Abstract
We recently identified a minireplicon of pBtoxis from Bacillus thuringiensis subsp. israelensis that contained an operon encoding two novel proteins (ORF156 and ORF157), both of which are required for replication. ORF157 contains a helix-turn-helix motif and shares no homology with known plasmid replication proteins (Rep), and ORF156 contains the signature motif present in FtsZ/tubulin proteins, the latter of which are known to function in cell division and chromosome segregation. Here we show that the minimal sequence composed of four 12-bp imperfect direct repeats (iterons) in the pBtoxis minireplicon was sufficient to replicate a reporter plasmid in B. thuringiensis subsp. israelensis when ORF156 and ORF157 functions were provided in trans. To further investigate the roles of ORF156 and ORF157 in pBtoxis replication, six-histidine-tagged recombinant rORF156 and rORF157 proteins were purified from Escherichia coli and used in electrophoretic mobility shift assays. Our results demonstrated that rORF157, but not rORF156, binds specifically to the pBtoxis iterons, suggesting that ORF157 functions as a Rep protein. Although rORF156 did not bind to the iteron sequence, we showed that it bound to rORF157-DNA complexes. In addition, we showed that rORF156 has GTPase activity characteristic of the FtsZ/tubulin superfamily of proteins. Taken together, these results suggest that the iterons compose the minimal replication origin (ori) of pBtoxis and that ORF157 and ORF156 are involved in the initiation of pBtoxis replication and possibly in the segregation and partitioning of this plasmid to daughter cells.
The large 128-kb plasmid, pBtoxis, in Bacillus thuringiensis subsp. israelensis encodes all major endotoxin proteins (Cyt1Aa, Cry4A, Cry4B, and Cry11A) responsible for the mosquito larvicidal activity of this strain (3, 7, 17). The molecular mechanism by which pBtoxis replicates has not been established, though preliminary evidence indicates it has unique features (18). For example, we recently identified a minireplicon of pBtoxis that contains an operon with two open reading frames (ORF), pBt156 and pBt157, coding for two novel proteins, respectively, ORF156 (54.4 kDa) and ORF157 (11.8 kDa) (18). The ORF157 protein contains a helix-turn-helix motif common in a wide variety of protein families, including those involved in transcription and DNA replication (1), whereas ORF156 contains a sequence motif present in the FtsZ/tubulin superfamily (3, 18) and ORF45 (RepX) encoded by the virulence plasmid, pXO1, in Bacillus anthracis (19). An apparent homologue of ORF157 is absent in pXO1. Despite the presence of conserved domains in ORF157, ORF156, and RepX, these proteins share no homology with proteins known to function in plasmid replication and partitioning (1, 19). Nevertheless, Tang et al. have shown that ORF156 and ORF157 are required for minireplicon replication (18). Indeed, Tinsley and Khan (19) have reported the RepX is also essential for replicating a pXO1-derived minireplicon and have proposed that the FtsZ-like proteins encoded by large plasmids in members of the Bacillus cereus sensu lato group (B. thuringiensis, B. anthracis, and Bacillus cereus) represent a novel family of initiator proteins involved in replicating large virulence plasmids. Though the specific functions of ORF156 and ORF157 in pBtoxis replication are unknown, in a more recent study Larsen et al. (12) showed that ORF156 fused to the green fluorescent protein (TubZ-GFP) assembles directional linear polymers characteristic of the tubulin superfamily of proteins and have also provided evidence that ORF157 (TubR) has a negative effect on ORF156 levels.
The pBtoxis minireplicon also contains two putative cis elements; a putative dnaA box motif (TTTTCCACT) similar to those found in plasmids in Escherichia coli and Bacillus subtilis, to which initiator DnaA proteins bind; and iterons known to be essential for initiating replication of large theta-replicons (6, 10, 11). Iterons function as binding sites for plasmid-encoded Rep proteins and in the formation of higher-order structures of iteron-initiator complexes that are required to initiate plasmid replication (4, 14, 15). Similar to other large plasmids with A+T-rich iterons, the pBtoxis iterons are composed of four 12-bp A+T-rich direct imperfect repeated sequences (T[A/T][A/T][C/A][G/A]GTTTA[A/C][A/C]), which together with the putative dnaA box are found upstream of the pBt157 gene (18).
