Abstract
Phosphopantetheine adenylyltransferase (PPAT) from Escherichia coli is an essential hexameric enzyme that catalyzes the penultimate step in coenzyme A (CoA) biosynthesis and is a target for antibacterial drug discovery. The enzyme utilizes Mg-ATP and phosphopantetheine (PhP) to generate dephospho-CoA (dPCoA) and pyrophosphate. When overexpressed in E. coli, PPAT copurifies with tightly bound CoA, suggesting a feedback inhibitory role for this cofactor. Using an enzyme-coupled assay for the forward-direction reaction (dPCoA-generating) and isothermal titration calorimetry, we investigated the steady-state kinetics and ligand binding properties of PPAT. All substrates and products bind the free enzyme, and product inhibition studies are consistent with a random bi-bi kinetic mechanism. CoA inhibits PPAT and is competitive with ATP, PhP, and dPCoA. Previously published structures of PPAT crystallized at pH 5.0 show half-the-sites reactivity for PhP and dPCoA and full occupancy by ATP and CoA. Ligand-binding studies at pH 8.0 show that ATP, PhP, dPCoA, and CoA occupy all six monomers of the PPAT hexamer, although CoA exhibits two thermodynamically distinct binding modes. These results suggest that the half-the-sites reactivity observed in PPAT crystal structures may be pH dependent. In light of previous studies on the regulation of CoA biosynthesis, the PPAT kinetic and ligand binding data suggest that intracellular PhP concentrations modulate the distribution of PPAT monomers between high- and low-affinity CoA binding modes. This model is consistent with PPAT serving as a “backup” regulator of pathway flux relative to pantothenate kinase.
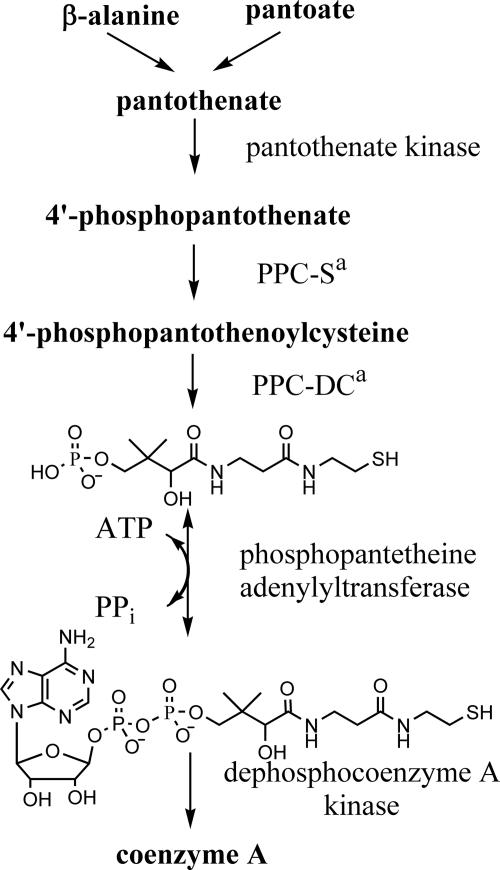
Coenzyme A (CoA) is the principal intracellular carrier of activated acyl groups. It is utilized in many metabolic reactions, including fatty acid metabolism and biosynthesis (3). CoA is synthesized from pantothenate in five enzymatic steps common to organisms from all taxa (Fig. 1). In Escherichia coli, the five enzymatic activities are catalyzed by four gene products (3), all of which are essential for growth in rich media (6, 8) (C. McPherson, unpublished data). E. coli phosphopantetheine adenylyltransferase (PPAT) catalyzes the penultimate step of this pathway and shows considerable homology to its eubacterial orthologs (7) but no significant sequence homology to the human ortholog (1, 4). PPAT has been implicated in metabolic regulation of the CoA biosynthetic pathway (17), as discussed in detail below. These findings indicate that the enzyme is a potential target for antibacterial drug discovery.
FIG. 1.
Biosynthesis of CoA in E. coli. In E. coli the enzymatic activities of phosphopantothenoylcysteine synthetase (PPC-S) and phosphopantothenoylcysteine decarboxylase (PPC-DC), indicated by a superscript “a” in the figure, are catalyzed by a single gene product.
PPAT catalyzes the magnesium-dependent adenosyl transfer from ATP to phosphopantetheine (PhP), generating dephospho-CoA (dPCoA) and pyrophosphate (7). The E. coli coaD (formerly kdtB) gene encoding PPAT has been cloned, and the overexpressed protein has been purified and characterized (7). PPAT is a homohexamer of 17.8-kDa subunits and overexpressed PPAT copurifies with approximately 0.5 equivalents of CoA per PPAT monomer. Characterization of the reverse reaction (generation of PhP) demonstrated steady-state kinetics consistent with a ternary complex mechanism. To our knowledge, no published study has described the forward direction reaction of the E. coli enzyme.
X-ray crystal structures of PPAT from several organisms have been reported. These include E. coli in complex with several ligands (9, 10, 25), Thermus thermophilus in complex with PhP (21), Bacillus subtilis in complex with ADP (2), and Mycobacterium tuberculosis (apo-protein) (15). Recently, crystals of Enterococcus faecalis (apo-protein) (13) and Helicobacter pylori (5) have been reported. PPAT structures from all five organisms exhibit a similar hexameric quaternary structure. In all but the E. coli structures the six monomers are structurally identical.
