Abstract
Neisseria meningitidis is a frequent commensal of the human nasopharynx causing severe invasive infections in rare cases. A functional two-partner secretion (TPS) system in N. meningitidis, composed of the secreted effector protein HrpA and its cognate transporter HrpB, is identified and characterized in this study. Although all meningococcal strains harbor at least one TPS system, the hrpA genes display significant C-terminal sequence variation. Meningococcal genes encoding the TPS effector proteins and their transporters are closely associated and transcribed into a single mRNA. HrpA proteins are translocated across the meningococcal outer membrane by their cognate transporters HrpB and mainly released into the environment. During this process, HrpA is proteolytically processed to a mature 180-kDa form. In contrast to other known TPS systems, immature HrpA proteins are stable in the absence of HrpB and accumulate within the bacterial cell. A small percentage of mature HrpA remains associated with the bacteria and contributes to the interaction of meningococci with epithelial cells.
Export of proteins to the surface and protein secretion are implicated in many aspects of bacterial life, including cell-to-cell communication, motility, and virulence. Highly sophisticated systems have evolved to ensure correct secretion of bacterial proteins, and many of them have been recognized as central virulence determinants. These include the type IV secretion system of Helicobacter pylori (2), type III secretion systems of members of the family Enterobacteriaceae (32), and bacterial autotransporters (18). Filamentous hemagglutinin (FHA) of Bordetella pertussis is the prototype of a family of large exoproteins in gram-negative bacteria that are secreted via the two-partner secretion (TPS) pathway (22, 23). FHA and related proteins, termed TpsA for two-partner secretion protein A, are translocated across the inner membrane via the universal Sec machinery (8). Transport across the outer membrane requires a specific transporter termed TpsB. In B. pertussis, this transporter is referred to as FhaC (23). The N-terminal region of the TpsA proteins contains a signal peptide for secretion via Sec and the adjacent TPS signal or secretion domain of approximately 245 amino acids (aa) that targets the TpsA protein to its cognate transporter, TpsB (8, 10, 22, 23). The genes encoding TpsA and TpsB are often found in close vicinity. Several TpsA proteins have been shown to contribute to virulence in plant and animal pathogens (5, 13, 38, 43, 48). B. pertussis FHA itself is both a major adhesion protein essential for establishing pathogen-host contact (29) and an important immunogen (26), representing the main constituent of the pertussis vaccine (35). By subtractive hybridization of DNA regions specific for Neisseria meningitidis strain Z2491 and absent in Neisseria gonorrhoeae, Klee et al. were the first to describe a homologue of FHA in meningococci (27). All three available meningococcal genomes contain open reading frames (ORFs) encoding putative TPS systems (4, 34, 44) (Table 1). According to their homology with FHA, the corresponding genes have been annotated Hrp (for hemagglutinin/hemolysin-related proteins) in strain MC58 and will be referred to as HrpA (TpsA homologue) and HrpB (TpsB homologue), respectively. In contrast to the other strains, the genome sequence of serogroup B strain MC58 (44) contains two almost identical copies of the TPS system and three additional putative hrpA genes that differ mainly in their C-terminal sequences (Table 1) (46). In addition, another putative hrpB gene is present in strain MC58. The highest degree of sequence similarity between B. pertussis FhaB and meningococcal HrpA exists within the N-terminal region of the predicted proteins (22 to 25% identity) comprising the domain necessary for binding of FhaB to its transporter and secretion by the TPS pathway. In contrast, the C-terminal sequences of the meningococcal HrpA proteins show no relevant homology to B. pertussis FhaB.
TABLE 1.
hrpA and hrpB genes in the sequenced meningococcal genomes
| Strain |
hrpA
|
hrpB
|
||||
|---|---|---|---|---|---|---|
| ORFa | Predicted protein length (aa) | % Identity (protein) to NMB1779 | ORF | Predicted protein length (aa) | % Identity (protein) to NMB1780 | |
| Z2491 | NMA0688 | 2,016 | 77.4 | NMA0687 | 580 | 98.6 |
| MC58 | NMB1779 | 1,995 | 100 | NMB1780 | 580 | 100 |
| NMB0497 | 1,976 | 93.5 | NMB0496b | 559 | 99.3 | |
| NMB0493 | 2,703 | 13.4 | −c | − | − | |
| NMB1214 | 2,274 | 16.0 | − | − | − | |
| NMB1768 | 2,514 | 13.9 | NMB1762 | 595 | 44.5 | |
| FAM18 | NMC0444 | 2,027 | 65.4 | NMC0443 | 580 | 99.0 |
NMA, NMB, and NMC denote the genes in strain Z2491, MC58, and FAM18 genomes (44), respectively, encoding proteins homologous to B. pertussis FhaB (HrpA in meningococci) and FhaC (HrpB in meningococci).
The protein encoded by NMB0496 lacks the first 21 aa at the N terminus and thus presumably contains no functional signal sequence.
−, not detected; genome of no associated putative hrpB gene could be detected in the MC58 for NMB0493 and NMB1214.
N. meningitidis colonizes the upper respiratory tract of approximately 10% of healthy individuals (6, 7, 12) and only occasionally invades the bloodstream, causing life-threatening septicemia and meningitis (39). Whereas invasive meningococci belong to a few clonal lineages in the vast majority of cases, isolates from healthy carriers are genetically highly diverse (12). The genetic background of the variable virulence potential in N. meningitidis is still unclear. However, a thorough comparison of the genetic composition of hyperinvasive strains with carrier strains might lead to a better understanding of virulence in N. meningitidis.
By analyzing a large collection of meningococcal carrier isolates, we show that TPS systems are present in all N. meningitidis strains. However, the effector proteins display a uniquely high degree of sequence variation. By constructing deletion mutants, a function of HrpB as a transporter for HrpA and the contribution of HrpA to meningococcal adherence to host epithelial cells could be established.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 822 N. meningitidis isolates obtained from healthy carriers were used for analysis, which have been described previously (11, 12). Meningococcal strains used for infection experiments are listed in Table S1 in the supplemental material. Strains were stored in glycerol stock at −80°C and plated on gonococcal (GC) agar (BD Difco, Heidelberg, Germany) supplemented with Poly-ViteX (bioMerieux, Marcy l'Etoile, France). When appropriate, chloramphenicol (7 μg/ml for N. meningitidis and 30 μg/ml for Escherichia coli), spectinomycin (125 μg/ml and 75 μg/ml, respectively), kanamycin (100 μg/ml and 30 μg/ml, respectively), and/or erythromycin (7 μg/ml) were added to the medium for selection.