To date, little is known about the molecular basis for DNA replication of the large virulence plasmids of the B. cereus sensu lato group. The novel features of the plasmid-encoded FtsZ tubulin-like proteins, ORF156 and RepX, and the helix-turn-helix-containing ORF157 suggest a conserved but unknown mechanism for propagating and restricting these plasmids within a narrow host range. Here, we demonstrate that the 48-bp iterons alone were sufficient to support replication of a reporter plasmid propagated in B. thuringiensis subsp. israelensis in which ORF156 and ORF157 functions were provided in trans by parental pBtoxis, demonstrating that the iterons compose the minimal replication origin (ori) of the pBtoxis minireplicon. Furthermore, we used electrophoretic mobility shift assays (EMSAs) to show that ORF157, but not ORF156, binds with high specificity to the iteron sequence, suggesting that this binding event initiates pBtoxis replication. Although ORF156 is not an iteron-binding protein, we show that it is an Mg2+-dependent GTPase that interacts directly with ORF157-DNA complexes, thereby implicating its role as a replication initiator protein.
MATERIALS AND METHODS
Bacterial strains, DNA manipulation, and transformation.
The bacterial strains described here are listed in Table 1. Luria-Bertani medium was used to culture Escherichia coli, B. thuringiensis subsp. israelensis 4Q7, and B. thuringiensis subsp. israelensis 4Q5. Antibiotic concentrations for bacterial selection were as follows: 100 μg/ml ampicillin and 10 μg/ml chloramphenicol. DNAs from B. thuringiensis and E. coli were extracted using the plasmid Midi and Maxi kits (QIAGEN) and the Wizard plus minipreps DNA purification system (Promega). DNA fragments were isolated from agarose gels using the QIAquick gel extraction kit (QIAGEN) and ligated using the Fast-Link DNA ligation kit (Epicenter). PCR was performed with the Expand Long Template PCR system (Roche Molecular Biochemicals) in a Px2 system thermocycler (Thermo Hybaid) for 30 cycles as follows: 94°C for 30 s, 55°C for 30 s, and 68°C for 2 min. Transformation of B. thuringiensis subsp. israelensis 4Q7 and B. thuringiensis subsp. israelensis 4Q5 with 5 μg plasmid DNA was performed as described previously (21, 22).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| B. thuringiensis | ||
| 4Q7 | Acrystalliferous strain of B. thuringiensis subsp. israelensis | Bacillus Genetic Stock Center |
| 4Q5 | Plasmid-cured mutant of B. thuringiensis subsp. israelensis; bears only pBtoxis plasmid | Bacillus Genetic Stock Center |
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | |
| Top10 | Invitrogen | |
| Plasmids | ||
| pC194 | Plasmid from Staphylococcus aureus containing a chloramphenicol resistance gene | 9 |
| pUC19 | High-copy-no. E. coli vector containing MCS; Ampr | 23 |
| pUCCl | pUC19 containing the chloramphenicol resistance gene from pC194 | This work |
| pUCSw | pUCCl containing 2.2-kb mini-replicon of pBtoxis | This work |
| pUCp | pUCCl containing 191-bp fragment upstream of ORF157 | This work |
| pUCID | pUCCl containing 97-bp fragment (48-bp iterons, 37-bp spacer, plus putative dnaA box) | This work |
| pUCIDM | pUCCl containing iterons, 37-bp spacer, and mutated dnaA box | This work |
| pUCIS | pUCCl containing iterons and 37-bp spacer | This work |
| pUCSD | pUCCl containing 37-bp spacer and putative dnaA box | This work |
| pUCI | pUCCl containing 48-bp iteron sequence | This work |
| pBtKp | pUCE containing 6.3-kb KpnI fragment of pBtoxis | 18 |
| pBAD/myc-HisB | Vector for expression of recombinant proteins containing C-terminal 6-His tag in E. coli | Invitrogen |
| pBAD156 | pBAD/myc-HisB containing pBt156 gene | This work |
| pBAD157 | pBAD/myc-HisB containing pBt157 gene | This work |
Construction of recombinant plasmids.
