Abstract
We describe purification and characterization of an oligopeptide permease protein (Hly-OppA) from Vibrio furnissii that has multifaceted functions in solute binding, in in vitro hemolysis, in antibiotic resistance, and as a virulence factor in bacterial pathogenesis. The solute-binding function was revealed by N-terminal and internal peptide sequences of the purified protein and was confirmed by discernible effects on oligopeptide binding, by accumulation of fluorescent substrates, and by fluorescent substrate-antibiotic competition assay experiments. The purified protein exhibited host-specific in vitro hemolytic activity against various mammalian erythrocytes and apparent cytotoxicity in CHO-K1 cells. Recombinant Hly-OppA protein and an anti-Hly-OppA monoclonal antibody exhibited and neutralized the in vitro hemolytic activity, respectively, which further confirmed the hemolytic activity of the gene product. In addition, a V. furnissii hly-oppA knockout mutant caused less mortality than the wild-type strain when it was inoculated into BALB/c mice, indicating the virulence function of this protein. Finally, the in vitro hemolytic activity was also confirmed with homologous ATP-binding cassette-type transporter proteins from other Vibrio species.
Members of the genus Vibrio are major causes of human gastroenteritis resulting from the consumption of contaminated marine products. Vibrio furnissii, first described as gas-producing biovar II of Vibrio fluvialis by Brenner et al. (4), like Vibrio parahaemolyticus, is thought to cause acute gastroenteritis with symptoms including diarrhea, abdominal cramps, nausea, and vomiting (4, 25, 39).
Hemolysin (Hly), in addition to other pathogenic Vibrio factors, such as proteases, hemagglutinins, and other hydrolytic exoenzymes, has been suggested to be an important virulence factor in the pathogenesis of many Vibrio species and is the most feared virulence factor involved in the gastrointestinal disorders caused by V. parahaemolyticus (3, 7, 23, 31, 33). However, little is known about V. furnissii and its possible production of toxins, which may be important in both pathogenesis and virulence. Several extracellular hemolysins have been isolated and characterized from various species of Vibrio, but not from V. furnissii (2, 5, 15, 19-21, 32, 40). Recently, a functional role of phosphomannomutase in the virulence of V. furnissii was reported (22).
Oligopeptide transport (Opp) systems in bacteria belong to the ATP-binding cassette (ABC) family of transporters. They are composed of five subunits: an extracellular oligopeptide-binding protein that specifically binds to incoming substrates and delivers them to the translocator, two transmembrane proteins that form the pore, and two ATP-binding proteins involved in ATP hydrolysis (13, 17, 18). The Opp systems have diverse functional roles, ranging from uptake of oligopeptides from growth media to various signaling processes (6, 9, 14, 16, 24, 27, 34, 38). Studies of this protein family have revealed that the oligopeptide permease A (OppA) of bacteria is one of the solute-binding proteins (SBPs) that play an important role in the transport of oligopeptides into the cell and in various signaling processes (24).
Although both Hly and OppA have been broadly characterized, no direct correlations between these two proteins have been reported. In this study, we report identification of an SBP (designated Hly-OppA), originally purified from extracellular media of V. furnissii, that has both a solute-binding function and an in vitro hemolytic activity, and we demonstrate its virulent effect in mice.
MATERIALS AND METHODS
Bacterial strains and materials.
V. furnissii strain ATCC 35016 was obtained in a freeze-dried form from the Culture Collection and Research Center (Hsin-Chu, Taiwan). This bacterium showed hemolysis on tryptic soy broth (TSB) agar plates containing 1.5% NaCl and 5% sheep blood. Phenyl Sepharose 6 Fast Flow, Mono Q, and Sepharose 4B columns were provided by Amersham Pharmacia Biotech (Piscataway, NJ). Protein molecular weight standards and a protein assay kit were obtained from Bio-Rad (Hercules, CA).
Purification of Hly-OppA from V. furnissii culture medium.
An Erlenmeyer flask containing 500 ml TSB was inoculated with V. furnissii and incubated at 37°C in a rotary shaker (180 cycles/min) for 50 h. The culture supernatant was first centrifuged at 6,483 × g for 30 min at 4°C and then subjected to ammonium sulfate precipitation at 60% saturation. The precipitated proteins were collected and resuspended in 10 mM Tris-HCl (pH 7.6). After dialysis in this buffer, the protein solution was loaded onto a Phenyl-Sepharose 6 Fast Flow column preequilibrated with 10 mM Tris-HCl-1 mM EDTA (pH 7.6) and eluted with a linear 0 to 50% ethylene glycol gradient. Fractions exhibiting hemolysis were pooled, dialyzed, and concentrated using a YM30 ultrafiltration membrane. The active sample was applied to a Mono Q column equilibrated with 10 mM Tris-HCl and then eluted with 4 void volumes of a step gradient consisting of 50, 100, 200, 300, and 500 mM and 1 M KCl. Protein eluted between 100 and 200 mM KCl. The active fractions represented partially purified protein and were used for preparation of a monoclonal antibody. The monoclonal antibody was conjugated onto Sepharose 4B and used for protein purification. Next, the protein-bound monoclonal antibody column was washed with 20 void volumes of 10 mM Tris-HCl buffer (pH 7.6) and eluted with 5 void volumes of 10 mM Tris-HCl (pH 2.8). The protein solution was neutralized with 80 μl of 1 M Tris-HCl (pH 10) and assayed to determine hemolytic activity and protein homogeneity.
Assay for hemolytic activity.
Hemolytic activity was determined using rabbit erythrocytes that were washed three times with 10 mM phosphate-buffered saline (PBS) (pH 7.6) and resuspended at a concentration of 4% (vol/vol). For hemolytic activity assays, 0.1 ml of 0.1% Triton X-100, which caused complete release of hemoglobin from erythrocytes and resulted in the maximum change in absorbance at 540 nm, was used as a positive control. The elution buffers, which caused negligible erythrocyte hemolysis compared with sample fractions, were used as negative controls. One hundred percent hemolysis was defined as the A540 of hemoglobin released from erythrocytes treated with 0.1% Triton X-100. One hemolytic unit was defined as the amount of Hly-OppA that caused release of 50% of the hemoglobin from the rabbit erythrocytes.
Molecular cloning and DNA sequencing.
