Abstract
Several serious diseases are caused by biofilm-associated Staphylococcus aureus. Colonial variants occur in biofilms of other bacterial species, and S. aureus variants are frequently isolated from biofilm-associated infections. Thus, we studied the generation of variants with altered expression of virulence factors in S. aureus biofilms. We observed that the number of variants found in biofilms, as measured by hemolytic activity, varied for different strains. Further study of hemolytic activity and signaling by the accessory gene regulator (Agr) quorum-sensing system in one S. aureus strain revealed three primary biofilm subpopulations: nonhemolytic (Agr deficient), hemolytic (Agr positive), and hyperhemolytic (also Agr positive). The nonhemolytic variant became the numerically dominant subpopulation in the biofilm. The nonhemolytic variant phenotype was stable and heritable, indicating a genetic perturbation, whereas the hyperhemolytic phenotype was unstable, suggesting a phase variation. Transcription profiling revealed that expression of the agr locus and many extracellular virulence factors was repressed in the nonhemolytic variant. Expression of the agr-activating gene, sarU, was also repressed in the nonhemolytic variant, suggesting one potential regulatory pathway responsible for the Agr-deficient phenotype. We suggest that the development of these variants in biofilms may have important clinical implications.
Biofilms are microbial communities embedded in a self-produced extracellular matrix (reviewed in reference 8). Many infections caused by Staphylococcus aureus, a common human pathogen, appear to be associated with, or closely resemble, biofilms. These include infections of implanted medical devices or foreign bodies, endocarditis, osteomyelitis, and even some skin infections (1, 8, 13, 22, 31, 33). Biofilm-associated infections are of particular concern, since they remain difficult to control with standard antibiotics (6) or to clear by host defenses (8, 20, 30).
There is increasing recognition that biofilm growth gives rise to a significant population of bacteria with a diverse set of phenotypes, often termed “variants.” These bacterial subpopulations often have characteristics that may benefit the overall community. For instance, some variants found in Pseudomonas aeruginosa biofilms demonstrate either increased ability to disseminate or accelerated biofilm formation (5). Of particular interest is the application of the “insurance hypothesis” to these complex communities of microorganisms (5). Under this hypothesis, the presence of diverse subpopulations enables the overall community to survive in a broader range of conditions and potential challenges than would a community comprised of a single genetic or phenotypic population. In P. aeruginosa biofilms, some variants have been demonstrated to have increased resistance to oxidative stress (5). Stable variants either isolated from biofilms or directly affecting biofilm growth have also been described in several other microorganisms, including Streptococcus pneumoniae (2), Staphylococcus epidermidis (15), Pseudomonas putida (16), Vibrio cholerae (36), Vibrio vulnificus (14), and even Candida spp. (17).
The generation of some variants, such as small colony variants (SCVs; reviewed in reference 26), during growth and infection by S. aureus is well described. Also, the generation of reversible phase variants, mediated by an insertion element, that are altered in their capacity to form biofilm has been studied in S. aureus and S. epidermidis (7, 15, 42). Less well understood are staphylococcal strains with stable defects in the accessory gene regulator (Agr) quorum-sensing system, often isolated from biofilm-associated or persistent infections (11, 12, 27, 38, 39). The Agr system has been assigned a central role in S. aureus pathogenesis (24, 25), and several Agr-regulated genes may also be important in biofilm development (reviewed in reference 41). The expression of surface-associated adhesins that mediate attachment to other cells and to biological surfaces, such as fibrinogen-binding protein, are generally repressed by Agr in response to increasing concentrations of the signal molecule, autoinducing peptide (AIP), encoded by the agr locus. In contrast, secreted virulence factors that may mediate detachment, such as delta-toxin (a molecule with surfactant-like properties) and tissue-degrading enzymes, such as the proteases and hemolysins, are generally induced by Agr. This is consistent with the observation that under certain conditions Agr mutants in staphylococcal species have been shown to form more robust biofilms both in vitro and in vivo than wild-type strains (37-40).
Given the frequency of S. aureus biofilm-associated infections and the common isolation of variants from patients, we sought to determine whether the growth of S. aureus in a biofilm resulted in the accumulation of variants with defects in the production of virulence factors, particularly those controlled by the Agr system.
MATERIALS AND METHODS
Growth media and strains.
