Abstract
In archaea, RNA endonucleases that act specifically on RNA with bulge-helix-bulge motifs play the main role in the recognition and excision of introns, while the eukaryal enzymes use a measuring mechanism to determine the positions of the universally positioned splice sites relative to the conserved domain of pre-tRNA. Two crystallographic structures of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728 (EndATa) have been solved to 2.5-Å and 2.7-Å resolution by molecular replacement, using the 2.7-Å resolution data as the initial model and the single-wavelength anomalous-dispersion phasing method using selenomethionine as anomalous signals, respectively. The models show that EndATa is a homodimer and that it has overall folding similar to that of other archaeal tRNA endonucleases. From structural and mutational analyses of H236A, Y229F, and K265I in vitro, we have demonstrated that they play critical roles in recognizing the splice site and in cleaving the pre-tRNA substrate.
Although RNA splicing is a required step for gene expression in all three taxonomic kingdoms, the eukarya, the archaea, and the bacteria, at least four different mechanisms of splicing have evolved. In bacteria, tRNA introns are self-splicing group I introns and the splicing mechanism is autocatalytic (3, 6). In eukarya, tRNA introns are small and invariably interrupt the anticodon loop 1 base 3′ to the anticodon. They are removed by the stepwise actions of an endonuclease, a ligase, and a phosphotransferase (7, 27). In Saccharomyces cerevisiae, there are three distinct enzymes required for tRNA splicing. An endonuclease acts first and is responsible for the recognition and excision of introns. A ligase subsequently joins the two exons in a multinucleotide-dependent reaction, leaving a 2′-phosphate group at the spliced junction. Finally a 2′-phosphotransferase catalyzes the transfer of the 2′-phosphate group to its cofactor, NAD (1). The yeast endonuclease is a heterotetramer consisting of Sen15, Sen2, Sen34, and Sen54 subunits, where Sen34 and Sen2 share sequence homology over a region of ∼115 amino acid residues. This conserved region defines a family of tRNA-splicing endonucleases (EndA) in both the archaea and eukarya (16, 21, 22, 38).
In archaea, the introns often reside in the same location as for eukaryal tRNA introns. The splicing mechanism in archaea is similar to eukaryal splicing but differs in some respects (15, 26). In archaea, EndA plays a key role in ensuring the correct removal of the intron from the pre-tRNA, pre-rRNA, and pre-mRNA (1, 13-15, 34, 41). EndAs from both eukarya and archaea cleave the pre-tRNA substrate, leaving 5′-hydroxyl and 2′,3′ cyclic phosphate termini, but these enzymes recognize their substrates differently. The eukaryal enzyme uses a measuring mechanism to determine the positions of the universally positioned splice sites relative to the conserved domain of pre-tRNA (20, 29). In contrast, archaeal splice site recognition appears to rely on the presence of a specific RNA structure. The majority of archaeal exon-intron boundaries form a folded RNA structure, termed the bulge-helix-bulge (BHB) motif, consisting of 2- or 3-nucleotide (nt) bulges separated by a 4-bp helix (5, 8, 9, 15, 20, 37). This motif is mainly found in the anticodon loop but can also be seen in various locations on precursor tRNAs (23, 27). Additionally, the BHB motif is recognized by the endonuclease for processing of other functional RNAs, such as rRNA and mRNA (13, 32, 35, 39). Although the BHB motif has some resemblance to yeast RNA exon-intron boundaries, archaeal enzymes are unable to process yeast RNA precursors, suggesting an RNA recognition mechanism peculiar to archaea (2, 16, 29, 38).
Some crystal structures of archaeal RNA-processing enzymes have already been identified and published (5, 19, 20, 40). Recently, the structure of EndA from Archaeoglobus fulgidus (EndAAf; Protein Data Bank [PDB] accession no. 2GJW) complexed with RNA substrate was reported (40). The archaeal endonucleases can be distinctly grouped into three subfamilies; homodimers, homotetramers, and heterotetramers. The crystal structure of splicing endonuclease from Methanococcus jannaschii (EndAMj; PDB accession no. 1A79) belongs to the homotetramers, while those from A. fulgidus (EndAAf; PDB accession no. 1RLV) and Sulfolobus solfataricus (EndASs; PDB accession no. 2CV8) are homodimers, although the structures from Sulfolobus species are for nonfunctional dimers of the catalytic subunit (5, 36, 41). The endonuclease from Thermoplasma acidophilum (EndATa) was predicted to be a dimeric enzyme, but it was not confirmed empirically (23). Here, we report the crystal structure of EndATa as two crystallographic forms. The predicted active site of EndA was formed from a Tyr-His-Lys catalytic triad near the C terminus. We have also confirmed the involvement of the active sites of these residues in EndATa by assaying the biochemical activities of the mutant proteins.