Molecular cloning, plasmids, and primers.
Plasmids and primers are listed in Table S2 in the supplemental material. Recombinant DNA techniques were performed according to standard laboratory procedures. Plasmids were introduced into E. coli by chemical transformation. Meningococcal deletion mutants were constructed by integrating a selection marker into the gene of interest, simultaneously deleting part of the gene. Transformation of N. meningitidis was performed as described previously (15), and mutants were selected on appropriate media (see above). Detailed information regarding mutant construction can be found in the supplemental material. Meningococcal mutants used in this study are listed in Table S1 in the supplemental material. All mutant strains were checked for lipopolysaccharide (LPS) immunotype (L3 and L8) in a standard enzyme-linked immunosorbent assay using antibodies kindly provided by W. Zollinger and expression of major surface markers (Opc, Opa, and pili). Capsule expression was assayed using monoclonal antibodies MAb 735 (serogroup B) and MAb 924 (serogroup C) (14). MAb SM1 (anti-class 1 pili) was kindly provided by M. Virji (Bristol, United Kingdom), MAb AD211 (anti-class 2 pili), MAb B306 (anti-Opc), and MAb 4B12/C11 (anti-Opa) were kindly provided by Mark Achtman (Berlin, Germany).
Chromosomal DNA isolation, PCR, Southern blotting, and DNA sequencing.
Chromosomal DNA was isolated from N. meningitidis using the Genomic-Tip system (QIAGEN, Hilden, Germany). Restriction enzymes and Taq polymerase were purchased from NEB (Frankfurt, Germany). For amplification of DNA fragments larger than 4 kb, a high-fidelity Taq polymerase was used (Platinum Taq DNA polymerase high fidelity; Invitrogen). Southern blot analysis was performed as previously described (19). Automated DNA sequencing was performed with the dye deoxy terminator cycle method on an Applied Biosystems 377 sequencer.
Dot blot hybridization.
Chromosomal DNA was prepared with the QIAmp DNA mini kit (QIAGEN), and 200-ng samples were spotted onto nylon membranes (Macherey-Nagel). Dot blot hybridization was performed as described previously (19). Probes specific for the meningococcal hrpA genes NMB0493, NMB1214, and NMB1768 were generated by PCR using the primer pairs OK99/OK100 (nucleotides 2050 to 4071), OK103/OK104 (nucleotides 2498 to 4526), and OK105/OK106 (nucleotides 1999 to 4004), respectively. A single probe detecting both NMB0497 and NMB1779 was generated by PCR using the primer pair OK90/OK91 (nucleotides 2482 to 4537). A probe specific for NMC0443 was amplified with the primer pair OK423/OK424 (nucleotides 752 to 1168). A probe specific for NMB1762 was amplified with primer pair OK489/OK490 (nucleotides 2 to 1779). Probes were labeled using a random primer digoxigenin system (Roche, Mannheim, Germany) and detected with the chemiluminescence detection kit (Roche).
RNA extraction and RT-PCR.
RNA was prepared using RNeasy kits (QIAGEN), and reverse transcription-PCR was performed with the One Step reverse transcriptase PCR (RT-PCR) kit (QIAGEN). To exclude DNA contamination of the samples, PCR without RT was performed simultaneously. In strain MC58, expression of the hrpA genes was analyzed with primer pair OK301/OK302 (NMB0493), OK307/OK308 (NMB1768), OK305/OK306 (NMB1214), and OK303/OK304 (NMB0497/NMB1779), each primer pair amplifying internal fragments of the respective genes. In strain 2120, expression of the hrpA gene NMC0444 was analyzed with primer pair OK442/OK443. To analyze the expression of hrpB genes homologous to NMB1780 and to simultaneously verify the operon organization of hrpA and hrpB genes, primer pairs were designed with the forward primer recognizing sequences at the 3′ end of hrpB and the reverse primer recognizing a conserved region at the 5′ end of the associated hrpA gene. Strains were analyzed with primer pair OK474/OK475 or OK338/OK339. Expression of hrpB genes homologous to NMB1762 was analyzed with primer pair OK491/OK492 (primer sequences are listed in Table S2 in the supplemental material).
Expression in E. coli and purification of NMB1779.
E. coli strain SCS1 containing pQE-NMB1779 (kindly provided by S. Klee) was grown overnight in 10 ml of LB medium containing 100 μg/ml ampicillin and 12 μg/ml kanamycin. The following day, 1 ml of the overnight culture was used to inoculate 30 ml of fresh medium. This culture was allowed to grow to an optical density at 600 nm (OD600) of 0.5 to 0.6 before the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.5 mM. After 3 h, the cells were harvested, and the recombinant protein was affinity purified under denaturing conditions by using a Ni-nitrilotriacetic acid spin kit (QIAGEN) according to the manufacturer's instructions.
Rabbit polyclonal antibodies against NMB1779.
Rabbit polyclonal antibodies to NMB1779 (RαNMB1779) were raised against the denatured affinity-purified recombinant protein in a New Zealand White female rabbit as previously described (1). RαNMB1779 was used in immunoblots at a dilution of 1:100.
Western blot detection of HrpA proteins.
Antibody RαNMB1779 was used to investigate the presence of HrpA proteins. For that purpose, an overnight culture of N. meningitidis was inoculated into 20 ml Dulbecco's modified Eagle medium (Biochrom) supplemented with Poly-ViteX (bioMerieux) and grown for 1 h at 37°C and 200 rpm. Cultures were adjusted to an OD600 of 0.1 in 20 ml and grown until they reached an OD600 of 1.2. For generation of concentrated supernatants, bacteria were pelleted by centrifugation, and the supernatant was passed through a 0.2-μm filter (Whatman). Supernatant proteins were concentrated with an Amicon Ultra-15 centrifugal filter device (Millipore). For generation of whole-cell lysates, cultures were adjusted to an OD600 of 0.6 in 1 ml. Bacteria were harvested by centrifugation, and the resulting pellet was resuspended in 50 μl of sample solution. Portions (25 μl) of the respective samples were used for electrophoresis, blotted, and incubated with the respective antibody. Detection was performed with the SuperSignal West Pico chemiluminescent substrate (Pierce).
Infection experiments.