A new growth medium (staphylococcal biofilm medium [SBM]) used for biofilm and some chemostat experiments consisted of 5% porcine serum (Sigma) in phosphate-buffered saline-0.1% glucose (pH 7.2). Porcine serum was chosen from among several other animal sera since it most closely resembled human serum with respect to expression of superantigens by S. aureus when added to complex media (data not shown). Enriched medium for chemostat experiments contained SBM with 20 g of Casamino Acids and 10 g of yeast extract/liter. Bacteria were grown in brain heart infusion broth (BHI; Difco) for the quantitative studies of hemolytic activity, AIP activity, and gene expression. For initial screens of variant production, colonies were plated on tryptic soy broth agar (Difco) and then transferred by using sterile toothpicks onto 5% sheep blood agar plates (Becton Dickinson). The S. aureus strains used were FRI1169, isolated from a patient with toxic shock syndrome (3); 15981, a clinical isolate described as biofilm positive (35); RF122, a bovine mastitis isolate (10); and 8325-4, a “prototypical” S. aureus strain (23).
Biofilm and chemostat experiments.
Drip reactor experiments were based in part on those conducted by Boles et al. (5). Biofilms were grown on stainless steel plates within stainless steel reactor chambers. Drip reactor chambers were initially filled with SBM containing a 1:100 dilution of an overnight culture of S. aureus and incubated 24 h at 37°C. Reactors were then set at a 15° angle, and SBM was dripped over the plates at a rate of 4.5 ml/h. After the indicated times, biofilms were removed from the plates by scraping into 10 ml of PBS.
Chemostats were filled with 150 ml of SBM, which was then inoculated with 1.5 ml of an overnight culture of S. aureus. After 24 h with stirring at 37°C, fresh medium was continuously added to the chemostats to achieve a volume change approximately every 4 h.
Cells from both the biofilm and chemostat cultures were dispersed by using a Branson Digital Sonifier 250 (40% power output, 10-s pulse), and the dispersed cells were diluted and plated as described above. Hemolytic activity was scored after overnight incubation of the plates at 37°C, followed by 1 h of incubation at 4°C. Hemolytic, nonhemolytic, and hyperhemolytic isolates were scored as such based on comparison to wild-type colonies.
Measurement of hemolytic activity.
Isolates from biofilms and chemostats were grown overnight in 5 ml of BHI broth. A 100-μl portion of this culture was then transferred to 5 ml of fresh BHI broth, followed by incubation with shaking for 8 h at 37°C. Cells were removed by centrifugation, and the hemolytic activity of the supernatant fluid was measured as described by Bernheimer (4). Incubation of supernatant fluids together with sheep blood was for 30 min at 37°C, followed by 30 min at 4°C. The relative hemolytic activity (hemolytic units measured in the test culture divided by hemolytic units measured in wild-type FRI1169 culture) was calculated based on the absorbance of the centrifuged (5 min at 150 × g) blood and supernatant mixture at an absorbance of 545 nm (Genesys 6; Thermo Electron Corp).
Agr AIP activity measurements.
S. aureus strains can be classified into at least four different Agr groups based on the identity of the AIP expressed by the agr locus (24). Generally, AIP from one Agr group inhibits Agr expression by a different group. Strain MN8 belongs to a different Agr group than does FRI1169, and AIP produced by FRI1169 inhibits Agr expression by MN8. We exploited this property to develop a bioassay to measure relative levels of AIP produced by the biofilm variants. An overnight culture of MN8 carrying an agr P3-gfp reporter (40) was diluted 1:100 into BHI containing 5 μg of chloramphenicol/ml; 50 μl of this suspension was added to each well of 96-well optical plates. The plates were incubated 2.5 h, after which various amounts (1, 5, 10, and 25 μl) of thawed supernatants from test strains were added to the wells. These supernatants were taken from the same cultures as used in the quantitative hemolytic activity assay described above and were stored at −80°C prior to assay. The plates were incubated an additional 22 h, and the fluorescence in each well was measured (Spectra Max M5; Molecular Devices). All incubations were performed at 37°C with shaking at 250 rpm. Normalized AIP activities were calculated based on the ability of the supernatants to inhibit agr P3-gfp activity versus the supernatants from cultures of wild-type FRI1169.
Sequencing agr.