The plasmids described here are listed in Table 1. To construct pUCCl, a vector that does not replicate in B. thuringiensis, the 1.0-kb Sau3AI/HpaII fragment containing the chloramphenicol resistance gene from pC194 (9) was cloned into the blunted NdeI site of pUC19. Plasmid pUCSw, which contained the minireplicon of pBtoxis (18), was constructed by cloning the 2.2-kb SwaI fragment from pBtKp (Table 1) in the SmaI site of pUCCl. Plasmid pUCP (Fig. 1), which contained the 191-bp sequence upstream from pBt157 was constructed by inserting a PCR fragment amplified with primers pP-F and pP-R (Table 2) into the HindIII and PstI sites in pUCCl. Plasmid pUCID (Fig. 1), which contained the four 12-bp-iteron sequence and the 37-bp spacer followed by the putative dnaA box motif (5′-TTTTCCACT-3′), was constructed by inserting a 97-bp PCR fragment amplified with primers pIDna-F and pIDna-R (Table 2) into the HindIII and PstI sites in pUCCl. Plasmid pUCIDM, which was identical to pUCID except, for having the putative dnaA box motif replaced with 5′-CCCCAAGAC-3′, was constructed using plasmid pUCID as the template and primers pIDM-F and pIDM-R; the ScaI/PstI-digested 1.8-kb amplicon was ligated to the larger ScaI/PstI fragment of pUCCl to generate pUCIDM. Plasmid pUCIS, which contained the iteron sequence and the 37-bp spacer sequence, but lacked the putative dnaA box motif, was constructed by ligating the ScaI/PstI-digested 1.8-kb fragment amplified with primers pIDM-F and pIS-R, using pUCID as the template, with the larger ScaI/PstI fragment of pUCCl. Plasmid pUCSD, which contained the 37-bp spacer sequence and dnaA box motif, but not the iterons, was constructed by ligating the NcoI/HindIII-digested 1.0-kb PCR fragment, which was amplified with primers pSD-F and pSD-R and using plasmid pUCID as the template, with the larger NcoI/HindIII fragment of pUCCl. To construct pUCI (Fig. 1), a synthetic double-stranded oligonucleotide prepared by annealing oligonucleotides 5′-CCCAAGCTTTAAAGGTTTAAATAACAGTTTAAATTTAAGTTTAACTTTCAGTTTACACTGCAGTT-3′ and 5′-AACTGCAGTGTAAACTGAAAGTTAAACTTAAATTTAAACTGTTATTTAAACCTTTAAAGCTTGGG-3′, which contained the 48-bp direct-repeat iteron sequence, was digested with HindIII and PstI (sites are underlined in sequences above) and inserted into the same sites in pUCCl. The nucleotide integrity of each construct was confirmed by DNA sequencing and restriction enzyme analyses.
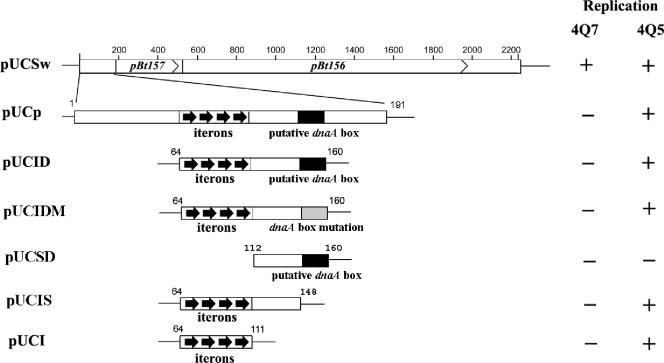
FIG. 1.
Schematic diagram of the plasmids used to define the replication origin of the minireplicon of pBtoxis. pUCSw contains the 2.2-kb minireplicon of pBtoxis with pBt157 and pBt156 coding for, respectively, ORF157 and ORF156; pUCp contains the 191-bp upstream sequence from pBt157; pUCID contains a 97-bp sequence which includes the iteron sequence, 37-bp spacer, and putative dnaA box motif; pUCIDM is identical to pUCID, except that the putative dnaA box motif was replaced with 5′-CCCCAAGAC-3′; pUCIS contains the iteron sequence and 37-bp spacer; pUCSD contains 37-bp spacer and putative dnaA box motif; and pUCI contains only the iterons. The arrows indicate the 12-bp A+T-rich direct repeats (iterons). The black box and gray box represent, respectively, the putative dnaA box motif and the mutation of the putative dnaA box motif. pUCSw, pUCp, pUCID, and pUCI are all derivatives of pUCCl, the parental plasmid incapable of replicating in Bacillus thuringiensis subsp. israelensis. The ability (+) or inability (−) of each plasmid to replicate in B. thuringiensis subsp. israelensis strain 4Q7 cured of native plasmids or strain 4Q5, which contains parental pBtoxis, is also shown. Gene terminology follows that used by Berry et al. (3).
