Abstract
We provide experimental and modeling evidence that the hydrodynamic environment can impact quorum sensing (QS) in a Pseudomonas aeruginosa biofilm. The amount of biofilm biomass required for full QS induction of the population increased as the flow rate increased.
Quorum sensing (QS), or intercellular signaling, is used by a wide range of bacterial species to coordinate gene expression in a population (8, 26). One of the best-studied QS mechanisms is the acyl homoserine-lactone (AHL) system used by many different gram-negative bacterial species (7, 8). QS has been characterized primarily in planktonic batch culture, where its onset corresponds to a specific population density at which an inducing concentration of signal occurs. This has promoted the description of QS as a cell density-dependent signaling mechanism.
In the environment, bacteria rarely encounter conditions similar to a well-mixed planktonic culture. Unlike the steady signal accumulation in batch cultures, signal levels in environmental systems might be affected by signal diffusion between bacteria and the surrounding environment. Therefore, an interesting question is this: where can QS-relevant concentrations of signal be found in the environment? One potential answer is in surface-associated communities called biofilms. Microorganisms in the natural environment often live in biofilms, where they grow in close proximity to each other (5, 15). McLean et al. demonstrated that biofilms present on rocks submerged in flowing aquatic systems produce measurable amounts of AHLs (14), supporting the idea that physiologically relevant concentrations of signal might be present in some environmental biofilm systems.
Several physical, biological, and chemical factors have the potential to influence QS in biofilm systems (12). The hydrodynamic environment is one such factor with the potential to influence QS in several ways. First, signal produced in the biofilm will diffuse into the bulk fluid and will be washed away (i.e., advected). Thus, not all of the signal will remain in the local environment due to mass transfer effects. Second, mass transfer also can produce nutrient gradients, and the nutritional environment has been shown to influence QS (1, 6). For example, iron availability can modulate QS in Pseudomonas aeruginosa (1, 6), while other components of the nutritional environment can delay the onset of QS (28). Third, the hydrodynamic environment can affect biofilm density and thickness (2, 18, 25), which in turn can affect signal gradients.
The common soil and aquatic bacterium P. aeruginosa is a paradigm organism for the study of AHL-based QS and biofilms. In this study, we examined the relationship between hydrodynamic environment and QS induction in a developing P. aeruginosa biofilm. We hypothesized that as fluid flow rate in a biofilm system increased, the amount of biofilm biomass required for full QS induction of the population also would increase.
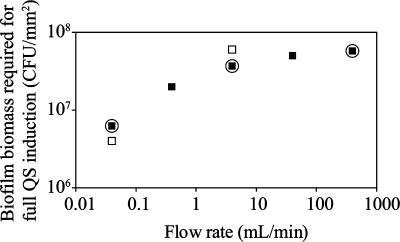
We used predictive modeling to gauge how flow conditions might affect the amount of biomass required for full QS induction of the population in a simple laboratory biofilm. This model focuses primarily on the effect of signal washout from the system. Several parameters were included in the model (see Table S1 in the supplemental material), including estimated per-cell signal production rate, biofilm biomass accumulation, Fickian diffusion of signal in the biofilm, and assumed geometry of a flat, two-dimensional biofilm. Based on past studies, we assumed the signal concentration required for full lasB induction to be 23 nM 3-oxo-dodecanoyl homoserine-lactone (3, 4, 17). We used a Stokes flow model for the fluid flow over the biofilm, which induced a diffusion boundary layer. The only nutrient gradient accounted for was dissolved oxygen, which was used as a measure of metabolic activity for estimating signal production at different depths in the biofilm. In our model, as flow rate increased, the predicted amount of biofilm biomass (CFU per attachment surface area) required for full QS induction of the population increased as well (Fig. 1).
FIG. 1.
Comparison of the amounts of biofilm biomass required for full QS induction of the population from model simulations (▪) and from tube biofilm reactor experiments (□). The three modeled data points that were tested experimentally are circled.