A primer walking-based approach was used to sequence both strands of the agr locus amplified from the genomes of FRI1169 and the nonhemolytic variant using high-fidelity PCR with the primers 5′-ATGATGATTAACTCATCC-3′ and 5′-CACGCGTCATATTTAA-3′. Sequencing was performed by the University of Iowa DNA Facility using an Applied Biosystems Model 3730 sequencer.
Transcriptome analysis.
Cultures of the S. aureus FRI1169 wild-type and nonhemolytic variant were inoculated to an optical density at 600 nm of 0.01 and incubated with shaking at 37°C. Cells were harvested at an optical density at 600 nm of 2 (early stationary phase), and mRNA was isolated and cDNA was prepared as described elsewhere (28). Labeled cDNA was hybridized to Affymetrix S. aureus microarrays at 45°C using the manufacturer's prokaryotic sample and assay processing protocol. Washing, staining, and scanning of microarrays were performed with an Affymetrix fluidic station. Analysis of expression profiling experiments was done as described elsewhere (28).
The original data files for the microarray experiments are deposited at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE8482.
RESULTS
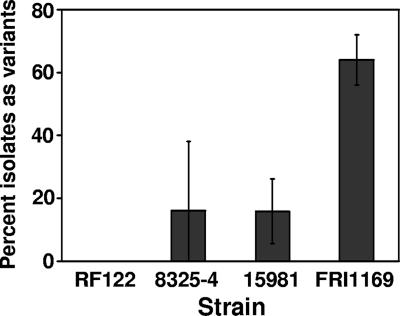
To assess the ability of S. aureus to generate variants during growth in a biofilm, we used the drip reactor system similar to that used by Boles et al. (5). One important modification (see Materials and Methods) was the use of a serum-containing medium that promotes the growth of staphylococcal biofilms under several growth conditions versus standard dilutions of complex media (data not shown). After 2 weeks of growth in these drip reactors, hemolytic variants were observed at various frequencies in cultures of three of four S. aureus strains tested (Fig. 1). Variants were also observed at various frequencies using other biofilm culture systems after growth for extended periods of time (data not shown). The strain that exhibited the highest number of variants, FRI1169, was chosen for further study.
FIG. 1.
Hemolytic variant formation in isolates of various S. aureus strains grown in drip reactor biofilms for 2 weeks as determined by plating on sheep blood agar. Values represent averages and standard deviations of variant percentages in four biofilm cultures of each strain. The value for FRI1169 is significantly higher than for the other strains (P < 0.01, Student t test).
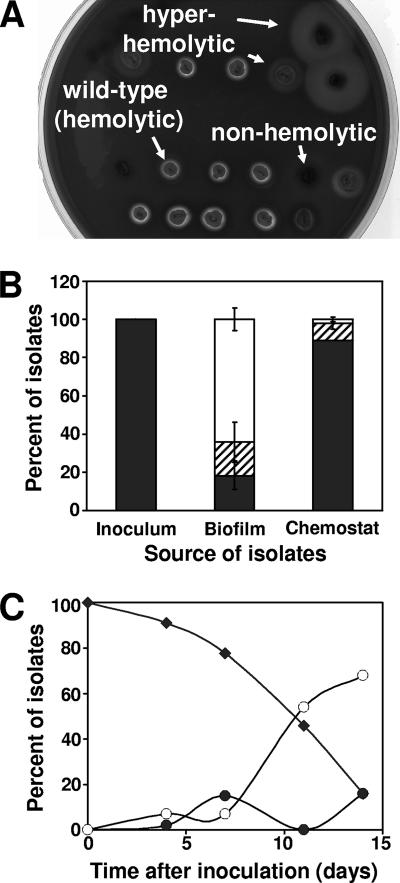
Colonies of FRI1169 isolated from drip reactors exhibited three primary phenotypes when plated on blood agar (Fig. 2A). These included being low or nonhemolytic, hemolytic (comparable to the wild type), and hyperhemolytic. The hemolyzed zone around the hyperhemolytic variants varied in size and demonstrated only partial hemolysis. When plated on complex medium, the nonhemolytic isolates were white, unlike the golden color of the parent strain. When plated on blood agar, only hemolytic colonies were observed among 100 isolates from the planktonic cultures used to inoculate the drip reactors (Fig. 2B). After 2 weeks, nearly two-thirds of the isolates from the drip reactors were nonhemolytic, with the remaining isolates equally divided between the hemolytic and hyperhemolytic populations. In contrast, nearly 90% of the isolates from a 2-week chemostat of planktonic cells exhibited normal hemolytic activity. Most of the remaining chemostat isolates were hyperhemolytic. Although variants were present within 4 days of the inoculation of the biofilm cultures (Fig. 2C), the ratio of variants to hemolytic isolates increased much more quickly between 7 and 11 days and appeared to stabilize by about 14 days. Cultures incubated beyond 14 days did not change appreciably in the total number of variants (data not shown). The percentage of isolates exhibiting hyperhemolysis varied over time. These variations in the amount of hemolytic activity, the type of hemolytic activity, and colony color of isolates from S. aureus biofilms suggest that multiple cellular systems are being affected in the variants.
