers) show better sequence conservation than structural subunits and thus are superior for generating trees that illustrate the phylogenetic relationships between divergent members of the chaperone/usher assembly class. This and a previous study (367) defined fimbrial clades by nodes on an FUP family tree (Fig. 2) because all chaperone/usher systems contain only a single usher gene, while multiple genes encoding chaperones may be present. An FUP-based definition of the α-, β-, γ-, κ-, π-, and σ-fimbrial clades is supported by (i) similarities in operon structures within each group (Fig. 3 to 9), (ii) the presence of distinct conserved protein domains in major subunits of different clades (Fig. 10), (iii) the definition of almost identical fimbrial clades using a chaperone tree (Fig. 11), and (iv) the formation of clusters of operons encoding similar surface structures.
The construction of FUP trees similar to the one shown in Fig. 2 is not a rapid and convenient method for classifying new fimbrial operons identified by whole-genome sequencing. However, our analysis suggests that results from a simple BLAST search may be sufficient for the placement of a putative fimbrial operon in our classification scheme (Table 1). This method may not always succeed in distinguishing between members of clades within the γ-fimbriae, because some groups (particularly γ1- and γ4-fimbriae) share similar characteristics. However, Table 1 illustrates that information on gene order and conserved protein domains found in fimbrial subunits will be sufficient for assigning a new operon to either the α-, β-, γ-, κ-, π-, or σ-fimbrial clades. In conclusion, the classification scheme proposed in this article (Table 1) delineates for the first time a simple and convenient method for assigning newly discovered chaperone/usher systems to one of six major phylogenetic clades (Fig. 2).
TABLE 1.
FUP-based classification scheme for fimbrial operons
| Major division | Clade | Subclade | Fig. | Alternate designation | Domain(s) in subunit | Domain(s) in chaperone | Core operon structurea | Other feature(s) |
|---|---|---|---|---|---|---|---|---|
| Alternate chaperone/usher family | α | 3 | Class 5 fimbriae, FUP cluster 8 | PFAM04449, PFAM07434 | COG3121 or no domain | CMUT | None | |
| Classical chaperone/ | β | 4a | FUP cluster 7 | PFAM06551 | COG3121, PFAM00345, | MCU | No tip adhesin | |
| usher family | γ | γ3 | 5 | FGL chaperone subfamily, Class 2 fimbriae, FUP clusters 4 and 5 | PFAM04619, PFAM06443, or PFAM05775 | and PFAM02753 | (M)CU(M) | No tip adhesin |
| γ1 | 5 | FUP cluster 1 | PFAM00419, COG3539 | MCUT | None | |||
| γ2 | 5 | FUP cluster 2 | MCCU | Three chaperones | ||||
| γ4 | 6 | FUP cluster 10 | MCUT | None | ||||
| π | 8 | FUP cluster 3 | MUCT | None | ||||
| κ | 7 | FUP cluster 6 | PFAM00419, COG3539, or PFAM02432 | MUC | No tip adhesin | |||
| Archaic chaperone/usher systems | σ | 9 | FUP cluster 9 | COG5430 | PFAM00345 and/or COG3121 | MCUT | None |
M, major subunit; C, chaperone; U, usher; T, tip adhesin.
Acknowledgments
Work in the laboratory of A.J.B. was supported by USDA/NRICGP grant no. 2002-35204-12247 and Public Health Service grants AI040124, AI044170, and AI065534.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Abe, N., K. Moriishi, M. Saito, and M. Naiki. 1993. Confirmed nucleotide sequence of fanF of Escherichia coli K99 fimbriae. Jpn. J. Vet. Res. 41:97-99. [PubMed] [Google Scholar]
- 2.Adams, L. M., C. P. Simmons, L. Rezmann, R. A. Strugnell, and R. M. Robins-Browne. 1997. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect. Immun. 65:5222-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmer, B. M., M. Tran, and F. Heffron. 1999. The virulence plasmid of Salmonella typhimurium is self-transmissible. J. Bacteriol. 181:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahrens, R., M. Ott, A. Ritter, H. Hoschutzky, T. Buhler, F. Lottspeich, G. J. Boulnois, K. Jann, and J. Hacker. 1993. Genetic analysis of the gene cluster encoding nonfimbrial adhesin I from an Escherichia coli uropathogen. Infect. Immun. 61:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, B. L., G. F. Gerlach, and S. Clegg. 1991. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 173:916-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 7.Altboum, Z., M. M. Levine, J. E. Galen, and E. M. Barry. 2003. Genetic characterization and immunogenicity of coli surface antigen 4 from enterotoxigenic Escherichia coli when it is expressed in a Shigella live-vector strain. Infect. Immun. 71:1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anantha, R. P., A. L. McVeigh, L. H. Lee, M. K. Agnew, F. J. Cassels, D. A. Scott, T. S. Whittam, and S. J. Savarino. 2004. Evolutionary and functional relationships of colonization factor antigen I and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect. Immun. 72:7190-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson, K. L., J. Billington, D. Pettigrew, E. Cota, P. Simpson, P. Roversi, H. A. Chen, P. Urvil, L. du Merle, P. N. Barlow, M. E. Medof, R. A. Smith, B. Nowicki, C. Le Bouguenec, S. M. Lea, and S. Matthews. 2004. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol. Cell 15:647-657. [DOI] [PubMed] [Google Scholar]
- 10.Baddour, L. M., G. D. Christensen, W. A. Simpson, and E. H. Beachey. 1990. Microbial adherence, p. 9-25. In G. L. Mandell, R. G. Douglas, Jr., and J. E. Bennet (ed.), Principles and practice of infectious disease, 3rd ed., vol. 2. Churchill Livingston, New York, NY. [Google Scholar]
- 11.Badger, J. H., T. R. Hoover, Y. V. Brun, R. M. Weiner, M. T. Laub, G. Alexandre, J. Mrazek, Q. Ren, I. T. Paulsen, K. E. Nelson, H. M. Khouri, D. Radune, J. Sosa, R. J. Dodson, S. A. Sullivan, M. J. Rosovitz, R. Madupu, L. M. Brinkac, A. S. Durkin, S. C. Daugherty, S. P. Kothari, M. G. Giglio, L. Zhou, D. H. Haft, J. D. Selengut, T. M. Davidsen, Q. Yang, N. Zafar, and N. L. Ward. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 188:6841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baga, M., M. Norgren, and S. Normark. 1987. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49:241-251. [DOI] [PubMed] [Google Scholar]
- 13.Bahrani, F. K., S. Cook, R. A. Hull, G. Massad, and H. L. Mobley. 1993. Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infect. Immun. 61:884-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahrani, F. K., G. Massad, C. V. Lockatell, D. E. Johnson, R. G. Russell, J. W. Warren, and H. L. Mobley. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 62:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahrani, F. K., and H. L. Mobley. 1993. Proteus mirabilis MR/P fimbriae: molecular cloning, expression, and nucleotide sequence of the major fimbrial subunit gene. J. Bacteriol. 175:457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahrani, F. K., and H. L. Mobley. 1994. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J. Bacteriol. 176:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahrani-Mougeot, F. K., S. Pancholi, M. Daoust, and M. S. Donnenberg. 2001. Identification of putative urovirulence genes by subtractive cloning. J. Infect. Dis. 183(Suppl. 1):S21-S23. [DOI] [PubMed] [Google Scholar]
- 18.Baker, E. E., H. Sommer, L. E. Foster, E. Meyer, and K. F. Meyer. 1952. Studies on immunization against plague. I. The isolation and characterization of the soluble antigen of Pasteurella pestis. J. Immunol. 68:131-145. [PubMed] [Google Scholar]
- 19.Bakermans, C., A. I. Tsapin, V. Souza-Egipsy, D. A. Gilichinsky, and K. H. Nealson. 2003. Reproduction and metabolism at −10 degrees C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 5:321-326. [DOI] [PubMed] [Google Scholar]
- 20.Bakker, D., C. E. Vader, B. Roosendaal, F. R. Mooi, B. Oudega, and F. K. de Graaf. 1991. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol. Microbiol. 5:875-886. [DOI] [PubMed] [Google Scholar]
- 21.Bakker, D., P. T. Willemsen, L. H. Simons, F. G. van Zijderveld, and F. K. de Graaf. 1992. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol. Microbiol. 6:247-255. [DOI] [PubMed] [Google Scholar]
- 22.Bakker, D., P. T. Willemsen, R. H. Willems, T. T. Huisman, F. R. Mooi, B. Oudega, F. Stegehuis, and F. K. de Graaf. 1992. Identification of minor fimbrial subunits involved in biosynthesis of K88 fimbriae. J. Bacteriol. 174:6350-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardwell, J. C. A. 1994. Building bridges: disulphide bond formation in the cell. Mol. Microbiol. 14:199-205. [DOI] [PubMed] [Google Scholar]
- 25.Barnhart, M. M., J. S. Pinkner, G. E. Soto, F. G. Sauer, S. Langermann, G. Waksman, C. Frieden, and S. J. Hultgren. 2000. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl. Acad. Sci. USA 97:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnhart, M. M., F. G. Sauer, J. S. Pinkner, and S. J. Hultgren. 2003. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. J. Bacteriol. 185:2723-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bäumler, A. J., A. J. Gilde, R. M. Tsolis, A. W. van der Velden, B. M. Ahmer, and F. Heffron. 1997. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J. Bacteriol. 179:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bäumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell, K. S., M. Sebaihia, L. Pritchard, M. T. Holden, L. J. Hyman, M. C. Holeva, N. R. Thomson, S. D. Bentley, L. J. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. A. Quail, M. Sanders, D. Walker, S. Whitehead, G. P. Salmond, P. R. Birch, J. Parkhill, and I. K. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101:11105-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger, C. N., O. Billker, T. F. Meyer, A. L. Servin, and I. Kansau. 2004. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC). Mol. Microbiol. 52:963-983. [DOI] [PubMed] [Google Scholar]