To experimentally examine our hypothesis, we used P. aeruginosa strains harboring fluorescence-based transcriptional reporters for the well-studied QS-regulated gene lasB (see Table S2 in the supplemental material) (9, 10, 17). These reporters were characterized initially in planktonic batch culture, and green fluorescent protein (GFP) fluorescence was monitored as a function of culture density. The chromosomal reporter strain (P. aeruginosa PAO1 lasB::gfp) was found to induce rapidly between mid- and late logarithmic phase (see Fig. S1 in the supplemental material), and the plasmid reporter strain (P. aeruginosa PAO1 plasB::gfp) showed similar results (data not shown). As expected, the reporters failed to fluoresce in their associated QS-deficient (lasI rhlI) strains (data not shown). All of these observations are consistent with previously published lasB expression data. Using an AHL bioassay (16) on a planktonic batch culture (Luria-Bertani [LB] medium at 23°C), the plasB::gfp reporter strain was induced maximally at a 3-oxo-dodecanoyl homoserine-lactone concentration of 17 nM (data not shown). For the rest of this study, QS was said to be fully induced in a reporter biofilm once the biofilm reached 100% of the maximum normalized fluorescence (i.e., relative fluorescence units divided by optical density at 600 nm) of the reporter strain in a planktonic batch culture.
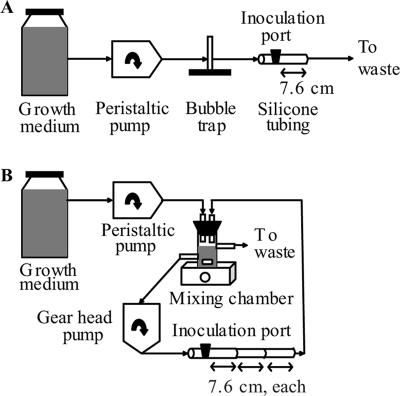
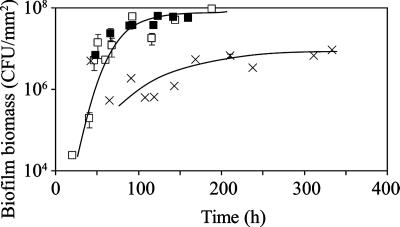
Three modeled data points (Fig. 1), corresponding to flow rates of 0.04 ml/min, 4.0 ml/min, and 380 ml/min, were chosen for analysis in laboratory biofilms. These flow rates correspond to velocities of 0.15 mm/s, 15 mm/s, and 1,400 mm/s, respectively, and to Reynold's numbers of 0.3, 30, and 3,000, respectively. For the two lower flow rates, biofilms were cultured in a once-through silicone tube biofilm reactor similar to those described previously (13, 20, 22) (Fig. 2A), where the flow was driven by a peristaltic pump. For the highest flow rate, to avoid the large volumes of growth medium necessary to run the reactor in a once-through format, biofilms were cultured in a recycle reactor similar to that described previously (23) (Fig. 2B). In the recycle reactor, fresh medium was pumped via a peristaltic pump from an influent reservoir to a mixing chamber, where it was mixed with recycled medium. To achieve the high flow rate (380 ml/min), a gear pump was used to pump the medium in the recycle loop. The hydraulic residence time in the system was approximately 35 min, which minimized planktonic accumulation. For both types of reactors, biofilms of P. aeruginosa PAO1 lasB::gfp were cultured at room temperature in LB medium on sections of 7.6-cm silicone tubing with a 2.4-mm inner diameter. At various time points, the biofilm was harvested by centrifugation and was resuspended in 1× phosphate-buffered saline by several cycles of sonication and vortexing; the absence of cell clumps in the suspension was verified by microscopy. Total biofilm biomass was determined by plate counts for each harvested sample. Biofilms grown at the two lower flow rates showed similar biofilm accumulation kinetics (Fig. 3), in which a steady-state level of biofilm biomass (∼5 × 107 CFU/mm2) was reached after approximately 100 h. Biofilms grown at the highest flow rate showed a lower steady-state level of biofilm biomass (∼5 × 106 CFU/mm2), which was reached after approximately 170 h. Scanning electron microscopy was performed on the biofilms grown at the two lower flow rates. The biofilms appeared to be fairly flat and uniform, forming a confluent mat on the tube surface (see Fig. S2 in the supplemental material); thus, the experimental biofilms formed in the tube biofilm reactors were similar in structure to the modeled two-dimensional biofilms.
