Abstract
The objective of this study was to determine whether the temperate Yersinia enterocolitica phage PY54 may interact with the related Escherichia coli phage N15 during both the lysogenic and the lytic cycle in the same cell. The PY54 and N15 prophages are linear plasmids which have been shown to be compatible and stably replicating in E. coli and Yersinia. In E. coli, the PY54 prophage does not restrict N15 propagation. In contrast, N15 reduces by use of its cor gene the susceptibility of Yersinia strains to PY54. Doubly lysogenic E. coli strains release PY54 virions, some of which apparently contain the N15 genome. Further experiments with replicative miniplasmid derivatives of PY54, N15, and the related Klebsiella oxytoca phage φKO2 demonstrated that the φKO2 and N15 plasmid prophages belong to the same incompatibility group.
Escherichia coli phage N15 (16), PY54 isolated from Yersinia enterocolitica (10), and the Klebsiella oxytoca phage φKO2 (5) are the only known temperate phages whose prophages are linear plasmids with terminal hairpins (telomeres). Virions of the three phages contain a linear genome with cohesive ends. Each phage and its respective plasmid prophage are circularly permuted molecules with sizes of 46.3 kb (N15 and PY54) or 51.6 kb (φKO2). Sequence analyses revealed similar genome organizations of the three phages (5). The left arm of the genomes carries mainly genes for virion assembly and plasmid partitioning (sop and spy). The right arm contains the protelomerase gene (tel) essential for the generation of the telomeres, a large gene encoding a multifunctional replication protein (RepA), immunity and anti-immunity genes (e.g., cB, cro, and antA), and genes contributing to host cell lysis. Overall the left arm of PY54 is more closely related to φKO2 than to N15, whereas N15 and φKO2 show the strongest similarities in the right arm. As the genomes of all three phages are mosaically related to λ and due to homologies to lambda-like phages, it has been suggested to include the telomere phages as a subgroup of the lambdoid phage family (5). Members of this family can interact with each other or with unrelated phages. Phage P22, for example, encodes an antirepressor (Ant) which is capable of inducing resident lambdoid phages by neutralizing the repressor already present in the cell (3). On the other hand, lambda rex genes prevent the growth of rII mutants of phage T4 (2), whereas the HK022 transcription termination factor Nun prematurely terminates early λ transcripts (13). Besides these interactions, some lambdoid phages can easily recombine with each other, producing functional hybrids (7). Campbell and Botstein (4) defined lambdoid phages as being capable of productive genetic recombination with λ. Horizontal gene exchange is therefore suggested to be a major component of evolution for these viruses (8).
In this work, we studied possible interactions between the telomere phages during their life cycles. The experiments were carried out to answer the question whether the detected DNA similarities between the phages may lead to any incompatibility, immunity, or recombination. Here we show that the PY54 and N15 prophages are compatible plasmids. The PY54 prophage is also compatible with a replicative φKO2 miniplasmid, while the N15 prophage cannot coexist with this miniplasmid in the same cell. We further demonstrate that the N15 cor gene reduces the susceptibility of Yersinia strains against PY54 infection and that doubly lysogenic E. coli strains release three types of virions, (i) N15, (ii) PY54, and (iii) PY54 particles harboring the N15 genome.
PY54 and N15 prophages are compatible plasmids.