Based on the N-terminal sequence obtained and the ABC-type oligopeptide transporter conserved gene sequence, we designed a set of degenerate primers (sense primer YKW-ABC-N1 [5′-CAAGAGTTCGTTCGTGGTAAC-3′] and antisense primer YKW-ABC-C1 [5′-TTATTGAGCTTTGATGTAAAG-3′]) in order to obtain the core region of the V. furnissii hly-oppA gene. PCR was carried out under the following conditions: denaturation at 94°C for 5 min and then 35 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min.
The rapid amplification of cDNA ends method was used for amplification of both the 5′ and 3′ ends (12). The internal sequence primers VF-ABC-R2 (5′-CGTTTCCCAACTTTCAGCAAC-3′) and VF-ABC-F2 (5′-GCGTGATATGCCAATCGCACC-3′) were used for amplification of the N-terminal and C-terminal DNA fragments, respectively. To obtain the 5′ end of the gene, a single antisense strand was amplified by PCR using genomic DNA and the specific antisense primer VF-ABC-R2. The resulting DNA was incubated with terminal transferase and dATP. The sense strand was then generated with a specific poly(dT) primer, using the antisense strand PCR product as the template. For 3′ end amplification, two anchor primers, VF-ABC-F2 and poly(dT), specific for the central portion and the 3′ nontranslated region, respectively, were used for PCR amplification of the total DNA, as previously described. The amplified DNA fragment was sequenced using an ABI PRISM 3100 autosequencer according to the manufacturer's protocol (Applied Biosystems, Foster City, CA).
Analysis of DNA and amino acid sequences.
The nucleotide sequence analysis and protein sequence comparisons were performed with the BLAST and ClustalW programs, using the BLAST network services at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Expression and purification of recombinant Hly-OppA protein.
The hly-oppA gene was first subcloned into two expression plasmids, pET3a and pET28a(+), for Hly-OppA protein expression and antibiotic susceptibility tests. The hly-oppA PCR fragment was amplified from V. furnissii genomic DNA with oligonucleotide primers YKW-V.fur-ABC-express-N1 (5′-GGAATTCCATATGCAGTTGTTCCAGCTGGCACC-3′; NdeI site underlined) and YKW-V.fur-ABC-express-C1 (5′-CCGGAATTCTTATT GCGCTTTGATGTAAAG-3′; EcoRI site underlined) to obtain a 1.56-kbp DNA fragment. This DNA fragment was digested with the NdeI and EcoR I restriction enzymes and ligated into the pET28a(+) and pET3a(+) vectors, which were predigested with the same restriction enzymes, to obtain recombinant expression plasmids pYKW1 and pYKW3, respectively. pYKW1 and pYKW3 were transformed into Escherichia coli BL21(DE3)(pLysS) cells to generate E. coli strains YKWEC1 and YKWEC3, respectively, for protein expression and antibiotic susceptibility tests.
YKWEC1 was grown in 300 ml of LB broth supplemented with 50 μg/ml kanamycin at 37°C to an A600 of 0.6. Expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the culture was incubated at 37°C for an additional 5 h before centrifugation. The cells were harvested and resuspended in 15 ml of 20 mM Tris-HCl (pH 7.6) buffer. The mixture was sonicated, and the cell debris was removed by centrifugation at 12,000 × g for 30 min at 4°C.
Preparation and characterization of monoclonal antibody.
A 0.5-ml solution containing equal parts of Freund's complete adjuvant and Mono Q chromatography-purified Hly-OppA protein (50 μg) was injected into female BALB/c mice. This was followed by three booster injections consisting of 50 μg of the protein emulsified with Freund's incomplete adjuvant at 10-day intervals; the animals were bled for hybridization 4 days after the last injection. A myeloma cell line (FO) was fused with spleen cells from immunized BALB/c mice at a ratio of 1:5. The culture medium (obtained between days 14 and 21 after fusion) was assayed for the production of specific antibodies by a solid-phase enzyme-linked immunosorbent assay, using purified protein as the antigen. Each monoclonal antibody was established by limiting dilutions at least twice. The monoclonal antibody preparation was used with purified and crude V. furnissii Hly-OppA protein. The specificity of the antibody was confirmed by Western blotting.
For immunoblot experiments, proteins were electrophoretically separated and transferred onto polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were washed with the PBS buffer (pH 7.6) containing 0.05% Tween 20 (PBST) for 10 min, and the immunoblot experiments were carried out by following the procedure for the ECL Western blotting system, using monoclonal antibody raised against Hly-OppA (1:500) and anti-mouse immunoglobulin-horseradish peroxidase (HRP) conjugate (1:5,000). Excess ligand was removed by washing preparations with PBST for 30 min, and detection of the proteins was performed according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, NJ). Membranes were exposed to Hyperfilm ECL (Amersham Pharmacia Biotech, Piscataway, NJ) for different times or until a suitable signal was obtained.
Construction of the V. furnissii hly-oppA knockout mutant and a strain with hly-oppA restored.
An hly-oppA knockout mutant of V. furnissii (VFYKW1) was constructed by the allelic exchange method (10). A 467-bp fragment, which included 162 bp of the 5′ end and 305 bp of the 3′ end of the hly-oppA gene, was amplified by PCR with primers YKW-ABC-N1 and YKW-VF-ABC-knockout-RF1 (antisense primer; 5′-AAGTTGGGAAACGGTGGTGACTACAACAA-3′) and primers YKW-VF-ABC-knockout-F1 (sense primer; 5′-AAGTTGGGAAACGGTGGTGACTACAACAA-3′) and YKW-ABC-C1. The PCR product was cloned into an allelic exchange suicide vector, pCVD442, to generate a recombinant plasmid, pVF-hly-oppA-K, which was transformed into a conjugal donor, E. coli S17-1 λpir (8). The transformed E. coli S17-1 λpir strain and V. furnissii were grown in LB and TSB, respectively, overnight at 37°C before transconjugation was performed. The pVF-hly-oppA-K vector was then transferred from E. coli to V. furnissii. The transconjugants were selected on thiosulfate citrate bile salts sucrose agar plates containing ampicillin (200 μg/ml) and tested for sensitivity to 10% sucrose. The sucrose-sensitive transconjugants were grown in LB containing 10% sucrose overnight and spread onto a plate containing LB with 10% sucrose for selection of sucrose-resistant clones, and they were further tested for ampicillin sensitivity. The sucrose-resistant, ampicillin-sensitive strains were then screened by PCR for a 467-bp product that represented the hly-oppA deletion. The presence or absence of the hly-oppA fragment was validated both by using a standard biochemical substrate assay kit (Microgen GN-ID Identification; Microgen Bioproducts Ltd., Camberley, United Kingdom) and by direct sequencing. Genomic DNA was purified from VFYKW1 and sequenced to confirm successful construction of the knockout strain. For construction of a strain with hly-oppA restored (VFYKW2), the hly-oppA gene containing the signal sequence was amplified by PCR using primers YKW-ABC-SP-N1 (sense primer; 5′-ATGTATAAAAATAAGATCACA-3′) and YKW-ABC-C1.