FIG. 2.
Hemolytic variant generation in cultures of S. aureus FRI1169. (A) Isolates from drip reactors plated onto sheep blood agar. (B) Distribution of hemolytic (▪), hyperhemolytic (▒), and nonhemolytic (□) isolates in overnight culture used to inoculate drip reactors, 2-week-old drip reactor biofilms, and 2-week-old chemostat (planktonic) cultures. Values represent averages and standard deviations of three independent cultures. Changes in the hemolytic and nonhemolytic (but not the hyperhemolytic) populations between the biofilm and chemostat cultures are significant (P < 0.001, Student t test). (C) Hemolytic (⧫), hyperhemolytic (•), and nonhemolytic (○) isolates from drip reactor biofilms sampled at 0, 4, 11, and 14 days after inoculation.
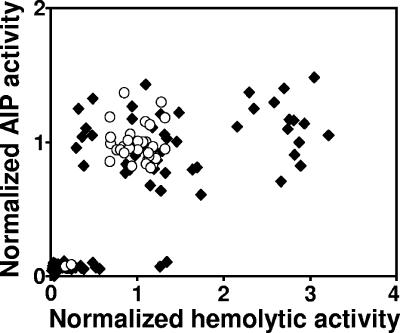
The expression of hemolysins is positively regulated by the S. aureus Agr system (24). To determine whether hemolytic activity in biofilm variants reflected signaling activity by Agr, we measured both hemolytic and AIP activity of 30 isolates from each of four 2-week drip reactor biofilms (total of 120 isolates) and 30 isolates from a 2-week chemostat culture of FRI1169. The data indicate that hemolytic activity generally reflected Agr function (Fig. 3). Isolates with wild-type levels of hemolytic activity generally expressed wild-type levels of AIP. Similarly, nonhemolytic isolates did not produce measurable AIP. In contrast, isolates that expressed two- to threefold hemolytic activity as wild-type isolates did not have a measurable increase in AIP expression. These results indicate that not only is Agr function altered in some variants, but additional regulatory systems controlling the expression of hemolysins are also likely affected.
FIG. 3.
Hemolytic and AIP-signaling activity of isolates from biofilm (⧫) and chemostat (○) cultures. Values are normalized to activities measured in wild-type FRI1169.
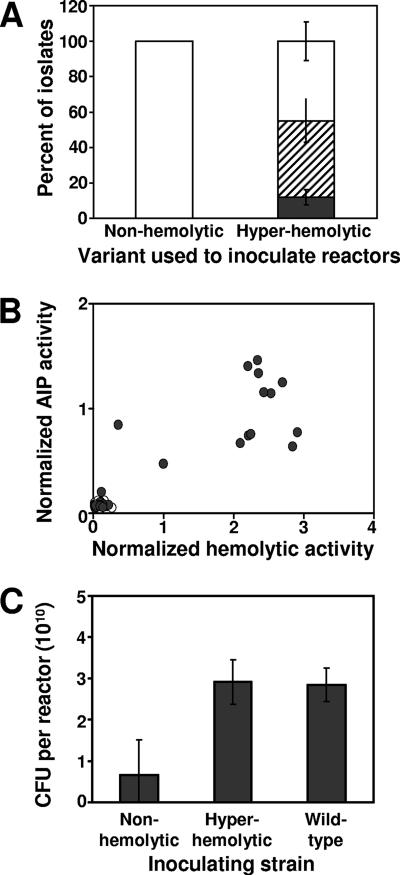
To determine whether the variant phenotypes observed were heritable and stable, one of the nonhemolytic variants and one of the hyperhemolytic variants isolated from a 2-week drip reactor biofilm were used to inoculate new drip reactor biofilms. Whereas the hyperhemolytic variant gave rise to multiple phenotypes, including hyperhemolytic, nonhemolytic, and a small number of hemolytic variants, the nonhemolytic variant gave rise only to other nonhemolytic variants (Fig. 4A). Correspondingly, none of the isolates from the nonhemolytic variant-inoculated culture regained AIP activity, whereas a range of AIP activities was observed in isolates from the hyperhemolytic variant-inoculated biofilm (Fig. 4B). Thus, the nonhemolytic, Agr-deficient phenotypes appear to reflect a heritable genotypic alteration, whereas the hyperhemolytic phenotype is less stable and may reflect some form of phase variation in biofilms. Interestingly, the biofilms inoculated with the hyperhemolytic variant and the wild-type strains resulted in biofilms with approximately four times as many recoverable cells than biofilms inoculated with the nonhemolytic variant (Fig. 4C).