- 32.Bernier, C., P. Gounon, and C. Le Bouguenec. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70:4302-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Colladovides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 34.Bock, K., M. E. Breimer, A. Brignole, G. C. Hansson, K.-A. Karlsson, G. Larson, H. Leffler, B. E. Samuelsson, N. Strömberg, C. S. Edén, and J. Thurin. 1985. Specificity of binding of a strain of uropathogenic Escherichia coli to Galα1→Gal-containing glycosphingolipids. J. Biol. Chem. 260:8545-8551. [PubMed] [Google Scholar]
- 35.Bonci, A., A. Chiesurin, P. Muscas, and G. M. Rossolini. 1997. Relatedness and phylogeny within the family of periplasmic chaperones involved in the assembly of pili or capsule-like structures of gram-negative bacteria. J. Mol. Evol. 44:299-309. [DOI] [PubMed] [Google Scholar]
- 36.Bouwman, C. W., M. Kohli, A. Killoran, G. A. Touchie, R. J. Kadner, and N. L. Martin. 2003. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J. Bacteriol. 185:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyd, E. F., and D. L. Hartl. 1997. Recent horizontal transmission of plasmids between natural populations of Escherichia coli and Salmonella enterica. J. Bacteriol. 179:1622-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd, E. F., C. W. Hill, S. M. Rich, and D. L. Hartl. 1996. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics 143:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boylan, M., C. J. Smyth, and J. R. Scott. 1988. Nucleotide sequence of the gene encoding the major subunit of CS3 fimbriae of enterotoxigenic Escherichia coli. Infect. Immun. 56:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley, D. E. 1985. Transfer systems of K88 and K99 plasmids. Plasmid 13:118-128. [DOI] [PubMed] [Google Scholar]
- 41.Brinton, C. C., Jr. 1959. Non-flagellar appendages of bacteria. Nature 183:782-786. [DOI] [PubMed] [Google Scholar]
- 42.Brinton, C. C., Jr. 1965. The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans. N. Y. Acad. Sci. 27:1003-1054. [DOI] [PubMed] [Google Scholar]
- 43.Bruant, G., N. Gousset, R. Quentin, and A. Rosenau. 2002. Fimbrial ghf gene cluster of genital strains of Haemophilus spp. Infect. Immun. 70:5438-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buckles, E. L., F. K. Bahrani-Mougeot, A. Molina, C. V. Lockatell, D. E. Johnson, C. B. Drachenberg, V. Burland, F. R. Blattner, and M. S. Donnenberg. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72:3890-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bullitt, E., and L. Makowski. 1995. Structural polymorphism of bacterial adhesion pili. Nature 373:164-167. [DOI] [PubMed] [Google Scholar]
- 47.Buts, L., J. Bouckaert, E. De Genst, R. Loris, S. Oscarson, M. Lahmann, J. Messens, E. Brosens, L. Wyns, and H. De Greve. 2003. The fimbrial adhesin F17-G of enterotoxigenic Escherichia coli has an immunoglobulin-like lectin domain that binds N-acetylglucosamine. Mol. Microbiol. 49:705-715. [DOI] [PubMed] [Google Scholar]
- 48.Cantey, J. R., R. K. Blake, J. R. Williford, and S. L. Moseley. 1999. Characterization of the Escherichia coli AF/R1 pilus operon: novel genes necessary for transcriptional regulation and for pilus-mediated adherence. Infect. Immun. 67:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao, J., A. S. Khan, M. E. Bayer, and D. M. Schifferli. 1995. Ordered translocation of 987P fimbrial subunits through the outer membrane of Escherichia coli. J. Bacteriol. 177:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capitani, G., O. Eidam, and M. G. Grutter. 2006. Evidence for a novel domain of bacterial outer membrane ushers. Proteins 65:816-823. [DOI] [PubMed] [Google Scholar]
- 51.Cassels, F. J., and M. K. Wolf. 1995. Colonization factors of diarrheagenic E. coli and their intestinal receptors. J. Ind. Microbiol. 15:214-226. [DOI] [PubMed] [Google Scholar]
- 52.Castano, I., F. Bastarrachea, and A. A. Covarrubias. 1988. gltBDF operon of Escherichia coli. J. Bacteriol. 170:821-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castano, I., N. Flores, F. Valle, A. A. Covarrubias, and F. Bolivar. 1992. gltF, a member of the gltBDF operon of Escherichia coli, is involved in nitrogen-regulated gene expression. Mol. Microbiol. 6:2733-2741. [DOI] [PubMed] [Google Scholar]
- 54.Chain, P. S., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agullo, V. L. Reyes, L. Hauser, M. Cordova, L. Gomez, M. Gonzalez, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 103:15280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 103:5977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi, B. K., and D. M. Schifferli. 2001. Characterization of FasG segments required for 987P fimbria-mediated binding to piglet glycoprotein receptors. Infect. Immun. 69:6625-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi, B. K., and D. M. Schifferli. 1999. Lysine residue 117 of the FasG adhesin of enterotoxigenic Escherichia coli is essential for binding of 987P fimbriae to sulfatide. Infect. Immun. 67:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 60.Clark, C. A., M. W. Heuzenroeder, and P. A. Manning. 1992. Colonization factor antigen CFA/IV (PCF8775) of human enterotoxigenic Escherichia coli: nucleotide sequence of the CS5 determinant. Infect. Immun. 60:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clouthier, S. C., K. H. Muller, J. L. Doran, S. K. Collinson, and W. W. Kay. 1993. Characterization of three fimbrial genes, sefABC, of Salmonella enteritidis. J. Bacteriol. 175:2523-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 63.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Connor, E. M., and M. R. Loeb. 1983. A hemadsorption method for detection of colonies of Haemophilus influenzae type b expressing fimbriae. J. Infect. Dis. 148:855-860. [DOI] [PubMed] [Google Scholar]
- 65.Cota, E., C. Jones, P. Simpson, H. Altroff, K. L. Anderson, L. du Merle, J. Guignot, A. Servin, C. Le Bouguenec, H. Mardon, and S. Matthews. 2006. The solution structure of the invasive tip complex from Afa/Dr fibrils. Mol. Microbiol. 62:356-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daraselia, N., D. Dernovoy, Y. Tian, M. Borodovsky, R. Tatusov, and T. Tatusova. 2003. Reannotation of Shewanella oneidensis genome. Omics 7:171-175. [DOI] [PubMed] [Google Scholar]
- 67.Darfeuille-Michaud, A., D. Aubel, G. Chauviere, C. Rich, M. Bourges, A. Servin, and B. Joly. 1990. Adhesion of enterotoxigenic Escherichia coli to the human colon carcinoma cell line Caco-2 in culture. Infect. Immun. 58:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das, M., A. Hart-Van Tassell, P. T. Urvil, S. Lea, D. Pettigrew, K. L. Anderson, A. Samet, J. Kur, S. Matthews, S. Nowicki, V. Popov, P. Goluszko, and B. J. Nowicki. 2005. Hydrophilic domain II of Escherichia coli Dr fimbriae facilitates cell invasion. Infect. Immun. 73:6119-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 70.de Almeida, D. F., M. Hungria, C. T. Guimaraes, R. V. Antonio, F. C. Almeida, L. G. P. de Almeida, R. de Almeida, J. A. Alves-Gomes, E. M. Andrade, J. Araripe, M. F. F. de Araujo, S. Astolfi, V. Azevedo, A. J. Baptista, L. A. M. Bataus, J. D. Batista, A. Belo, C. van den Berg, M. Bogo, S. Bonatto, J. Bordignon, M. M. Brigido, C. A. Brito, M. Brocchi, H. A. Burity, A. A. Camargo, D. D. Cardoso, N. P. Carneiro, B. S. Cavada, L. M. O. Chueire, T. B. Creczynski-Pasa, N. C. da Cunha, N. Fagundes, C. L. Falcao, F. Fantinatti, L. P. Farias, M. S. S. Felipe, L. P. Ferrari, J. A. Ferro, M. T. Ferro, G. R. Franco, N. S. A. de Freitas, L. R. Furlan, R. T. Gazzinelli, E. A. Gomes, P. R. Goncalves, T. B. Grangeiro, D. Grattapaglia, E. C. Grisard, E. S. Hanna, S. N. Jardim, J. Laurino, L. C. T. Leoi, L. F. A. Lima, M. D. Loureiro, M. D. C. P. de Lyra, H. M. F. Madeira, G. P. Manfio, A. Q. Maranhao, W. S. Martins, S. M. Z. di Mauro, S. R. B. de Medeiros, R. D. Meissner, M. A. M. Moreira, F. F. do Nascimento, M. F. Nicolas, J. G. Oliveira, S. C. Oliveira, R. F. C. Paixao, J. A. Parente, F. D. P. Pedrosa, S. D. J. Pena, J. O. Pereira, M. Pereira, L. S. C. Pinto, L. D. Pinto, J. I. R. Porto, D. P. Potrich, C. E. Ramalho-Neto, A. M. M. Reis, L. U. Rigo, E. Rondinelli, E. B. P. do Santos, F. R. Santos, M. P. C. Schneider, H. N. Seuanez, A. M. R. Silva, A. L. D. da Silva, D. W. Silva, R. Silva, I. D. Simoes, D. Simon, C. M. D. Soares, R. D. A. Soares, E. M. Souza, K. R. L. de Souza, R. C. Souza, M. B. R. Steffens, M. Steindel, S. R. Teixeira, et al. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100:11660-11665.14500782 [Google Scholar]
- 71.Dean, E. A. 1990. Comparison of receptors for 987P pili of enterotoxigenic Escherichia coli in the small intestines of neonatal and older pig. Infect. Immun. 58:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dodson, K. W., J. S. Pinkner, T. Rose, G. Magnusson, S. J. Hultgren, and G. J. Waksman. 2001. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105:733-743. [DOI] [PubMed] [Google Scholar]
- 76.Dong, Z., Z. Zhang, S. Li, and C. Huang. 1998. Study on the genetic determinants responsible for expression and assembly of CS3 fimbriae. Chin. J. Biotechnol. 14:67-74. [PubMed] [Google Scholar]