HEp-2 epithelial cells were grown in RPMI 1640 medium supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal calf serum in a 96-well microtiter plate until confluent growth. FaDu epithelial cells (ATCC HTB-43) were grown in Eagle's minimum essential medium (Cambrex) with 2 mM l-glutamine and Earle's balanced salt solution adjusted to contain 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids (Cambrex), 1.0 mM sodium pyruvate (Cambrex), and 10% heat-inactivated fetal calf serum in a 96-well microtiter plate until confluent growth. Prior to infection experiments, cells were washed with phosphate-buffered saline, and infection experiments were performed in RPMI 1640 medium without serum. Infection experiments were performed as described previously (28). The number of cell-associated bacteria was calculated as a percentage of total bacteria present in the well after 5 h of infection. All experiments were performed at least three times unless indicated otherwise. Two-tailed Student's t test was used to calculate statistical significance (P values).
Protein and nucleic acid sequence analyses.
Public databases of published protein and nucleotide sequences were searched using the BLAST programs available at the http://www.ncbi.nih.gov/BLAST/ and http://www.tigr.org websites. The genome databases of meningococcal strain Z2491, MC58, and FAM18 were searched using the BLAST servers available at TIGR (http://www.tigr.org) and the Sanger Institute (http://www.sanger.ac.uk). Conserved domain searches were performed using SwissPfam available at http://www.sanger.ac.uk, InterProScan available at http://us.expasy.org/prosite/, and the NCBI Conserved Domain Search available at http://www.ncbi.nih.gov/. Protein sequences were aligned using the multiple-sequence alignment ClustalW available at http://clustalw.ddbj.nig.ac.jp/top-e.html. Phylogenetic analysis was performed with MEGA (version 3.1) available at www.megasoftware.net.
RESULTS
Presence and distribution of hrpA genes among N. meningitidis carrier strains.
A panel of 822 N. meningitidis isolates from healthy individuals was analyzed for the presence of hrpA genes. Genetic characteristics of these strains including multilocus sequence typing analysis have been described previously (12). The presence and distribution of hrpA genes were analyzed by dot blotting with specific probes for meningococcal hrpA genes NMB0493, NMB1214, NMB1768, and NMB0497/1779. A total of 687 strains that gave unambiguous results were analyzed (see Table S3 in the supplemental material). Ninety-seven percent (210/216) of the strains belonging to one of the known hypervirulent lineages were found to be positive for at least one of the probes: 112/114 (98%) isolates belonging to the sequence type 41 and 44 (ST-41/44) complex, all 34 (100%) isolates belonging to the ST-32 complex, 59/63 (94%) isolates belonging to the ST-23 complex, all 3 (100%) isolates belonging to the ST-11 complex, and both (100%) isolates belonging to the ST-8 complex (see Table S3 in the supplemental material). The majority of the remaining strains (334/471 [71%]) not belonging to hypervirulent lineages also carried at least one hrpA gene. Among those strains that did not hybridize with the probes, 113/137 (82%) belonged to four of five clonal complexes harboring the capsule null locus (cnl) (11) including all strains of the ST-53 complex (52 strains), the ST-198 complex (42 strains), the ST-1117 complex (17 strains), and the ST-1136 complex (2 strains). The majority of hrpA-positive strains (255/544 [47%]) hybridized only with the probe specific for NMB0497/NMB1779 (see Table S3 in the supplemental material). Of 544 hrpA-positive strains, 159 (29%) strains hybridized with the probes specific for NMB0493, NMB1768, and NMB0497/NMB1779 but not with the probe for NMB1214. More than 50% of these strains were members of the ST-41/44 complex (86 isolates). A positive signal with all four probes could be demonstrated in 76/544 strains (14%), including most isolates of the ST-32 complex (33/34), all isolates of the ST-269 complex (12 isolates), and a set of strains belonging to the ST-35 complex (10/32 [31%]). Interestingly, the presence of a hrpA gene homologous to either NMB0493, NMB1214, or NMB1768 was always associated with a positive signal for NMB0497/NMB1779. Though there were no isolates representing the three serogroup A-related hypervirulent lineages among the carrier strains analyzed, the presence of one hrpA gene in the published genome sequence of strain Z2491 suggests that hrpA genes are also distributed among the serogroup A strains.
Presence and distribution of hrpB genes among N. meningitidis carrier strains.
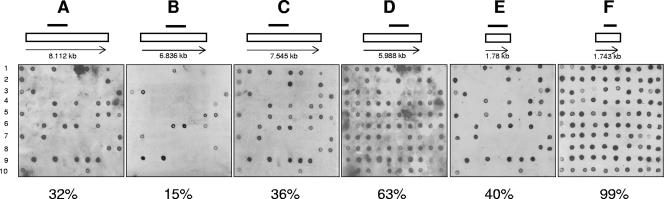
Given the high sequence variability of known hrpA genes in N. meningitidis, strains that did not react with any of the four probes could either completely lack a hrpA homologue or contain hrpA genes not detected by the probes due to sequence variation. Therefore, additional dot blots were performed with a probe specific for the highly conserved meningococcal hrpB gene NMC0443 (Table 1). Almost all strains (801/822 [97.4%]) hybridized with the probe specific for the hrpB gene (Fig. 1). As the genes for cognate exoprotein and transporter protein are closely associated in known TPS systems, the presence of genes coding for possible transporter proteins suggests that strains which were negative with the MC58 hrpA probes harbored undetected hrpA genes with nucleotide sequences different from those present in strain MC58. This hypothesis was tested by PCR of a selection of 30 strains representing hypervirulent clonal complexes (ST-8 complex, ST-11 complex, ST-23 complex, and ST-41/44 complex) and nonhypervirulent clonal complexes (ST-22 complex, ST-60 complex, and ST-162 complex) including those harboring the capsule null locus (ST-53 complex, ST-198 complex, ST-845 complex, and ST-1117 complex). Additionally, the 21 strains which did not hybridize with the hrpB probe were tested. The forward primer recognized the 3′ terminus of the highly conserved hrpB gene (corresponding to nucleotides 1641 to 1663 in NMC0443). The reverse primer recognized a conserved motif within the secretion domain present in all known meningococcal hrpA genes (corresponding to nucleotides 501 to 520 in NMC0444). A DNA fragment with the expected size was amplified in all strains with the exception of one isolate belonging to the ST-774 complex (data not shown). NMB1762 has been hypothesized to encode the cognate transporter protein for HrpANMB1768 (46). By dot blot hybridization with a probe specific for NMB1762, 328 out of the 822 strains analyzed (40%) gave a positive result (Fig. 1). Almost all strains harboring NMB1768-homologous genes hybridized also with the probe for NMB1762 (288/294 [97.9%]). In all six negative strains, PCR with primers specific for NMB1762 revealed the presence of this gene. Similarly, the strains that hybridized with the probe for NMB1762 but did not hybridize with the probe for NMB1768 or gave ambiguous results (34/328 [10.3%]) were shown to harbor both genes by PCR with primers specific for NMB1768. The only exception was one isolate of the ST-167 complex that was negative with both the probe and the PCR for NMB1768.