Oligopeptide binding tests.
For oligopeptide binding experiments, 4 μg/ml each of the Hly-OppA protein and bovine serum albumin (BSA) and different concentrations (0.1, 1, 4, 16, and 64 μg/ml) of a 9-mer oligopeptide library were dot blotted onto a PVDF membrane. The unbound area on the membrane was blocked with 5% nonfat milk and washed three times with PBS containing 0.05% Tween 20. The membrane was incubated with anti-Hly-OppA antibody (1:500) and then with anti-mouse immunoglobulin-HRP conjugate (1:5,000). Membranes were exposed to a 3,3-diaminobenzidine tetrahydrochloride (DAB) solution (10 μl of 30% H2O2 and 100 μl of DAB [20 mg/ml DAB in dimethyl sulfoxide] in 10 ml of 50 mM Tris buffer [pH 7.6]) until a suitable signal was obtained.
Antibiotic susceptibility tests.
Individual plates were inoculated with V. furnissii and E. coli containing recombinant hly-oppA expression plasmids. Filter paper disks that were 6.0 mm in diameter were impregnated with ampicillin (10 μg), carbenicillin (100 μg), cephalothin (30 μg), chloramphenicol (30 μg), colistin sulfate (10 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), penicillin G (10 U), polymyxin B (300 U), streptomycin (10 μg), or tetracycline (30 μg) and were placed on the surface of the plates. The plates were incubated for 16 h at 37°C under microaerophilic conditions. The resulting inhibition zone diameters were expressed in millimeters.
Fluorescent substrate binding and antibiotic competition of the V. furnissii Hly-OppA protein.
The solute-binding efficiency of fluorescent substrates was determined using a Thermo Labsystems fluorometer. The excitation and emission wavelengths were 544 and 590 nm for ethidium bromide (EtBr) and 485 and 538 nm for SYBR green, respectively. To demonstrate binding of EtBr in intact bacteria, wild-type V. furnissii, VFYKW1, YKWEC1, and YKWEC2 were grown overnight in LB (supplemented with 25 μg/ml kanamycin for strains YKWEC1 and YKWEC2) and were diluted into fresh LB. When the cultures reached an A600 of 1 at 37°C, the bacteria were centrifuged at 825 × g for 2 min and resuspended in buffer (50 mM potassium phosphate [pH 7.2], 25 mM glucose, 5 mM MgSO4). The process was repeated twice, and the resulting pellets were resuspended in 1 ml buffer. For the solute-binding activity assay, either EtBr (10 μM) or SYBR green (100 U) was added to a fluorometry plate containing 200 μl of cells. The resulting solution was monitored at 25°C for fluorescence excitation and emission for 20 min at 20-s intervals. When the antibiotic competition assay was performed, different concentrations (500 μg/ml and 2 mg/ml) of ampicillin and 10 μM EtBr were mixed and then added to a fluorometry plate containing 200 μl of cells. For SYBR green competition assays of EtBr influx, different concentrations (5 and 50 U) of SYBR green were added, and the binding efficiency of EtBr was recorded as previously described.
Biofilm assay.
The biofilm formation assay used was based on a modified method (37). Cells from overnight colonies were grown on tryptic soy agar (TSA) plates and in 3 ml TSB at 37°C overnight. Cells from the plates were freshly inoculated into 3 ml TSB and grown to an A600 of 0.5. Three microliters of each cell suspension was added to 1 ml TSB in borosilicate glass tubes. The cultures were then incubated at 37°C without shaking for the time required. The tubes were rinsed with distilled water to remove nonadherent cells. Biofilms were stained by addition of 1.2 ml of 1% crystal violet for 25 min, followed by a distilled water rinse. The cell-associated dye was solubilized in 1.2 ml dimethyl sulfoxide and quantified by measuring the A570 of the resulting solution. Each assay was performed at least in triplicate.
Thermostability of the purified protein.
The effect of temperature on the hemolytic activity of purified Hly-OppA was determined by incubating 2 hemolytic units of the purified protein in PBS for 30 min at different temperatures (16, 25, 30, 37, 40, 42, 45, 50, 52, 55, 57, 60, 65, 70, and 75°C) and then assaying the residual hemolytic activity with 4% rabbit erythrocytes, as previously described.
Detection of cytotoxic effects on CHO-K1 cells.
The purified Hly-OppA was assayed to determine its activity against CHO-K1 cells. Cells were grown in an F-12 medium supplemented with 10% fetal bovine serum and were maintained in a humidified atmosphere consisting of 5% CO2 in air at 37°C. Single-cell suspensions were obtained from ∼90% confluent cultures by harvesting cells with trypsin-EDTA and then seeded into six-well plates. For morphological studies, 1 μg/ml of Hly-OppA was added to a cell culture, and the plates were incubated for 30 min at 37°C. Cells were stained with 50% trypan blue exclusion stain and then visualized by microscopy. In parallel, cells treated with BSA and PBS were used as negative controls.
Cell viability was assessed with a cell counting kit, and the assays were performed in F-12 medium supplemented with 10% fetal bovine serum. The CHO-K1 cells were treated with serial dilutions of Hly-OppA (0, 0.125, 0.25, 0.5, 1, and 2 μg/ml) and maintained in a humidified atmosphere consisting of 5% CO2 in air at 37°C for 24 h. After trypsinization and trypan blue exclusion of the cells, the cells were counted and the results were compared with the numbers of cells in the untreated cultures. The viability data were expressed as means and standard deviations from the three independent experiments.
Scanning electron microscopic analysis of cell morphology.