FIG. 4.
Variant generation in 2-week biofilms inoculated with nonhemolytic and hyperhemolytic variants. (A) Distribution of hemolytic (▪), hyperhemolytic (▒), and nonhemolytic (□) isolates from drip reactors inoculated with indicated variant as determined by plating on sheep blood agar. Values represent averages and standard deviations of variant percentages in four biofilm cultures inoculated with each variant. (B) Hemolytic and AIP-signaling activity of isolates from biofilms inoculated with nonhemolytic (○) or hyperhemolytic (•) variants. (C) Viable cells recovered from reactors inoculated with indicated variant. Values represent averages and standard deviations of variant percentages in four biofilm cultures inoculated with each variant. Differences between the values for the hyperhemolytic or wild-type strain and the nonhemolytic strain are significant (P < 0.01, Student t test), while there is no significant difference between the values for the hyperhemolytic and wild-type strains (P = 0.87, Student t test).
If the function of Agr is disrupted in the nonhemolytic variants, this should be reflected in the expression of other important virulence factors in this variant. This was confirmed by using microarrays to compare expression of certain virulence factors in planktonic cultures of wild-type FRI1169 versus the nonhemolytic variant. Expression of several secreted virulence factor genes, including those encoding hemolysins, was repressed in the nonhemolytic variant (Table 1). The array experiments also confirmed that expression of both primary transcripts of the agr locus was repressed. These are the P2 transcript, encoding the signal generation and recognitions systems (agrACDB), and the P3 transcript, RNAIII, which is thought to be the regulatory effector molecule of the Agr system. Interestingly, expression of sarU was also repressed in the nonhemolytic variant. SarU is through to activate both P2 and P3 transcription (21), and its repression in the variant likely contributed to the repression of agr.
TABLE 1.
Change in expression of staphylococcal extracellular accessory protein and regulatory genes in a nonhemolytic variant
| Gene | Virulence factor | Fold changea |
|---|---|---|
| agrACDB | Accessory gene regulator | −9 |
| cap5a | Polysaccharide capsule type 5 | −6 |
| clfB | Clumping factor B | 4 |
| geh | Glycerol ester hydrolase | −3 |
| hlb | Beta-hemolysin | −18 |
| hld/RNAIII | Delta-hemolysin/toxin | −33 |
| hlgBC | Gamma-hemolysin (components B and C) | −11 |
| lip | Butyryl lipase | −8 |
| lukS/F | P-V leukocidin | −6 |
| plc | Phosphatidylinositol-specific phospholipase C | −10 |
| sak | Staphylokinase | −4 |
| sarU | SarU (SarA homolog) | −6 |
| scp | Staphopain | −8 |
| splA-F | Serine protease-like | −41 |
| sspA | Serine protease (V8 protease) | −4 |
| sspB | Cysteine protease | −3 |
| tst | Toxic shock syndrome toxin-1 | −5 |
Value represents the average fold change in gene expression as determined from two independent experiments comparing a nonhemolytic variant to wild-type FRI1169. Only genes identified in reference 25 as staphylococcal extracellular accessory proteins or regulatory genes whose expression was significantly altered are shown.