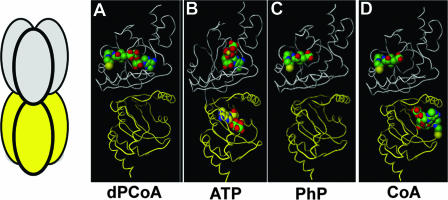
In the published E. coli PPAT structures, the dyad axis of the hexamer divides the protein into two asymmetrical trimers (Fig. 2). The three component monomers within each trimer are structurally identical. However, comparison of any two monomers across the dyad axis shows distinct differences. Cocrystallization of E. coli PPAT with dPCoA or PhP shows density for these ligands in only one trimer of the hexamer (11). Cocrystallization with Mn-ATP results in ligand density in all six monomers, but the ligand was well resolved in only one trimer. Surprisingly, the CoA-bound structure shows occupancy in all six monomers but different binding modes across the dyad axis. One trimer shows density for only the PhP moiety of CoA bound in a conformation similar to that of the dPCoA and PhP costructures (upper monomer of Fig. 2D). The CoA conformation in the other trimer shows the PhP moiety is reorganized within the PhP binding site and the adenosine portion occupies a novel binding site (lower monomer of Fig. 2D) (10).
FIG. 2.
Cocrystal structures of E. coli PPAT. The leftmost panel shows a diagram depicting hexameric PPAT. Similarly colored monomers are structurally equivalent. In the remaining panels, previously published costructures of PPAT showing bound ligands—dPCoA (PDB:1B6T [11]), ATP (PDB:1GN8 [9]), PhP (PDB:1QJC [9]), and CoA (PDB:1H1T [10])—are presented.
To facilitate our efforts to identify novel inhibitors of PPAT, we investigated the steady-state kinetic mechanism and ligand binding properties of PPAT, including its inhibition by CoA. Our data are consistent with a random Bi-Bi mechanism and show that CoA is a competitive inhibitor versus ATP, PhP, and dPCoA. Isothermal titration calorimetry (ITC) binding studies performed at pH 8.0 indicate that all PPAT monomers bind ATP, PhP, dPCoA, and CoA. These results are discussed in light of the published crystal structures and the regulatory role of PPAT in CoA biosynthesis.
MATERIALS AND METHODS
General.
All chemicals were obtained from Sigma Aldrich (St. Louis, MO) unless otherwise noted. PhP was synthesized as described previously (16) by Naeja Pharmaceuticals (Edmonton, Alberta, Canada) and 7-methyl-8-thioguanosine (MESG) was purchased from Berry & Associates (Dexter, MI). Primers were synthesized by Sigma Genosys (Woodlands, TX). Inorganic pyrophosphatase from Staphylococcus aureus was provided by Z. Xu (Pfizer, Inc., Groton, CT). E. coli strain JL4 was a gift of J. Liu (Pfizer, Inc., Ann Arbor, MI).
Production of E. coli PPAT.
The PPAT open reading frame was amplified from genomic DNA of strain JL4, a derivative of E. coli K-12, using the primers 5′-CATTAATATGCAAAAACGGGCGATTTATCC-3′ and 5′-CGGATCCCTACGCTAACTTCGCCATCAGCG-3′. The PCR products were subcloned into pET9a or pET14b to give the wild-type and N-terminal His6-tagged constructs, respectively. Both clones were sequenced to confirm the integrity of the PPAT open reading frame. His6-tagged PPAT was expressed from the E. coli BL21(DE3) clone using the pET14b plasmid. The expression was initiated from a 20% glycerol stock stored at −80°C. A portion (1 ml) of the glycerol stock was used to inoculate 200 ml of LB medium containing 10 mg of ampicillin/ml in a 500-ml shake flask. The flask was incubated at 30°C in a shaking incubator at 250 rpm for 6 to 8 h, until the culture became cloudy. An aliquot (0.5 ml) from the shake flask culture was used to inoculate a 20-liter working volume Braun fermentor which had been prepared with sterile media composed of 400 g of yeast extract (Difco 0127), 400 g of Acidicase Peptone (BBL 211843), 400 g of Casitone (Difco 0259), 400× gelysate peptone (BBL 211870), 40 g of KH2PO4 (anhydrous), 40 g of K2HPO4 (anhydrous), 40 g of Na2HPO4·7H2O, and 10 g of ampicillin/liter (pH 6.5 to 6.8). The fermentation temperature was set at 30°C, the fermentor impeller speed was set at 800 rpm, and air was sparged into the fermentor at about 8 liters per min. The pH was maintained at 6.8 + 0.2 by the addition of 85% lactic acid (USB 18140), and foam was controlled by addition of Antifoam 289 (Sigma) as needed. After 17 h of growth the turbidity reached an optical density at 600 nm of 11. The fermentor was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 3.2 mM. At 3 h after induction the fermentor was harvested, and the cell paste was collected by centrifugation.
The cell paste (600 g, wet weight) was resuspended to 1.8 liters in 25 mM HEPES (pH 7.5), 1% Triton X-100, 250 mM NaCl, 5 mM imidazole, 5 mM magnesium chloride, and 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). Benzonase (80 μl; EM Industries) was added, and the cells were lysed by two passages through a Dyno-Mill Type KDL (Glenn Mills) using a 600-ml chamber and 500 ml of 0.25- to 0.5-mm glass beads. The impeller speed was 4,200 rpm, and the flow rate was 100 ml/min.
The lysate was clarified by centrifugation and was batch bound to 100 ml of nickel-NTA agarose (QIAGEN) overnight at 4°C. The next day the resin was packed into a 5-cm-diameter column and washed sequentially with the following buffers: (i) 25 mM HEPES (pH 7.5)-1 M NaCl, 20 mM imidazole-5 mM TCEP and (ii) 25 mM HEPES (pH 7.5)-500 mM NaCl-40 mM imidazole-5 mM TCEP. The washes were carried out until a baseline at 280 nm was reached as determined by UV absorbance monitor. The protein was eluted from the column using 25 mM HEPES (pH 7.5), 250 mM NaCl, 200 mM imidazole, and 5 mM TCEP. After elution, the protein was concentrated to 300 ml and loaded onto a Superdex 200 resin column (14 by 100 cm). The column was run at 40 ml/min using 10 mM HEPES (pH 8.0)-150 mM NaCl-1 mM TCEP. Fractions were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the PPAT fractions were pooled.