TABLE 2.
Primers used for PCR
| Primer | Sequence (5′ to 3′)a |
|---|---|
| pIDna-F | CCCAAGCTTTAAAGGTTTAAATAACAGTTTAAA |
| pIDna-R | AACTGCAGGAGAGTGGAAAAATTCACCTAACC |
| pP-F | CCCAAGCTTAAATTTAATTTTAGGTTTAAATTT |
| pP-R | AACTGCAGAACTCCCATCTGTTTAATTAATTC |
| pIDM-F | AAAAGTACTCACCAGTCACAGAAAAGCATCTT |
| pIDM-R | CGGAATTCGTCTTGGGGATTCACCTAACCTATTTATATCA |
| pIS-R | CGGAATTCATTCACCTAACCTATTTATATCAC |
| pSD-F | CCCAAGCTTTACCATTCCCAGTGTGATATAAAT |
| pSD-R | CATGCCATGGAATAATAGAAAGAGAAAAAGCATT |
| pBAD156-F | CATGCCATGGCAATGTTATTAAACAGTAATGAACTA |
| pBAD156-R | GCTCTAGACCACGTTTTTTAAATGGATTTGA |
| pBAD157-F | CATGCCATGGCAATGAATAGGGATCACTTTTATACG |
| pBAD157-R | GCTCTAGACCTAAATTACGCATTTCCATTGC |
Restriction endonuclease cleavage sites are typed in boldface, and the mutated nucleotide bases are underlined.
Expression and purification of recombinant ORF156 and ORF157 proteins.
The l-arabinose-based pBAD/myc-his expression system (Invitrogen) was used for synthesis of recombinant six-histidine-tagged ORF156 and ORF157 proteins, respectively, rORF156 and rORF157. The pBt156 and pBt157 open reading frames were amplified with primers pBAD156-F and pBAD156-R and pBAD157-F and pBAD157-R (Table 2), respectively, digested with NcoI and XbaI, and cloned in the same sites in pBAD/myc-hisB for expression in E. coli TOP10 (Invitrogen). Recombinant proteins were purified using imidazole-based buffers and Ni-nitrilotriacetic acid resin columns, according to the manufacturer's protocol (QIAGEN). The purified proteins were desalted and concentrated using Centricon centrifugal filters (Millipore). Protein concentration was assayed using the Coomassie protein assay kit (Pierce), and Western blot analysis using anti-His C-terminal antibody (Invitrogen) was performed to confirm purification of rORF156 and rORF157.
EMSA.
Synthetic oligonucleotides 5′-TAAAGGTTTAAATAACAGTTTAAATTTAAGTTTAACTTTCAGTTTACA-3′ and 5′-TGTAAACTGAAAGTTAAACTTAAATTTAAACTGTTATTTAAACCTTTA-3′, containing the 48-bp iteron sequence, were boiled for 5 min and allowed to anneal by slowly cooling to room temperature. The annealed double-stranded DNA was purified from a 15% polyacrylamide gel and then labeled with [γ-32P]ATP (Amersham) using T4 polynucleotide kinase (Promega), eluted from a Micro Bio-Spin P-30 Tris chromatograph column (Bio-Rad), and quantified by scintillation. For EMSA, reaction mixtures containing 2 μl 5× gel shift binding buffer (Promega), 30 fmol of labeled iterons (25,000 cpm), and 30 pmol of rORF157 or 360 pmol of rORF156, were incubated at room temperature for 20 min, and the DNA-protein complexes were resolved in a 5% polyacrylamide gel (Bio-Rad) by electrophoresis in 0.5× TBE (0.045 M Tris-borate, 0.001 M EDTA) buffer at 100 V for 2 h. Gels were dried on Whatman paper and exposed to X-ray film.