FIG. 1.
DNA dot blot analysis of a selection of carrier strains with probes for the hrpA genes NMB0493 (A), NMB1214 (B), NMB1768 (C), and NMB0497/NMB1779 (D) and the hrpB genes NMB1762 (E) and NMB0496/NMB1780 (F). Each blot (A to F) shows the results of a selection of 100 carrier isolates (alpha 201 to 300). The order of bacterial strains is the same in all panels. The number below each panel is the percentage of strains out of the 822 strains analyzed that hybridized with the respective probe. The locations of the probes used for hybridization are indicated by black bars above the open rectangles representing the respective hrpA and hrpB ORFs. The numbers below the open rectangles indicate the sizes (in kilobases) of the ORFs.
Sequence analysis of hrpAalpha 14.
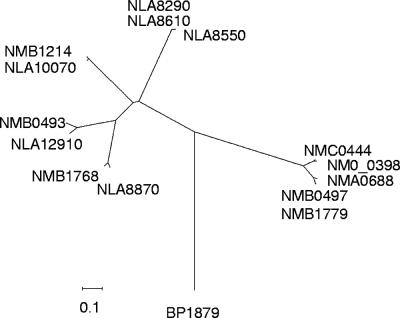
To determine whether strains that did not hybridize with the MC58 hrpA probes but displayed a positive PCR result with the primer pair for hrpA and hrpB harbor a full-length hrpA gene, the complete nucleotide sequence of the single hrpA gene of strain alpha 14 (cnl:ST-53) was determined from contigs of a whole-genome sequencing project (C. Schoen et al., unpublished data). The 5,949-bp hrpA gene from strain alpha 14 (hrpAalpha 14) (NM00398) encodes a protein with a predicted molecular mass of approximately 207 kDa displaying an overall identity to NMB1779 of 50.8%, with the highest degree of similarity at the N terminus (98.8% identity within the first 321 aa) including the TPS domain. Motifs critical for secretion of FHA in B. pertussis and conserved in strain MC58 hrpA genes are also present in NM00398 (see Fig. S1 in the supplemental material). In contrast, the C terminus was more diverse, displaying only 41.5% sequence identity, which accounts for the failure of the probes derived from strain MC58 to detect hrpAalpha 14. hrpAalpha 14 is most closely related to NMC0444 with an overall nucleotide identity of 51.8% and an overall amino acid identity of 78.6%. A phylogenetic tree was constructed to compare HrpAalpha 14 with the other known meningococcal HrpA proteins. Accordingly, meningococcal HrpA proteins can clearly be separated into two groups (Fig. 2). HrpAalpha 14 clustered into group I, comprising the single HrpA proteins of meningococcal strains Z2491 and FAM18 together with the two HrpA proteins of strain MC58 encoded by NMB0497 and NMB1779. Group II comprised the three HrpA proteins of strain MC58 encoded by NMB0493, NMB1214, and NMB1768 which have no close homologues in the other two available N. meningitidis sequences (Fig. 2). Even HrpA proteins within one of these groups display significant C-terminal sequence variation (Fig. 2). The protein sequences encoded by six putative hrpA genes identified in the preliminary whole-genome sequence of Neisseria lactamica ST640 (available at www.sanger.ac.uk) were included in the analysis. Only genes encoding HrpA homologous to group II, but not to group I, meningococcal HrpA proteins were detected in N. lactamica (Fig. 2).
FIG. 2.
Phylogenetic tree based on the deduced amino acid sequences of the proteins encoded by the hrpA genes of strains MC58, Z2491, FAM18, and alpha 14. In addition, the protein sequences encoded by six putative hrpA genes present in the preliminary sequence of Neisseria lactamica ST640 were included in the analysis. The meningococcal HrpA proteins clearly cluster into two separate groups. The HrpA protein identified in strain alpha 14 (denoted by NM0_0398 in the figure) is closely related to the group of HrpA proteins encoded by genes homologous to NMB1779. In contrast, the six HrpA proteins identified in N. lactamica (denoted by NLA8870, NLA12910, NLA10070, NLA8290, NLA8610, and NLA8550 in the figure) are closely related to the second group of meningococcal HrpA proteins encoded by NMB0493, NMB1214, and NMB1768. BP1879 denotes the ORF encoding the filamentous hemagglutinin of B. pertussis. Alignments were performed with ClustalW. Tree construction was performed with MEGA (version 3.1) using the neighbor-joining method. Regions of the alignment containing gaps were excluded from the analysis. The scale bar indicates linkage distance.
Expression analysis of hrpA and hrpB genes in a selection of meningococcal strains.
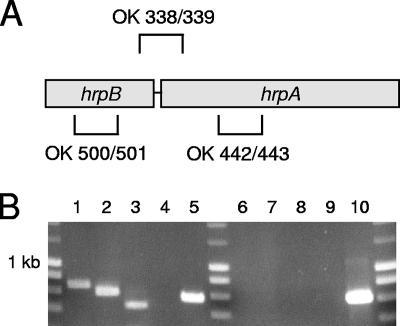
RT-PCR was performed to detect the presence of hrpA and hrpB transcripts in total RNA isolated from different meningococcal strains. In strain MC58, expression was confirmed for the hrpA genes NMB0493, NMB1214, NMB1768, and NMB0497/NMB1779 and for the hrpB genes NMB1780/NMB0496 and NMB1762. As NMB0497 and NMB1779 and their associated putative transporters NMB0496 and NMB1780 display almost 100% sequence identity, differentiation between the genes was not achieved. Using a forward primer recognizing the 3′ end of the hrpB gene NMB1780 and a reverse primer recognizing a conserved region within the 5′ end of the hrpA gene NMB1779, it could be shown that both genes are organized in an operon and transcribed into a single mRNA. Similar results were obtained with strain 2120 harboring only one hrpA gene (Fig. 3). Therefore, the typical operon structure of other TPS systems is conserved in N. meningitidis. The expression of hrpA genes was also analyzed in a subset of the carrier strains representing 11 clonal complexes using the primer pair for hrpA and hrpB. In all strains, a DNA fragment of the expected size was amplified (data not shown). Similarly, a selection of nine carrier strains representing nine different clonal complexes that had been shown to harbor NMB1762 homologues could be shown to express the respective gene. Therefore, all hrpA and hrpB genes were found to be transcribed in all meningococcal isolates tested.