MATERIALS AND METHODS
Cloning and purification.
The endA gene encoding site-specific recombinase from T. acidophilum strain DSM1728 was amplified from purified T. acidophilum genomic DNA by PCR. The sequences of the forward and reverse oligonucleotide primers designed from the published genomic sequence were 5′-AAT TGA ATT CAT GGA ACA GGG TAT CTG TGG ATC TCA-3′ and 5′-AAT TCT CGA GTT AAA TAA TAT CTT TCA CCC TTT TCA GGG-3′, respectively. (The bases in boldface represent the EcoRI and XhoI digestion sites, respectively.) The amplified DNA was inserted into the digested expression vector, a modified pET-21b (Novagen). This vector encodes a hexahistidine tag ahead of a thrombin recognition site just before the multicloning site. The vector therefore facilitates expression of N-terminally His-tagged proteins from which the tag can be removed with thrombin. The protein was overexpressed in Escherichia coli BL21(DE3) cells. The recombinant protein in the supernatant fraction was purified by two chromatographic steps. The first purification step utilized the N-terminal histidine tag in an Ni2+-chelated Hi-Trap HP column (Amersham Pharmacia). The histidine tag was removed from the protein by dialysis with thrombin overnight at 291 K before gel filtration on a HiLoad 26/60 Superdex-200 prep grade column (Amersham Pharmacia) using 50 mM Tris-HCl (pH 8.5), 100 mM NaCl, 5% (vol/vol) glycerol, 10 mM dithiothreitol (DTT). The homogeneity of the purified protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The purified protein was concentrated to 9.1 mg/ml using Centri-prep (Millipore) in buffer A. The protein concentration was estimated by the Bradford method (Bio-Rad) using bovine serum albumin (1 mg/ml) as a standard. A similar procedure was followed to express and purify the mutant proteins H236A, Y229F, and K265I. The purified selenomethionyl protein was also obtained from E. coli B834(DE3) cells.
Crystallization and data collection.
Initial crystallization was performed at 317 K by the sitting-drop method using a Hydra II Plus One crystallization robot (Matrix Technology) with approximately 1,500 conditions and a ratio of 200 nl precipitants to 200 nl protein solution. Crystallization trials were established using screening kits from Hampton Research, Jena Biosciences, and Emerald Biostructures. Initially, two types of crystal were obtained under conditions of the same salt and different buffers. The crystals were then optimized by a hanging-drop vapor diffusion method using 24-well plates by mixing 2 μl protein solution with 2 μl reservoir solution. Each hanging drop was placed over 0.5 ml reservoir solution. Finally, tetragonal crystals and monoclinic crystals were obtained using 2.9 M NaCl-0.1 M citric acid (pH 5.6) and 2.4 M NaCl-0.1 M Na/K phosphate (pH 6.2), respectively. Crystals of EndATa were mounted in nylon CryoLoops (Hampton Research), immersed in cryoprotectant (30% [vol/vol] glycerol in reservoir solution) for a few seconds, and then flash-cooled in a cold nitrogen stream. X-ray diffraction data from the monoclinic form were collected from a cryo-cooled crystal using an ADSC Q210 charge-coupled device detector at beamline 4A at Pohang Light Source, Pohang, South Korea. X-ray diffraction data from the tetragonal form were collected using an ADSC Q315 charge-coupled device detector at Photon Factory, Tsukuba, Japan (beamline BL-5A). The wavelength of the synchrotron X-rays was 1,000 Å. The raw data were processed and scaled using DENZO and SCALEPACK (25).
Structure determination and refinement.
The structure of EndATa was determined by the single-wavelength anomalous-diffraction (SAD) method using peak data collected with selenomethionyl protein crystals (33). Phases were calculated with SOLVE and RESOLVE (31) and had a mean figure of merit of 0.45 at 2.7-Å resolution. The SAD phasing and refinement statistics are given in Table 1. The model was completed by iterative cycles of model building with Coot (10) and refinement with CNS (4). The final model contained 300 amino acids and 100 water molecules for EndATa, yielding an Rfactor and Rfree of 21.5% and 27.8%, respectively (4).
TABLE 1.