PY54 and N15 are temperate phages that specifically infect Y. enterocolitica and E. coli, respectively. To ascertain whether, regardless of the host range of the phages, the plasmid prophages are able to replicate in both species, we used standard transformation procedures to introduce PY54 DNA into E. coli C-1a (18) and N15 DNA into the Y. enterocolitica strain 83/88 (10, 17). Selection of transformants containing the respective prophage was achieved by use of the PY54-A and -C mutants, harboring a kanamycin resistance gene (11), and the N15-C02 cat (chloramphenicol acetyltransferase) mutant (see Fig. 2A). While the mutants PY54-A and N15-C02 have a phenotype similar to that of the wild-type phages, mutant PY54-C is exclusively lysogenic. Restriction analyses documented the linear prophages in the transformants (data not shown), which indicates that both phage plasmids are functional in E. coli and Yersinia. To examine compatibility, the E. coli C-1a (PY54) strain and the Y. enterocolitica 83/88 (N15) strain were lysogenized with the mutants of N15 and PY54, respectively. The single lysogenic strains could readily be infected with the second phage, and both prophages coexisted in E. coli and Yersinia. We have determined the compatibility for approximately 100 generations under nonselective conditions. The doubly lysogenic strains showed the same stability as single lysogenic strains containing either phage. Thus, the N15 and PY54 prophages are compatible plasmids. As the group of telomere phages comprises three members, the question arises whether the N15 and PY54 prophages are also compatible with the K. oxytoca prophage φKO2.
FIG. 2.
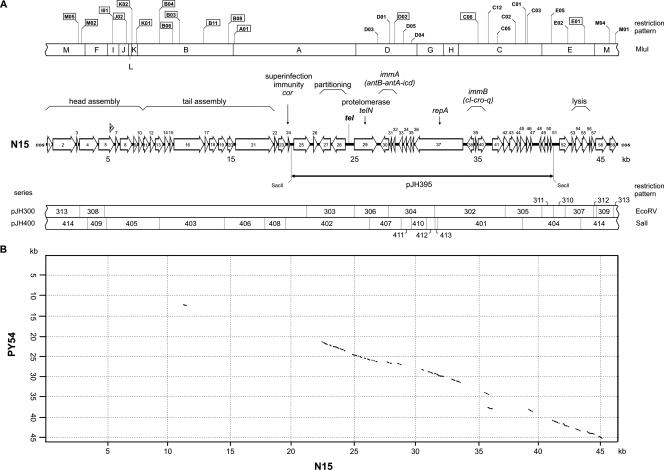
N15 genome and its homologies to PY54. (A) The top bar shows the MluI restriction pattern of N15. Indicated are the insertion sites of cat. Insertions leading to defective phage particles are depicted in boxes. In the middle, the N15 gene map, including a functional assignment according to Ravin et al. (15), is shown. The bar indicates the position of the linear miniplasmid pJH395. The bottom panel shows the EcoRV and SalI restriction fragments used for the construction of an N15 DNA fragment library. (B) Dot plot alignment of N15 and PY54. The alignment was performed with MacVector (version 8.0; Oxford Molecular Group). The numbers on the axes give the scale for the genomes in base pairs.
N15 and φKO2 plasmid prophages belong to the same incompatibility group.
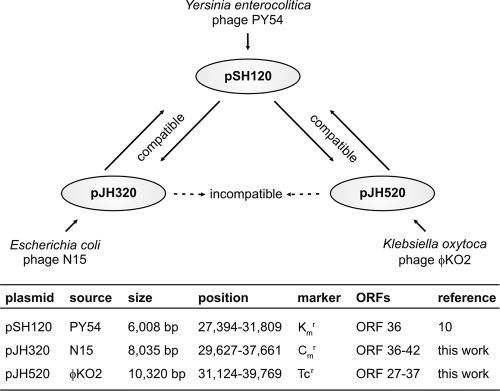
While N15 and PY54 can be propagated in hosts, for phage φKO2 a suitable indicator strain could not yet be identified (5). To allow compatibility studies with the φKO2 prophage, we made attempts to introduce a marker gene into the φKO2 genome and to transform its host strain K. oxytoca CCUG 15788 (Culture Collection, University of Göteborg, Sweden) with PY54 and N15 DNA. Since all of these trials failed, compatibility was studied using circular miniplasmid derivatives of the prophages retaining replicative competence (Fig. 1). Plasmid pSH120 is a derivative of a PY54 mutant and is essentially composed of repA and a kanamycin resistance gene located downstream from repA (11). The N15 miniplasmid pJH320 was obtained by digestion of N15 mutant D04 DNA (Fig. 2A) with AatII and subsequent religation. This miniplasmid contains the N15 genes for RepA, cB, Cro, Q, and a cat gene. The φKO2 miniplasmid pJH520 was constructed by digesting the phage DNA with DraI and ligating the restriction fragments with the tet resistance gene of the transposon Tn5. Among its 11 open reading frames, this plasmid harbors the predicted genes for RepA, the prophage repressor CB, and the antirepressors Cro and AntA.