FIG. 2.
Schematics depicting tube biofilm reactors. (A) Once-through reactor for flow in the laminar regime. (B) Recycle reactor for flow in the transitional regime between laminar and turbulent.
FIG. 3.
Accumulation of biofilm biomass in the tube biofilm reactor. Flow in the laminar regime, 0.04 ml/min (□) and 4.0 ml/min (▪); flow in the transitional regime between laminar and turbulent, 380 ml/min (×). Lines on the plot show the trend of the data. For simplicity, error bars representing the standard deviations are shown only for the data points that are based on the averages of at least two separate harvested biofilms. The data points without error bars are based on results from a single harvested biofilm. The biofilm biomass for each harvested biofilm was measured by triplicate plate counts.
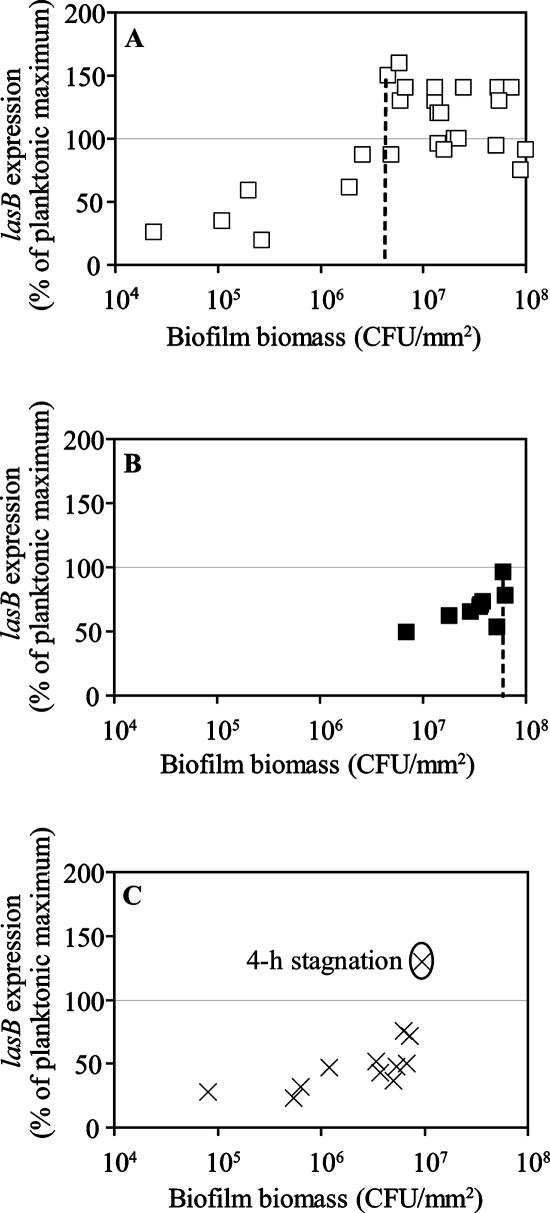
Expression of lasB in the biofilm population was monitored over time using fluorimetry (Fig. 4) and microscopy (data not shown). At the lowest flow rate (0.04 ml/min), the amount of biofilm biomass required for full QS induction of the population was estimated to be 4 × 106 CFU/mm2 (Fig. 4A). At the intermediate flow rate (4.0 ml/min), the amount of biofilm biomass required for full QS induction was estimated to be 6 × 107 CFU/mm2 (Fig. 4B). The highest flow rate was achieved using the recycle reactor. Signal accumulation in the recycled medium was monitored and found to be below the detection limit of the AHL bioassay (data not shown). Full QS induction of the biofilm population grown at the highest flow rate was never observed, even after 200 h of reactor operation, although an upper limit of 80% induction of the population was seen (Fig. 4C). However, QS was fully induced in this population after the flow was stagnated for 4 h (Fig. 4C).
FIG. 4.