To remove bound CoA from the protein, the protein solution was buffer exchanged into 10 mM citrate (pH 5.0)-1 mM TCEP by using an S1Y10 spiral cartridge (Millipore). After buffer exchange the protein was concentrated to 300 ml and run on a column of Superdex 200 resin (14 by 100 cm) which had been equilibrated with 10 mM HEPES (pH 8.0), 150 mM NaCl, and 1 mM TCEP. The column was run as described above, and the fractions were checked by SDS-PAGE. PPAT fractions were pooled and concentrated to >10 mg/ml. The pooled protein was divided into aliquots and stored at −80°C.
Throughout the manuscript, PPAT concentrations represent the concentration of monomers unless otherwise noted.
Production of E. coli purine nucleoside phosphorylase (PNP).
The deoD gene of E. coli strain JL4 was cloned, expressed, and purified as previously described (14).
Forward direction steady-state kinetic assays.
PPAT was assayed in the forward direction by using a modification of the enzyme-coupled phosphate detection system described by Webb (24). Typical assays were conducted at 25°C in 500 μl (cuvette) or 70 μl (microplate). In addition to 14 nM PPAT, each reaction contained 50 mM Tris (pH 8.0), 6 mM magnesium chloride, 1 mM TCEP, 20 μg of S. aureus pyrophosphatase/ml, 0.1 U of E. coli PNP/ml, and 150 μM MESG. In this variation of the PNP-MESG assay, the pyrophosphate produced by PPAT turnover is cleaved by inorganic pyrophosphatase to two molecules of orthophosphate. The orthophosphate is then consumed by PNP, resulting in phosphorolysis of MESG and an increase in absorbance at 360 nm. Various amounts of ATP and PhP were added, but magnesium was maintained at a minimum of 6 mM (or twofold greater than the highest ATP concentration tested). Reaction rates were determined as described above for PNP except that the observed rates were halved to account for pyrophosphate cleavage.
In addition to the coupled PNP-MESG assay, PPAT was also assayed in the forward direction by using high-pressure liquid chromatography (HPLC). Reaction mixtures were as described above; however, PNP and MESG were omitted. Pyrophosphatase was included in forward-direction reactions to maintain a favorable equilibrium for dPCoA formation. When necessary, reactions (60 μl) were quenched by the addition of trichloroacetic acid to a final concentration of 1% (wt/vol). Aliquots (20 μl) of each reaction mix were injected onto a Zorbax XDB-C18 column (4.6 by 50 mm) in a-1 ml/min mobile phase of 20 mM potassium phosphate (pH 5.5)-8% (vol/vol) methanol with detection at 260 nm. Under these conditions, ATP elutes in the void volume and dPCoA elutes approximately 3 min after injection. dPCoA was quantified by using a standard curve made from authentic dPCoA.
Steady-state kinetic and inhibition data were fit by using the enzyme kinetics module of Sigma Plot (Jandel Scientific).
Reverse direction steady-state assay.
PPAT was assayed in the reverse direction by using a coupled hexokinase/glucose-6-phosphate dehydrogenase system as described previously (7). In this assay system, the ATP synthesized by PPAT is consumed by hexokinase producing ADP and glucose-6-phosphate. Glucose-6-phosphate dehydrogenase oxidizes glucose-6-phosphate to gluconolactone with a concomitant reduction of NADP+. The reduction of NADP+ is monitored at 340 nm.
Equilibrium constant determination.
The equilibrium constant of PPAT was approximated by measuring the approach to equilibrium (18) at various ratios of dPCoA and pyrophosphate (products) to ATP and PhP (substrates). Reactions were conducted as described above for the forward-direction HPLC assay; however, pyrophosphatase was excluded from the reaction mix. The change in dPCoA concentration over time was used to determine net flux.
ITC.
Ligand binding affinities and stoichiometries were determined by using a VP-ITC (Microcal, Northampton, MA). Solutions of PPAT (typically 200 μM in volumes of 10 ml or less) were dialyzed against 2 liters of 50 mM Tris (pH 8.0)-0.5 mM TCEP-5 mM magnesium chloride. Ligands to be titrated were solvated in dialysis buffer. Concentrated ligands were used to titrate solutions of PPAT (typically 30 μM concentrations of PPAT monomers or less) at 25°C. The data were collected and analyzed by using software provided with the instrument (Origin). The data were fit to either one or two independent-site binding models as described below. In the case of PPAT, the one independent site model assumes that the heat produced by binding to each monomer is equal to and independent of other monomers. Similarly, the two-site model assumes that all PPAT monomers contributing to each binding “site” behave identically.
Determination of solution ligand binding stoichiometries.
PPAT (50 μM in 50 mM Tris [pH 8.0]-0.5 mM TCEP-5 mM magnesium chloride or 50 μM in 100 mM sodium acetate [pH 5.0]-1.1 M ammonium sulfate-200 mM sodium chloride-0.5 mM TCEP) was incubated with a 10-fold molar excess of ligand for 10 min and then dialyzed for 2 h against a 2,000-fold excess of buffer without ligand. Dialysis was conducted at room temperature using a 10,000 molecular weight cutoff Slidealyzer (Pierce). The protein was then repeatedly passed over a 5-ml PD-10 desalting column (Amersham Pharmacia Biotech) equilibrated in the above buffer until no free ligand was detected in the small molecule fraction and the amount of bound ligand remained unchanged. If the amount of bound ligand did not stabilize, the experiment was terminated after four passages had been completed. For PPAT samples equilibrated with ATP, CoA, or dPCoA, bound ligand was determined as follows. An electronic absorbance spectrum of the protein was obtained, and the protein was precipitated by incubation at 95°C for 3 min, followed by centrifugation. The electronic absorbance spectrum of the supernatant (small molecules) was collected and used to quantify the amount of bound ligand. The difference spectrum (before and after boiling and centrifugation) was used to estimate free PPAT (ɛa = 8.44 mM−1 cm−1 at 280 nm based on the tyrosine and tryptophan content). For PPAT samples equilibrated with PhP, the amount of bound PhP was determined as described above except that the small molecule fraction was assayed using the standard PNP-pyrophosphatase-MESG-coupled assay described previously. An excess of ATP was added (200 μM), and the total change in the absorbance at 360 nm was determined upon addition of the PhP-containing supernatant. After correction for dilution, this value was used to determine the concentration of bound PhP.