Data collection and refinement statistics
| Statistic | Value for data setb
|
|
|---|---|---|
| Native | Se-Met peak | |
| X-ray diffraction data | ||
| Space group | P43212 | C2 |
| Wavelength (Å) | 1.12714 | 0.97949 |
| Cell dimension | ||
| a, b, c (Å) | 57.17, 57.17, 459.91 | 164.91 46.09 58.00 |
| α, β, γ (°) | 90.00 90.00 90.00 | 90.00 92.61 90.00 |
| Resolution (Å) | 50-2.5 (2.54-2.50) | 50-2.7 (2.90-2.70) |
| No. of reflections | 263,396 (14,582) | 104,483 (10,735) |
| No. of unique reflections | 49,839 (2,514) | 11,669 (1,887) |
| Rsyma | 0.062 (0.34) | 0.066 (0.29) |
| I/ΣI | 16.68 (4.44) | 26.25 (3.69) |
| Completeness (%) | 98.2 (99.9) | 98.0 (97.6) |
| Overall figure of merit | 0.45 | |
| No. of molecules in asymmetric unit | 2 | 1 |
| Crystallographic refinement statistics | ||
| Resolution (Å) | 20-2.7 | 20-2.5 |
| Rworkc/Rfree (%) | 22.5/28.5 | 21.5/27.8 |
| Avg B factor (Å2) | 44.5 | 45.1 |
| RMS | ||
| Bond length (Å) | 0.009 | 0.008 |
| Bond angles (°) | 1.4 | 1.3 |
| Ramachandran plot by PROCHECK (%) | ||
| Most favorable region | 85.1 | 83.1 |
| Additional allowed region | 14.9 | 16.5 |
| Generously allowed region | 0 | 0.4 |
Rsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
Values in parentheses are for the highest-resolution shell.
Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where all reflections belong to a test set of 5% randomly selected data.
The dimeric structure of EndATa was also determined by molecular replacement with the monomeric structure, having an Rfactor and Rfree of 22.4% and 28.5%, respectively.
Analytical ultracentrifugation measurements.
Sedimentation velocity experiments were performed at 20°C in a Beckman XL-I centrifuge using an eight-slot rotor (An-50Ti) at 40,000 rpm. EndA was analyzed at concentrations of 0.63, 0.32, and 0.13 mg/ml (17.6 μM, 9.5 μM, and 3.8 μM, respectively) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl. Twelve-millimeter double-sector Epon charcoal-filled centerpieces and sapphire windows were used with 400 μl of protein solution in the sample cell and 420 μl of buffer in the reference cell. Absorbance measurements at 280 nm were taken in 0.002-cm radial steps. Absorbance and refractive-index scans were measured from each cell every 5 min for 10 h. The partial specific volume of EndA was estimated to be 0.7349 ml/g, and the solvent density was calculated to be 1.00499 g/ml, using the program SEDENTERP v1.08 (17, 18). Analysis of the sedimentation velocity data for EndA was performed with SEDFIT (version 8.9) (17, 28).
Construction of the pre-tRNAArchEuka gene.
For the intron cleavage assay and binding assay, we chose as the substrate the hybrid pre-tRNAArchEuka (11, 24), which consists of yeast tRNAPhe with an archaeal BHB region. The pre-tRNAArchEuka gene, with an upstream NcoI restriction site and hammerhead ribozyme sequences followed by a HindIII restriction site at the end, was created by PCR using six partially overlapping synthetic DNA oligomers as a template for PCR. The amplified DNA fragment was digested with NcoI and HindIII and then ligated into pUC19 downstream of a T7 RNA polymerase promoter. The resulting plasmid was prepared on a large scale and then linearized with HindIII and purified with phenol-chloroform-isoamyl alcohol, followed by ethanol precipitation.
Preparation of pre-tRNA.
In vitro transcription was performed by an established method (30) using linearized plasmid. The transcript of the hammerhead ribozyme and pre-tRNAArchEuka tandem sequences autocleaved into 94-nt pre-tRNAArchEuka (GCGGATTTAGCTCAGTTGGAGAGCGCCAGACTCCAGAGGCTTACACCGGAGATATCTGGAGGTCCTGTGTTCGATCCACAGA ATTCGCAAGCT) during the transcription reaction. The RNA was purified with a 15% (vol/vol) acrylamide, 7 M urea, 1× Tris-borate-EDTA (TBE [10.8 g of Tris base, 5.5 g of boric acid, and 4 ml of 0.5 M EDTA, pH 8.0]) denaturing gel (40 by 20 cm). The pre-tRNA was eluted from the excised band, ethanol precipitated, and dissolved in Milli Q water.
Intron cleavage assay.