The V. furnissii wild-type strain and the hly-oppA knockout mutant were incubated in broth medium for the time required. Cell morphology was determined by scanning electron microscopy. Briefly, cell-containing coverslips were fixed in a 2.5% glutaraldehyde solution for 1 h at room temperature. The coverslips were then treated with a 1% osmium tetroxide solution for 30 min, dehydrated with a graded acetone series, washed with an ethanol series, and dried with hexamethyldisilazane (HMDS). The coverslips were then mounted onto stubs using colloidal silver and were sputter coated with gold-palladium.
Virulence of wild-type V. furnissii and hly-oppA knockout mutant strains in mice.
BALB/c mice ranging from 8 to 12 weeks old were used as previously described (38). Mice were anesthetized and injected subcutaneously with various amounts (108 to ∼1012 CFU) of the wild-type V. furnissii and VFYKW1 strains. The numbers of surviving mice inoculated with various numbers of CFU were then monitored for 1 month to determine the 50% lethal dose. Three mice were used in each of the 50% lethal dose experiments, and the data represent three independent experiments.
Nucleotide sequence accession number.
The nucleotide sequence of the V. furnissii hly-oppA gene has been deposited in the GenBank database under accession number DQ777764.
RESULTS
Purification and determination of the sequence of the V. furnissii protein with hemolytic activity.
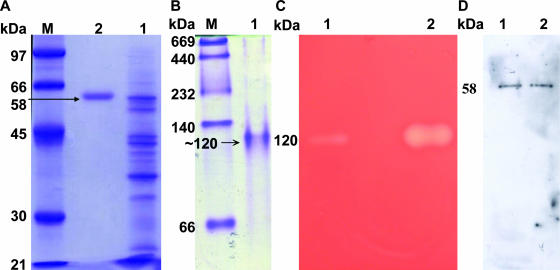
We obtained 877-fold enrichment of the specific in vitro hemolytic activity of Hly-OppA from growth media of V. furnissii (see Table S1 in the supplemental material). Electrophoresis of the homogeneous protein revealed a molecular mass of ∼58 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) (Fig. 1A). A single band at approximately 120 kDa was found using nondenaturing PAGE, and the hemolytic activity of this protein band suggested that it is a dimeric protein under physiological conditions (Fig. 1B and 1C). Immunoblot analysis revealed that both crude and purified proteins produced single bands (Fig. 1D).
FIG. 1.
Purification and characterization of the hemolytic activity of the Hly-OppA protein from V. furnissii. (A) The extracellular medium from a V. furnissii culture (lane 1) was passed through Phenyl Sepharose 6 Fast Flow, Mono Q, and antibody-conjugated Sepharose 4B columns to obtain a homogeneous protein (lane 2) with a molecular mass of ∼58 kDa, as shown by sodium dodecyl sulfate-PAGE. (B) Native PAGE of purified Hly-OppA, showing a molecular mass of ∼120 kDa. (C) Hemolytic activity detected when the ammonium sulfate-precipitated protein fraction (lane 1) and antibody-conjugated purified Hly-OppA (lane 2) from native PAGE were embedded in a blood agar plate. (D) Immunoblot analysis with antiserum against Hly-OppA, revealing that both the crude (lane 1) and purified (lane 2) proteins yielded a single band. Lane M contained markers.
To determine the protein's identity, the purified protein was subjected to both N-terminal determination and internal amino acid sequence determination, and the corresponding gene was cloned and sequenced. The N-terminal sequence was AVVPAGTRLADVQEFVRNC. Three internal peptide sequences were IATAIQSMWK, VTYLPIENQVAEMNR, and SNPLNFTLLYNTSENHK. The gene coding sequence was a 1,551-bp open reading frame that encodes a 516-amino-acid polypeptide with a calculated molecular mass of 58,516 Da; this molecular mass is comparable to the size of the protein purified from the culture medium. A database search of these sequences showed high sequence similarity to the oligopeptide ABC transporter proteins which belong to bacterial extracellular SBP family 5 (35). The deduced amino acid sequence had 91, 82, 73, and 71% identity to V. fluvialis OppA and the ABC-type oligopeptide transporter sequences from Vibrio cholerae, Vibrio vulnificus, and V. parahaemolyticus, respectively. In addition, a conserved AESWETTDNKTFIFHLRKNAKW sequence motif, homologous to the consensus sequence of bacterial extracellular SBP family 5, was identified between amino acids 61 and 82. Compared to the sequence of V. fluvialis OppA, however, a 27-amino-acid region of the signal sequence was absent from the translated N-terminal sequence of the purified protein, but the corresponding sequence was present in the DNA sequence. The extensive sequence homology of the purified protein to bacterial extracellular SBP family 5 but not to the expected hemolysin sequence reported so far is a very interesting finding. These results prompted us to further investigate the solute binding, in vitro hemolytic activity, morphological effects, and role in virulence of the hly-oppA gene product.
Identification of the solute-binding function of the V. furnissii Hly-OppA protein.
To identify the solute-binding function of Hly-OppA, we performed oligopeptide-binding, accumulation of fluorescent substrate, and fluorescent substrate-antibiotic competition experiments. A 9-mer oligopeptide library was first immunoblotted with the anti-Hly-OppA monoclonal antibody and anti-mouse immunoglobulin-HRP conjugate to exclude the possibility of nonspecific binding. The library was then incubated with purified Hly-OppA protein and detected with the above-mentioned primary and secondary antibodies. The results showed that the 9-mer oligopeptide library exhibited concentration-dependent binding, whereas no binding affinity was observed for the BSA protein (Fig. 2).
FIG. 2.
Dot blots demonstrating binding of the 9-mer oligopeptide library to purified Hly-OppA protein in a concentration-dependent manner (lanes 3 to 7). Lanes 1 and 2 contained Hly-OppA protein and BSA, which were positive (P) and negative (N) controls, respectively.