Traber and Novick (34) have described a slipped-mispairing mutation in AgrA that can result in delayed activation of agr and failure to translate alpha- and delta-hemolysins. This is caused by the addition of an extra A residue to a run of seven A's at the 3′ end of agrA. To determine whether this mutation might contribute to the hemolysin-deficient phenotype that we have observed, we sequenced the agr locus of wild-type FRI1169 and the nonhemolytic variant used to inoculate secondary drip reactors. Both the wild type and the variant had a run of seven A's at the 3′ end of agrA as found in prototypical strains NCTC 8325, COL, and N315 and did not carry the extra A residue as observed by Traber and Novick. Instead, a single mutation was observed in the agr locus of the nonhemolytic variant, a C-to-A conversion in agrA (corresponding to nucleotide 4102 in GenBank accession number X52543) resulting in the conversion of a threonine residue to a lysine residue. This mutation seems insufficient to explain the phenotypes and gene expression patterns observed in the nonhemolytic variant.
DISCUSSION
Our study demonstrates that variants with altered expression of virulence factors are generated during biofilm growth of S. aureus and that for strain FRI1169, the nonhemolytic/Agr-deficient variants become the numerically dominant subpopulation. This nonhemolytic/Agr-deficient phenotype appears to be the result of a heritable genetic event(s). We have identified one potential regulatory pathway, the repression of sarU, that could contribute to agr repression in the nonhemolytic variant. However, we have not yet discovered the genetic event(s) that might be occurring in S. aureus during biofilm growth leading to variant generation. The multiple phenotypes and gene expression patterns observed suggest that several genetic events may be occurring, including events outside the agr locus, and the combination of mutations may vary from isolate to isolate. We also have not distinguished among the possible reasons for the accumulation of nonhemolytic/Agr-deficient variants in the FRI1169 biofilms. It could be that in biofilms, nonhemolytic variants are generated at a higher rate, selection favors them, or they detach less frequently than the hemolytic/Agr-positive cells. One clue might be the observation that when the medium used in the chemostat cultures was enriched to increase the cell density by 5- to 10-fold, the generation of variants increased to ca. 50% of the overall population (data not shown). Thus, cell density and/or the by-products of metabolism may contribute to mechanisms within S. aureus that generate phenotypic and genotypic diversity. Another clue might be the observation that when drip reactor cultures are inoculated with a mixture of wild-type strains 8325-4 or 15981 and their corresponding nonhemolytic variants (95:5 ratio, respectively), the resulting 14-day-old biofilms contain a significantly higher percentage of variants than observed in the experiment in Fig. 1, where only the wild-type strains were used as the inoculum (data not shown). Thus, once the biofilm colony is established, the nonhemolytic variants may have some selective advantage within the biofilm or are more likely to be retained within the biofilm. It should be noted that changes in Agr activity would also be expected to alter the expression of many genes not directly associated with pathogenesis (9), and the impact of these gene products on biofilm development has yet to be fully understood.
Agr-deficient variants may be better suited to biofilm formation and long-term, chronic infection since (i) they tend to express the surface adhesins that mediate cell-to-cell and cell-to-surface interactions (e.g., fibrinogen-binding protein), while repressing factors that may facilitate detachment (e.g., delta-toxin), and (ii) they tend to express more immuno-evasive factors (e.g., protein A) than immunostimulatory ones (e.g., superantigen) (18, 24). In addressing the first idea, our laboratory has confirmed here that Agr-deficient variants become the predominant form in biofilms grown in a serum-based medium. Also, we have some evidence that the Agr-positive population is not completely lost from the biofilm, suggesting that there may be some mechanism to retain the capability to express invasive factors at an appropriate stage of infection. Indeed, we have detected the frequent detachment of cells expressing Agr from the biofilm (40). This may have important clinical implications, as detaching cells expressing Agr are also likely to be expressing extracellular virulence factors important in causing acute infection. Interestingly, drip reactor biofilms inoculated with Agr-positive cells, either wild type or hyperhemolytic, contained more viable bacteria than did biofilms formed from a hemolytic/Agr-deficient variant. Thus, at least under these conditions, the Agr-positive strains may be more adept at colonization.