The splicing assays for endoribonuclease activity were performed with the in vitro-transcribed pre-tRNAArchEuka. The pre-tRNAArchEuka was incubated with both wild-type and mutant EndA in cleavage buffer (20 mM HEPES-Na, pH 8.0, 5 mM DTT, 1 mM spermidine, 10 mM MgCl2) for 30 min at 55°C. The reaction solutions were mixed with 2× denaturing loading dye (90% [vol/vol] formamide, 10 mM Tris, pH 7.4, 2 mM EDTA, 0.1% [wt/vol] bromophenol blue, 0.1% [wt/vol] xylene cyanol) and run on denaturing gel (20% [vol/vol] polyacrylamide, 7 M urea, 1× TBE), followed by toluidine blue staining.
Binding assay for EndA and pre-tRNAArchEuka.
The binding of EndA (wild type, H236A, Y229F, and K265I) to pre-tRNAArchEuka was monitored using a gel mobility shift assay. A constant concentration of pre-tRNAArchEuka (2.4 M) was incubated with several concentrations of EndA in 20 mM HEPES, pH 8.0, 10 mM MgCl2, 100 mM NaCl, 5 mM DTT, 1 mM spermidine, 20% (vol/vol) glycerol at 37°C for 5 min. The samples were loaded on 1% (wt/vol) agarose, 1× TBE buffer and run at 20 mA for 50 min. The RNA bands were stained with ethidium bromide.
Protein structure accession numbers.
The coordinates of EndATa have been deposited in the Protein Data Bank (accession no. 2OHC for the tetragonal form [dimer] and no. 2OHE for the monoclinic form [monomer]).
RESULTS AND DISCUSSION
Functional characterization of EndATa.
The EndA open reading frame in T. acidophilum encodes an ortholog of the M. jannaschii EndA responsible for the splicing of the BHB motif in pre-tRNA. To test the splicing endo-RNase activity, assays were performed with in vitro-transcribed pre-tRNAArchEuka (Fig. 1A and B). The pre-tRNAArchEuka was incubated with both wild-type and mutant EndA in cleavage buffer. Figure 1B shows that the purified EndATa clearly displayed splicing activity and cleaved the substrate completely, reducing the full-length pre-tRNA to two fragments, a 5′ fragment (39 nt) and a 3′ fragment (37 nt). In contrast to the wild type, the mutant enzymes showed dramatically reduced splicing activities (Fig. 1B). These sites exhibited 50-fold less activity than the wild type, implying a significant role in catalysis. Mutation of Tyr-229 to alanine reduced the enzymatic activity by about 50-fold, while mutations of H256A and K265I completely abolished the activity. The formation of a specific complex between EndATa and the pre-tRNAArchEuka substrate was tested by electrophoretic mobility shift assays (Fig. 1C). Figure 1C clearly shows the formation of a stable complex between EndATa and the pre-tRNAArchEuka substrate in the transition state. As can be seen in Fig. 1C, a band shift was observed, indicative of complex formation. While two mutants (H236A and Y229F) showed mobilities similar to that of the wild type, K265I showed significantly weaker binding, in which Lys-265 probably played an important role in making the triad of enzyme activity for the tRNA substrate (Fig. 1C). Recently, the structure of EndAAf (PDB accession no. 2GJW) complexed with RNA substrate was reported (40). Therefore, in this process, the enzyme-substrate complex could form a very stable transient complex.
FIG. 1.
In vitro cleavage and binding assays with a substrate containing BHB by T. acidophilum endonuclease. (A) Secondary structure of the pre-tRNAArchEuka, which is prepared with two kinds of ribozyme sequence unit added to the head and tail of a pre-tRNA sequence. The splice site is indicated by the arrow. (B) Splicing activity assay of wild-type and mutant EndATa with full-length pre-tRNA substrate. The control (no added enzyme) shows no spontaneous cleavage. Wild-type EndATa cleaved the substrate completely; the full-length pre-tRNA was cut to give a 5′ fragment (39 nt) and a 3′ fragment (37 nt) in the agarose gel. All mutants showed reduced activity, especially H236A and K265I, which were essentially inactive. (C) Electrophoretic mobility of pre-tRNA incubated with EndATa wild-type and mutant forms. H236A and Y229F showed unaltered binding to pre-tRNA, but K265I bound to the substrate much more weakly than the wild type.
Overall structure of EndATa.