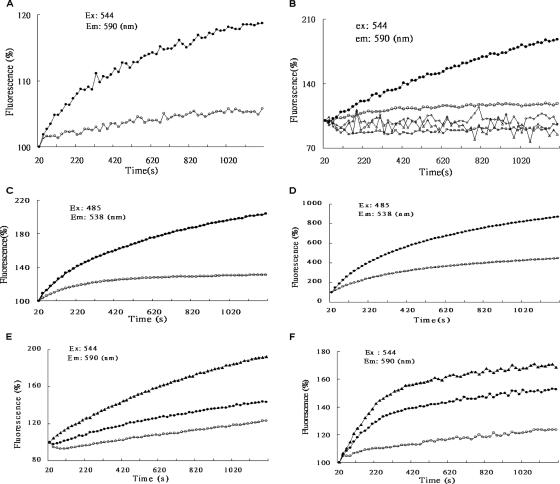
We then investigated the accumulation of the fluorescent substrate of the hly-oppA gene product, using the wild-type V. furnissii strain and the hly-oppA gene knockout mutant (VFYKW1), as well as the strain with the hly-oppA gene transformed into E. coli (YKWEC1) (Fig. 3). We assessed the binding of EtBr and SYBR green (26) in a substrate competition assay by adding different concentrations of ampicillin to EtBr and measuring the effect on the fluorescence emission intensity. The results show that both EtBr and SYBR green accumulation increased with time for both wild-type V. furnissii and YKWEC1, but little effect was seen with VFYKW1 and YKWEC2 (Fig. 4A to D). Moreover, adding different concentrations of ampicillin or SYBR green to the EtBr resulted in a decrease in fluorescence intensity in a concentration-dependent manner compared to the results with EtBr alone (Fig. 4E and 4F), demonstrating the solute-binding activity of the hly-oppA gene product.
FIG. 3.
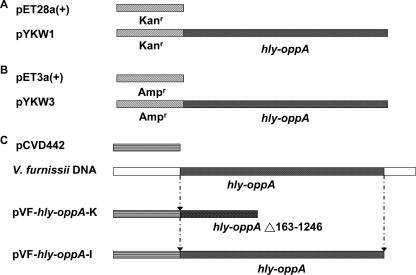
Vector construction for expression and knockout of the hly-oppA gene. (A) The full-length hly-oppA gene was treated with the NdeI and EcoRI restriction enzymes and inserted into a kanamycin-resistant pET28a(+) vector predigested with the same enzymes to generate pYKW1. (B) The same hly-oppA open reading frame was inserted into the NdeI/EcoRI sites of ampicillin-resistant pET3a(+) to generate pYKW3. (C) V. furnissii chromosomal DNA was used as a template for amplification of 162 bp of the 5′ end and 305 bp of the 3′ end of the hly-oppA gene. The 467-bp PCR fragment was cloned into an allelic exchange suicide vector, pCVD442, to generate the knockout plasmid pVF-hly-oppA-K. The pVF-hly-oppA-K plasmid was transferred from E. coli to V. furnissii to generate the V. furnissii hly-oppA knockout strain VFYKW1. The full-length hly-oppA gene was cloned into pCVD442 to generate pVF-hly-oppA-I and transferred to VFYKW1 to generate the V. furnissii VFYKW2 strain with hly-oppA restored.
FIG. 4.
Solute-binding activity and competitive inhibition tests using the fluorescent substrates EtBr (10 μM) and SYBR green (100 U) and various concentrations of ampicillin. The excitation (Ex) and emission (Em) wavelengths were 544 and 590 nm for EtBr and 485 and 538 nm for SYBR green, respectively. (A) Transport of EtBr in wild-type V. furnissii (•) or VFYKW1 (○). (B) Transport of EtBr in YKWEC1 or YKWEC2. □, EtBr alone; ▵, pET28a(+) alone; ⋄, pYKW1 alone; ○, pET28a(+) plus EtBr; •, pYKW1 plus EtBr. (C) Transport of SYBR green in wild-type V. furnissii (•) or VFYKW1 (○). (D) Transport of SYBR green in YKWEC1 or YKWEC2. ○, pET28a(+) plus SYBR green; •, pYKW1 plus SYBR green. (E) Ampicillin competitively inhibited transport of EtBr in YKWEC1 in a concentration-dependent manner. ▴, pYKW1 plus EtBr; •, pYKW1 plus EtBr plus 0.5 mg/ml ampicillin; ○, pYKW1 plus EtBr plus 2 mg/ml ampicillin. (F) SYBR green competitively inhibited transport of EtBr in YKWEC1 in a concentration-dependent manner. ▴, pYKW1 plus EtBr; •, pYKW1 plus EtBr plus 5 U SYBR green; ○, pYKW1 plus EtBr plus 50 U SYBR green.
Characterization of the in vitro hemolytic function and cytotoxicity of the V. furnissii Hly-OppA protein.
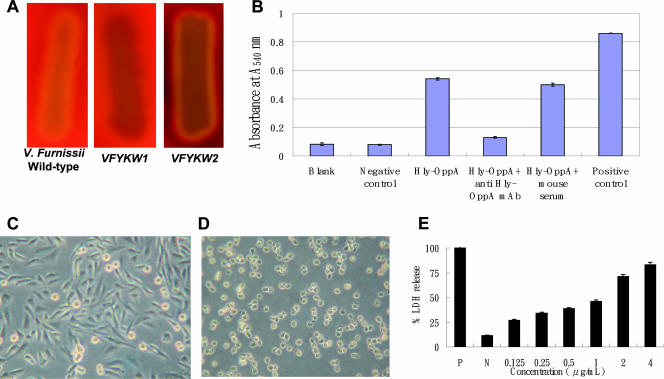
To clearly demonstrate the in vitro hemolytic activity and cytotoxicity of V. furnissii Hly-OppA, we carried out blood agar and erythrocyte lysis assays, mammalian cell cytotoxicity experiments, and anti-Hly-OppA monoclonal antibody protection experiments. Hemolytic activity was present on TSA plates incubated at 37°C with wild-type V. furnissii and VFYKW2 within 11 and 12 h, respectively, but was not present with VFYKW1 even after 36 h of incubation, as shown in Fig. 5A. Incubation with the purified protein also led to significant lysis of sheep erythrocytes and release of hemoglobin as measured by a change in absorbance at 540 nm (Fig. 5B). A decrease in erythrocyte lysis was observed when the protein was neutralized with anti-Hly-OppA monoclonal antibody, suggesting that the monoclonal antibody could block hemolytic activity (Fig. 5B). When mammalian CHO-K1 cells were treated with various concentrations of purified Hly-OppA, morphological changes, including cell detachment, loss of cell cytoplasm with cell shrinkage, and reduction in nucleus size, were observed (Fig. 5C and D). Cytotoxicity was also determined by measuring the amount of lactate dehydrogenase (LDH) activity released into the medium following destabilization of the plasma membrane and the accumulation of LDH in the extracellular compartment after addition of Hly-OppA. The amount of LDH found in the culture medium after addition of 4 μg/ml Hly-OppA was 80% of the amount released after the addition of lysis buffer (Fig. 5E). Purified Hly-OppA exhibited host-specific activities against various erythrocytes, with the highest hemolytic activity against rabbit erythrocytes (100%) and lower activity against mouse (61%), pig (60%), and human erythrocytes (11%), demonstrating the in vitro hemolytic activity and cytotoxicity of the Hly-OppA protein.