- 77.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 80.Duguid, J. P. 1959. Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 21:271-286. [DOI] [PubMed] [Google Scholar]
- 81.Duguid, J. P., E. S. Anderson, and I. Campbell. 1966. Fimbriae and adhesive properties in salmonellae. J. Pathol. Bacteriol. 92:107-138. [DOI] [PubMed] [Google Scholar]
- 82.Duguid, J. P., I. W. Smith, G. Dempster, and P. N. Edmunds. 1955. Non-flagellar filamentous appendages (fimbriae) in Bacterium coli. J. Pathol. Bacteriol. 70:335-348. [DOI] [PubMed] [Google Scholar]
- 83.Duthy, T. G., P. A. Manning, and M. W. Heuzenroeder. 2001. Characterization of the CsfC and CsfD proteins involved in the biogenesis of CS5 pili from enterotoxigenic Escherichia coli. Microb. Pathog. 31:115-129. [DOI] [PubMed] [Google Scholar]
- 84.Duthy, T. G., P. A. Manning, and M. W. Heuzenroeder. 2002. Identification and characterization of assembly proteins of CS5 pili from enterotoxigenic Escherichia coli. J. Bacteriol. 184:1065-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duthy, T. G., L. H. Staendner, P. A. Manning, and M. W. Heuzenroeder. 1999. CS5 pilus biosynthesis genes from enterotoxigenic Escherichia coli O115:H40. J. Bacteriol. 181:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edwards, R. A., J. Cao, and D. M. Schifferli. 1996. Identification of major and minor chaperone proteins involved in the export of 987P fimbriae. J. Bacteriol. 178:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eisenbrandt, R., M. Kalkum, E. M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 88.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evans, D. G., and D. J. Evans, Jr. 1978. New surface-associated heat-labile colonization factor antigen (CFA/II) produced by enterotoxigenic Escherichia coli of serogroups O6 and O8. Infect. Immun. 21:638-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Evans, D. G., D. J. Evans, Jr., S. Clegg, and J. A. Pauley. 1979. Purification and characterization of the CFA/I antigen of enterotoxigenic Escherichia coli. Infect. Immun. 25:738-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evans, D. G., T. K. Satterwhite, D. J. Evans, Jr., and H. L. DuPont. 1978. Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect. Immun. 19:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 94.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Folkesson, A., A. Advani, S. Sukupolvi, J. D. Pfeifer, S. Normark, and S. Lofdahl. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol. Microbiol. 33:612-622. [DOI] [PubMed] [Google Scholar]
- 96.Friedrich, M. J., N. E. Kinsey, J. Vila, and R. J. Kadner. 1993. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol. Microbiol. 8:543-558. [DOI] [PubMed] [Google Scholar]
- 97.Froehlich, B., E. Holtzapple, T. D. Read, and J. R. Scott. 2004. Horizontal transfer of CS1 pilin genes of enterotoxigenic Escherichia coli. J. Bacteriol. 186:3230-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Froehlich, B., J. Parkhill, M. Sanders, M. A. Quail, and J. R. Scott. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Froehlich, B. J., A. Karakashian, L. R. Melsen, J. C. Wakefield, and J. R. Scott. 1994. CooC and CooD are required for assembly of CS1 pili. Mol. Microbiol. 12:387-401. [DOI] [PubMed] [Google Scholar]
- 100.Froehlich, B. J., A. Karakashian, H. Sakellaris, and J. R. Scott. 1995. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect. Immun. 63:4849-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gaastra, W., H. Sommerfelt, L. van Dijk, J. G. Kusters, A. M. Svennerholm, and H. M. Grewal. 2002. Antigenic variation within the subunit protein of members of the colonization factor antigen I group of fimbrial proteins in human enterotoxigenic Escherichia coli. Int. J. Med. Microbiol. 292:43-50. [DOI] [PubMed] [Google Scholar]
- 102.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 103.Garcia, M. I., P. Gounon, P. Courcoux, A. Labigne, and C. Le Bouguenec. 1996. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol. Microbiol. 19:683-693. [DOI] [PubMed] [Google Scholar]
- 104.Garcia, M. I., M. Jouve, J. P. Nataro, P. Gounon, and C. Le Bouguenec. 2000. Characterization of the AfaD-like family of invasins encoded by pathogenic Escherichia coli associated with intestinal and extra-intestinal infections. FEBS Lett. 479:111-117. [DOI] [PubMed] [Google Scholar]
- 105.Garcia, M. I., A. Labigne, and C. Le Bouguenec. 1994. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J. Bacteriol. 176:7601-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gerlach, G. F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 171:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gillies, R. R., and J. P. Duguid. 1958. The fimbrial antigens of Shigella flexneri. J. Hyg. (London) 56:303-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gilsdorf, J. R., K. W. McCrea, and C. F. Marrs. 1997. Role of pili in Haemophilus influenzae adherence and colonization. Infect. Immun. 65:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilsdorf, J. R., M. Tucci, and C. F. Marrs. 1996. Role of pili in Haemophilus influenzae adherence to, and internalization by, respiratory cells. Pediatr. Res. 39:343-348. [DOI] [PubMed] [Google Scholar]
- 110.Gohl, O., A. Friedrich, M. Hoppert, and B. Averhoff. 2006. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl. Environ. Microbiol. 72:1394-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong, M., and L. Makowski. 1992. Helical structure of P pili from Escherichia coli. Evidence from X-ray fiber diffraction and scanning transmission electron microscopy. J. Mol. Biol. 228:735-742. [DOI] [PubMed] [Google Scholar]
- 114.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 115.Goss, T. J., A. Perez-Matos, and R. A. Bender. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183:6607-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gounon, P., M. Jouve, and C. Le Bouguenec. 2000. Immunocytochemistry of the AfaE adhesin and AfaD invasin produced by pathogenic Escherichia coli strains during interaction of the bacteria with HeLa cells by high-resolution scanning electron microscopy. Microbes Infect. 2:359-365. [DOI] [PubMed] [Google Scholar]
- 117.Gousset, N., A. Rosenau, P. Y. Sizaret, and R. Quentin. 1999. Nucleotide sequences of genes coding for fimbrial proteins in a cryptic genospecies of Haemophilus spp. isolated from neonatal and genital tract infections. Infect. Immun. 67:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grassl, G., B. Bufe, B. Muller, M. Rosel, and D. Kleiner. 1999. Characterization of the gltF gene product of Escherichia coli. FEMS Microbiol. Lett. 179:79-84. [DOI] [PubMed] [Google Scholar]
- 119.Greenberg, D. E., L. Ding, A. M. Zelazny, F. Stock, A. Wong, V. L. Anderson, G. Miller, D. E. Kleiner, A. R. Tenorio, L. Brinster, D. W. Dorward, P. R. Murray, and S. M. Holland. 2006. A novel bacterium associated with lymphadenitis in a patient with chronic granulomatous disease. PLoS Pathog. 2:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grewal, H. M., H. Valvatne, M. K. Bhan, L. van Dijk, W. Gaastra, and H. Sommerfelt. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect. Immun. 65:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guignot, J., M.-F. Bernet-Camard, C. Poüs, L. Plançon, C. Le Bouguenec, and A. L. Servin. 2001. Polarized entry of uropathogenic Afa/Dr diffusely adhering Escherichia coli strain IH11128 into human epithelial cells: evidence for α5β1 integrin recognition and subsequent internalization through a pathway involving caveolae and dynamic unstable microtubules. Infect. Immun. 69:1856-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hahn, E., P. Wild, U. Hermanns, P. Sebbel, R. Glockshuber, M. Haner, N. Taschner, P. Burkhard, U. Aebi, and S. A. Muller. 2002. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323:845-857. [DOI] [PubMed] [Google Scholar]
- 123.Hahn, E., P. Wild, E. M. Schraner, H. U. Bertschinger, M. Haner, S. A. Muller, and U. Aebi. 2000. Structural analysis of F18 fimbriae expressed by porcine toxigenic Escherichia coli. J. Struct. Biol. 132:241-250. [DOI] [PubMed] [Google Scholar]
- 124.Hales, B. A., H. Beverley-Clarke, N. J. High, K. Jann, R. Perry, J. Goldhar, and G. J. Boulnois. 1988. Molecular cloning and characterisation of the genes for a non-fimbrial adhesin from Escherichia coli. Microb. Pathog. 5:9-17. [DOI] [PubMed] [Google Scholar]
- 125.Hamers, A. M., H. J. Pel, G. A. Willshaw, J. G. Kusters, B. A. van der Zeijst, and W. Gaastra. 1989. The nucleotide sequence of the first two genes of the CFA/I fimbrial operon of human enterotoxigenic Escherichia coli. Microb. Pathog. 6:297-309. [DOI] [PubMed] [Google Scholar]
- 126.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haslam, D. B., T. Boren, P. Falk, D. Ilver, A. Chou, Z. Xu, and S. Normark. 1994. The amino-terminal domain of the P-pilus adhesin determines receptor specificity. Mol. Microbiol. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 128.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 129.He, H., H. A. Snyder, and S. Forst. 2004. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology 150:1439-1446. [DOI] [PubMed] [Google Scholar]
- 130.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 131.Helander, A., H. M. Grewal, W. Gaastra, and A. M. Svennerholm. 1997. Detection and characterization of the coli surface antigen 6 of enterotoxigenic Escherichia coli strains by using monoclonal antibodies. J. Clin. Microbiol. 35:867-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Helander, A., G. C. Hansson, and A. M. Svennerholm. 1997. Binding of enterotoxigenic Escherichia coli to isolated enterocytes and intestinal mucus. Microb. Pathog. 23:335-346. [DOI] [PubMed] [Google Scholar]