FIG. 3.
RT-PCR analysis of hrpA and hrpB expression in strain 2120. (A) Schematic representation of the primer pairs used in panel B, primers OK338/OK339 (OK 338/339), OK500/OK501 (OK 500/501), and OK442/OK443 (OK 442/443). (B) Transcripts of hrpA and hrpB were detected in total RNA preparations of strain 2120. Additionally, it could be shown that both genes are transcribed into a single mRNA. The following templates were included in the samples: total RNA of strain 2120 (lanes 1 to 3, 5, and 6 to 8), no template (negative control) (lanes 4 and 9), and genomic DNA of strain 2120 (positive control) (lane 10). The primer sets included in the samples were as follows: hrpB-specific primers (OK500/OK501 primers [see panel A]) (lanes 1 and 6), hrpA-specific primers (OK442/OK443 primers [see panel A]) (lanes 2, 7, and 10), primer pair with the forward primer recognizing the 3′ end of the hrpB gene and the reverse primer recognizing a conserved region within the 5′ end of the adjacent hrpA (OK338/OK339 [see panel A]) (lanes 3 and 8), and primers specific for lgtA (OK487/OK488, positive control) (lanes 4, 5, 8, and 9). The samples loaded in lanes 6 to 10 were not subjected to the reverse transcription step of the RT-PCR procedure and served as controls to rule out DNA contamination of the RNA templates.
Meningococcal HrpA proteins are expressed, proteolytically processed, and secreted in an HrpB-dependent manner.
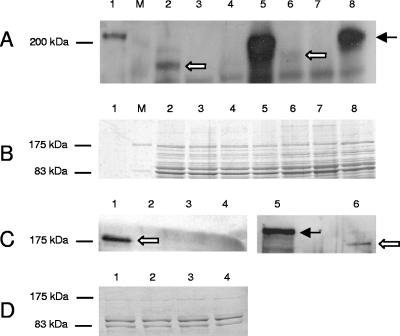
To investigate the expression of meningococcal HrpA proteins, a polyclonal rabbit antiserum was raised against the HrpA protein encoded by NMB1779. For that purpose, His-tagged NMB1779 was expressed and purified in E. coli. This antiserum was able to recognize recombinant HrpA protein in a Western blot; the protein migrated to a position of approximately 210 kDa (Fig. 4). However, when Western blotting was performed with whole-cell lysates of strain MC58, no band corresponding in size to the recombinant protein could be detected (Fig. 4). Instead, the antiserum did detect small amounts of a protein of approximately 180 kDa which was absent in ΔNMB0497 ΔNMB1779 mutants. Interestingly, analysis of a ΔhrpB mutant (simultaneous deletion of NMB0496 and NMB1780) revealed a prominent band corresponding in size to the full-length recombinant HrpA protein. Therefore, proteolytic processing of meningococcal HrpA proteins might occur during or after translocation across the outer membrane. To determine whether HrpA proteins are secreted into the environment, whole-cell lysates of strain 2517 and its ΔhrpA and ΔhrpB deletion mutants and a ΔhrpA ΔhrpB double deletion mutant were compared with concentrated supernatants of the same strains (Fig. 4). Strain 2517 was chosen for these experiments, as only a single hrpA gene and its associated hrpB gene are present in this strain. Similar to the results obtained with the ΔhrpB mutant of strain MC58, a prominent band of approximately 210 kDa was detected in whole-cell lysates of strain 2517 ΔhrpB (Fig. 4). This protein could not be detected in both the ΔhrpA mutant and the ΔhrpA ΔhrpB mutant and most likely represents the unprocessed HrpA protein accumulating in the bacterial cell due to the deletion of its transporter HrpB. As for strain MC58, a band of approximately 180 kDa was detected in whole-cell lysates of strain 2517. In addition, a protein of the same size was detected in the culture supernatants (Fig. 4). This form of HrpA was absent from supernatants and cell lysates of the ΔhrpA and ΔhrpA ΔhrpB mutants. These data provide evidence that HrpA proteins are effectively released into the culture supernatant in the presence of HrpB. Furthermore, HrpA proteins are processed during or after secretion as indicated by the lower molecular mass of the mature HrpA. A small proportion of the processed HrpA remains associated with the bacterial cell.
FIG. 4.
Expression analysis of HrpA proteins by Western immunoblot assays. Proteins were separated on a 7.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with polyclonal antibody RαNMB1779. The arrows indicate the processed (white arrows) and unprocessed (black arrow) HrpA proteins. (A) Western blot of whole-cell lysates of strain MC58, strain 2517, and their respective ΔhrpA and ΔhrpB mutants. Lanes: 1, purified HrpA; M, molecular mass standard (in kilodaltons); 2, MC58 (wild type); 3, MC58 ΔsiaD ΔhrpA; 4, MC58 ΔgalE ΔhrpA; 5, MC58 ΔsiaD ΔlgtA ΔhrpB; 6, 2517; 7, 2517 ΔhrpA; 8, 2517 ΔhrpB. In strain MC58, ΔhrpA denotes simultaneous deletion of NMB0497 and NMB1779, whereas ΔhrpB denotes simultaneous deletion of NMB0496 and NMB1780. (B) Coomassie blue-stained gel of whole-cell lysates used in panel A. (C) Western blot of culture supernatants of strain 2517 and its respective ΔhrpA and ΔhrpB deletion mutants as well as the double deletion mutant ΔhrpA ΔhrpB in comparison to whole-cell lysate of strain 2517 ΔhrpB. Lanes: 1, 2517; 2, 2517 ΔhrpA; 3, 2517 ΔhrpB; 4, 2517 ΔhrpA ΔhrpB; 5, whole-cell lysate of strain 2517 ΔhrpB; 6, 2517. (D) Coomassie blue-stained gel of culture supernatants used in panel C.
Contribution of hrpA to the interaction of meningococci with host cells.