EndATa crystallized in two crystal forms, one tetragonal and the other monoclinic. The monoclinic structure of EndATa was determined by the SAD method to 2.7-Å resolution using selenomethionine as an anomalous scatterer (Fig. 2A). The tetragonal crystal structure was solved by molecular replacement to 2.5-Å resolution using the monoclinic structure as a search model (Fig. 2B). Phasing and refinement statistics are shown in Table 1. The monoclinic and tetragonal forms have one and two molecules in the asymmetric unit, respectively. Purified EndATa appeared at the dimer size on a gel filtration column, which had the enzymatic activity from the monomer with a molecular mass of 35 kDa. Analytical ultracentrifugation and dynamic light scattering also showed EndATa to exist as a dimer in solution (data not shown). The dimer appears to be physiological, because protein contacts in the asymmetric unit are identical in the two crystal forms and they involve conserved amino acids. The monomer of EndATa has overall dimensions of about 90 by 35 by 30 Å and consists of two major domains, the N-terminal domain and the catalytic domain, with a linker region between them. The overall structures of EndATa are illustrated in Fig. 2. The N-terminal domain consists of a mixed antiparallel/parallel β-sheet and three α-helices. The linker domain consists of α4 and three consecutive β-strands (β6 to β8). The catalytic domain consists of four α-helices and 11 β strands (β9 to β19). The protein has similar conformations in both crystal forms, but there are structural differences between the dimer and the monomer caused by crystal packing. The Cα root mean square deviations (RMSD) are 0.94 Å for the monomer and one molecule (A) in the dimer and 1.00 Å for the monomer and the other molecule (B) in the dimer (Fig. 2C). The 2.5-Å tetragonal structure is shown as a ribbon model in Fig. 2B. Superposition of the Cα atoms of the two monomers in the tetragonal crystal gives an RMSD of 0.94 Å for residues 2 to 289. Significant differences were observed in loops (94 to 102, 230 to 237, and 244 to 249) (Fig. 2C). When the loop segment alone was compared with those of other protomers (monomer and molecules A and B in the dimer), the RMSD for Cα atoms were 1.2 Å. The electron density in the loop region is well defined for each protomer, and the average temperature factor in this region is similar to that of the whole molecule. Nevertheless, the notable conformational differences indicate that these loops are the most flexible regions in the molecule.
FIG. 2.
Overall structures of the two crystallographic forms of the T. acidophilum endonuclease. (A) The structure of the monoclinic form. (B) The structure of the tetragonal form. (C) Structural variations of the two crystallographic forms. Red is the monoclinic form (monomer), green is the tetragonal form (protomer A), and yellow is the tetragonal form (protomer B).
Dimer interface.
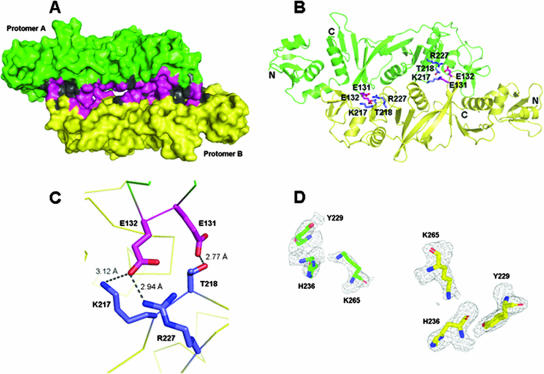
In the tetragonal crystal, molecules A and B form two apparent dimers along the noncrystallographic twofold axes. The extensive dimeric interface is formed by the packing of portions of α8, L22 (residues 217 to 224 between β15 and β16), L14 (residues 131 to 132 between β9 and β10), L16 (residues 162 to 163 between β11 and β12), and the groove between the N terminus (residues 60 to 76) and the C terminus (residues 159 to 164 and 177 to 186). As shown in Fig. 3, it is a broad region that is 14.0% of the total accessible surface of the dimer and buries with a surface area of 3,764 Å2 (Fig. 3A). In total, 90 residues are involved in the dimerization, including several conserved hydrophobic residues (Met, Leu, Gly, Phe, and Val), as cited in reports published previously (20). Overall, there are nonpolar (39.9%), polar (40%), and polar charged (21.1%) residue fractions in the dimer interface. There are three main ionic contacts across the dimeric interface (Fig. 3C). Sequence alignment of EndAs from various species shows Glu-131 and Glu-132 of EndATa to be well conserved. These residues sit on a loop that packs closely against the partner chain and appear to be essential for dimerization (Fig. 3B). Glu-131 forms a hydrogen bond with Thr-218′ of the other subunit (prime indicates the neighboring chain), and its distance is 2.77 Å. The Oɛ1 of Glu-132 makes two hydrogen bonds with Lys-217′ and Arg-227′, and its distances are 3.12 Å and 2.94 Å, respectively (Fig. 3C). Dimerization of two monomers could establish sufficient space for the enzymatic activity and tRNA substrate binding with three catalytic residues containing His-236, Tyr-229, and Lys-265. Archaeal endonucleases can be distinctly grouped into three subfamilies, that of homodimers, that of homotetramers, and that of heterotetramers (5, 36, 41). Dimerization of the EndA family appears to be important for stability and splicing activity. Overall, dimer formation by broad and strong hydrophilic and hydrophobic interactions helps to stabilize substrate binding for enzymatic reactions with EndATa.