- 133.Hess, P., A. Altenhofer, A. S. Khan, N. Daryab, K. S. Kim, J. Hacker, and T. A. Oelschlaeger. 2004. A Salmonella fim homologue in Citrobacter freundii mediates invasion in vitro and crossing of the blood-brain barrier in the rat pup model. Infect. Immun. 72:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 135.Hochhut, B., C. Wilde, G. Balling, B. Middendorf, U. Dobrindt, E. Brzuszkiewicz, G. Gottschalk, E. Carniel, and J. Hacker. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61:584-595. [DOI] [PubMed] [Google Scholar]
- 136.Holden, M. T., R. W. Titball, S. J. Peacock, A. M. Cerdeno-Tarraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thomson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davis, D. DeShazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Barrell, P. C. Oyston, and J. Parkhill. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Holmgren, A., and C. Brändén. 1989. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature 342:248-251. [DOI] [PubMed] [Google Scholar]
- 138.Honarvar, S., B. K. Choi, and D. M. Schifferli. 2003. Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol. Microbiol. 48:157-171. [DOI] [PubMed] [Google Scholar]
- 139.Houwink, A. L., and W. van Iterson. 1950. Electron microscopical observations on bacterial cytology; a study on flagellation. Biochim. Biophys. Acta 5:10-44. [DOI] [PubMed] [Google Scholar]
- 140.Hudault, S., O. B. Spiller, B. P. Morgan, and A. L. Servin. 2004. Human diffusely adhering Escherichia coli expressing Afa/Dr adhesins that use human CD55 (decay-accelerating factor) as a receptor does not bind the rodent and pig analogues of CD55. Infect. Immun. 72:4859-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Bäumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 142.Hung, C. S., J. Bouckaert, D. Hung, J. Pinkner, C. Widberg, A. DeFusco, C. G. Auguste, R. Strouse, S. Langermann, G. Waksman, and S. J. Hultgren. 2002. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44:903-915. [DOI] [PubMed] [Google Scholar]
- 143.Hung, D. L., S. D. Knight, R. M. Woods, J. S. Pinkner, and S. J. Hultgren. 1996. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 15:3792-3805. [PMC free article] [PubMed] [Google Scholar]
- 144.Hurst, M. R., T. R. Glare, T. A. Jackson, and C. W. Ronson. 2000. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182:5127-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hurst, M. R., M. O'Callaghan, and T. R. Glare. 2003. Peripheral sequences of the Serratia entomophila pADAP virulence-associated region. Plasmid 50:213-229. [DOI] [PubMed] [Google Scholar]
- 146.Ideses, D., D. Biran, U. Gophna, O. Levy-Nissenbaum, and E. Z. Ron. 2005. The lpf operon of invasive Escherichia coli. Int. J. Med. Microbiol. 295:227-236. [DOI] [PubMed] [Google Scholar]
- 147.Imberechts, H., H. De Greve, C. Schlicker, H. Bouchet, P. Pohl, G. Charlier, H. Bertschinger, P. Wild, J. Vandekerckhove, J. Van Damme, et al. 1992. Characterization of F107 fimbriae of Escherichia coli 107/86, which causes edema disease in pigs, and nucleotide sequence of the F107 major fimbrial subunit gene, fedA. Infect. Immun. 60:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Imberechts, H., P. Wild, G. Charlier, H. De Greve, P. Lintermans, and P. Pohl. 1996. Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb. Pathog. 21:183-192. [DOI] [PubMed] [Google Scholar]
- 149.Inoue, O. J., J. H. Lee, and R. E. Isaacson. 1993. Transcriptional organization of the Escherichia coli pilus adhesin K99. Mol. Microbiol. 10:607-613. [DOI] [PubMed] [Google Scholar]
- 150.Iriarte, M., J. C. Vanooteghem, I. Delor, R. Diaz, S. Knutton, and G. R. Cornelis. 1993. The Myf fibrillae of Yersinia enterocolitica. Mol. Microbiol. 9:507-520. [DOI] [PubMed] [Google Scholar]
- 151.Isaacson, R. E. 1977. K99 surface antigen of Escherichia coli: purification and partial characterization. Infect. Immun. 15:272-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Isaacson, R. E., B. Nagy, and H. W. Moon. 1977. Colonization of porcine small intestine by Escherichia coli: colonization and adhesion factors of pig enteropathogens that lack K88. J. Infect. Dis. 135:531-539. [DOI] [PubMed] [Google Scholar]
- 153.Isaacson, R. E., and P. Richter. 1981. Escherichia coli 987P pilus: purification and partial characterization. J. Bacteriol. 146:784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jacob-Dubuisson, F., J. Heuser, K. Dodson, S. Normark, and S. J. Hultgren. 1993. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 12:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jacob-Dubuisson, F., J. Pinkner, Z. Xu, R. Striker, A. Padmanhaban, and S. J. Hultgren. 1994. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc. Natl. Acad. Sci. USA 91:11552-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jacobs, A. A., B. H. Simons, and F. K. de Graaf. 1987. The role of lysine-132 and arginine-136 in the receptor-binding domain of the K99 fibrillar subunit. EMBO J. 6:1805-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jagnow, J., and S. Clegg. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397-2405. [DOI] [PubMed] [Google Scholar]
- 158.Jalajakumari, M. B., C. J. Thomas, R. Halter, and P. A. Manning. 1989. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol. Microbiol. 3:1685-1695. [DOI] [PubMed] [Google Scholar]
- 159.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. Deboy, A. S. Durkin, M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 161.Jones, C. H., P. Dexter, A. K. Evans, C. Liu, S. J. Hultgren, and D. E. Hruby. 2002. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 184:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jones, C. H., F. Jacob-Dubuisson, K. Dodson, M. Kuehn, L. Slonim, R. Striker, and S. J. Hultgren. 1992. Adhesin presentation in bacteria requires molecular chaperones and ushers. Infect. Immun. 60:4445-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 92:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jordi, B. J., G. A. Willshaw, B. A. van der Zeijst, and W. Gaastra. 1992. The complete nucleotide sequence of region 1 of the CFA/I fimbrial operon of human enterotoxigenic Escherichia coli. DNA Seq. 2:257-263. [DOI] [PubMed] [Google Scholar]
- 165.Jouve, M., M. I. Garcia, P. Courcoux, A. Labigne, P. Gounon, and C. Le Bouguenec. 1997. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect. Immun. 65:4082-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 167.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 168.Keegan, S. J., C. Graham, D. E. Neal, G. Blum-Oehler, J. N′Dow, J. P. Pearson, and D. L. Gally. 2003. Characterization of Escherichia coli strains causing urinary tract infections in patients with transposed intestinal segments. J. Urol. 169:2382-2387. [DOI] [PubMed] [Google Scholar]
- 169.Kerneis, S., M. F. Bernet, M. H. Coconnier, and A. L. Servin. 1994. Adhesion of human enterotoxigenic Escherichia coli to human mucus secreting HT-29 cell subpopulations in culture. Gut 35:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kerneis, S., G. Chauviere, A. Darfeuille-Michaud, D. Aubel, M. H. Coconnier, B. Joly, and A. L. Servin. 1992. Expression of receptors for enterotoxigenic Escherichia coli during enterocytic differentiation of human polarized intestinal epithelial cells in culture. Infect. Immun. 60:2572-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Khan, A. S., N. C. Johnston, H. Goldfine, and D. M. Schifferli. 1996. Porcine 987P glycolipid receptors on intestinal brush borders and their cognate bacterial ligands. Infect. Immun. 64:3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Khan, A. S., B. Kniep, T. A. Oelschlaeger, I. Van Die, T. Korhonen, and J. Hacker. 2000. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect. Immun. 68:3541-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Khan, A. S., and D. M. Schifferli. 1994. A minor 987P protein different from the structural fimbrial subunit is the adhesin. Infect. Immun. 62:4233-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kisiela, D., A. Sapeta, M. Kuczkowski, T. Stefaniak, A. Wieliczko, and M. Ugorski. 2005. Characterization of FimH adhesins expressed by Salmonella enterica serovar Gallinarum biovars Gallinarum and Pullorum: reconstitution of mannose-binding properties by single amino acid substitution. Infect. Immun. 73:6187-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Klemm, P., B. J. Jorgensen, B. Kreft, and G. Christiansen. 1995. The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J. Bacteriol. 177:621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Klemm, P., B. J. Jorgensen, I. van Die, H. de Ree, and H. Bergmans. 1985. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Gen. Genet. 199:410-414. [DOI] [PubMed] [Google Scholar]