For competition assays, binding reactions were first performed at room temperature for 10 min in binding buffer containing purified proteins and a 1,000× molar excess of either unlabeled iterons as the specific competitor or an unlabeled 30-bp DNA fragment of Herves (2), a transposable element from the mosquito, as the nonspecific competitor (Fig. 2B). After incubation, the 32P-labeled iteron was added and the reaction was continued for another 20 min at room temperature. For protein-protein interaction studies with rORF156 and rORF157-iteron complexes, both proteins were added simultaneously.
FIG. 2.
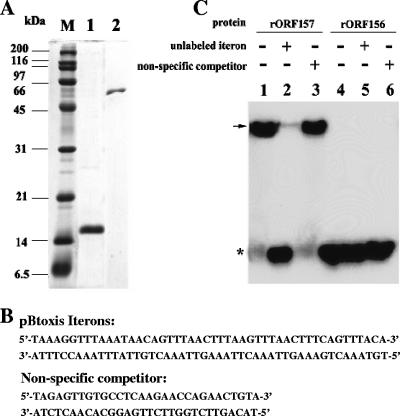
Recombinant protein purification and EMSAs showing that ORF157, but not ORF156, is an iteron-binding protein. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) showing purification of histidine-tagged ORF157 and ORF156, respectively. Lane 1, rORF157; lane 2, rORF156; and lane M, protein molecular mass markers. (B) Nucleotide sequence of the pBtoxis iterons and nonspecific DNA substrates used in EMSAs. (C) EMSA using the 48-bp γ-32P-labeled pBtoxis iterons as the probe, with 30 pmol of rORF157 (lanes 1, 2, and 3) and 360 pmol of rORF156 (lanes 4, 5, and 6). The locations of DNA-protein complexes (arrow) and unbound labeled iterons (asterisk) are shown. Reaction mixtures were incubated in the presence (+) or absence (−) of nonspecific competitor and unlabeled iterons as a specific competitor to determine the iteron-specific binding by rORF157 and rORF156.
GTPase assay.
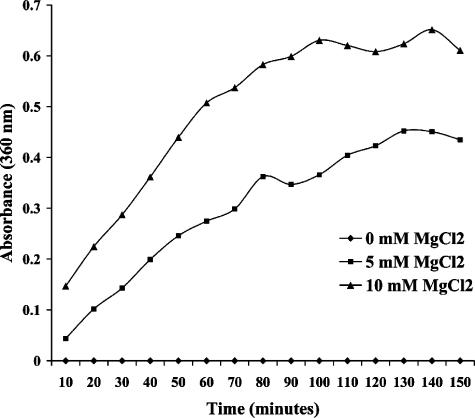
The GTPase assay was performed using the EnzChek phosphate assay kit (Molecular Probes) with 2-amino-6-mercapto-7-methylpurine ribose (MESG) substrate, according to manufacturer's protocol, with and without 5 mM MgCl2 or 10 mM MgCl2. Reactions were initiated by adding GTP (Invitrogen) to a final concentration of 0.5 mM. The continuous absorbance trace at a wavelength of 360 nm reflected Pi released from rORF156-GTP complexes followed by phosphorylase coupling reactions with MESG. Absorption at 360 nm was measured at 10-min intervals.
RESULTS AND DISCUSSION
Replication origin of pBtoxis minireplicon.
To identify the minimal sequences required to replicate the pBtoxis minireplicon (18), different fragments from the minireplicon were cloned into the vector pUCC1 to generate the reporter plasmids pUCSw, pUCp, pUCID, pUCIDM, pUCIS, pUCSD, and pUCI (Fig. 1). The reporter plasmid pUCSw contained the 2.2-kb pBtoxis minireplicon that included the pBt157 and pBt156 ORFs coding for, respectively, ORF157 and ORF156, and the iterons and putative dnaA box motif (18); pUCp contained the 191-bp sequence upstream from pBt157 with the iterons and putative dnaA box; pUCID contained the iterons and the 37-bp spacer followed by the putative dnaA box; pUCIDM contained the iterons, the 37-bp spacer, and the mutated dnaA box; pUCIS contained the iterons and 37-bp spacer; pUCSD contained the 37-bp spacer and putative dnaA box; and pUCI contained only the iterons. When these plasmids were introduced into B. thuringiensis subsp. israelensis 4Q7, a strain cured of native plasmids including pBtoxis, pUCSw, but not pUCp, pUCID, pUCIDM, pUCIS, pUCSD and pUCI, replicated under selection with chloramphenicol. The inability of the latter six plasmids to replicate in strain 4Q7 supports our previous observation that both ORF157 and ORF156 are required for plasmid replication (18).