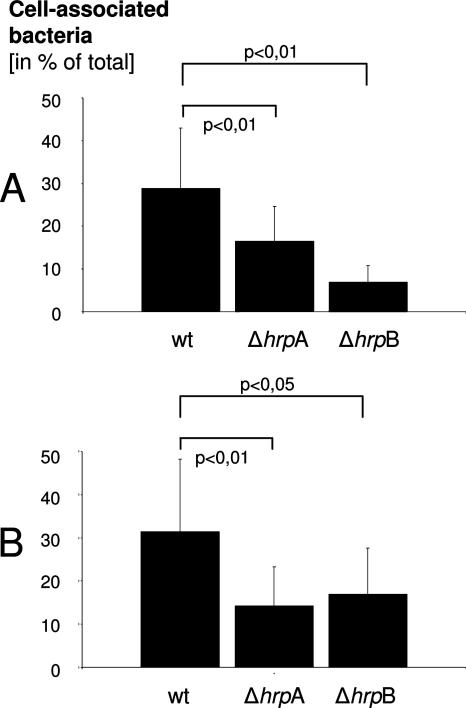
To investigate whether the presence of different hrpA genes in strain MC58 contributes to adherence, mutant NMB0493, NMB1214, and NMB1768 genes were deleted in the unencapsulated strain MC58 ΔsiaD and the unencapsulated and truncated-LPS strain MC58 ΔsiaD ΔlgtA by insertional inactivation of these genes. As NMB0497 represents an almost identical copy of NMB1779, both genes were inactivated simultaneously in strain MC58 ΔsiaD and in the unencapsulated and truncated-LPS strain MC58 ΔsiaABCD ΔgalE. Inactivation of hrpA genes was confirmed by PCR and Southern blot analysis (data not shown). Unencapsulated and truncated-LPS parental strains were chosen, because capsule expression and the presence of the LPS alpha-chain shields meningococci from interaction with epithelial cells (O. Kurzai et al., unpublished data). Infection experiments were performed to quantify the adherence of the ΔhrpA mutants to epithelial cells in comparison to their parental strains. The proportion of cell-associated bacteria (adherent and viable intracellular) was quantified 5 h postinfection (p.i.). No difference could be detected in the proportion of cell-associated bacteria between the hrpA mutants and their parental strains (data not shown). These data indicate that adherence to epithelial cells is not mediated by an individual hrpA present in MC58. As the possibility that the proteins encoded by the different hrpA genes share a common function in the interaction with host cells cannot be excluded, further infection experiments were performed using mutants of serogroup C strain 2120 harboring only a single hrpA gene. hrpA mutants were constructed in the unencapsulated strain 2120 ΔsiaD and in the unencapsulated and truncated-LPS strain 2120 ΔsiaD ΔlgtA by insertional inactivation. Irrespective of the parental strain genotype, deletion of hrpA or hrpB did not influence growth kinetics (data not shown). Whereas no difference in the number of HEp-2 cell-associated bacteria could be detected for strain 2120 ΔsiaD and its respective hrpA mutant, deletion of the hrpA gene in the strain with truncated LPS led to a significant decrease in the proportion of cell-associated bacteria 5 h p.i. (16.8% versus 34.4% cell-associated bacteria; P < 0.01) (Fig. 5). Similar results were obtained with FaDu epithelial cells (Fig. 5), indicating that HrpA contributes to meningococcal adherence to different epithelial cell lines in an unencapsulated and LPS-truncated background and might be functionally redundant in strains carrying more than one allele. The same results were obtained with three additional independent hrpA mutants of strain 2120 ΔsiaD ΔlgtA (data not shown). Additionally, deletion of the hrpB gene in strain 2120 ΔsiaD ΔlgtA also led to a significant decrease in the proportion of cell-associated bacteria 5 h p.i. (Fig. 5), thus providing further evidence for the functional relevance of HrpA secretion by its associated HrpB. In contrast to epithelial cells, no difference could be detected in the number of cell-associated bacteria in infection experiments with human dendritic cells (data not shown), suggesting that the adhesive properties of HrpA are cell type specific.
FIG. 5.
Adhesion of a ΔhrpA deletion mutant and a ΔhrpB deletion mutant of strain 2517 ΔlgtA to HEp2 (A) and FaDu epithelial cells (B) 5 h p.i. Compared to the wild-type (wt) parental strain, the proportion of cell-associated bacteria was significantly lower in the ΔhrpA mutant and in the ΔhrpB mutant. Means plus standard deviations (error bars) for 10 independent experiments are indicated. Two-tailed Student's t test was used to calculate statistical significance (P values).
DISCUSSION
Several large exoproteins in various gram-negative bacteria are predicted to be secreted via the TPS system (23). However, relatively few of these proteins have been investigated in detail. FHA, the most extensively characterized of these exoproteins, is an important virulence factor of Bordetella and constitutes the main component of the pertussis vaccine (29). In this study, meningococcal hrpA genes encoding high-molecular-weight proteins related to FHA were analyzed. The sequence similarities between FHA and HrpA are restricted to the N terminus comprising the TPS domain which is the hallmark of proteins secreted by the TPS pathway. In contrast, the C-terminal portions of the proteins that have been shown to contain the functional domains in Bordetella FHA (31) show no relevant homology between the two species. The conserved TPS domain contains secretion determinants mediating translocation of the respective protein across the outer membrane by its associated transporter termed FhaC in B. pertussis (21, 23, 46). The presence of the TPS domain including conserved NPNGI and NPNL motifs in the derived protein sequence of the hrpA genes of strains MC58, Z2491, FAM18, and alpha 14 together with the fact that more than 99% of the strains investigated in this study contain genes homologous to fhaC suggests the presence of a functional TPS in N. meningitidis.
Klee et al. were able to demonstrate the presence of genes homologous to the hrpA gene of strain Z2491 in 13 strains mainly representing the known hypervirulent lineages of N. meningitidis (27). In this study, by analyzing a large collection of carrier isolates comprising 51 clonal complexes, we provide evidence that hrpA genes are ubiquitously present in N. meningitidis and are not restricted to strains of the hypervirulent lineages. We propose to divide the meningococcal hrpA genes into two groups according to their degree of sequence similarity. Group I comprises the two almost identical genes NMB0497 and NMB1779 together with the single hrpA genes present in strain Z2491 (NMA0688), strain FAM18 (NMC0444), and strain alpha 14 (NM00398). Group II comprises NMB1768, NMB0493, and NMB1214. As no genes closely related to group II had until now been identified in meningococcal strains other than strain MC58, our data for the first time provide evidence that they are not restricted to MC58 or its close relatives. From an evolutionary point of view, it is interesting to notice that only hrpA genes homologous to group II were detected in N. lactamica strain ST640. These findings are congruent with those of a recent study by Snyder and Saunders (41). The authors were able to demonstrate that genes homologous to NMB0493 and NMB1214 are present in all 13 N. lactamica strains investigated using comparative genome hybridization to a pan-Neisseria microarray (41). Genes homologous to NMB1768 were detected in a subset of N. lactamica strains, whereas genes homologous to NMB1779 could not be detected in N. lactamica (41). Similarly, the genome of N. gonorrhoeae strain FA 1090 contains an ORF disrupted by multiple stop codons, which otherwise would encode a TpsA protein with homology to NMB1768 (46). Even hrpA genes from the same group can show considerable sequence variation in the C terminus, which might be due to a direct recombination-based mechanism for C-terminal exchange between hrpA genes and a number of silent cassettes encoding alternative C termini present in the meningococcal genome (4, 46). For example, NMB1779 and hrpAalpha 14, both group I meningococcal hrpA genes, show a sequence diversity of >49%. It is intriguing to speculate that C-terminal sequence variation might have arisen from selective pressure and could alter the functions of these proteins and modulate the virulence potential of different clonal lineages.