- 178.Klemm, P., I. Orskov, and F. Orskov. 1982. F7 and type 1-like fimbriae from three Escherichia coli strains isolated from urinary tract infections: protein chemical and immunological aspects. Infect. Immun. 36:462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Knutton, S., D. R. Lloyd, D. C. Candy, and A. S. McNeish. 1985. Adhesion of enterotoxigenic Escherichia coli to human small intestinal enterocytes. Infect. Immun. 48:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Knutton, S., M. M. McConnell, B. Rowe, and A. S. McNeish. 1989. Adhesion and ultrastructural properties of human enterotoxigenic Escherichia coli producing colonization factor antigens III and IV. Infect. Immun. 57:3364-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Korhonen, T. K., V. Vaisanen-Rhen, M. Rhen, A. Pere, J. Parkkinen, and J. Finne. 1984. Escherichia coli fimbriae recognizing sialyl galactosides. J. Bacteriol. 159:762-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Korotkova, N., E. Cota, Y. Lebedin, S. Monpouet, J. Guignot, A. L. Servin, S. Matthews, and S. L. Moseley. 2006. A subfamily of Dr adhesins of Escherichia coli bind independently to decay-accelerating factor and the N-domain of carcinoembryonic antigen. J. Biol. Chem. 281:29120-29130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kroncke, K. D., I. Orskov, F. Orskov, B. Jann, and K. Jann. 1990. Electron microscopic study of coexpression of adhesive protein capsules and polysaccharide capsules in Escherichia coli. Infect. Immun. 58:2710-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kubiet, M., R. Ramphal, A. Weber, and A. Smith. 2000. Pilus-mediated adherence of Haemophilus influenzae to human respiratory mucins. Infect. Immun. 68:3362-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kuehn, M. J., J. Heuser, S. Normark, and S. J. Hultgren. 1992. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356:252-255. [DOI] [PubMed] [Google Scholar]
- 186.Kuehn, M. J., S. Normark., and S. J. Hultgren. 1991. Immunoglobulin-like PapD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc. Natl. Acad. Sci. USA 88:10586-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Labigne-Roussel, A., M. A. Schmidt, W. Walz, and S. Falkow. 1985. Genetic organization of the afimbrial adhesin operon and nucleotide sequence from a uropathogenic Escherichia coli gene encoding an afimbrial adhesin. J. Bacteriol. 162:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Labigne-Roussel, A. F., D. Lark, G. Schoolnik, and S. Falkow. 1984. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect. Immun. 46:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lalioui, L., M. Jouve, P. Gounon, and C. Le Bouguenec. 1999. Molecular cloning and characterization of the afa-7 and afa-8 gene clusters encoding afimbrial adhesins in Escherichia coli strains associated with diarrhea or septicemia in calves. Infect. Immun. 67:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lalioui, L., and C. Le Bouguénec. 2001. afa-8 gene cluster is carried by a pathogenicity island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Langstraat, J., M. Bohse, and S. Clegg. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 193.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, et al. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li, H., L. Qian, Z. Chen, D. Thahbot, G. Liu, T. Liu, and D. G. Thanassi. 2004. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 344:1397-1407. [DOI] [PubMed] [Google Scholar]
- 196.Li, X., D. E. Johnson, and H. L. Mobley. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67:2822-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li, X., and H. L. Mobley. 1998. MrpB functions as the terminator for assembly of Proteus mirabilis mannose-resistant Proteus-like fimbriae. Infect. Immun. 66:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li, X., H. Zhao, L. Geymonat, F. Bahrani, D. E. Johnson, and H. L. Mobley. 1997. Proteus mirabilis mannose-resistant, Proteus-like fimbriae: MrpG is located at the fimbrial tip and is required for fimbrial assembly. Infect. Immun. 65:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lindler, L. E., M. S. Klempner, and S. C. Straley. 1990. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect. Immun. 58:2569-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lindler, L. E., and B. D. Tall. 1993. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol. Microbiol. 8:311-324. [DOI] [PubMed] [Google Scholar]
- 201.Locht, C., M. C. Geoffroy, and G. Renauld. 1992. Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J. 11:3175-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Low, A. S., F. Dziva, A. G. Torres, J. L. Martinez, T. Rosser, S. Naylor, K. Spears, N. Holden, A. Mahajan, J. Findlay, J. Sales, D. G. Smith, J. C. Low, M. P. Stevens, and D. L. Gally. 2006. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 74:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Low, A. S., N. Holden, T. Rosser, A. J. Roe, C. Constantinidou, J. L. Hobman, D. G. Smith, J. C. Low, and D. L. Gally. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033-1047. [DOI] [PubMed] [Google Scholar]
- 204.Low, D., B. Braaten, and M. van der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 205.Ludi, S., J. Frey, D. Favre, and M. H. Stoffel. 2006. Assessing the expression of enterotoxigenic Escherichia coli-specific surface antigens in recombinant strains by transmission electron microscopy and immunolabeling. J. Histochem. Cytochem. 54:473-477. [DOI] [PubMed] [Google Scholar]
- 206.Lugering, A., I. Benz, S. Knochenhauer, M. Ruffing, and M. A. Schmidt. 2003. The Pix pilus adhesin of the uropathogenic Escherichia coli strain X2194 (O2:K(−):H6) is related to Pap pili but exhibits a truncated regulatory region. Microbiology 149:1387-1397. [DOI] [PubMed] [Google Scholar]
- 207.Lund, B., F. Lindberg, B. I. Marklund, and S. Normark. 1987. The PapG protein is the alpha-D-galactopyranosyl-(1-4)-beta-D-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 84:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lymberopoulos, M. H., S. Houle, F. Daigle, S. Leveille, A. Bree, M. Moulin-Schouleur, J. R. Johnson, and C. M. Dozois. 2006. Characterization of Stg fimbriae from an avian pathogenic Escherichia coli O78:K80 strain and assessment of their contribution to colonization of the chicken respiratory tract. J. Bacteriol. 188:6449-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 210.Manting, E. H., and A. J. Driessen. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 211.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Massad, G., F. K. Bahrani, and H. L. Mobley. 1994. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect. Immun. 62:1989-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Massad, G., J. F. Fulkerson, Jr., D. C. Watson, and H. L. Mobley. 1996. Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the aft gene cluster. Infect. Immun. 64:4390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Massad, G., and H. L. Mobley. 1994. Genetic organization and complete sequence of the Proteus mirabilis pmf fimbrial operon. Gene 150:101-104. [DOI] [PubMed] [Google Scholar]
- 216.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 217.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 218.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 219.McConnell, M. M., H. Chart, A. M. Field, M. Hibberd, and B. Rowe. 1989. Characterization of a putative colonization factor (PCFO166) of enterotoxigenic Escherichia coli of serogroup O166. J. Gen. Microbiol. 135:1135-1144. [DOI] [PubMed] [Google Scholar]
- 220.McConnell, M. M., M. Hibberd, A. M. Field, H. Chart, and B. Rowe. 1990. Characterization of a new putative colonization factor (CS17) from a human enterotoxigenic Escherichia coli of serotype O114:H21 which produces only heat-labile enterotoxin. J. Infect. Dis. 161:343-347. [DOI] [PubMed] [Google Scholar]
- 221.McCrea, K. W., W. J. Watson, J. R. Gilsdorf, and C. F. Marrs. 1997. Identification of two minor subunits in the pilus of Haemophilus influenzae. J. Bacteriol. 179:4227-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Medigue, C., E. Krin, G. Pascal, V. Barbe, A. Bernsel, P. N. Bertin, F. Cheung, S. Cruveiller, S. D'Amico, A. Duilio, G. Fang, G. Feller, C. Ho, S. Mangenot, G. Marino, J. Nilsson, E. Parrilli, E. P. Rocha, Z. Rouy, A. Sekowska, M. L. Tutino, D. Vallenet, G. von Heijne, and A. Danchin. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Meslet-Cladiere, L. M., A. Pimenta, E. Duchaud, I. B. Holland, and M. A. Blight. 2004. In vivo expression of the mannose-resistant fimbriae of Photorhabdus temperata K122 during insect infection. J. Bacteriol. 186:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mooi, F. R., I. Claassen, D. Bakker, H. Kuipers, and F. K. de Graaf. 1986. Regulation and structure of an Escherichia coli gene coding for an outer membrane protein involved in export of K88ab fimbrial subunits. Nucleic Acids Res. 14:2443-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Morrow, B. J., J. E. Graham, and R. Curtiss III. 1999. Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect. Immun. 67:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Morschhauser, J., V. Vetter, L. Emody, and J. Hacker. 1994. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol. Microbiol. 11:555-566. [DOI] [PubMed] [Google Scholar]