When the reporter plasmids pUCSw, pUCp, and pUCID (each of which contained different fragments of the pBtoxis minireplicon but included the four 12-bp iterons and 37-bp spacer sequence followed by the putative dnaA box) were introduced into B. thuringiensis subsp. israelensis 4Q5, a strain that harbors only pBtoxis (Table 1), 110, 114, and 115 transformants, respectively, grew per 5 μg plasmid DNA under selection with chloramphenicol. When the 4Q5 strain was transformed with pUCIDM, which contained the iterons, 37-bp spacer sequence, and the mutated dnaA box motif, 110 Cmr transformants/5 μg plasmid DNA were obtained, indicating that the putative dnaA box motif was not required for minireplicon replication.
Reporter plasmids containing the iterons with (pUCIS) and without (pUCI) the 37-bp spacer sequence also replicated in the 4Q5 strains, yielding, respectively, 97 and 15 colonies per 5 μg of plasmid DNA. However, whereas transformants containing pUCIS were recovered by 24 h, those containing pUCI formed smaller colonies that appeared after 48 h of incubation. Finally, when the 4Q5 strains were electroporated with pUCSD, which contained the 37-bp spacer sequence and putative dnaA box but lacked the iterons, transformants were not recovered after repeated attempts.
In all of these experiments, the presence of pBtoxis in each recombinant strain resistant to chloramphenicol was inferred by the presence of the large crystalline inclusion composed of toxin proteins encoded by this plasmid, as observed by phase-contrast microscopy. This indicated that the autonomously replicating plasmids did not have a deleterious effect on the stability of parental pBtoxis. Importantly, to confirm that the reporter plasmids replicated in recombinant 4Q5 strains and were not propagated and selected for by recombination events with chromosomal DNA or pBtoxis, plasmids were extracted from recombinant 4Q5 strains and used to transform E. coli. These reporter plasmids replicated in E. coli under ampicillin selection. Restriction enzyme and PCR analyses showed that they were identical to those used in the autonomous replication assays in the 4Q5 strains. These results demonstrated that the iterons alone functioned as the minimal replication origin (ori) of the pBtoxis minireplicon.
ORF157 is an iteron-binding protein.
The structural organization and occurrence of the iterons, pBt157, and pBt156 in an operon and their essential role in propagating the pBtoxis minireplicon suggested that the ORF157 and ORF156 proteins interact cooperatively with the cis elements in initiating pBtoxis replication. To determine whether these proteins have strong iteron-binding specificity, six-histidine-tagged recombinant proteins, rORF157 and rORF156, synthesized in E. coli, were purified by affinity chromatography (Fig. 2A) and used in EMSAs with radiolabeled iterons or unlabeled specific (iterons) and nonspecific competitor DNA (Fig. 2B). Whether the six-histidine tags in the recombinant proteins influenced the results observed in our study is not known. Nevertheless, a marked electrophoretic mobility shift was observed when rORF157 was incubated with the substrate-specific labeled iteron probe in the absence or presence of the nonspecific competitor (Fig. 2C, lanes 1 and 3). When rORF157 was preincubated with unlabeled iterons, followed by the addition of labeled iterons, a shift in mobility was not observed due to saturation of the protein with the unlabeled specific probe (Fig. 2C, lane 2). These results demonstrate that the rORF157 binds to the pBtoxis iterons with high specificity. In similar assays, no complexes were observed with rORF156 and the labeled iterons in either the absence or presence of unlabeled specific and nonspecific competitors (Fig. 2C, lanes 4, 5, and 6). These results demonstrate that ORF156 is not an iteron-binding protein.
ORF156 binds to ORF157-iteron complexes.