Single-turnover experiments.
PPAT (315 μM in 1 ml of 50 mM Tris [pH 8.0], 5 mM magnesium chloride, and 0.5 mM TCEP) was incubated with a 10-fold molar excess of either PhP or dPCoA for 10 min at room temperature and then dialyzed twice in 4 liters of the above buffer for 2 h at room temperature to remove unbound ligand. The amount of bound dPCoA or PhP was determined as described above. To the PPAT-PhP complex, ATP (final concentration equal to that of PPAT monomers) and 5 μg of S. aureus pyrophosphatase were added. After a 10-min incubation, the mixture was heated to 95°C for 3 min, and precipitated protein was removed by centrifugation. The amount of dPCoA formed was determined by HPLC as described above. A similar procedure was followed for the PPAT-dPCoA complex except that pyrophosphate (final concentration equal to that of PPAT monomers) and hexokinase (5 μg) were added to initiate the reaction.
RESULTS
Purification of wild-type and N-terminal His6-tagged PPAT.
Wild-type PPAT was purified from an overexpressing strain of E. coli. As previously reported (7), the protein copurified as an apparent hexamer with roughly 0.5 equivalents of CoA tightly bound per PPAT monomer. Attempts to remove the contaminating CoA from the wild-type protein using the published procedure (7) were unsuccessful, although the CoA could be easily exchanged with dPCoA and PhP by thorough dialysis against buffer containing either ligand at 100 μM (data not shown). Dialysis against 2 mM ATP was also sufficient to displace the bound CoA (data not shown).
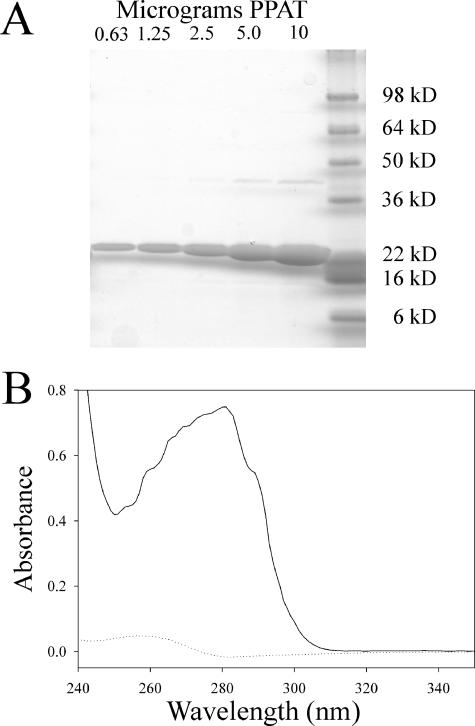
PPAT was also produced as an N-terminal fusion to a His6 tag with a thrombin cleavage site. This protein also exhibited 0.5 equivalents of tightly bound CoA per PPAT monomer, as confirmed by comigration with authentic CoA on HPLC (data not shown). Using the published procedure of dialysis at pH 5.0 and gel filtration (7), most of the contaminating CoA could be removed from the protein. After removal of copurifying CoA, typical preparations of His6-tagged PPAT were >95% pure and retained 0.05 to 0.1 equivalents of CoA per PPAT monomer (Fig. 3). Activity assays and ligand-binding studies using the tagged or wild-type protein showed no differences in kinetic parameters. Both wild-type and His6-tagged protein eluted from calibrated gel filtration columns with apparent molecular weights consistent with a homohexamer (data not shown). Attempts to remove the His6 tag by thrombin cleavage resulted in clipping of the tag, as evidenced by a decreased apparent molecular weight on reducing SDS-PAGE gels. However, the thrombin-digested protein was still readily bound by nickel-NTA resin and required 200 mM imidazole for elution, indicating that the clipped fusion tag remained tightly associated with the protein. Wild-type PPAT shows weak interaction with nickel-NTA resin but elutes in the presence of 10 to 20 mM imidazole. For routine use, the His6 tag was left intact.
FIG. 3.
Characterization of purified His6-tagged E. coli PPAT. (A) Reducing SDS-PAGE (8 to 20%, Tris-glycine) showing various amounts of purified PPAT. (B) Electronic absorbance spectrum of 90 μM PPAT after purification and removal of CoA (solid line) and spectrum of PPAT-associated small molecules released upon boiling the sample from panel B and centrifugation of the precipitated protein (dotted line). For consistency, spectra were normalized based on the absorbance at 350 nm. In samples with low absorbance, this normalization process can lead to slight negative absorbance values such as that seen around 280 nm in the dotted-line spectrum.
Steady-state kinetics.