HrpA proteins are expressed and released into the culture supernatant by a HrpB-dependent mechanism. However, in contrast to B. pertussis and other known TPS systems, where the exoprotein FHA is rapidly degraded intracellularly in the absence of the transporter FhaC (21) (9, 25), meningococcal HrpA accumulates in the bacteria and remains stable and undegraded if HrpB has been deleted. Our data strongly suggest that the HrpA protein is proteolytically processed during or after secretion. Similar processing has been described for B. pertussis (29) and Haemophilus influenzae (3). Details of the secretion and processing of HrpA will be addressed in further studies.
Until now, the role of FHA-related proteins in the interaction with the human host has been studied in detail in only a few bacterial species. B. pertussis FHA mediates adherence to epithelial cells (20, 37, 45) and macrophages (36, 40) and is required for tracheal colonization in vivo (26). Similarly, the FHA-related proteins of nontypeable H. influenzae, HMW1 and HMW2, were shown to mediate binding to macrophages (33) and human epithelial cells (42). Our data clearly show that meningococcal HrpA proteins contribute to adherence to host cells. A hrpA deletion mutant of strain 2120, which harbors only one hrpA gene, displayed significantly reduced adherence to different epithelial cells. This effect was observed in an unencapsulated background with short LPS (L8 immunotype). It has previously been shown that capsule expression as well as expression of the LPS alpha-chain can shield meningococci from interaction with host cells (28). Both capsule expression and expression of the LPS alpha-chain are subject to phase variation in meningococci (17, 24), and therefore, capsule-deficient as well as LPS-truncated variants may play a role during colonization and pathogenesis (17, 30). Indeed, the function of Opc, a well-known adhesin and invasin of N. meningitidis is also dependent on a lack of capsule expression and LPS truncation (47). Due to the fact that only small amounts of HrpA remain cell associated and can be detected in whole-cell lysates, the possibility that the effect of the hrpA or hrpB deletion on adhesion is indirect (e.g., by altering outer membrane composition), cannot be excluded. However, no gross changes in expression of outer membrane proteins could be detected in the deletion mutants. The roles of hrpA genes with homology to NMB1768, NMB0493, and NMB1214 have not been addressed selectively, as strains harboring one of these genes always possess additional hrpA genes homologous to NMB1779/NMB0497. In strain MC58, deletions of a single hrpA gene had no effect on adhesion, and although we were able to construct a double deletion of NMB1779 and NMB0497 in MC58, the mutant displayed the same adherence phenotype as the parental strain. This might indicate that hrpA genes of both groups are functionally redundant in interaction with epithelial cells despite considerable sequence difference.
In summary, FHA-like proteins (HrpA) as well as FhaC-like proteins (HrpB) are encoded and expressed by virtually all N. meningitidis strains belonging to both hypervirulent and nonhypervirulent clonal complexes. HrpA proteins are secreted via transporters encoded by hrpB genes and accumulate in the bacteria after deletion of this transporter. However, Western blot analysis as well as the observation that HrpA proteins play a role in adhesion to host cells indicates that a proportion of the secreted HrpA remains surface associated. A similar phenomenon has also been described for another meningococcal adhesin of the autotransporter family, App (16). By analogy to the situation in B. pertussis, this renders these proteins interesting candidates for designing potential vaccines against serogroup B meningococci.
Supplementary Material
Acknowledgments
This work would not have been possible without the excellent technical assistance of Julia Oechsner. Antibodies have been kindly donated by Wendell Zollinger, Sanjay Ram, Mark Achtman, and Mumtaz Virji. The NMB1779 expression construct was kindly donated by Silke Klee. Heike Claus and Ulrich Vogel contributed to this work in many helpful discussions and provided dot blots of the strains from the Bavarian carriage study. Christoph Schoen kindly provided data from the strain alpha 14 genome project.
This work was supported by the German Research Foundation (DFG) SFB479 (grant B2 to O.K. and M.F.).