FIG. 1.
Compatibility between the telomere phages PY54, N15, and φKO2 and their miniplasmid derivatives. (Top) Results of the compatibility studies. (Bottom) Sizes and compositions of the miniplasmids. ORF, open reading frame.
Transformation experiments revealed that the miniplasmids replicate in all three genera. Compatibility between the plasmids was investigated by either cotransformation or subsequent introduction into the same host strains. In E. coli and Yersinia, the PY54-derived plasmid pSH120 showed full compatibility with pJH320 (N15) and pJH520 (φKO2) (Fig. 1). Moreover, pSH120 could easily be introduced into the φKO2 host strain, where it stably replicated. By contrast, we were unable to introduce pJH320 into the Klebsiella strain CCUG 15788. N15 incompatibility with φKO2 was also evident by the fact that E. coli C-1a could not be cotransformed with pJH320 and pJH520 nor could the plasmids successively be introduced into an E. coli strain. The studies clearly showed that pSH120 is fully compatible with the N15 and φKO2 miniplasmids, while these two miniplasmids obviously do not coexist (Fig. 1).
It is likely that the incompatibility between the N15 miniplasmid pJH320 and the φKO2-derived plasmid pJH520 is caused by highly homologous RepA replication proteins of the phages, which are 92% identical. The repA genes harbor the prophage replication origins, which contain several conserved structure elements (e.g., AT-rich region and dam methylation sites). Hence, the initiation proteins probably have the same origin specificities, as was already surmised (5). In contrast, the PY54 and N15 RepA proteins are only 49% identical and show some deviations in the nucleotide binding motifs. Moreover, the PY54 origin differs slightly from those of N15 and φKO2 (23), which might be essential for the compatibility between pSH120 and the other two miniplasmids. Studies on interaction of the prophages have documented that the PY54 and N15 plasmids belong to different incompatibility groups while N15 and φKO2 are affiliated with the same group. We next wanted to learn whether there is any interaction between N15 and PY54 during their release from doubly lysogenic bacteria.
PY54 and N15 are inducible in E. coli and Y. enterocolitica.
For induction experiments in E. coli, the wild-type strain C-1a was chosen because it is suited for PY54 release, particularly at 28°C, the temperature used to induce the phage in Yersinia (Table 1). A PY54 lysogenic E. coli C-1a strain was infected by N15. The resulting doubly lysogenic strain as well as C-1a strains harboring either PY54 or N15 was treated with mitomycin C (17). N15 was released from the doubly lysogenic bacteria with nearly the same efficiency as from a single lysogenic strain (Table 1). In contrast, lysates prepared with the doubly lysogenic bacteria exhibited a reduction of the PY54 titer by 1 to 2 orders of magnitude compared to lysates prepared with the single lysogenic E. coli C-1a strain. We conclude that this effect is not caused by direct interaction between the phages but rather by the fact that N15 release in E. coli starts significantly earlier (30 to 60 min) than that of PY54, independent of the temperature (28°C or 37°C) used for induction (data not shown).
TABLE 1.