There is substantial supporting evidence that Agr variants arise commonly both in vitro and in vivo and may play an important role in staphylococcal pathogenesis, particularly in persistent and/or biofilm-associated infections. Somerville et al. (32) found that serial passage of S. aureus in planktonic culture resulted in the loss of Agr function in a large percentage of the cell population, along with hemolytic and aconitase activities. These authors hypothesized that frequent mutations of the agr locus create a mixed population of bacteria, with some cells expressing colonization factors and others expressing secreted exotoxins. Under a particular environment with specific ecological and/or immunological selection, the Agr variant best able to adapt would emerge. Interestingly, strain FRI1169 itself was shown in early studies of toxin production to generate variants under certain growth conditions with altered levels of toxic shock syndrome toxin expression (19). One can hypothesize that Agr expression might have also been affected in these variants.
agr mutants are frequently found among staphylococcal clinical isolates. Vuong et al. (39) found that 26% of 105 S. aureus isolates failed to produce delta-toxin, indicating that they were deficient in quorum-sensing-mediated regulation. When Goerke et al. (12) isolated staphylococci from the lungs of cystic fibrosis patients, not only did the strains generally express low levels of RNAIII, but several isolates were also found to be Agr negative. Fowler et al. (11) showed that the percentage of S. aureus isolates recovered from patients with persistent bacteremia defective in delta-toxin production (a consistent indicator of Agr activity) was higher than in isolates from patients with resolving bacteremia (71% versus 39%, respectively). Interestingly, staphylococci with intermediate resistance to glycopeptide antibiotics are frequently isolated from biomedical device-related infection, which are also likely to be biofilm associated. These same staphylococcal strains were shown to be predominantly Agr negative (27).
Similar findings regarding Agr and biofilms have been made in studies of S. epidermidis, which contains an Agr system with a high degree of similarity to that of S. aureus. Vuong et al. (38) determined that S. epidermidis agr mutants show increased colonization of indwelling devices in a rabbit model, as well as increased adhesion to epithelial cells. The same study also found that isolates from joint prostheses infections were more likely to carry a nonfunctional Agr system than isolates from healthy individuals.
Our observation of the eventual numerical dominance of the biofilm by the FRI1169 nonhemolytic variant is consistent with the results of Schwan et al. (29). This group examined the behavior of mixed populations of hyperhemolytic, hemolytic, and nonhemolytic variants in a murine abscess model of infection. They found that the percentage of nonhemolytic variants, likely representing Agr-negative bacteria, recovered from the wound increased over time, whereas the number of hyperhemolytic variants decreased dramatically over the same time period. A wound infection model demonstrated the same trend, although to a lesser degree. In contrast, in a model of systemic infection, hemolytic variants seemed to be favored in isolates recovered from murine livers and spleens. Thus, Agr activity likely facilitates survival and pathogenesis in some host environments, but not in others.
Our study indicates that the rate of variant generation may be very different among different clinical isolates (Fig. 1). This leads to several intriguing questions: namely, what are the implications of being infected by an isolate more likely to generate variants? Is it more likely that this will lead to persistent infection? With regard to the insurance hypothesis described by Boles et al. (5), do the clinical isolates with greater ability to generate variants demonstrate increased fitness in terms of ability to infect new patient populations or increased resistance to immune response or antibiotic therapies? Only correlation of clinical studies with assessment of variant generation will fully address the role that variants play in staphylococcal pathogenesis.
Acknowledgments
This study was funded in part by a gift from the Procter & Gamble Co. to the University of Iowa Foundation and College of Medicine. J.M.Y. performed much of this research while an NIH Ruth L. Kirschstein postdoctoral fellow.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Akiyama, H., T. Hamada, W. K. Huh, O. Yamasaki, T. Oono, W. Fujimoto, and K. Iwatsuki. 2003. Confocal laser scanning microscopic observation of glycocalyx production by Staphylococcus aureus in skin lesions of bullous impetigo, atopic dermatitis, and pemphigus foliaceus. Br. J. Dermatol. 148:526-532. [DOI] [PubMed] [Google Scholar]
- 2.Allegrucci, M., and K. Sauer. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 189:2030-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergdoll, M. S., B. A. Crass, R. F. Reiser, R. N. Robbins, and J. P. Davis. 1981. A new staphylococcal enterotoxin, enterotoxin F, associated with toxic-shock-syndrome Staphylococcus aureus isolates. Lancet 1:1017-1021. [DOI] [PubMed] [Google Scholar]