QS induction in the tube biofilm reactor. Flow rates were (A) 0.04 ml/min, (B) 4.0 ml/min, and (C) 380 ml/min (with and without stagnation). The vertical dashed line indicates the biofilm biomass at which QS was fully induced (i.e., lasB expression at 100% of planktonic maximum).
Our data and model suggest that signal washout might explain why QS fails to fully induce at the highest flow rate, although we cannot rule out mass transfer effects on nutrient gradients. These data also suggest that QS may not be fully operative at high flow rates and thus not play a significant role in biofilm formation. Supporting this point, Purevdorj et al. demonstrated that P. aeruginosa wild-type and QS mutant strains formed nearly identical biofilms under high flow conditions (19).
At the two lower flow rates, which are in the laminar flow region, our experimental values for QS induction were very close to the values predicted by the model (Fig. 1). At the highest flow rate, which is in the transitional regime between laminar and turbulent, full QS induction of the population was not seen experimentally; this was consistent with the model, since the maximum biofilm biomass in the flowing experimental system (7 × 106 CFU/mm2 [Fig. 4C]) never reached the level predicted by the model to be necessary for full QS induction (6 × 107 CFU/mm2 [Fig. 1]).
To demonstrate some of the same principles in a different format, we next examined lasB expression in a flow cell biofilm reactor (24). This system was inoculated with P. aeruginosa PAO1 plasB::gfp and operated with LB medium at room temperature. This system is very similar to that shown in Fig. 2A, except that the biofilm was grown in a 1-mm- by 1-mm-square glass capillary (Friedrick & Dimmock, Millville, NJ), with flow rates of 0.005 ml/min, 0.1 ml/min, and 1.0 ml/min. These flow rates correspond to velocities of 0.08 mm/s, 1.7 mm/s, and 17 mm/s, respectively, and to Reynold's numbers of 0.08, 1.7, and 17, respectively. The advantage of the flow cell biofilm reactor is that it can be interrogated by direct microscopy. Biofilm biomass and structural characteristics were measured using the image analysis software COMSTAT (11) (see Fig. S3 in the supplemental material). In this system, the biofilm was much more structured and heterogeneous than were the tube biofilms (Fig. 5). At earlier points in biofilm development, lasB was not expressed in the flow cell system although a significant amount of biofilm biomass was present (Fig. 5A and B). To determine if the mass transfer environment might be responsible, flow was stopped, and much of the population was observed to be expressing lasB within 6 h (Fig. 5C and D). Nearly uniform levels of expression were seen throughout the system, also demonstrating that oxygen is not limiting for proper GFP folding. To complement this experiment, another biofilm was grown to a point of development similar to that shown in Fig. 5, and inducing concentrations (10 μM) of the two primary AHLs, 3-oxo-dodecanoyl homoserine-lactone and butyryl homoserine-lactone, were supplied exogenously. The entire population was seen to rapidly induce within 10 min (data not shown). Taken together, the tube biofilm reactor data and the flow cell data suggest that mass transfer influences QS induction in a developing biofilm.
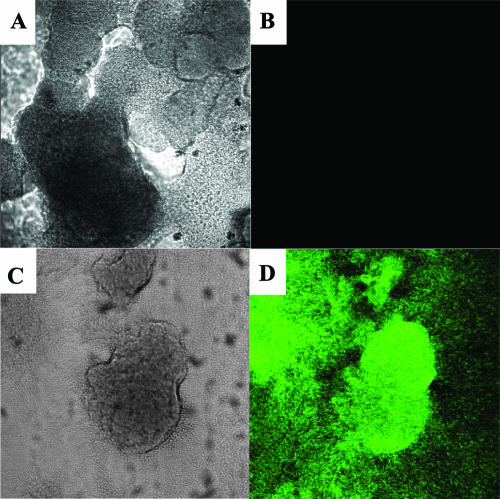
FIG. 5.
QS induction in the flow cell biofilm reactor. Transmitted light and epifluorescent maximum projections of the confocal scanning laser microscopy stacks are shown for a biofilm grown in the flow cell at 1 ml/min. (A) Transmitted light image and (B) corresponding epifluorescent image showing low lasB expression in the biofilm under flowing conditions. (C) Transmitted light image and (D) corresponding epifluorescent image showing full lasB expression after a 6-h stagnation period.