Efficiencies of plating of lysates of E. coli C-1a and Y. enterocolitica 83/88 harboring PY54, N15, or both phagesa
| Prophage(s) | Release (titer [PFU]) of indicated phage from:
|
|
|---|---|---|
| E. coli C-1a | Y. enterocolitica 83/88 | |
| PY54 | PY54 | |
| PY54-A | 1 × 107 | 5 × 107 |
| PY54-A/N15-C02 | 1 × 106 | 3 × 107 |
| PY54-C | 8 × 107* | 1 × 108* |
| PY54-C/N15-C02 | 7 × 105* | 8 × 107* |
| N15 | N15 | |
| N15-C02 | 1 × 109 | 5 × 108 |
| N15-C02/PY54-A | 8 × 108 | 3 × 108 |
| N15-C02/PY54-C | 7 × 108 | 3 × 108 |
Strains were grown in 10 ml LB at 28°C and 37°C to an A588 of 0.2 to 0.3. Mitomycin C (2.5 μg/ml) was added, and the cultures were further incubated until lysis occurred. After removal of cell debris by centrifugation at 10,000 × g for 10 min, phages were passed through 0.2-μm sterile filters. A portion (100 μl) of each diluted lysate was incubated for 10 min with 100 μl indicator strain. Then, 4.5 ml LB soft agar (0.8%) was added and the mixture was poured on LB agar plates. The phage titers were determined after incubation overnight at 28°C (PY54) or 37°C (N15). *, the PY54-C mutant is able to lysogenize Yersinia exclusively; for that reason, the efficiency of plating was determined by plating infected bacteria on agar containing kanamycin (see text for details).
Switching to Yersinia, we initially studied N15 release from the bacteria at different temperatures. Upon introduction by transformation, induction of the N15-C02 mutant at 28°C and 37°C gave lysates with nearly identical phage titers, approximately half as high as that from E. coli C-1a (Table 1). As with PY54 isolated from E. coli, N15 particles released from Yersinia showed an unchanged host range and were able to infect only E. coli. The Y. enterocolitica strain 83/88 containing N15 was lysogenized with PY54, followed by induction of the phages. Contrary to the situation with E. coli, the doubly lysogenic Yersinia strain released nearly as many PY54 and N15 particles as the single lysogenic strains (Table 1), which can be explained by the similar onsets of N15 and PY54 release in this species.
The similar genome organizations of PY54 and N15 inspired us to study changes in the induced phages that might arise by inaccurate packaging of the virus DNA or by recombination. Therefore, in the following experiments, phages released from doubly lysogenic strains were analyzed in detail.
Doubly lysogenic E. coli strains release PY54 particles harboring the N15 genome.
A dot plot alignment of PY54 and N15 discloses significant similarities in the right arms of their genomes (Fig. 2B). To elucidate if there is any DNA exchange of the prophages in doubly lysogenic bacteria, we designed recombination experiments using a series of N15 cat mutants and the PY54-A and -C mutants (see above). It is assumed that in the case of recombination some phages will carry both resistance genes on their genome. The N15 cat mutants were generated using a HyperMu <CHL-1> insertion kit (Epicenter, Madison, WI). Following mutagenesis, the phage DNA was introduced into the supercompetent E. coli strain GeneHogs (Invitrogen). The cat insertion sites were determined by molecular cloning of tagged MluI restriction fragments using the vector pLitmus38 (Apr; New England Biolabs) and subsequent sequencing.
Figure 2A illustrates that many cat insertions in the left arm of the N15 genome resulted in defective phage particles. To examine recombination also in this region, we cloned the MluI restriction fragments including cat of the defective N15 mutants by use of the vector pLitmus38. E. coli C-1a strains harboring the prophages of PY54-A or PY54-C were then either lysogenized with intact N15 cat mutants or transformed with the recombinant plasmids. The resulting strains were cultivated at 28°C and 37°C, supplemented with kanamycin (100 μg/ml) and chloramphenicol (12.5 μg/ml). After reaching an A588 of 0.2 to 0.3, phages were induced with mitomycin C (2.5 μg/ml), harvested by ultracentrifugation at 70,000 × g for 90 min, and resuspended in 500 μl SM buffer (17). A portion (100 μl) of each lysate containing approximately 109 to 1010 N15 and PY54 particles/ml and dilutions of the lysates were used to infect E. coli C-1a and Y. enterocolitica 83/88 with multiplicities of infection between 0.01 and 100. The infected indicator strains were plated on agar containing chloramphenicol, kanamycin, or both antibiotics.