- 227.Mu, X. Q., and E. Bullitt. 2006. Structure and assembly of P-pili: a protruding hinge region used for assembly of a bacterial adhesion filament. Proc. Natl. Acad. Sci. USA 103:9861-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mu, X. Q., E. H. Egelman, and E. Bullitt. 2002. Structure and function of Hib pili from Haemophilus influenzae type b. J. Bacteriol. 184:4868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 230.Nataro, J. P., D. Yikang, J. A. Giron, S. J. Savarino, M. H. Kothary, and R. Hall. 1993. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect. Immun. 61:1126-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Neeser, J. R., A. Chambaz, M. Golliard, H. Link-Amster, V. Fryder, and E. Kolodziejczyk. 1989. Adhesion of colonization factor antigen II-positive enterotoxigenic Escherichia coli strains to human enterocytelike differentiated HT-29 cells: a basis for host-pathogen interactions in the gut. Infect. Immun. 57:3727-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Newton, H. J., J. Sloan, V. Bennett-Wood, L. M. Adams, R. M. Robins-Browne, and E. L. Hartland. 2004. Contribution of long polar fimbriae to the virulence of rabbit-specific enteropathogenic Escherichia coli. Infect. Immun. 72:1230-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ng, T. W., L. Akman, M. Osisami, and D. G. Thanassi. 2004. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186:5321-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nishiyama, M., R. Horst, O. Eidam, T. Herrmann, O. Ignatov, M. Vetsch, P. Bettendorff, I. Jelesarov, M. G. Grutter, K. Wuthrich, R. Glockshuber, and G. Capitani. 2005. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Norgren, M., M. Baga, J. M. Tennent, and S. Normark. 1987. Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol. Microbiol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 236.Nowicki, B., J. P. Barrish, T. Korhonen, R. A. Hull, and S. I. Hull. 1987. Molecular cloning of the Escherichia coli O75X adhesin. Infect. Immun. 55:3168-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nowicki, B., A. Labigne, S. Moseley, R. Hull, S. Hull, and J. Moulds. 1990. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect. Immun. 58:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nowicki, B., J. Moulds, R. Hull, and S. Hull. 1988. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect. Immun. 56:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nowicki, B., C. Svanborg-Eden, R. Hull, and S. Hull. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect. Immun. 57:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nudleman, E., and D. Kaiser. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7:52-62. [DOI] [PubMed] [Google Scholar]
- 241.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Olasz, F., P. Z. Fekete, G. Blum-Oehler, Z. Boldogkoi, and B. Nagy. 2005. Characterization of an F18+ enterotoxigenic Escherichia coli strain from post weaning diarrhoea of swine, and of its conjugative virulence plasmid pTC. FEMS Microbiol. Lett. 244:281-289. [DOI] [PubMed] [Google Scholar]
- 243.Old, D. C., and R. A. Adegbola. 1982. Haemagglutinins and fimbriae of Morganella, Proteus and Providencia. J. Med. Microbiol. 15:551-564. [DOI] [PubMed] [Google Scholar]
- 244.Oremland, R. S., S. E. Hoeft, J. M. Santini, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Orskov, I., and F. Orskov. 1966. Episome-carried surface antigen K88 of Escherichia coli. I. Transmission of the determinant of the K88 antigen and influence on the transfer of chromosomal markers. J. Bacteriol. 91:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Orskov, I., and F. Orskov. 1990. Serologic classification of fimbriae. Curr. Top. Microbiol. Immunol. 151:71-90. [PubMed] [Google Scholar]
- 247.Orskov, I., F. Orskov, H. W. Smith, and W. J. Sojka. 1975. The establishment of K99, a thermolabile, transmissible Escherichia coli K antigen, previously called “Kco,” possessed by calf and lamb enteropathogenic strains. Acta Pathol. Microbiol. Scand. B 83:31-36. [DOI] [PubMed] [Google Scholar]
- 248.Ottow, J. C. 1975. Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 29:79-108. [DOI] [PubMed] [Google Scholar]
- 249.Ou, J. T., M.-Y. Lin, and H.-L. Chao. 1994. Presence of F-like ori-T base-pair sequences on the virulence plasmids of Salmonella serovars Gallinarum, Enteritidis, and Typhimurium, but absent in those of Choleraesuis and Dublin. Microb. Pathog. 17:13-21. [DOI] [PubMed] [Google Scholar]
- 250.Oudega, B., and F. K. De Graaf. 1988. Genetic organization and biogenesis of adhesive fimbriae of Escherichia coli. Antonie Leeuwenhoek 54:285-299. [DOI] [PubMed] [Google Scholar]
- 251.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 252.Paranchych, W., and L. S. Frost. 1988. The physiology and biochemistry of pili. Adv. Microb. Physiol. 29:53-114. [DOI] [PubMed] [Google Scholar]
- 253.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 254.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 255.Parkkinen, J., G. N. Rogers, T. Korhonen, W. Dahr, and J. Finne. 1986. Identification of the O-linked sialyloligosaccharides of glycophorin A as the erythrocyte receptors for S-fimbriated Escherichia coli. Infect. Immun. 54:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson III, L. S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pere, A., M. Leinonen, V. Vaisanen-Rhen, M. Rhen, and T. K. Korhonen. 1985. Occurrence of type-1C fimbriae on Escherichia coli strains isolated from human extraintestinal infections. J. Gen. Microbiol. 131:1705-1711. [DOI] [PubMed] [Google Scholar]
- 258.Pere, A., B. Nowicki, H. Saxen, A. Siitonen, and T. K. Korhonen. 1987. Expression of P, type-1, and type-1C fimbriae of Escherichia coli in the urine of patients with acute urinary tract infection. J. Infect. Dis. 156:567-574. [DOI] [PubMed] [Google Scholar]
- 259.Perez-Casal, J., J. S. Swartley, and J. R. Scott. 1990. Gene encoding the major subunit of CS1 pili of human enterotoxigenic Escherichia coli. Infect. Immun. 58:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pettigrew, D., K. L. Anderson, J. Billington, E. Cota, P. Simpson, P. Urvil, F. Rabuzin, P. Roversi, B. Nowicki, L. du Merle, C. Le Bouguenec, S. Matthews, and S. M. Lea. 2004. High resolution studies of the Afa/Dr adhesin DraE and its interaction with chloramphenicol. J. Biol. Chem. 279:46851-46857. [DOI] [PubMed] [Google Scholar]
- 261.Piatek, R., B. Zalewska, K. Bury, and J. Kur. 2005. The chaperone-usher pathway of bacterial adhesin biogenesis—from molecular mechanism to strategies of anti-bacterial prevention and modern vaccine design. Acta Biochim. Pol. 52:639-646. [PubMed] [Google Scholar]
- 262.Piatek, R., B. Zalewska, O. Kolaj, M. Ferens, B. Nowicki, and J. Kur. 2005. Molecular aspects of biogenesis of Escherichia coli Dr fimbriae: characterization of DraB-DraE complexes. Infect. Immun. 73:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Potter, E. Schwartz, A. Strittmatter, I. Voss, G. Gottschalk, A. Steinbuchel, B. Friedrich, and B. Bowien. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 264.Poole, S. T., A. L. McVeigh, R. P. Anantha, L. H. Lee, Y. M. Akay, E. A. Pontzer, D. A. Scott, E. Bullitt, and S. J. Savarino. 2007. Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol. Microbiol. 63:1372-1384. [DOI] [PubMed] [Google Scholar]
- 265.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Pouttu, R., B. Westerlund-Wikstrom, H. Lang, K. Alsti, R. Virkola, U. Saarela, A. Siitonen, N. Kalkkinen, and T. K. Korhonen. 2001. matB, a common fimbrillin gene of Escherichia coli, expressed in a genetically conserved, virulent clonal group. J. Bacteriol. 183:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pugsley, A. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Raina, S., D. Missiakas, L. Baird, S. Kumar, and C. Georgopoulos. 1993. Identification and transcriptional analysis of the Escherichia coli htrE operon which is homologous to pap and related pilin operons. J. Bacteriol. 175:5009-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Read, T. D., M. Dowdell, S. W. Satola, and M. M. Farley. 1996. Duplication of pilus gene complexes of Haemophilus influenzae biogroup aegyptius. J. Bacteriol. 178:6564-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Reis, M. H., M. Heloiza, T. Affonso, L. R. Trabulsi, A. J. Mazaitis, R. Maas, and W. K. Maas. 1980. Transfer of a CFA/I-ST plasmid promoted by a conjugative plasmid in a strain of Escherichia coli of serotype O128ac:H12. Infect. Immun. 29:140-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Riegman, N., R. Kusters, H. Van Veggel, H. Bergmans, P. Van Bergen en Henegouwen, J. Hacker, and I. Van Die. 1990. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J. Bacteriol. 172:1114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Rippinger, P., H. U. Bertschinger, H. Imberechts, B. Nagy, I. Sorg, M. Stamm, P. Wild, and W. Wittig. 1995. Designations F18ab and F18ac for the related fimbrial types F107, 2134P and 8813 of Escherichia coli isolated from porcine postweaning diarrhoea and from oedema disease. Vet. Microbiol. 45:281-295. [DOI] [PubMed] [Google Scholar]
- 273.Roberts, J. A., B.-I. Marklund, D. Ilver, D. Haslam, M. B. Kaack, G. Baskin, M. Louis, R. Mollby, J. Winberg, and S. Normark. 1994. The Gala(1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. USA 91:11889-11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Roos, V., M. A. Schembri, G. C. Ulett, and P. Klemm. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799-1806. [DOI] [PubMed] [Google Scholar]
- 275.Roosendaal, B., and F. K. de Graaf. 1989. The nucleotide sequence of the fanD gene encoding the large outer membrane protein involved in the biosynthesis of K99 fimbriae. Nucleic Acids Res. 17:1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Roosendaal, B., W. Gaastra, and F. K. de Graaf. 1984. The nucleotide-sequence of the gene encoding the K99 subunit of entero-toxigenic Escherichia-coli. FEMS Microbiol. Lett. 22:253-258. [Google Scholar]