Environmentally and clinically relevant biofilms are found in hydrodynamic environments that range from turbulent flow (e.g., biofilms on a rock in a river) to little or no flow (e.g., biofilms in the airways of people suffering from cystic fibrosis). In this report, we provide evidence that the hydrodynamic environment can impact QS induction in biofilms. Our study focused on a single, well-known QS-regulated gene. Other researchers have shown that there are multiple classes of QS-regulated genes (21, 27), and our future studies will examine if representatives of these other classes of QS-regulated genes also are subject to the effects of the hydrodynamic environment in biofilms. Another limitation of our study is that our simple model does not account for nutritional parameters known to influence QS. As the effect of the nutritional environment on QS becomes better understood, we will attempt to incorporate these nutritional parameters into our model and test the model experimentally. As researchers attempt to understand and control QS in environmentally and clinically relevant contexts, consideration of factors such as the hydrodynamic environment will become increasingly important in developing an accurate model of these complex situations.
Supplementary Material
Acknowledgments
This work was supported in part by grants from NSF (MCB 0133-833) to M.R.P., NIH (GM67248-01) to D.L.C., NIH (DC04173) to G. D. Ehrlich, NIH (GM60052) to P.S., and the W.M. Keck Foundation to P. Stewart.
Footnotes
Published ahead of print on 17 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bollinger, N., D. J. Hassett, B. H. Iglewski, J. W. Costerton, and T. R. McDermott. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183:1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Characklis, W. G. 1980. Biofilm development and destruction. Final report EPRI CS-1554. Electric Power Research Institute, Palo Alto, CA.
- 3.Chopp, D. L., M. J. Kirisits, B. Moran, and M. R. Parsek. 2003. The dependence of quorum sensing on the depth of a growing biofilm. Bull. Math. Biol. 65:1053-1079. [DOI] [PubMed] [Google Scholar]
- 4.Chopp, D. L., M. J. Kirisits, B. Moran, and M. R. Parsek. 2002. A mathematical model of quorum sensing in a growing bacterial biofilm. J. Ind. Microbiol. Biotechnol. 29:339-346. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Duan, K., and M. G. Surette. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189:4827-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 8.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 9.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, K. M., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1994. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 176:3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 12.Horswill, A. R., P. Stoodley, P. S. Stewart, and M. R. Parsek. 2007. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 387:371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean, R. J. C., M. Whiteley, D. J. Stickler, and W. C. Fuqua. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259-263. [DOI] [PubMed] [Google Scholar]
- 15.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 16.Parsek, M. R., A. L. Schaefer, and E. P. Greenberg. 1997. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 26:301-310. [DOI] [PubMed] [Google Scholar]
- 17.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira, M. O., M. Kuehn, S. Wuertz, T. Neu, and L. F. Melo. 2002. Effect of flow regime on the architecture of a Pseudomonas fluorescens biofilm. Biotechnol. Bioeng. 78:164-171. [DOI] [PubMed] [Google Scholar]
- 19.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68:4457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer, A. L., E. P. Greenberg, and M. R. Parsek. 2001. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay, p. 41-47. In R. J. Doyle (ed.), Microbial growth in biofilms, part A: developmental and molecular biological aspects, vol. 336. Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 21.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 23.Stoodley, P., R. Cargo, C. J. Rupp, S. Wilson, and I. Klapper. 2002. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 29:361-367. [DOI] [PubMed] [Google Scholar]
- 24.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ. Microbiol. 1:447-455. [DOI] [PubMed] [Google Scholar]
- 25.Villasenor, J. C., M. C. M. van Loosdrecht, C. Picioreanu, and J. J. Heijnen. 2000. Influence of different substrates on the formation of biofilms in a biofilm airlift suspension reactor. Water Sci. Technol. 41:323-330. [Google Scholar]
- 26.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 27.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarwood, J. M., E. M. Volper, and E. P. Greenberg. 2005. Delays in Pseudomonas aeruginosa quorum-controlled gene expression are conditional. Proc. Natl. Acad. Sci. USA 102:9008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.