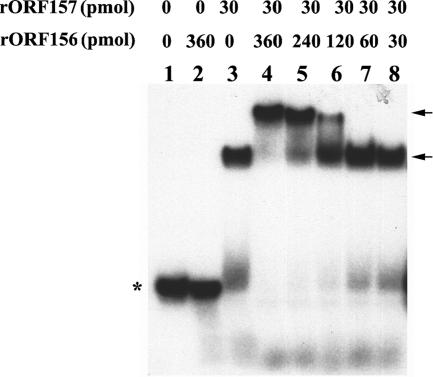
To further study the putative role of ORF156 in pBtoxis replication, EMSAs were performed to determine whether this protein could interact with rORF157-iteron complexes. These assays clearly demonstrated that rORF156 bound the rORF157-iteron complexes, probably through rORF156-rORF157 protein-protein interactions, resulting in a larger electrophoretic mobility shift (Fig. 3). The extent of the shift was dependent on the relative ratios of the two proteins used in the assay. For example, when the ratio of rORF157 was kept constant (30 pmol), a one- or twofold increase in rORF156 (30 or 60 pmol) had little effect on the shift of the rORF157-iteron complexes (Fig. 3, lanes 7 and 8). In contrast, by increasing the molar ratios of rORF156 to rORF157 to 4:1, 8:1, and 12:1, corresponding increases in mobility shifts of protein-DNA complexes were observed (Fig. 3, lanes 4, 5, and 6).
FIG. 3.
EMSAs showing that ORF156 binds to ORF157-iteron complexes. Thirty femtomoles of the 48-bp γ-32P-labeled pBtoxis iteron probe was added to each EMSA reaction mixture. Lane 1, negative control without protein; lane 2, 360 pmol purified rORF156 added to the reaction mixture; lanes 3 to 8, 30 pmol purified rORF157 added to reaction mixtures with different amounts of purified rORF156 (0, 360, 240, 120, 60, and 30 pmol). The locations of DNA-protein complexes (arrows) and unbound labeled iterons (asterisk) are shown.
Putative roles of ORF157 and ORF156 in pBtoxis replication.
Although proteins other than ORF157 (11.8 kDa) could interact with the pBtoxis iterons, the iteron-binding specificity of this protein suggests that its function is similar to those of other plasmid-encoded Rep proteins, including RepE (37 kDa) and RepA (29 kDa) (6, 8, 16). Rep proteins are known to interact directly with genome-encoded DnaA to form multiprotein complexes that catalyze the initiation of plasmid replication (4, 13). In addition, the binding of RepA to cis elements represses transcription from the repA promoter, thereby providing a mechanism for autoregulating plasmid replication and copy number. Whether ORF157 functions as an autoregulator of transcription or pBtoxis replication is unknown. However, recent work by Larsen et al. (12) showed that in a heterologous system using E. coli as a host, in the absence of ORF157, recombinant ORF156-GFP levels were 4.5-fold higher than in the presence of ORF157. Therefore, it is likely that ORF157 functions as a negative regulator of ORF156.
The ORF156 protein contains a motif (GGGVGTG) conserved in all members of the tubulin/FtsZ superfamily (PROSITE motif PS00227: [S/A/G]GGTG[S/A/T]G) that is involved in Mg2+-dependent GTP binding and hydrolysis (3, 5, 20), an enzymatic activity demonstrated for rORF156 in the present study (Fig. 4). Tinsley and Khan (19) have demonstrated that this motif is required for RepX function. However, as GTP was not included in our EMSAs, it is unlikely that the GTPase activity is required for the recruitment and assembly of ORF156 with the ORF157-iteron complex.
FIG. 4.
GTPase activity of the rORF156 protein in the absence or presence of MgCl2. Activity was measured without MgCl2 or with 0, 5, and 10 mM MgCl2 in 50 mM Tris-HEPES (1:1) buffer (pH 8.0) at 37°C at different time intervals by absorbance at 360 nm using the MESG substrate assay system as described in Materials and Methods.