- 277.Roosendaal, E., A. A. Jacobs, P. Rathman, C. Sondermeyer, F. Stegehuis, B. Oudega, and F. K. de Graaf. 1987. Primary structure and subcellular localization of two fimbrial subunit-like proteins involved in the biosynthesis of K99 fibrillae. Mol. Microbiol. 1:211-217. [DOI] [PubMed] [Google Scholar]
- 278.Ruer, S., S. Stender, A. Filloux, and S. de Bentzmann. 2007. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J. Bacteriol. 189:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Russell, P. W., and P. E. Orndorff. 1992. Lesions in two Escherichia coli type 1 pilus genes alter pilus number and length without affecting receptor binding. J. Bacteriol. 174:5923-5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rychlik, I., M. A. Lovell, and P. A. Barrow. 1998. The presence of genes homologous to the K88 genes faeH and faeI on the virulence plasmid of Salmonella gallinarum. FEMS Microbiol. Lett. 159:255-260. [DOI] [PubMed] [Google Scholar]
- 281.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sajjan, U. S., H. Xie, M. D. Lefebre, M. A. Valvano, and J. F. Forstner. 2003. Identification and molecular analysis of cable pilus biosynthesis genes in Burkholderia cepacia. Microbiology 149:961-971. [DOI] [PubMed] [Google Scholar]
- 283.Sakai, T., K. Kanai, K. Osatomi, and K. Yoshikoshi. 2004. Identification and characterization of a fimbrial gene cluster of Edwardsiella tarda expressing mannose-resistant hemagglutination. Fish Pathol. 39:87-93. [Google Scholar]
- 284.Sakai, T., K. Kanai, K. Osatomi, and K. Yoshikoshi. 2003. Identification of a 19.3-kDa protein in MRHA-positive Edwardsiella tarda: putative fimbrial major subunit. FEMS Microbiol. Lett. 226:127-133. [DOI] [PubMed] [Google Scholar]
- 285.Sakellaris, H., D. P. Balding, and J. R. Scott. 1996. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol. Microbiol. 21:529-541. [DOI] [PubMed] [Google Scholar]
- 286.Sakellaris, H., and J. R. Scott. 1998. New tools in an old trade: CS1 pilus morphogenesis. Mol. Microbiol. 30:681-687. [DOI] [PubMed] [Google Scholar]
- 287.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 288.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sauer, F. G., K. Fütterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 290.Sauer, F. G., H. Remaut, S. J. Hultgren, and G. Waksman. 2004. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta 1694:259-267. [DOI] [PubMed] [Google Scholar]
- 291.Saulino, E. T., E. Bullitt, and S. J. Hultgren. 2000. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. USA 97:9240-9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Saulino, E. T., D. G. Thanassi, J. S. Pinkner, and S. J. Hultgren. 1998. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17:2177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Savarino, S. J., P. Fox, Y. Deng, and J. P. Nataro. 1994. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J. Bacteriol. 176:4949-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Scaletsky, I. C., J. Michalski, A. G. Torres, M. V. Dulguer, and J. B. Kaper. 2005. Identification and characterization of the locus for diffuse adherence, which encodes a novel afimbrial adhesin found in atypical enteropathogenic Escherichia coli. Infect. Immun. 73:4753-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schifferli, D. M., and M. A. Alrutz. 1994. Permissive linker insertion sites in the outer membrane protein of 987P fimbriae of Escherichia coli. J. Bacteriol. 176:1099-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schifferli, D. M., E. H. Beachey, and R. K. Taylor. 1990. The 987P fimbrial gene cluster of enterotoxigenic Escherichia coli is plasmid encoded. Infect. Immun. 58:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Schmoll, T., H. Hoschutzky, J. Morschhauser, F. Lottspeich, K. Jann, and J. Hacker. 1989. Analysis of genes coding for the sialic acid-binding adhesin and two other minor fimbrial subunits of the S-fimbrial adhesin determinant of Escherichia coli. Mol. Microbiol. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 298.Schneiker, S., V. A. Martins dos Santos, D. Bartels, T. Bekel, M. Brecht, J. Buhrmester, T. N. Chernikova, R. Denaro, M. Ferrer, C. Gertler, A. Goesmann, O. V. Golyshina, F. Kaminski, A. N. Khachane, S. Lang, B. Linke, A. C. McHardy, F. Meyer, T. Nechitaylo, A. Puhler, D. Regenhardt, O. Rupp, J. S. Sabirova, W. Selbitschka, M. M. Yakimov, K. N. Timmis, F. J. Vorholter, S. Weidner, O. Kaiser, and P. N. Golyshin. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Servin, A. L. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 18:264-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shipley, P. L., C. L. Gyles, and S. Falkow. 1978. Characterization of plasmids that encode for the K88 colonization antigen. Infect. Immun. 20:559-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Siitonen, A., R. Martikainen, R. Ikaheimo, J. Palmgren, and P. H. Makela. 1993. Virulence-associated characteristics of Escherichia coli in urinary tract infection: a statistical analysis with special attention to type 1C fimbriation. Microb. Pathog. 15:65-75. [DOI] [PubMed] [Google Scholar]
- 303.Simons, B. L., P. T. Willemsen, D. Bakker, B. Roosendaal, F. K. De Graaf, and B. Oudega. 1990. Structure, localization and function of FanF, a minor component of K99 fibrillae of enterotoxigenic Escherichia coli. Mol. Microbiol. 4:2041-2050. [DOI] [PubMed] [Google Scholar]
- 304.Simons, L. H., P. T. Willemsen, D. Bakker, F. K. de Graaf, and B. Oudega. 1991. Localization and function of FanH and FanG, minor components of K99 fimbriae of enterotoxigenic Escherichia coli. Microb. Pathog. 11:325-336. [DOI] [PubMed] [Google Scholar]
- 305.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. Nhani, Jr., F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. Pereira, Jr., J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, M. A. J. de Rosa, V. E. de Rosa, Jr., R. G. de Sá, R. V. Santelli, H. E. Sawasaki, A. C. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-159. [DOI] [PubMed] [Google Scholar]
- 306.Slonim, L. N., J. S. Pinkner, C. I. Branden, and S. J. Hultgren. 1992. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 11:4747-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Smeds, A., K. Hemmann, M. Jakava-Viljanen, S. Pelkonen, H. Imberechts, and A. Palva. 2001. Characterization of the adhesin of Escherichia coli F18 fimbriae. Infect. Immun. 69:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Smith, H. R., S. M. Scotland, and B. Rowe. 1983. Plasmids that code for production of colonization factor antigen II and enterotoxin production in strains of Escherichia coli. Infect. Immun. 40:1236-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Sokurenko, E. V., H. S. Courtney, D. E. Ohman, P. Klemm, and D. L. Hasty. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Spears, P. A., L. M. Temple, D. M. Miyamoto, D. J. Maskell, and P. E. Orndorff. 2003. Unexpected similarities between Bordetella avium and other pathogenic bordetellae. Infect. Immun. 71:2591-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Stapleton, A. E., M. R. Stroud, S. I. Hakomori, and W. E. Stamm. 1998. The globoseries glycosphingolipid sialosyl galactosyl globoside is found in urinary tract tissues and is a preferred binding receptor in vitro for uropathogenic Escherichia coli expressing pap-encoded adhesins. Infect. Immun. 66:3856-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Steven, A. C., M. E. Bisher, B. L. Trus, D. Thomas, J. M. Zhang, and J. L. Cowell. 1986. Helical structure of Bordetella pertussis fimbriae. J. Bacteriol. 167:968-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.St. Geme, J. W., J. S. Pinkner III, G. P. Krasan, J. Heuser, E. Bullitt, A. L. Smith, and S. J. Hultgren. 1996. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc. Natl. Acad. Sci. USA 93:11913-11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Stirm, S., F. Orskov, I. Orskov, and A. Birch-Andersen. 1967. Episome-carried surface antigen K88 of Escherichia coli. 3. Morphology. J. Bacteriol. 93:740-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Stirm, S., F. Orskov, I. Orskov, and B. Mansa. 1967. Episome-carried surface antigen K88 of Escherichia coli. II. Isolation and chemical analysis. J. Bacteriol. 93:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 318.Striker, R., U. Nilsson, A. Stonecipher, G. Magnusson, and S. J. Hultgren. 1995. Structural requirements for the glycolipid receptor of human uropathogenic Escherichia coli. Mol. Microbiol. 16:1021-1029. [DOI] [PubMed] [Google Scholar]
- 319.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 320.Stromberg, N., B. I. Marklund, B. Lund, D. Ilver, A. Hamers, W. Gaastra, K. A. Karlsson, and S. Normark. 1990. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα(1-4)Gal-containing isoreceptors. EMBO J. 9:2001-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Svenson, S. B., H. Hultberg, G. Kallenius, T. K. Korhonen, R. Mollby, and J. Winberg. 1983. P-fimbriae of pyelonephritogenic Escherichia coli: identification and chemical characterization of receptors. Infection 11:61-67. [DOI] [PubMed] [Google Scholar]
- 322.Tacket, C. O., D. R. Maneval, and M. M. Levine. 1987. Purification, morphology, and genetics of a new fimbrial putative colonization factor of enterotoxigenic Escherichia coli O159:H4. Infect. Immun. 55:1063-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Tarkkanen, A. M., B. Westerlund-Wikstrom, L. Erkkila, and T. K. Korhonen. 1998. Immunohistological localization of the MrkD adhesin in the type 3 fimbriae of Klebsiella pneumoniae. Infect. Immun. 66:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 326.Tennent, J. M., F. Lindberg, and S. Normark. 1990. Integrity of Escherichia coli P pili during biogenesis: properties and role of PapJ. Mol. Microbiol. 4:747-758. [DOI] [PubMed] [Google Scholar]