FIG. 5.
Effect of the Hly-OppA protein on the V. furnissii hemolytic phenotype, erythrocyte lysis, morphology, and cytotoxicity in CHO-K1 cells. (A) Hemolytic phenotype of wild-type V. furnissii, VFYKW1, and VFYKW2 on TSA containing 5% sheep blood. (B) Erythrocyte lysis and hemoglobin release caused by purified Hly-OppA protein in the presence or absence of anti-Hly-OppA monoclonal antibody, as measured by the change in absorbance at 540 nm. Blank, PBS buffer; Negative control, BSA (0.5 μg/μl); Hly-OppA, 0.1 μg/μl Hly-OppA; Hly-OppA + anti Hly-OppA mAb, 0.1 μg/μl Hly-OppA plus 0.1 μg/μl anti-Hly-OppA monoclonal antibody; Hly-OppA + mouse serum, 0.1 μg/μl Hly-OppA plus 0.1 g/μl mouse serum; Positive control, 0.1% Triton X-100. (C and D) CHO-K1 cells were not exposed (C) or exposed (D) to the Hly-OppA protein (1 μg/ml) for 30 min at 37°C. (E) Dose-dependent cytotoxicity of the Hly-OppA protein in CHO-K1 cells. CHO-K1 cells were exposed to various concentrations of the Hly-OppA protein for 30 min, and the viability of the cells was determined using a commercial cytotoxicity assay kit. The data are the means and standard deviations from at least three independent experiments.
Determination of the morphology and biofilm production of the V. furnissii hly-oppA knockout mutant.
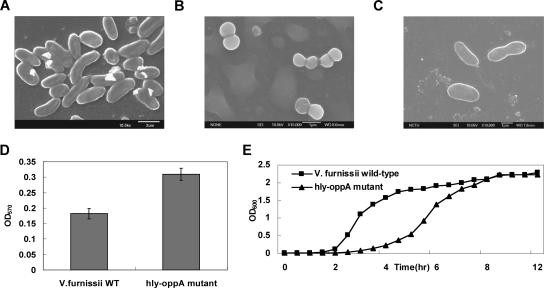
To investigate whether the hly-oppA gene product affects the phenotype of V. furnissii, we assessed morphology and biofilm production on TSB. Scanning electron microscopy examination revealed that the wild-type strain has a rod-shaped morphology, whereas the knockout mutant forms diplococci (Fig. 6A and B). As expected, the cell morphology of a V. furnissii strain with hly-oppA restored (VFYKW2) was the same as that of the wild-type strain (Fig. 6C). We then examined whether the growth rate and biofilm production of the V. furnissii hly-oppA mutant differed from the growth rate and biofilm production of the wild-type strain. The biofilm production of VFYKW1 was about twice that of the wild-type (Fig. 6D), whereas the planktonic cell numbers of the wild type were twice those of VFYKW1 (Fig. 6E). These data suggest that the absence of the hly-oppA gene affects both the cell growth rate and biofilm production, as well as cell morphology.
FIG. 6.
Scanning electron micrographs, biofilm productivity, and growth curves of wild-type V. furnissii and VFYKW1. (A) Micrograph of wild-type V. furnissii exhibiting a rod-shaped morphology. Magnification, ×10,000. (B) Diplococcus-shaped VFYKW1 with a “dehydrated string” morphology on the cellular surface. Magnification, ×10,000. (C) VFYKW2 exhibiting a rod-shaped morphology similar to that of the wild type. (D) Comparison of biofilm production by wild-type V. furnissii (V. furnissii WT) and VFYKW1 (hly-oppA mutant). OD570, optical density at 570 nm. (E) Comparison of growth ratios of wild-type V. furnissii and VFYKW1. OD600, optical density at 600 nm.
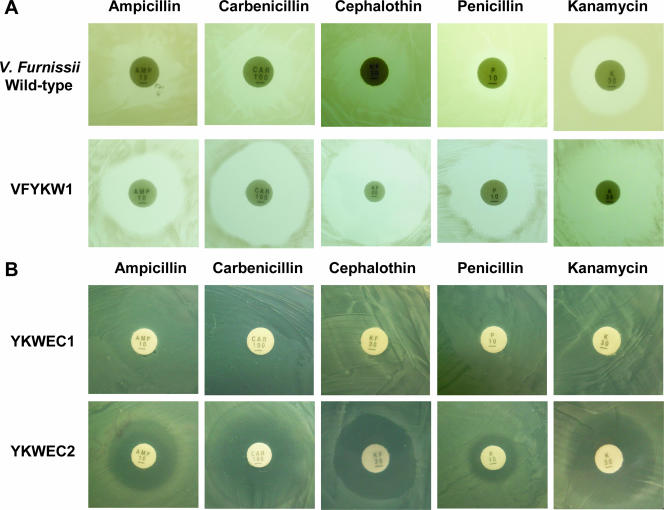
Functional roles of the hly-oppA gene product in antibiotic resistance and virulence.
We next investigated the contribution of the hly-oppA gene product to the development of multidrug resistance in both the wild-type V. furnissii strain and the VFYKW1 knockout strain, as well as in E. coli, and the virulence in mice. Discernible differences in antibiotic resistance between the wild-type V. furnissii and VFYKW1 strains were observed, with the latter exhibiting increased susceptibility, when several antibiotics were tested (Fig. 7A). In addition, two recombinant plasmids, pYKW1 and pYKW3, were constructed, transformed into E. coli BL21(DE3)(pLysS) cells, and used to investigate the effects of different antibiotics. Significant antibiotic resistance was also observed in E. coli cells expressing Hly-OppA recombinant protein but not in E. coli cells lacking this protein (Fig. 7B).