FIG. 3.
Dimer interface of the T. acidophilum endonuclease. (A) Schematic representation of the surface model in the dimer structure. The color coding of each protomer is as described in the legend to Fig. 2B. The hydrophilic interactions are shown in magenta, and the hydrophobic interactions are in dark gray. The ratio of hydrophilic to hydrophobic is 1.5:1. (B) The key residues in dimeric interfaces are shown as a ball-and-stick model. The residues E131 and E132 on loop L14 form hydrogen bonds with T218, R227, and K217, respectively, on the opposite protomer. (C) Close-up view of the key interactions on the dimer interface. The residues E131 and E132 of molecule A are in magenta, and K217, T218, and R227 of molecule B are in blue. (D) 2Fo-Fc electron density map covering the region around the catalytic residues (Y229, H236, and K265) of EndATa in the dimer structure at 1.0 σ.
The active site.
The active site has high structural similarity among members of the EndAs. Therefore, the location and mode of substrate binding in the EndA protein could be deduced by analogy with the structures of EndAAf in complex with the BHB substrate (40). On each enzyme surface, there is a cavity with a negative charge located along the center of the dimer interface, and this high negative potential is favorable for attracting substrates with a positive charge, such as the base ring of tRNA. A comparison of the overall structure and the electrostatic-potential surfaces of the EndA family is shown in Fig. 4A and B. The active site is formed close to the dimer interface. Three residues equivalent to His-236, Tyr-229, and Lys-265 in our models have been implicated in the enzymatic activity and tRNA substrate binding (20, 21, 40). The distances between the active sites are 20.4 Å for His-236, 30.6 Å for Tyr-229, and 19.7 Å for Lys-265. These distances correspond to an intron length of pre-tRNA (Fig. 5A).
FIG. 4.
Structural comparison of the tRNA endonucleases. (A) Schematic representation of the surface model in the dimer structure. Molecular surfaces are colored according to electrostatic potential (negative and positive in red and blue, respectively). The orientation is the same as in Fig. 2B. This figure was made with the Pymol program. (B) Comparison of the EndATa, EndAMj, and EndAAf active sites in stereoview. Red is EndAMj, yellow is EndAAf, and blue is EndATa.
FIG. 5.
Structural arrangement of catalytic residues and the flexibility of His236. (A) Arrangement of catalytic residues. The detailed distances between the active sites are given in the text. (B) The position of His-236. Green and yellow show protomers A and B of the dimer, respectively; red represents the monomer.
In the case of EndAMj, it has been suggested that His-125 (His-236 for EndATa) functions as the general base and Tyr-115 (Tyr-229) as the general acid, and Lys-156 (Lys-265) stabilizes the transition state (20, 21). Two residues (His-236 and Tyr-229) occur on short loops between β-strands, except that Lys-265 lies between the β-strand and α-helix. Mutagenesis was undertaken to replace each of these side chains to observe the effects on the behavior of the enzyme and whether these residues are involved in substrate binding or the enzyme action mechanism. The in vitro enzymatic activities of these mutants are shown in Fig. 1C. H236A and Y229F show the same affinity as wild-type EndA for substrate RNA, but K265I showed significantly weaker binding, which probably accounts for the low level of enzyme activity. Comparisons of the EndATa dimer (tetragonal form) with the EndATa monomer (monoclinic form) showed that the His-236 residue is more flexible than the other two catalytic residues (Fig. 5B). When EndA binds to the tRNA substrate, His-236 in these regions can adapt to the suitable sites in the substrate. Lys-265 is critical for binding to the tRNA substrate. These results all clearly support the major mechanistic conclusions reached in previous studies (19, 20, 21).
Comparison with related enzymes.