We did not detect any colony on doubly selective agar. As expected, the infected E. coli indicator strain gave colonies exclusively in the presence of chloramphenicol (N15). In contrast, colonies of the infected Y. enterocolitica strain 83/88 appeared on agar containing not only kanamycin (PY54) but also chloramphenicol. Up to 40 chloramphenicol-resistant Yersinia colonies were visible after infection of the bacteria with 109 phages. The chloramphenicol-resistant Yersinia lysogens were analyzed further. They harbored the N15 prophage and released N15 particles infecting E. coli exclusively. Southern hybridization did not exhibit any PY54 DNA in the N15 lysogenic Yersinia strains (data not shown). Furthermore, the complete sequence determination of one N15 mutant (M04) recovered from Yersinia did not exhibit any nucleotide alterations. How could N15 gain access to Y. enterocolitica? It has never been possible to infect Yersinia with N15, even with concentrated lysates (1010 phages/ml). Even coinfection with PY54 gave no Yersinia N15 lysogens. It can therefore only be concluded that some N15 genomes were packed into PY54 particles present in the same E. coli cells. Both phages have nearly identical genome sizes. Although their cos sites are rather different regarding the sequences and overhangs, it cannot be excluded that in analogy to the process of general transduction, some N15 genomes were unspecifically encapsidated by PY54.
Recombination of the prophages was also investigated in Yersinia. As with E. coli we could not isolate any phage containing both resistance genes. In contrast to the former experiments, doubly lysogenic Yersinia strains released PY54 and N15 particles infecting exclusively their natural host. There was no indication that encapsidation of one phage genome into the virion of the other phage had occurred.
The studies of recombination suggest that in E. coli and Yersinia, an exchange of DNA between PY54 and N15 did not take place. PY54 is able to encapsidate DNA of at least 50 kb, significantly more than its genome size including two resistance genes (data not shown). Thus, it is conceivable that the stretches of DNA sequences identical in both phages are too short for efficient homologous recombination. The longest identical stretch located in the partitioning gene sopB (spyB) has a length of 18 nucleotides. It has been reported previously that segments of approximately 20 bp or more are sufficient for significant recombination in E. coli (19, 22). Therefore, N15 and φKO2 might be more capable of exchanging DNA because these phages share stretches of up to 50 identical nucleotides in some of their genes (repA and sopA). However, recombination studies with N15 and φKO2 are restricted because of the incompatibility of the prophages. Nevertheless, compared to other lambdoid phages that harbor genes 97% and more identical (e.g., cI, cro, and int), the overall homologies between the telomere phages are rather low. Hence, it is doubtful whether the three known telomere phages can exchange DNA by homologous recombination. However, as it is conceivable that other similar telomere phages can be found in the environment, gene exchange might occur vigorously also between members of this group. Functional phage hybrids of course can also arise by nonhomologous recombination, but these recombination events presumably occur very infrequently and over a long period of time (9).
Since we have analyzed interactions between PY54 and N15 at the lysogenic stage and during induction, the last section will focus on the interplay of the phages during lytic growth.
PY54 does not restrict N15 propagation in E. coli, whereas the N15 cor gene attenuates PY54 infection in Yersinia.