Footnotes
Published ahead of print on 14 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ait-Tahar, K., K. G. Wooldridge, D. P. Turner, M. Atta, I. Todd, and D. A. Ala'Aldeen. 2000. Autotransporter A protein of Neisseria meningitidis: a potent CD4+ T-cell and B-cell stimulating antigen detected by expression cloning. Mol. Microbiol. 37:1094-1105. [DOI] [PubMed] [Google Scholar]
- 2.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and J. W. St. Geme III. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., S. Hobbie, and R. Ondraczek. 1992. Serratia marcescens forms a new type of cytolysin. FEMS Microbiol. Lett. 79:299-305. [DOI] [PubMed] [Google Scholar]
- 6.Cartwright, K. A., J. M. Stuart, D. M. Jones, and N. D. Noah. 1987. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99:591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caugant, D. A., E. A. Hoiby, P. Magnus, O. Scheel, T. Hoel, G. Bjune, E. Wedege, J. Eng, and L. O. Froholm. 1994. Asymptomatic carriage of Neisseria meningitidis in a randomly sampled population. J. Clin. Microbiol. 32:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier, N., M. Moser, H. G. Koch, K. L. Schimz, E. Willery, C. Locht, F. Jacob-Dubuisson, and M. Muller. 2004. Membrane targeting of a bacterial virulence factor harbouring an extended signal peptide. J. Mol. Microbiol. Biotechnol. 8:7-18. [DOI] [PubMed] [Google Scholar]
- 9.Choi, P. S., A. J. Dawson, and H. D. Bernstein. 2007. Characterization of a novel two-partner secretion system in Escherichia coli O157:H7. J. Bacteriol. 189:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clantin, B., H. Hodak, E. Willery, C. Locht, F. Jacob-Dubuisson, and V. Villeret. 2004. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc. Natl. Acad. Sci. USA 101:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813-1819. [DOI] [PubMed] [Google Scholar]
- 12.Claus, H., M. C. Maiden, D. J. Wilson, N. D. McCarthy, K. A. Jolley, R. Urwin, F. Hessler, M. Frosch, and U. Vogel. 2005. Genetic analysis of meningococci carried by children and young adults. J. Infect. Dis. 191:1263-1271. [DOI] [PubMed] [Google Scholar]
- 13.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frosch, M., I. Gorgen, G. J. Boulnois, K. N. Timmis, and D. Bitter-Suermann. 1985. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc. Natl. Acad. Sci. USA 82:1194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frosch, M., E. Schultz, E. Glenn-Calvo, and T. F. Meyer. 1990. Generation of capsule-deficient Neisseria meningitidis strains by homologous recombination. Mol. Microbiol. 4:1215-1218. [DOI] [PubMed] [Google Scholar]
- 16.Hadi, H. A., K. G. Wooldridge, K. Robinson, and D. A. Ala'Aldeen. 2001. Identification and characterization of App: an immunogenic autotransporter protein of Neisseria meningitidis. Mol. Microbiol. 41:611-623. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt, S., A. Müller, H. Sillmann, M. Mühlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilse, R., S. Hammerschmidt, W. Bautsch, and M. Frosch. 1996. Site-specific insertion of IS1301 and distribution in Neisseria meningitidis strains. J. Bacteriol. 178:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishibashi, Y., D. A. Relman, and A. Nishikawa. 2001. Invasion of human respiratory epithelial cells by Bordetella pertussis: possible role for a filamentous hemagglutinin Arg-Gly-Asp sequence and alpha5beta1 integrin. Microb. Pathog. 30:279-288. [DOI] [PubMed] [Google Scholar]
- 21.Jacob-Dubuisson, F., C. Buisine, E. Willery, G. Renauld-Mongenie, and C. Locht. 1997. Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J. Bacteriol. 179:775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 23.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 24.Jennings, M. P., Y. N. Srikhanta, E. R. Moxon, M. Kramer, J. T. Poolman, B. Kuipers, and P. van der Ley. 1999. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology 145:3013-3021. [DOI] [PubMed] [Google Scholar]
- 25.Julio, S. M., and P. A. Cotter. 2005. Characterization of the filamentous hemagglutinin-like protein FhaS in Bordetella bronchiseptica. Infect. Immun. 73:4960-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura, A., K. T. Mountzouros, D. A. Relman, S. Falkow, and J. L. Cowell. 1990. Bordetella pertussis filamentous hemagglutinin: evaluation as a protective antigen and colonization factor in a mouse respiratory infection model. Infect. Immun. 58:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klee, S. R., X. Nassif, B. Kusecek, P. Merker, J. L. Beretti, M. Achtman, and C. R. Tinsley. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurzai, O., C. Schmitt, H. Claus, U. Vogel, M. Frosch, and A. Kolb-Mäurer. 2005. Carbohydrate composition of meningococcal lipopolysaccharide modulates the interaction of Neisseria meningitidis with human dendritic cells. Cell. Microbiol. 7:1319-1334. [DOI] [PubMed] [Google Scholar]
- 29.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 30.Mackinnon, F. G., R. Borrow, A. R. Gorringe, A. J. Fox, D. M. Jones, and A. Robinson. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb. Pathog. 15:359-366. [DOI] [PubMed] [Google Scholar]
- 31.Mazar, J., and P. A. Cotter. 2006. Topology and maturation of filamentous haemagglutinin suggest a new model for two-partner secretion. Mol. Microbiol. 62:641-654. [DOI] [PubMed] [Google Scholar]
- 32.Mota, L. J., I. Sorg, and G. R. Cornelis. 2005. Type III secretion: the bacteria-eukaryotic cell express. FEMS Microbiol. Lett. 252:1-10. [DOI] [PubMed] [Google Scholar]
- 33.Noel, G. J., S. J. Barenkamp, J. W. St. Geme III, W. N. Haining, and D. M. Mosser. 1994. High-molecular-weight surface-exposed proteins of Haemophilus influenzae mediate binding to macrophages. J. Infect. Dis. 169:425-429. [DOI] [PubMed] [Google Scholar]
- 34.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 35.Patel, S. S., and A. J. Wagstaff. 1996. A cellular pertussis vaccine (Infanrix-DTPa; SB-3). A review of its immunogenicity, protective efficacy and tolerability in the prevention of Bordetella pertussis infection. Drugs 52:254-275. [DOI] [PubMed] [Google Scholar]
- 36.Relman, D., E. Tuomanen, S. Falkow, D. T. Golenbock, K. Saukkonen, and S. D. Wright. 1990. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell 61:1375-1382. [DOI] [PubMed] [Google Scholar]
- 37.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 40.Saukkonen, K., C. Cabellos, M. Burroughs, S. Prasad, and E. Tuomanen. 1991. Integrin-mediated localization of Bordetella pertussis within macrophages: role in pulmonary colonization. J. Exp. Med. 173:1143-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder, L. A., and N. J. Saunders. 2006. The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics 7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatum, F. M., A. G. Yersin, and R. E. Briggs. 2005. Construction and virulence of a Pasteurella multocida fhaB2 mutant in turkeys. Microb. Pathog. 39:9-17. [DOI] [PubMed] [Google Scholar]
- 44.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 45.Tuomanen, E., A. Weiss, R. Rich, F. Zak, and O. Zak. 1985. Filamentous hemagglutinin and pertussis toxin promote adherence of Bordetella pertussis to cilia. Dev. Biol. Stand. 61:197-204. [PubMed] [Google Scholar]
- 46.van Ulsen, P., and J. Tommassen. 2006. Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol. Rev. 30:292-319. [DOI] [PubMed] [Google Scholar]
- 47.Virji, M., K. Makepeace, I. R. Peak, D. J. Ferguson, M. P. Jennings, and E. R. Moxon. 1995. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol. Microbiol. 18:741-754. [DOI] [PubMed] [Google Scholar]
- 48.Ward, C. K., J. L. Latimer, J. Nika, M. Vakevainen, J. R. Mock, K. Deng, R. J. Blick, and E. J. Hansen. 2003. Mutations in the lspA1 and lspA2 genes of Haemophilus ducreyi affect the virulence of this pathogen in an animal model system. Infect. Immun. 71:2478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.