EndATa shows high sequence identity with the C-terminal regions, having a catalytic activity toward EndAMj and EndAAf (47.3% and 44.0%, respectively) (http://www.ncbi.nlm.nih.gov/BLAST) (Fig. 6). Structural comparisons and a database search using DALI (12) revealed various structures that have similar folding patterns. Three of these structures, EndAMj (PDB entry 1A79; Z score = 11.7; RMSD = 2.5 Å; 131 residues), EndAAf (PDB entry 1RLV; Z score = 21.8; RMSD = 3.1 Å for 261 residues), and tRNA-splicing endonuclease subunit Sen (PDB entry 2GW6; Z score = 5.8; RMSD = 3.3 Å for 91 residues), are particularly useful for an understanding of RNA substrate recognition by EndA. Interestingly, DALI suggests that Holiday junction resolvase from Pyrococcus furiosus shows the third-highest Z score (6.0; PDB entry 1GEF; RMSD = 2.7 Å for 82 residues). The recently published structure of EndAAf complexed with a BHB motif provides structural information about the cleavage mechanism of the EndA family (40). While the overall fold of the catalytic C-terminal domain is similar that of the other archaeal EndAs, the N-terminal domain is notably different from that of EndAMj. Comparisons of the EndAMj homotetramer with the EndATa homodimer showed that most of the conserved residues are located on the catalytic domain. The backbone structure of EndATa could be superimposed on EndAMj and EndAAf, with RMSD of 3.78 Å and 3.09 Å, respectively, for the Cα atoms of the overall structures (each RMSD value was calculated with CNS [4]). Therefore, EndATa is more similar to EndAAf than to EndAMj.
FIG. 6.
Structure-assisted sequence alignment of the endonucleases. Alignments were performed using CLUSTALW and ESPript (http://www.ebi.ac.uk/clustalw; http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Protein designations are as follows: Tacido, T. acidophilum EndA; Afulgi, A. fulgidus EndA; Mjannasi, M. jannaschii EndA; Hvolcan, Halobacterium volcanii EndA; Sen2 and Sen34, S. cerevisiae endonuclease subunits Sen2 and Sen34. Strictly conserved residues have a red background, and residues that are conserved between groups are boxed in blue. Catalytic residues that are well conserved are indicated by triangles.
Conclusions.
We report that the two crystal structures of EndATa strongly demonstrate that the enzyme belongs to a homodimer EndA subfamily. From structural and mutational analyses of H236A, Y229F, and K265I in vitro, we have demonstrated that they play a critical role in recognizing the splice site and in cleaving the pre-tRNA substrate. Our results clearly support the major mechanistic conclusions reached for other members of the EndA family.
Acknowledgments
We thank H. S. Lee and his staff at beamline 4A, Pohang Accessory Laboratory, for assistance in data collection and J. Tame for reading the manuscript. We also thank S. Wakatsuki and his staff for their kind support during X-ray data collection at beamline BL-5A of the Photon Factory, Tsukuba, Japan.
K.Y.H. is supported by KU grants, and Y.K.K. is supported by the Seoul Fellowship. This experiment was supported by the Functional Proteomics Center, 21C Frontier Program, of the Korea Ministry of Science and Technology.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Abelson, J., C. R. Trotta, and H. Li. 1998. tRNA splicing. J. Biol. Chem. 273:12685-12688. [DOI] [PubMed] [Google Scholar]
- 2.Belfort, M., and A. Weiner. 1997. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell 89:1003-1006. [DOI] [PubMed] [Google Scholar]
- 3.Biniszkiewicz, D., E. Cesnaviciene, and D. A. Shub. 1994. Self-splicing group-I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 13:4629-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Calvin, K., D. M. Hall, F. Xu, S. Xue, and H. Li. 2005. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J. Mol. Biol. 353:952-960. [DOI] [PubMed] [Google Scholar]
- 6.Cech, T. 1993. Structure and mechanism of the large catalytic RNAs: group I and group II introns and ribonuclease P subunit, p. 239-270. In R. F. Gesteland and J. F. Atkins (ed.), RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Culbertson, M. R., and M. Winey. 1989. Split tRNA genes and their products: a paradigm for the study of cell function and evolution. Yeast 5:405-427. [DOI] [PubMed] [Google Scholar]
- 8.Diener, J. L., and P. B. Moore. 1998. Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: the bulge-helix-bulge motif. Mol. Cell 1:883-894. [PubMed] [Google Scholar]
- 9.Di Nicola Negri, E., S. Fabbri, E. Bufardeci, M. I. Baldi, D. Gandini Attardi, E. Mattoccia, and G. P. Tocchini-Valentini. 1997. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89:859-866. [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 11.Fabbri, S., P. Fruscoloni, E. Bufardeci, E. Di Nicola Negri, M. I. Baldi, D. Gandini Attardi, E. Mattoccia, and G. P. Tocchini-Valentini. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonuclease. Science 280:284-286. [DOI] [PubMed] [Google Scholar]
- 12.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 13.Kjems, J., and R. A. Garrett. 1988. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54:693-703. [DOI] [PubMed] [Google Scholar]
- 14.Kjems, J., and R. A. Garrett. 1991. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc. Natl. Acad. Sci. USA 88:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjems, J., J. Jensen, T. Olesen, and R. A. Garrett. 1989. Comparison of tRNA and rRNA intron splicing in the extreme thermophile and Archaebacterium Desulfurococcus mobilis. Can. J. Microbiol. 35:210-214. [DOI] [PubMed] [Google Scholar]
- 16.Kleman-Leyer, K., D. W. Armbruster, and C. J. Daniels. 1997. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell 89:839-847. [DOI] [PubMed] [Google Scholar]
- 17.Lave, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, J. C. Horton, and A. J. Rowe (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, London, United Kingdom.