FIG. 7.
Susceptibility to various antibiotics of (A) the wild-type V. furnissii and VFYKW1 knockout strains and (B) recombinant strains YKWEC1 and YKWEC2. The concentrations of various antibiotics utilized in the experiment are described in Materials and Methods.
To investigate the virulence of hly-oppA in vivo, BALB/c mice were injected with wild-type V. furnissii and VFYKW1 and the mortality rates were determined (11, 38). No pathogenic effects were observed when 8 × 1012 CFU of VFYKW1 was injected into mice, but the lethal dose of the V. furnissii wild-type strain was 5 × 109 CFU. These results indicate that the hly-oppA gene is required for V. furnissii lethality in mice.
Determination of in vitro hemolytic activity of homologous SBPs from other Vibrio species.
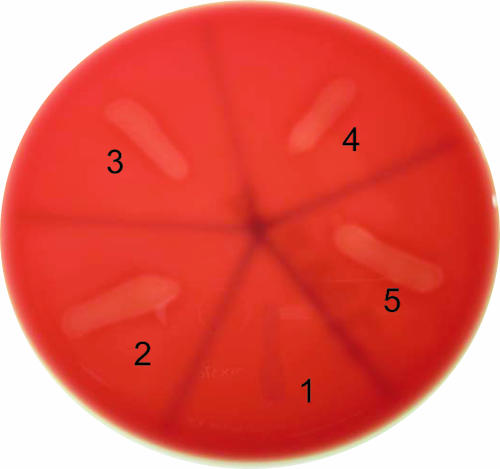
Is the Hly-OppA protein sequence with in vitro hemolytic activity unique to V. furnissii, or is it also present in homologous proteins from other Vibrio species? To answer this question, the V. furnissii hly-oppA gene and the homologous genes encoding ABC-type oligopeptide transporter sequences from V. fluvialis, V. vulnificus, and V. parahaemolyticus were each cloned into a chitinase signal sequence-derived pRSET vector. Apparent in vitro hemolytic phenotypes were observed for all four pRSET-derived recombinant plasmids but not for the control plasmid (Fig. 8), suggesting that the in vitro hemolytic activity may be prevalent in Vibrio species with gene sequences homologous to hly-oppA.
FIG. 8.
Characterization of in vitro hemolytic phenotypes of various Vibrio homologous ABC transporter proteins cloned and overexpressed in E. coli. Section 1, pRSET (negative control); section 2, pRSET-Hly-oppA from V. furnissii; section 3, pRSET-Hly-oppA from V. fluvialis; section 4, pRSET-Hly-oppA from V. vulnificus; section 5, pRSET-Hly-oppA from V. parahaemolyticus.
DISCUSSION
There is increasing evidence for a connection between ABC-type transporter proteins and bacterial virulence. A lipoprotein that confers a hemolytic phenotype to E. coli and shows sequence homology to periplasmic siderophore-binding proteins was identified in Campylobacter jejuni (29, 30). Disruption of an oppB gene in Bacillus thuringiensis was reported to abolish expression of the plcR regulon, a pleiotropic regulator of virulence factors in both B. thuringiensis and Bacillus cereus, and resulted in a nonhemolytic phenotype of B. thuringiensis (14). Further, a mutational study of the oppA gene of group A streptococci also suggested that Opp plays an important role in the pathogenesis of group A streptococcus infection, suggesting that it has dual roles in the regulation of several virulence genes and regulatory genes (38). However, no evidence directly indicating the involvement of OppA in virulence was reported.
Acosta et al. reported that an OppA protein purified from an E. coli K-12 strain exhibited resistance to various aminoglycoside antibiotics (1). Moreover, nine potential drug transporters belonging to resistance-nodulation-division-type systems, drug efflux systems of the major facilitator, multidrug and toxic compound extrusion systems, or ABC superfamilies in Salmonella enterica have been hypothesized to play important roles in multidrug resistance and virulence phenotypes (28). Finally, a recently reported in vivo challenge of plague with the Yersinia pestis OppA protein also suggested that antibody to OppA was responsible for protection, although the functional role of OppA in the virulence of Y. pestis and the mechanism by which immunization with OppA provides protection against Y. pestis remain unclear (36).
In the present study we report that a protein with in vitro hemolytic activity containing an amino acid sequence that is highly homologous to the ABC-type oligopeptide transporter protein from other Vibrio species was purified from the V. furnissii extracellular medium. The solute-binding function and in vitro hemolytic activity of this protein, as well as its cytotoxicity, were characterized using purified and recombinant proteins, as well as anti-Hly-OppA monoclonal antibody protection experiments. An hly-oppA knockout mutant showed a difference in fluorescent substrate accumulation and attenuation of multiple drug resistance to various classes of antibiotics. Both scanning electron microscopy investigation and determination of the mortality rate in BALB/c mice inoculated with both wild-type V. furnissii and the knockout mutant revealed effects on cellular morphology and pathogenesis. Since Hly-OppA has multiple roles in allocrite transport, antibiotic resistance, in vitro hemolysis, changes in morphology, and virulence in mice, the role of this protein is obviously complicated. Further studies are in progress to determine whether the virulence and pathogenesis upon V. furnissii infection and the mode of regulation affect Hly-OppA binding and subsequent molecular interactions.
Supplementary Material
Acknowledgments
We thank National Chiao Tung University and the MOE ATU Program for financially supporting this research.
We are grateful to Yaw-Kuen Li and Michael Donnenberg for supplying the expression vector, the pRSET vector, and the knockout vector pCVD442. We also thank Yu-Ju Chen and Hsin-Kai Liao (Academia Sinica Institute of Chemistry, Taipei, Taiwan, Republic of China) for kindly performing the internal amino acid sequence analyses.