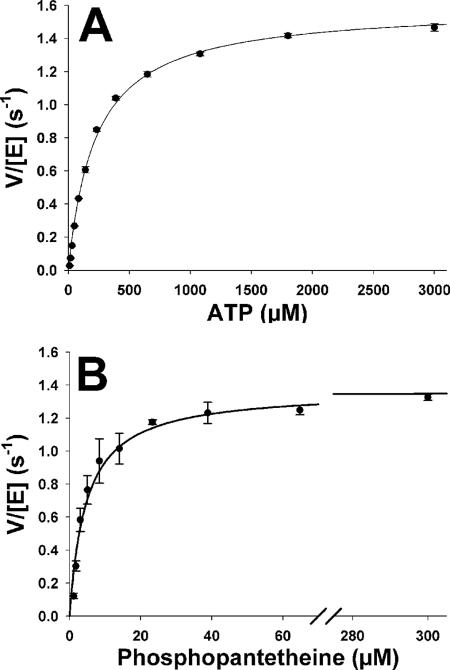
The steady-state kinetics of the forward reaction were investigated by using an enzyme-coupled assay that utilizes inorganic pyrophosphatase from S. aureus to cleave the pyrophosphate generated by PPAT turnover. The resulting orthophosphate is utilized by PNP for phosphorolysis of a chromogenic nucleoside (7-methyl-6-thioguanosine, MESG), resulting in an absorbance increase at 360 nm (24). Initial rates were determined at fixed saturating concentrations of PhP (200 μM) and varied ATP concentrations. These data were fit to the Michaelis-Menten equation with the following results: Km(ATP) = 220 ± 10 μM and kcat = 1.59 ± 0.01 s−1 (Fig. 4A). PhP concentrations were varied at a constant ATP concentration of 2.5 mM. Fitting the initial rates as described above gave values of Km(PhP) = 4.7 ± 0.5 μM and kcat = 1.37 ± 0.07 s−1 (Fig. 4B). The kinetic constants for the reverse reaction were consistent with those previously reported (7), although the kcat value was slightly lower. These data are summarized in Table 1.
FIG. 4.
Steady-state kinetic analysis of the PPAT forward-direction reaction. The rates of PPAT reaction measured at various concentrations of ATP and saturating PhP (A) and PhP and saturating ATP (B) are depicted. The assay was performed as described in Materials and Methods, and the data were globally fit to the Michaelis-Menten equation.
TABLE 1.
Steady-state kinetic parameters for the PPAT forward- and reverse-direction reactions
| Substrate | Assay direction | Mean ± SEMa
|
||
|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s) | ||
| PhP | Forward | 4.7 ± 0.5 | 1.37 ± 0.03 | (2.9 ± 0.1) × 105 |
| ATP | Forward | 220 ± 10 | 1.59 ± 0.01 | (7.1 ± 0.2) × 103 |
| dPCoA | Reverse | 17 ± 2 | 1.4 ± 0.1 | (4.4 ± 0.3) × 104 |
| Pyrophosphate | Reverse | 230 ± 10 | 1.4 ± 0.1 | (6.1 ± 0.4) × 103 |
Assays were conducted as described in Materials and Methods and represent the averages from at least three determinations.
Product inhibition studies.
Geerlof et al. reported kinetic data for the reverse reaction that were consistent with a ternary complex mechanism (7). We attempted to conduct similar experiments on the forward reaction in which either ATP or PhP concentrations were varied at subsaturating concentrations of the other substrate. We were unable to obtain satisfactory data due to the low Km for PhP and the sensitivity limits of the assay. Product inhibition studies were conducted as an alternative method to probe the kinetic mechanism. In the forward direction, dPCoA was competitive with both ATP and PhP. Since pyrophosphate is a substrate for the coupling enzymes, it was tested as a product inhibitor of ATP using the HPLC assay above described. In this assay, which monitors dPCoA formation, pyrophosphate was competitive with ATP. We were unable to establish a conclusive product inhibition profile of pyrophosphate versus PhP due to the low Km for PhP and the insensitivity of the assay. Methylene diphosphate (PCP) was investigated as a pyrophosphate analog, but no inhibition was observed at PCP concentrations up to 2 mM. When the reaction was run in the reverse direction, PhP was competitive with dPCoA and noncompetitive with pyrophosphate. Since ATP is a substrate for the coupling enzymes, several ATP analogs were investigated as surrogate product inhibitors. No inhibition of PPAT was observed with AMPCPP, ATP-γ-S, ADPPNP, or ADP at concentrations up to 1 mM. We also attempted to use the HPLC assay to monitor dPCoA consumption. However, the very small amount of dPCoA consumed in a typical assay relative to the starting amount resulted in unacceptable errors. These data are summarized in Table 2.
TABLE 2.
Product and CoA inhibition results for E. coli PPATa
| Variable substrate | Cosubstrate (concn) | Product or inhibitor | Reaction direction | Mean ± SEM
|
||
|---|---|---|---|---|---|---|
| Ki (μM) | α | Pattern of inhibition | ||||
| ATP | PhP (300 μM) | dPCoA | Forward | 192 ± 33 | ND | C |
| ATP | PhP (300 μM) | Pyrophosphate | Forwardb | 120 ± 15 | ND | C |
| PhP | ATP (3.0 mM) | dPCoA | Forward | 121 ± 16 | ND | C |
| dPCoA | Pyrophosphate (1.6 mM) | PhP | Reverse | 8.4 ± 0.7 | ND | C |
| Pyrophosphate | dPCoA (500 μM) | PhP | Reverse | 131 ± 15 | 1.64 ± 0.32 | NC |
| ATP | PhP (300 μM) | CoA | Forward | 70 ± 8 | ND | C |
| PhP | ATP (3.0 mM) | CoA | Forward | 14 ± 2 | ND | C |
| Pyrophosphate | dPCoA (300 μM) | CoA | Reverse | 117 ± 18 | 1.98 ± 0.52 | NC |
| dPCoA | Pyrophosphate (1.6 mM) | CoA | Reverse | 11 ± 2 | ND | C |
Assays were conducted as described in Materials and Methods unless otherwise noted. Abbreviations: C, competitive; NC, noncompetitive; ND, not determined (the parameter was not fit in the best inhibition model).