At first, we investigated the influence of the PY54 prophage on N15 propagation in E. coli. Infection of the E. coli C-1a strain containing PY54 with N15 gave comparable numbers of plaques as with the nonlysogenic C-1a control strain. Similarly, lysogenization of the strains by use of the N15-C02 cat mutant occurred with the same efficiencies. Thus, the PY54 prophage does not affect N15 infection and propagation in E. coli. To determine the influence of the N15 prophage on PY54 propagation in Yersinia, the N15 lysogenic Y. enterocolitica 83/88 strain was infected with PY54. Compared to a plasmidless control strain, we observed 1 to 2 orders of magnitude of reduced efficiency of plating and a similarly reduced ability of PY54 to lysogenize the Yersinia strain containing N15. By comparison, the linear N15 miniplasmid pJH395, composed of the 21.3-kb SacII fragment (N15 positions 19964 to 41294 [Fig. 2A]) and a cat gene inserted by transposon mutagenesis at position 29671, did not hamper PY54 propagation. This was unexpected because this large plasmid carries genes for, e.g., the protelomerase, partitioning proteins, and the prophage repressor that might cause an incompatibility with or immunity to PY54. To test the influence of further gene products, an N15 DNA fragment library in pBR329 covering the whole genome was constructed (Fig. 2A). First, a high-titer N15 phage lysate was purified by CsCl step gradients (17). The phage DNA was isolated as described previously (12). Circular DNA was obtained by ligating the cohesive ends of the N15 DNA with T4 DNA ligase. Thereafter, EcoRV and SalI restriction fragments were inserted into the corresponding sites of pBR329 (Apr Cmr Tcr) (6). Plasmids containing EcoRV restriction fragments resulted in the pJH300 series, and those harboring SalI restriction fragments gave the pJH400 series. Except for the EcoRV fragment of 16.4 kb, all other restriction fragments could be obtained by molecular cloning in E. coli.
The constructs were introduced into Yersinia. All transformants were used for infection experiments with PY54, but only plasmid pJH402, containing a 6.8-kb SalI restriction fragment (N15 positions 19423 to 26204), mediated the observed inhibition of PY54 infection (Fig. 2A). Besides other genes, this DNA fragment carries the N15 cor gene, for which counterparts in the PY54 and φKO2 genomes could not be found. cor is known to prevent superinfection of an N15 lysogenic E. coli strain with N15, φ80, and T1 by interacting with the phage receptor (21). To elucidate whether cor may also affect PY54 infection, we amplified the gene by PCR. The forward and reverse primers contained restriction sites for NdeI and HindIII, respectively. To generate a defective cor gene as a control, a frameshift mutation (additional G) was introduced into the forward primer. The digested products were ligated to the corresponding sites of the expression vector pMS470Δ8cat, yielding plasmids pJH340 and pJH340X (defective cor gene). Plasmid pMS470Δ8cat is based on the vector pMS470Δ8 (1), containing an inducible tac promoter. In this plasmid, a cat gene was introduced by transposon mutagenesis (see above) to allow for selection in Yersinia. Upon transformation with the resulting plasmid, pJH340, cor expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
While strains harboring pMS470Δ8cat or pJH340X, containing a defective cor gene, gave the same phage titers as the wild-type strain, the intact cor gene reduced the infectivity of PY54 by 1 to 2 orders of magnitude. Moreover, in E. coli the cloned cor gene conferred complete resistance against N15 infection, as already reported by Vostrov et al. (21). Which role does the Cor protein play in Yersinia? We determined the number of phages that had not adsorbed to the host strains expressing cor or Δcor and found that supernatants of the strain expressing cor contained approximately twice as many phages as supernatants of the Δcor strain. This implies that cor diminishes PY54 adsorption. It has been suggested that in E. coli the Cor protein interacts with FhuA, a receptor protein essential for the uptake of ferrichrome and other substances as well as for N15 adsorption (20, 21). An analogous protein (FcuA) with slightly different substrate specificities was also detected in Y. enterocolitica (14). FcuA might be implicated in PY54 attachment by binding to a tail protein (open reading frame 22 product) that is 27% identical to the lambda host specificity protein J (11). Cor might interact with FcuA as well, albeit to a lower extent than with the FhuA receptor protein. Nevertheless, since PY54 infects only Y. enterocolitica O:5 and O:5,27 strains, it is likely that an O-antigen component is also important for adsorption.
Conclusions.