- 18.Lebowitz, J., M. S. Lewis, and P. Schuck. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, H., and J. Abelson. 2000. Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 302:639-648. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., C. R. Trotta, and J. Abelson. 1998. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280:279-284. [DOI] [PubMed] [Google Scholar]
- 21.Lykke-Anderson, J., and R. A. Garret. 1997. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 16:6290-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marck, C., and N. H. Grosjean. 2003. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA 9:1513-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, M. J., and C. C. Query. 2000. Joining of RNAs by splinted ligation. Methods Enzymol. 317:109-123. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 26.Palmer, J. R., T. Baltrus, J. N. Reeve, and C. J. Daniels. 1992. Transfer RNA genes from the hyperthermophilic archaeon, Methanopyrus kandleri. Biochim. Biophys. Acta 1132:315-318. [DOI] [PubMed] [Google Scholar]
- 27.Phizicky, E. M., and C. L. Greer. 1993. Pre-tRNA splicing: variation on a theme or exception to the rule? Trends Biochem. Sci. 18:ix. [DOI] [PubMed] [Google Scholar]
- 28.Randau, L., K. Calvin, M. Hall, J. Yuan, M. Podar, H. Li, and D. Söll. 2005. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl. Acad. Sci. USA 102:17934-17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes, V. M., and J. Abelson. 1988. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55:719-730. [DOI] [PubMed] [Google Scholar]
- 30.Sampson, J. R., and O. C. Uhlenbeck. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 85:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck, P., M. S. Perugini, N. R. Gonzales, G. J. Howlett, and D. Schubert. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82:1096-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, T. H., T. S. Rozhdestvensky, B. C. d'Orval, M. L. Bortolin, H. Huber, B. Charpentier, C. Branlant, J. P. Bachellerie, J. Brosius, and A. Huttenhofer. 2002. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, L. D., and C. J. Daniels. 1988. A tRNA (Trp) intron endonuclease from Halobacterium volcanii: unique substrate recognition properties. J. Biol. Chem. 263:17951-17959. [PubMed] [Google Scholar]
- 35.Thompson, L. D., and C. J. Daniels. 1990. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem. 265:18104-18111. [PubMed] [Google Scholar]
- 36.Tocchini-Valentini, G. D., P. Fruscoloni, and G. P. Tocchini-Valentini. 2005. Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl. Acad. Sci. USA 102:8933-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tocchini-Valentini, G. D., P. Fruscoloni, and G. P. Tocchini-Valentini. 2005. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. USA 102:15418-15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotta, C. R., F. Miao, E. A. Arn, C. K. Ho, R. Rauhut, and J. N. Abelson. 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89:849-858. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, Y., S. Yokobori, T. Inaba, A. Yamagishi, T. Oshima, Y. Kawarabayasi, H. Kikuchi, and K. Kita. 2002. Introns in protein-coding genes in Archaea. FEBS Lett. 510:27-30. [DOI] [PubMed] [Google Scholar]
- 40.Xue, S., K. Calvin, and H. Li. 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312:906-910. [DOI] [PubMed] [Google Scholar]
- 41.Yoshinari, S., S. Fujita, R. Masui, S. Kuramitsu, S. Yokobori, K. Kita, and Y. Watanabe. 2005. Functional reconstitution of a crenarchaeal splicing endonuclease in vitro. Biochem. Biophy. Res. Commun. 334:1254-1259. [DOI] [PubMed] [Google Scholar]