Reaction progress was monitored by HPLC assay.
Inhibition of PPAT by CoA species.
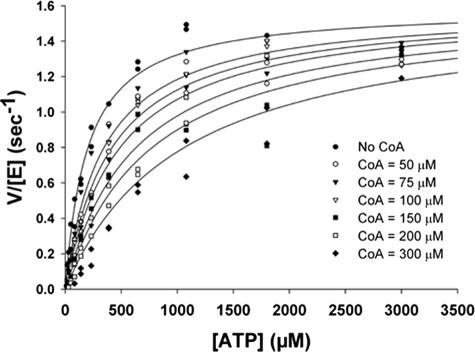
CoA was investigated as an inhibitor of PPAT. When ATP, PhP, or dPCoA were varied at fixed saturating concentrations of the second substrate, CoA is competitive with all three (Table 2). The inhibition of PPAT by CoA at variable concentrations of ATP is shown in Fig. 5. CoA is a noncompetitive inhibitor of pyrophosphate at saturating dPCoA concentrations (Table 2). Acetyl-CoA, desulfo-CoA, and palmitoyl-CoA were similarly tested, but significant inhibition of PPAT was not observed even at concentrations up to 500 μM (data not shown).
FIG. 5.
Competitive inhibition of PPAT by CoA. Rate of PPAT reaction measured at various concentrations of ATP at different fixed concentrations of CoA (indicated in the legend). The assay was performed as described in Materials and Methods, and the data were globally fit to a variety of inhibition models. The fit shown in the figure is to a simple Michaelis-Menten competitive inhibition model which provided the best fit as gauged by corrected Akaike's information criterion.
Equilibrium constant determination.
Various ratios of substrates (ATP and PhP) to products (pyrophosphate and dPCoA) were incubated with catalytic quantities of PPAT in the absence of pyrophosphatase. The approach to equilibrium (18) was followed by using HPLC to determine the consumption (reverse reaction) or generation (forward reaction) of dPCoA. The data shown in Table 3 indicate that the equilibrium constant lies between 1.25 and 1.95, slightly favoring the forward reaction.
TABLE 3.
Estimation of PPAT equilibrium constant at pH 8.0 and 25°Ca
| Combination | Concn (μM) prior to the addition of enzyme
|
Ratio of products to substratesb | PPAT fluxc | |||
|---|---|---|---|---|---|---|
| ATP | PhP | Pyrophosphate | dPCoA | |||
| 1 | 400 | 400 | 200 | 200 | 0.25 | Forward |
| 2 | 400 | 160 | 400 | 200 | 1.3 | Forward |
| 3 | 400 | 400 | 500 | 500 | 1.6 | Reverse |
| 4 | 400 | 40 | 200 | 200 | 2.5 | Reverse |
The approach to equilibrium was measured as described in Materials and Methods.
That is, the ratio of [dPCoA] and [pyrophosphate] to [ATP] and [PhP].
Flux was determined by monitoring an increase (forward reaction) or decrease (reverse reaction) in [dPCoA] over a fixed period of time.
ITC experiments.
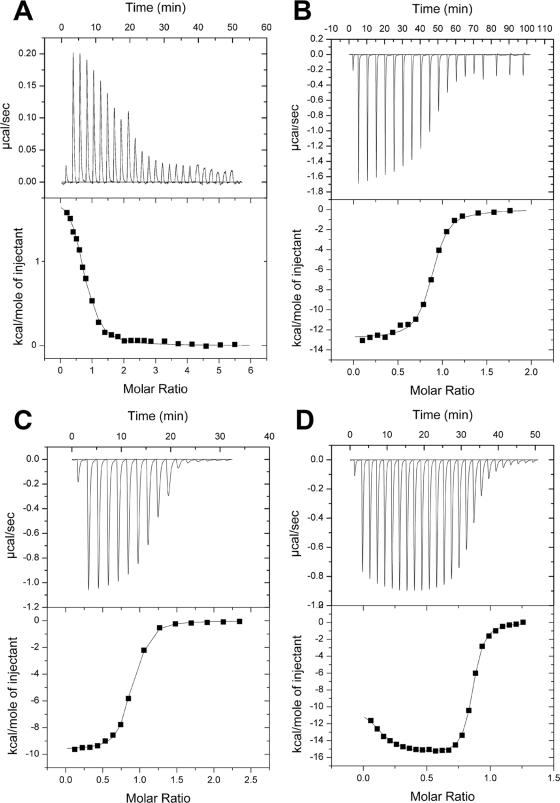
The binding of ligands at physiological pH was investigated by using ITC. For uniformity, all titration experiments contained 5 mM magnesium chloride and 0.5 mM TCEP (to prevent oxidation of PhP, CoA, and dPCoA). All ligands, with the exception of ATP, were observed to show similar binding constants in the presence or absence of magnesium chloride (data not shown). Titrations of PPAT with ATP, PhP, and dPCoA are shown in Fig. 6A, B, and C, respectively. In each case the data were best fit to a single-site model of ligand binding. The resulting thermodynamic values and binding stoichiometries are shown in Table 4. All three ligands showed at least 0.8 mol of ligand bound per PPAT monomer. When pyrophosphate was titrated into PPAT, small exothermic heats of binding were observed. Pyrophosphate did not saturate PPAT even at 100-fold excess ligand, suggesting a dissociation constant greater than 0.5 mM (data not shown). Titration of PPAT with CoA (Fig. 6D), desulfo-CoA (data not shown), and acetyl-CoA (data not shown) gave binding isotherms with two distinct phases that were best fit by a two independent sites binding model. The first phase accounts for approximately 0.1 ligands per monomer (0.6 per hexamer), while the second phase accounts for 0.76 ligands per monomer (4.6 per hexamer).