Our data underline that the phages PY54 and N15 are more distantly related than N15 and φKO2. This can be inferred from the following results. (i) The PY54 and N15 plasmid replicons are diverse and as a result compatible, while the φKO2 and N15 prophages belong to the same incompatibility group. (ii) Studies of immunity revealed no influence of PY54 on N15 propagation and only a weak cor-mediated inhibition of N15 on PY54 propagation. The data suggest that only limited interactions between DNA binding proteins of PY54 and N15 and the target sequences of the respective other phages exist. The in vivo observations are backed by in silico data (5). The N15 genome reveals stronger overall homologies to φKO2 than to PY54. It is therefore conceivable that unlike with N15 and PY54, some N15 and φKO2 DNA regions would have the potential to exchange DNA by homologous recombination, as it is often found with other lambdoid phages.
Acknowledgments
We thank Eckhard Strauch and Ben Davies for critical reading of the manuscript and Erich Lanka for constructive discussions.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to E. Lanka.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Balzer, D., G. Ziegelin, W. Pansegrau, V. Kruft, and E. Lanka. 1992. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 20:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benzer, S. 1955. Fine structure of a genetic region in bacteriophage. Proc. Natl. Acad. Sci. USA 41:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, A., and D. Botstein. 1983. Evolution of the lambdoid phages, p. 365-380. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 5.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covarrubias, L., and F. Bolivar. 1982. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 7.Gemski, P., Jr., L. S. Baron, and N. Yamamoto. 1972. Formation of hybrids between coliphage lambda and Salmonella phage P22 with a Salmonella typhimurium hybrid sensitive to these phages. Proc. Natl. Acad. Sci. USA 69:3110-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrix, R. W., G. F. Hatfull, and M. C. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154:253-257. [DOI] [PubMed] [Google Scholar]
- 9.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 10.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48:989-1003. [DOI] [PubMed] [Google Scholar]
- 11.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 12.Hertwig, S., A. Popp, B. Freytag, R. Lurz, and B. Appel. 1999. Generalized transduction of small Yersinia enterocolitica plasmids. Appl. Environ. Microbiol. 65:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King, R. A., P. L. Madsen, and R. A. Weisberg. 2000. Constitutive expression of a transcription termination factor by a repressed prophage: promoters for transcribing the phage HK022 nun gene. J. Bacteriol. 182:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koebnik, R., K. Hantke, and V. Braun. 1993. The TonB-dependent ferrichrome receptor FcuA of Yersinia enterocolitica: evidence against a strict co-evolution of receptor structure and substrate specificity. Mol. Microbiol. 7:383-393. [DOI] [PubMed] [Google Scholar]
- 15.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 16.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Sasaki, I., and G. Bertani. 1965. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 40:365-376. [DOI] [PubMed] [Google Scholar]
- 19.Shen, P., and H. V. Huang. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uc-Mass, A., E. J. Loeza, M. de la Garza, G. Guarneros, J. Hernandez-Sanchez, and L. Kameyama. 2004. An orthologue of the cor gene is involved in the exclusion of temperate lambdoid phages. Evidence that Cor inactivates FhuA receptor functions. Virology 329:425-433. [DOI] [PubMed] [Google Scholar]
- 21.Vostrov, A. A., O. A. Vostrukhina, A. N. Svarchevsky, and V. N. Rybchin. 1996. Proteins responsible for lysogenic conversion caused by coliphages N15 and φ80 are highly homologous. J. Bacteriol. 178:1484-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watt, V. M., C. J. Ingles, M. S. Urdea, and W. J. Rutter. 1985. Homology requirements for recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:4768-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegelin, G., N. Tegtmeyer, R. Lurz, S. Hertwig, J. Hammerl, B. Appel, and E. Lanka. 2005. The repA gene of the linear Yersinia enterocolitica prophage PY54 functions as a circular minimal replicon in Escherichia coli. J. Bacteriol. 187:3445-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]