While favoring a model that ORF156 binds ORF157-iteron complexes, we cannot exclude the possibility that other proteins encoded by pBtoxis have a similar function. However, our observation that rORF156 physically interacts with the rORF157-iteron complexes in vitro suggests a functional role for ORF156 based on what is known about the function of the tubulins and FtsZ proteins. The polymer-forming GTPases that compose the tubulin/FtsZ superfamily are similar to eukaryotic tubulins and are essential for cell division in prokaryotes (20). Tubulins assemble directional linear polymers that participate in a number of cytoskeletal processes in eukaryotes, including DNA segregation (12). In contrast, FtsZ proteins assemble into a ring structure, the so-called “Z-ring,” in a GTPase-dependent manner on the inner surface of the cytoplasmic membrane and recruit cytoplasmic proteins that mediate progressive septum invagination and subsequent cell division (20). Interesting, Larsen et al. (12) have demonstrated the recombinant ORF156-GFP (TubZ-GFP) forms dynamic linear polymers, suggesting that ORF156 is a novel type of tubulin-based cytoskeletal component in bacteria. Taken together, it is tempting to suggest that the interaction of ORF156 and the ORF157-iteron complex facilitates the segregation and partitioning of pBtoxis in daughter cells during cell division. Additionally, the role of ORF156 could be to mediate interactions between iteron-bound ORF157 and other proteins involved in pBtoxis replication.
Acknowledgments
We thank Jeffrey J. Johnson for technical assistance during this study and P. W. Atkinson (Department of Entomology, University of California, Riverside) for providing the nonspecific competitor DNA used in this study.
This research was supported by a grant to B.A.F. from the U.S. National Institutes of Health (AI 145817).
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 2.Arensburger, P., Y. J. Kim, J. Orsetti, C. Aluvihare, D. A. O'Brochta, and P. W. Atkinson. 2005. An active transposable element, Herves, from the African malaria mosquito Anopheles gambiae. Genetics 169:697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, C., S. O'Neil, E. Ben-Dov, A. F. Jones, L. Murphy, M. A. Quail, M. T. G. Holden, D. Harris, A. Zaritsky, and J. Parkhill. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68:5082-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 5.de Boer, P., R. Crossley, and L. Rothfield. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254-256. [DOI] [PubMed] [Google Scholar]
- 6.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federici, B. A. 1995. The future of microbial insecticides as vector control agents. J. Am. Mosq. Control Assoc. 11:260-268. [PubMed] [Google Scholar]
- 8.Giraldo, R., and M. E. Fernandez-Tresguerres. 2004. Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids. Plasmid 52:69-83. [DOI] [PubMed] [Google Scholar]
- 9.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. Freeman and Company, New York, NY.
- 11.Krause, M., B. Ruckert, R. Lurz, and W. Messer. 1997. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274:365-380. [DOI] [PubMed] [Google Scholar]
- 12.Larsen, R. A., C. Cusumano, A. Fujioka, G. Lim-Fong, P. Patterson, and J. Pogliano. 2007. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 21:1340-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, Y. B., H. J. Datta, and D. Bastia. 1998. Mechanistic studies of initiator-initiator interaction and replication initiation. EMBO J. 17:5192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee, S., I. Patel, and D. Bastia. 1985. Conformational changes in a replication origin induced by an initiator protein. Cell 43:189-197. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay, G., and D. K. Chattoraj. 1993. Conformation of the origin of P1 plasmid replication. Initiator protein induced wrapping and intrinsic unstacking. J. Mol. Biol. 231:19-28. [DOI] [PubMed] [Google Scholar]
- 16.Sharma, S., B. K. Sathyanarayana, J. G. Bird, J. R. Hoskins, B. Lee, and S. Wickner. 2004. Plasmid P1 RepA is homologous to the F plasmid RepE class of initiators. J. Biol. Chem. 279:6027-6034. [DOI] [PubMed] [Google Scholar]
- 17.Stein, C., G. W. Jones, T. Chalmers, and C. Berry. 2006. Transcriptional analysis of the toxin-coding plasmid pBtoxis from Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 72:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang, M., D. K. Bideshi, H.-W. Park, and B. A. Federici. 2006. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 72:6948-6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tinsley, E., and S. A. Khan. 2006. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 188:2829-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughan, S., B. Wickstead, K. Gull, and S. G. Addinall. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58:19-29. [DOI] [PubMed] [Google Scholar]
- 21.Wu, D., X. L. Cao, Y. Y. Bai, and A. I. Aronson. 1991. Sequence of an operon containing a novel delta-endotoxin gene from Bacillus thuringiensis. FEMS Microbiol. Lett. 65:31-35. [DOI] [PubMed] [Google Scholar]
- 22.Wu, D., and B. A. Federici. 1993. A 20-kilodalton protein preserves cell viability and promotes CytA crystal formation during sporulation in Bacillus thuringiensis. J. Bacteriol. 175:5276-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]