FIG. 6.
ITC titrations of PPAT with various ligands. In each panel the top half shows the raw thermogram. The bottom half of each panel shows the integrated heats (corrected for heats of dilutions due to slight differences in buffer) as a function of the added titrant. (A) Titration of 20 μM PPAT with 660 μM ATP; (B) titration of 21.1 μM PPAT with 600 μM PhP; (C) titration of 20 μM PPAT with 580 μM dPCoA; (D) titration of 30 μM PPAT with 300 μM CoA.
TABLE 4.
Binding parameters of ligands to PPAT as determined by ITC at pH 8.0 and 298°Ka
| Ligand | Mean ± SEM (phase 1, phase 2)
|
TΔS (kcal/mol) (phase 1, phase 2) | ||
|---|---|---|---|---|
| Stoichiometry (per PPAT monomer) | Kd (μM) | ΔH (kcal/mol) | ||
| ATP | 0.79 ± 0.01 | 1.85 ± 0.22 | 1.90 ± 0.03 | 10.0 |
| PhP | 0.93 ± 0.01 | 0.27 ± 0.04 | −11.6 ± 0.1 | −2.4 |
| dPCoA | 0.76 ± 0.01 | 0.17 ± 0.01 | −9.43 ± 0.1 | 0.02 |
| CoA | 0.08 ± 0.002, 0.76 ± 0.01 | ND, 0.06 ± 0.004 | ND†, −16.9 ± 0.3 | ND*, −6.8 |
| Desulfo-CoA | 0.08*, 0.80 ± 0.01 | ND, 0.15 ± 0.03 | ND†, −6.4 ± 0.3 | ND*, 3.1 |
| Acetyl- CoA | 0.08*, 0.76 ± 0.01 | ND, 0.55 ± 0.08 | ND†, −8.0 ± 0.6 | ND*, 0.6 |
Where applicable, phase 1 and phase 2 values are given, separated by a comma. ND, the errors associated with these values exceeded the value itself. *, The stoichiometry of the first phase was fixed based on the results of the CoA titration. †, The enthalpy of the first stage of CoA, acetyl-CoA, and desulfo-CoA binding was endothermic.
The results of product inhibition studies and the finding that pyrophosphate binds free PPAT suggested that a dead-end complex of PPAT, PhP, and pyrophosphate is kinetically relevant. To test this hypothesis, the apparent dissociation constants of ATP and PhP from PPAT were measured by ITC in the presence of pyrophosphate. Pyrophosphate at 0.5 mM had no statistically significant effect on the dissociation constant of PhP but reduced the apparent affinity of ATP by a factor of 1.9 ± 0.3 (data not shown). This allows an estimation of the pyrophosphate dissociation constant as approximately 0.6 mM. In both the presence and the absence of pyrophosphate, the ATP binding isotherm was best fit by a single binding site model in which all binding sites are equivalent and independent. The experiment could not be conducted at greater pyrophosphate concentrations due to precipitation of buffer components. In addition, the enthalpy of pyrophosphate binding was too weak for ITC to accurately measure the effects of either ATP or PhP on pyrophosphate binding to PPAT.
Solution binding experiments.
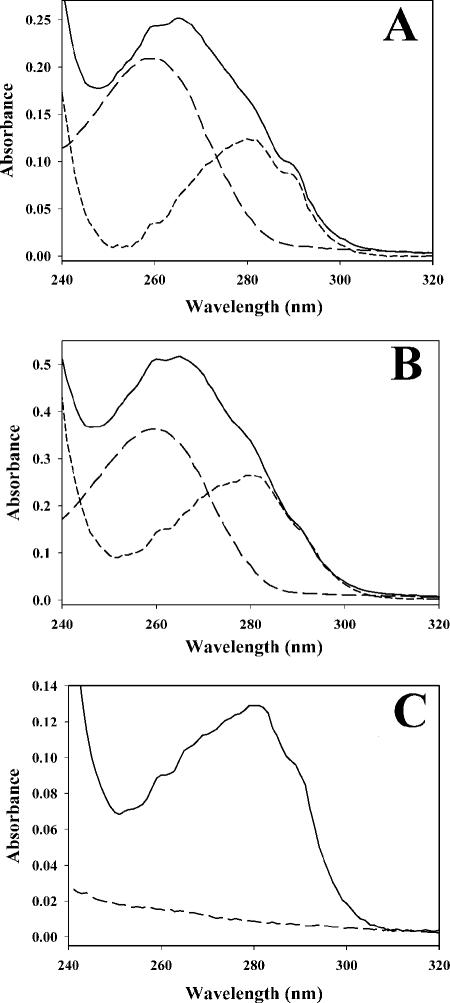
As previously reported (7), we found that PPAT copurifies with CoA tightly bound even after multiple chromatographic steps. This property was utilized as a tool to examine binding stoichiometries. A 10-fold molar excess of CoA was incubated with ligand-free PPAT for 10 min, followed by dialysis against buffer (pH 8.0) lacking ligand. The PPAT sample was then repeatedly concentrated and passed over disposable 5-ml PD-10 gel filtration columns (Amersham-Pharmacia Biotech) until the amount of bound ligand remained unchanged and no free ligand was detectable in the small molecule fraction of the column. The gel filtration process required approximately 1 h to complete. After this treatment, the amount of bound CoA was determined by electronic absorbance spectroscopy (Fig. 7A). Similar experiments were attempted with other ligands, including dPCoA and PhP (Fig. 7B and C). Since PhP has no chromophore, it was quantified by analytical enzyme assay. PhP, dPCoA, and CoA were bound by PPAT at stoichiometries of approximately one molecule of ligand per PPAT monomer (Table 5). ATP slowly dissociated upon repeated passages over the desalting column. After four passages, only 0.6 mol of ATP was bound per PPAT monomer (electronic absorbance spectra not shown). Identical experiments were conducted with dPCoA, CoA, and PhP under the reported crystallization conditions (100 mM sodium acetate [pH 5.0], 1.1 M ammonium sulfate, 200 mM NaCl) (11). Under these conditions, the ligands did not remain associated with the protein after dialysis and gel filtration, indicating a greatly reduced affinity under these conditions.