- 4.Bernheimer, A. W. 1988. Assay of hemolytic toxins. Methods Enzymol. 165:213-217. [DOI] [PubMed] [Google Scholar]
- 5.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2000. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J. Appl. Microbiol. 88:1028-1037. [DOI] [PubMed] [Google Scholar]
- 11.Fowler, V. G., Jr., G. Sakoulas, L. M. McIntyre, V. G. Meka, R. D. Arbeit, C. H. Cabell, M. E. Stryjewski, G. M. Eliopoulos, L. B. Reller, G. R. Corey, T. Jones, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbiocidal protein. J. Infect. Dis. 190:1140-1149. [DOI] [PubMed] [Google Scholar]
- 12.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 14.Grau, B. L., M. C. Henk, and G. S. Pettis. 2005. High-frequency phase variation of Vibrio vulnificus 1003: isolation and characterization of a rugose phenotypic variant. J. Bacteriol. 187:2519-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53:367-374. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, S. K., J. A. Haagensen, M. Gjermansen, T. M. Jorgensen, T. Tolker-Nielsen, and S. Molin. 2007. Characterization of a Pseudomonas putida rough variant evolved in a mixed species biofilm with Acinetobacter sp. strain C6. J. Bacteriol. 189:4932-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, J. J., R. J. Turner, and H. Ceri. 2007. A subpopulation of Candida albicans and Candida tropicalis biofilm cells are highly tolerant to chelating agents. FEMS Microbiol. Lett. 272:172-181. [DOI] [PubMed] [Google Scholar]
- 18.Kong, K. F., C. Vuong, and M. Otto. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296:133-139. [DOI] [PubMed] [Google Scholar]
- 19.Lee, A. C., and M. S. Bergdoll. 1985. Spontaneous occurrence of Staphylococcus aureus mutants with different pigmentation and ability to produce toxic shock syndrome toxin 1. J. Clin. Microbiol. 22:308-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leid, J. G., M. E. Shirtliff, J. W. Costerton, and A. P. Stoodley. 2002. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 70:6339-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayberry-Carson, K. J., B. Tober-Meyer, J. K. Smith, D. W. Lambe, Jr., and J. W. Costerton. 1984. Bacterial adherence and glycocalyx formation in osteomyelitis experimentally induced with Staphylococcus aureus. Infect. Immun. 43:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 24.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 25.Novick, R. P. 2006. Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation, p. 496-516. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 26.Proctor, R. 2006. Respiration and small-colony variants of Staphylococcus aureus, p. 434-442. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, DC.
- 27.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwan, W. R., M. H. Langhorne, H. D. Ritchie, and C. K. Stover. 2003. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed infections in murine abscesses and wounds. FEMS Immunol. Med. Microbiol. 38:23-28. [DOI] [PubMed] [Google Scholar]
- 30.Shiau, A. L., and C. L. Wu. 1998. The inhibitory effect of Staphylococcus epidermidis slime on the phagocytosis of murine peritoneal macrophages is interferon-independent. Microbiol. Immunol. 42:33-40. [DOI] [PubMed] [Google Scholar]
- 31.Shirtliff, M. E., J. T. Mader, and A. K. Camper. 2002. Molecular interactions in biofilms. Chem. Biol. 9:859-871. [DOI] [PubMed] [Google Scholar]
- 32.Somerville, G. A., S. B. Beres, J. R. Fitzgerald, F. R. DeLeo, R. L. Cole, J. S. Hoff, and J. M. Musser. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoodley, P., S. Kathju, L. Nistico, L. Hall-Stoodley, A. Gieseke, J. M. Coticchia, M. Baratz, J. E. Kerschner, Y. Liu, J. C. Post, and G. D. Ehrlich. 2007. Bacterial biofilms: standards for clinical diagnostics. Are we there yet?, abstr. S2:2, p. 21. Abstr. Biofilms 2007. American Society for Microbiology, Washington, DC.
- 34.Traber, K., and R. Novick. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ- and α-haemolysins. Mol. Microbiol. 59:1519-1530. [DOI] [PubMed] [Google Scholar]
- 35.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 36.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vuong, C., F. Gotz, and M. Otto. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect. Immun. 68:1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vuong, C., S. Kocianova, Y. Yao, A. B. Carmody, and M. Otto. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190:1498-1505. [DOI] [PubMed] [Google Scholar]
- 39.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 40.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarwood, J. M. 2006. Quorum-sensing-dependent regulation of staphylococcal virulence and biofilm development, p. 199-231. In D. R. Demuth and R. J. Lamont (ed.), Bacterial cell-to-cell communication: role in virulence and pathogenesis. Cambridge University Press, Cambridge, United Kingdom.
- 42.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]