- 327.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 328.Thanassi, D. G., E. T. Saulino, M.-J. Lombardo, R. Roth, J. Heuser, and S. J. Hultgren. 1998. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl. Acad. Sci. USA 95:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klösgen, T. Patschkowski, C. Rückert, O. Rupp, S. Schneiker, S. C. Schuster, F.-J. Vorhölter, E. Weber, A. Pühler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Thompson, B. G., and R. G. Murray. 1981. Isolation and characterization of the plasma membrane and the outer membrane of Deinococcus radiodurans strain Sark. Can. J. Microbiol. 27:729-734. [DOI] [PubMed] [Google Scholar]
- 331.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 332.Tomich, M., and C. D. Mohr. 2004. Transcriptional and posttranscriptional control of cable pilus gene expression in Burkholderia cenocepacia. J. Bacteriol. 186:1009-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Tomoeda, M., M. Inuzuka, and T. Date. 1975. Bacterial sex pili. Prog. Biophys. Mol. Biol. 30:23-56. [DOI] [PubMed] [Google Scholar]
- 334.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333-344. [DOI] [PubMed] [Google Scholar]
- 336.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Bäumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Valent, Q. A., J. Zaal, F. K. de Graaf, and B. Oudega. 1995. Subcellular localization and topology of the K88 usher FaeD in Escherichia coli. Mol. Microbiol. 16:1243-1257. [DOI] [PubMed] [Google Scholar]
- 339.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.van Die, I., B. van Geffen, W. Hoekstra, and H. Bergmans. 1985. Type 1C fimbriae of a uropathogenic Escherichia coli strain: cloning and characterization of the genes involved in the expression of the 1C antigen and nucleotide sequence of the subunit gene. Gene 34:187-196. [DOI] [PubMed] [Google Scholar]
- 341.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 342.Van Loy, C. P., E. V. Sokurenko, and S. L. Moseley. 2002. The major structural subunits of Dr and F1845 fimbriae are adhesins. Infect. Immun. 70:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Van Loy, C. P., E. V. Sokurenko, R. Samudrala, and S. L. Moseley. 2002. Identification of amino acids in the Dr adhesin required for binding to decay-accelerating factor. Mol. Microbiol. 45:439-452. [DOI] [PubMed] [Google Scholar]
- 344.Vetsch, M., D. Erilov, N. Moliere, M. Nishiyama, O. Ignatov, and R. Glockshuber. 2006. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 7:734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Viboud, G. I., N. Binsztein, and A. M. Svennerholm. 1993. A new fimbrial putative colonization factor, PCFO20, in human enterotoxigenic Escherichia coli. Infect. Immun. 61:5190-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Viboud, G. I., M. M. McConnell, A. Helander, and A. M. Svennerholm. 1996. Binding of enterotoxigenic Escherichia coli expressing different colonization factors to tissue-cultured Caco-2 cells and to isolated human enterocytes. Microb. Pathog. 21:139-147. [DOI] [PubMed] [Google Scholar]
- 347.Voegele, K., H. Sakellaris, and J. R. Scott. 1997. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 94:13257-13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Ward, N., O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methe, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. K. Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M. Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Watson, W. J., J. R. Gilsdorf, M. A. Tucci, K. W. McCrea, L. J. Forney, and C. F. Marrs. 1994. Identification of a gene essential for piliation in Haemophilus influenzae type b with homology to the pilus assembly platform genes of gram-negative bacteria. Infect. Immun. 62:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Weber, A., K. Harris, S. Lohrke, L. Forney, and A. L. Smith. 1991. Inability to express fimbriae results in impaired ability of Haemophilus influenzae b to colonize the nasopharynx. Infect. Immun. 59:4724-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Weening, E. H., J. D. Barker, M. C. Laarakker, A. D. Humphries, R. M. Tsolis, and A. J. Bäumler. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Wennerås, C., J.-R. Neeser, and A.-M. Svennerholm. 1995. Binding of the fibrillar CS3 adhesin of enterotoxigenic Escherichia coli to rabbit intestinal glycoproteins is competitively prevented by GalNAcβ1-4Gal-containing glycoconjugates. Infect. Immun. 63:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Willems, R. J., C. Geuijen, H. G. van der Heide, M. Matheson, A. Robinson, L. F. Versluis, R. Ebberink, J. Theelen, and F. R. Mooi. 1993. Isolation of a putative fimbrial adhesin from Bordetella pertussis and the identification of its gene. Mol. Microbiol. 9:623-634. [DOI] [PubMed] [Google Scholar]
- 356.Willems, R. J., C. Geuijen, H. G. van der Heide, G. Renauld, P. Bertin, W. M. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]
- 357.Willems, R. J., H. G. van der Heide, and F. R. Mooi. 1992. Characterization of a Bordetella pertussis fimbrial gene cluster which is located directly downstream of the filamentous haemagglutinin gene. Mol. Microbiol. 6:2661-2671. [DOI] [PubMed] [Google Scholar]
- 358.Willshaw, G. A., M. M. McConnell, H. R. Smith, and B. Rowe. 1990. Structural and regulatory genes for coli surface associated antigen 4 (CS4) are encoded by separate plasmids in enterotoxigenic Escherichia coli strains of serotype 0.25.H42. FEMS Microbiol. Lett. 56:255-260. [PubMed] [Google Scholar]
- 359.Wittig, W., R. Prager, M. Stamm, W. Streckel, and H. Tschape. 1994. Expression and plasmid transfer of genes coding for the fimbrial antigen F107 in porcine Escherichia coli strains. Zentralbl. Bakteriol. 281:130-139. [DOI] [PubMed] [Google Scholar]
- 360.Wolf, M. K., G. P. Andrews, D. L. Fritz, R. W. Sjogren, Jr., and E. C. Boedeker. 1988. Characterization of the plasmid from Escherichia coli RDEC-1 that mediates expression of adhesin AF/R1 and evidence that AF/R1 pili promote but are not essential for enteropathogenic disease. Infect. Immun. 56:1846-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wolf, M. K., G. P. Andrews, B. D. Tall, M. M. McConnell, M. M. Levine, and E. C. Boedeker. 1989. Characterization of CS4 and CS6 antigenic components of PCF8775, a putative colonization factor complex from enterotoxigenic Escherichia coli E8775. Infect. Immun. 57:164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wolf, M. K., and E. C. Boedeker. 1990. Cloning of the genes for AF/R1 pili from rabbit enteroadherent Escherichia coli RDEC-1 and DNA sequence of the major structural subunit. Infect. Immun. 58:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wolf, M. K., L. A. de Haan, F. J. Cassels, G. A. Willshaw, R. Warren, E. C. Boedeker, and W. Gaastra. 1997. The CS6 colonization factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEMS Microbiol. Lett. 148:35-42. [DOI] [PubMed] [Google Scholar]
- 364.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 365.Xu, Z., C. H. Jones, D. Haslam, J. S. Pinkner, K. Dodson, J. Kihlberg, and S. J. Hultgren. 1995. Molecular dissection of PapD interaction with PapG reveals two chaperone-binding sites. Mol. Microbiol. 16:1011-1020. [DOI] [PubMed] [Google Scholar]
- 366.Yang, X., Z. Zhang, C. Liu, B. Zhang, T. Chen, and C. Huang. 1995. Cloning of a cluster of genes encoding coli-surface antigen 6 (CS6) of human enterotoxigenic E. coli. Wei Sheng Wu Xue Bao 35:390-393. (In Chinese.) [PubMed] [Google Scholar]
- 367.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier. 2002. Protein-translocating outer membrane porins of gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]
- 368.Zalewska, B., R. Piatek, K. Bury, A. Samet, B. Nowicki, S. Nowicki, and J. Kur. 2005. A surface-exposed DraD protein of uropathogenic Escherichia coli bearing Dr fimbriae may be expressed and secreted independently from DraC usher and DraE adhesin. Microbiology 151:2477-2486. [DOI] [PubMed] [Google Scholar]
- 369.Zavialov, A. V., J. Berglund, A. F. Pudney, L. J. Fooks, T. M. Ibrahim, S. MacIntyre, and S. D. Knight. 2003. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587-596. [DOI] [PubMed] [Google Scholar]
- 370.Zav'yalov, V. P., T. V. Chernovskaya, E. V. Navolotskaya, A. V. Karlyshev, S. MacIntyre, A. M. Vasiliev, and V. M. Abramov. 1995. Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett. 371:65-68. [DOI] [PubMed] [Google Scholar]
- 371.Zav'yalov, V. P., G. A. Zav'yalova, A. I. Denesyuk, and T. Korpela. 1995. Modelling of steric structure of a periplasmic molecular chaperone Caf1M of Yersinia pestis, a prototype member of a subfamily with characteristic structural and functional features. FEMS Immunol. Med. Microbiol. 11:19-24. [DOI] [PubMed] [Google Scholar]
- 372.Zhu, G., H. Chen, B. K. Choi, F. Del Piero, and D. M. Schifferli. 2005. Histone H1 proteins act as receptors for the 987P fimbriae of enterotoxigenic Escherichia coli. J. Biol. Chem. 280:23057-23065. [DOI] [PubMed] [Google Scholar]
- 373.Zunino, P., L. Geymonat, A. G. Allen, C. Legnani-Fajardo, and D. J. Maskell. 2000. Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol. Med. Microbiol. 29:137-143. [DOI] [PubMed] [Google Scholar]