Footnotes
Published ahead of print on 14 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Acosta, M. B. R., R. C. C. Ferreira, G. Padilla, L. C. S. Ferreira, and S. O. P. Costa. 2000. Altered expression of oligopeptide-binding protein (OppA) and aminoglycoside resistance in laboratory and clinical Escherichia coli strains. J. Med. Microbiol. 49:409-413. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., U. H. Stroeher, and P. A. Manning. 1988. Extracellular proteins of Vibrio cholerae: nucleotide sequence of the structural gene (hlyA) for the haemolysin of the haemolytic El Tor strain 017 and characterization of the hlyA mutation in the non-haemolytic classical strain 569B. Mol. Microbiol. 2:481-488. [DOI] [PubMed] [Google Scholar]
- 3.Bhakdi, S., M. Roth, A. Sziegoleit, and J. Tranum-Jensen. 1984. Isolation and identification of two hemolytic forms of streptolysin-O. Infect. Immun. 46:394-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner, D. J., F. W. Hickman-Brenner, J. V. Lee, A. G. Steigerwalt, G. R. Fanning, D. G. Hollis, J. J. Farmer III, R. E. Weaver, S. W. Joseph, and R. J. Seidler. 1983. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 18:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y. C., M. C. Chang, Y. C. Chuang, and C. L. Jeang. 2004. Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr. Microbiol. 49:175-179. [DOI] [PubMed] [Google Scholar]
- 6.Dassa, E., and P. Bouige. 2001. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152:211-229. [DOI] [PubMed] [Google Scholar]
- 7.Datta-Roy, K., K. Banerjee, S. P. De, and A. C. Ghose. 1986. Comparative study of expression of hemagglutinins, hemolysins, and enterotoxins by clinical and environmental isolates of non-O1 Vibrio cholerae in relation to their enteropathogenicity. Appl. Environ. Microbiol. 52:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detmers, F. J., F. C. Lanfermeijer, and B. Poolman. 2001. Peptides and ATP binding cassette peptide transporters. Res. Microbiol. 152:245-258. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, J. J., C. P. Shao, Y. C. Ho, C. K. Yu, and L. I. Hor. 2001. Isolation and characterization of a Vibrio vulnificus mutant deficient in both extracellular metalloprotease and cytolysin. Infect. Immun. 69:5943-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garmory, H. S., and R. W. Titball. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 72:6757-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 15.Han, J. H., J. H. Lee, Y. H. Choi, J. H. Park, T. J. Choi, and I. S. Kong. 2002. Purification, characterization and molecular cloning of Vibrio fluvialis hemolysin. Biochim. Biophys. Acta 1599:106-114. [DOI] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 18.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 19.Hirono, I., T. Masuda, and T. Aoki. 1996. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathog. 21:173-182. [DOI] [PubMed] [Google Scholar]
- 20.Honda, T., Y. X. Ni, and T. Miwatani. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, G. T., J. Y. Lee, S. H. Huh, J. H. Yu, and L. S. Kong. 1997. Nucleotide sequence of the vmhA gene encoding hemolysin from Vibrio mimicus. Biochim. Biophys. Acta 1360:102-104. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. H., S. H. Ahn, J. H. Lee, E. M. Lee, N. H. Kim, K. J. Park, and L. S. Kong. 2003. Genetic analysis of phosphomannomutase/phosphoglucomutase from Vibrio furnissii and characterization of its role in virulence. Arch. Microbiol. 180:240-250. [DOI] [PubMed] [Google Scholar]
- 23.Kreger, A., and D. Lockwood. 1981. Detection of extracellular toxin(s) produced by Vibrio vulnificus. Infect. Immun. 33:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, E. M., S. H. Ahn, J. H. Park, J. H. Lee, S. C. Ahn, and I. S. Kong. 2004. Identification of oligopeptide permease (opp) gene cluster in Vibrio fluvialis and characterization of biofilm production by oppA knockout mutation. FEMS Microbiol. Lett. 240:21-30. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. V., P. Shread, A. L. Furniss, and T. N. Bryant. 1981. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6). J. Appl. Bacteriol. 50:73-94. [DOI] [PubMed] [Google Scholar]
- 26.Mata, M. T., F. Baquero, and J. C. Perez-Diaz. 2000. A multidrug efflux transport in Listeria monocytogenes. FEMS Microbiol. Lett. 187:185-188. [DOI] [PubMed] [Google Scholar]
- 27.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino, K., T. Latifi, and E. A. Groisman. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126-141. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. F., and P. T. Richardson. 1995. Molecular characterization of a Campylobacter jejuni lipoprotein with homology to periplasmic siderphore-binding proteins. J. Bacteriol. 177:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payne, S. M., and K. M. Lawlor. 1990. Molecular studies on iron acquisition by non-Escherichia coli species, p. 225-248. In B. H. Iglewski and V. L. Clark (ed.), The molecular basis of bacterial pathogenesis, vol. XI. Academic Press Ltd., London, United Kingdom. [Google Scholar]
- 31.Raimondi, F., J. P. Kao, C. Fiorentini, A. Fabbri, G. Donelli, N. Gasparini, A. Rubino, and A. Fasano. 2000. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect. Immun. 68:3180-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinoda, S., K. Ishida, E. G. Oh, K. Sasahara, S. Miyoshi, M. A. Chowdhury, and T. Yasuda. 1993. Studies on hemolytic action of a hemolysin produced by Vibrio mimicus. Microbiol. Immunol. 37:405-409. [DOI] [PubMed] [Google Scholar]
- 33.Shirai, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lutticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe, M., H. S. Atkins, D. N. Harland, S. J. Elvin, A. J. Stagg, O. Mirza, R. W. Titball, B. Byrne, and K. A. Brown. 2006. The ABC transporter protein OppA provides protection against experimental Yersinia pestis infection. Infect. Immun. 74:3687-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wai, S. N., Y. Mizunoe, and S. Yoshida. 1999. How Vibrio cholerae survive during starvation. FEMS Microbiol. Lett. 180:123-131. [DOI] [PubMed] [Google Scholar]
- 38.Wang, C. H., C. Y. Lin, Y. H. Luo, P. J. Tsai, Y. S. Lin, M. T. Lin, W. J. Chuang, C. C. Liu, and J. J. Wu. 2005. Effects of oligopeptide permease in group A streptococcal infection. Infect. Immun. 73:2881-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, P. A., P. R. Brayton, T. N. Bryan, and R. R. Colwell. 1986. Numerical taxonomy of vibrios isolated from aquatic environments. Int. J. Syst. Bacteriol. 36:531-543. [Google Scholar]
- 40.Yoh, M., T. Honda, T. Miwatani, S. Tsunasawa, and F. Sakiyama. 1989. Comparative amino acid sequence analysis of hemolysins produced by Vibrio hollisae and Vibrio parahaemolyticus. J. Bacteriol. 171:6859-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.