FIG. 7.
Nonequilibrium study of PPAT ligand binding. PPAT was mixed with an excess of substrate, and unbound ligand was removed by repeated rapid gel filtration chromatography until the amount of bound ligand stabilized. Bound ligand was quantified as described in Materials and Methods. In each case the solid line represents the electronic absorbance spectrum of the protein with ligand bound. The long dashes show the electronic absorbance spectrum of PPAT-associated ligands released upon boiling and centrifugation of precipitated protein. The short-dash line is the difference spectrum from the previous two datasets (i.e., apo-PPAT). The ligands depicted are CoA (A), dPCoA (B), and PhP (C).
TABLE 5.
PPAT ligand binding stoichiometries at pH 8.0 after repeated gel filtration in the absence of liganda
| Ligand | Stoichiometry (per PPAT monomer) |
|---|---|
| PhP | 0.91 ± 0.11 |
| ATP | 0.58 ± 0.07b |
| dPCoA | 0.96 ± 0.09 |
| CoA | 0.94 ± 0.16 |
Solution binding studies were conducted as described in Materials and Methods and represent the average ± the standard deviation of at least two independent determinations.
ATP stoichiometries continued to decrease after each successive round of gel filtration, whereas the other ligands stabilized after the second round of gel filtration and showed no change after at least two additional passages.
Single-turnover experiments.
The experiment described above established that PPAT binds one molecule of ATP, PhP, and dPCoA per monomer. To show that the fully ligand-bound enzyme is catalytically competent, dPCoA-bound PPAT or PhP-bound PPAT was incubated with one equivalent of either pyrophosphate or ATP. The results discussed above suggest that ATP and pyrophosphate were unlikely to remain tightly associated with the enzyme after turnover. Therefore, appropriate coupling enzymes were added to drive the reaction to completion by consuming any pyrophosphate (forward reaction) or ATP (reverse reaction) released. For every molar equivalent of PPAT-PhP complex, 0.91 ± 0.02 equivalents of dPCoA were generated upon addition of ATP (data not shown). Similarly, each molar equivalent of PPAT:dPCoA generated 0.88 ± 0.02 mol of ATP after pyrophosphate addition.
DISCUSSION
Bacterial CoA biosynthesis is an attractive target for antibacterial drug development for several reasons. These include the essentiality of the pathway for bacterial growth, the high conservation of several members of the pathway across diverse bacterial species, and a general lack of sequence homology to human orthologs. In the present study, we describe the kinetic and ligand binding properties of PPAT, the penultimate enzyme in this pathway.
Purification of PPAT from an overexpressing strain of E. coli results in protein with 0.5 equivalents of tightly bound CoA, which was removed from the His6-tagged protein by gel filtration at pH 5.0 but not from the wild-type protein (7). Easier removal of CoA from the His-tagged protein may be due to destabilization of the enzyme by the His6 tag which leads to a lower affinity for CoA. Despite these differences between wild-type PPAT and N-terminal His6-tagged PPAT at pH 5.0, the proteins behaved identically in kinetic and ligand-binding assays conducted at pH 8.0.
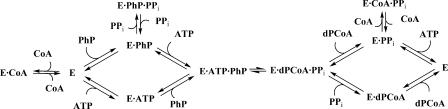
We have investigated the steady-state kinetics of the E. coli PPAT forward reaction at pH 8.0 by using an enzyme-coupled pyrophosphate detection assay. PPAT follows Michaelis-Menten kinetics with respect to ATP and PhP and shows no evidence of allosteric behavior. Product inhibition patterns and the observation that the free enzyme is capable of binding all substrates and products are consistent with a random-order Bi-Bi mechanism in which a ternary complex of PPAT, PhP, and ATP is formed. These findings are consistent with the ternary complex proposed by Geerlof et al. (7) in their investigation of the reverse reaction. The noncompetitive (α > 1) behavior of pyrophosphate versus PhP indicates pyrophosphate binds free PPAT and the PPAT-PhP complex. Although this is consistent with the placement of the substrates in the crystal structures shown in Fig. 2, we sought additional confirmation with soluble PPAT. We expected that the apparent dissociation constant of ATP but not that of PhP would be affected by the presence of pyrophosphate. ITC measurements confirmed that inclusion of 500 μM pyrophosphate in ITC titrations reduced the apparent affinity of ATP by approximately twofold but had no effect on the dissociation constant of PhP. These results are consistent with no physical overlap of these substrates in the X-ray crystal costructures and indicate that a dead-end ternary complex of PPAT-PhP-pyrophosphate could form under appropriate conditions. These results are summarized in the kinetic scheme shown in Fig. 8.
FIG. 8.
Putative kinetic mechanism of E. coli PhP including inhibition by CoA. Although not shown, the E·PPi complex can also bind PhP to form the dead end E·PhP·PPi complex.
The proposed role of CoA as a feedback regulator of PPAT implies that CoA binding affects PPAT activity. In our assay, CoA inhibits PPAT and is competitive with ATP, PhP, and dPCoA. CoA is noncompetitive (α > 1) with pyrophosphate. As discussed above, this is likely due to the lack of physical overlap of the pyrophosphate and CoA binding sites. Interestingly, the dissociation constant of CoA from the free enzyme is 65 nM as measured by ITC, whereas the kinetic inhibition constant (Ki) in the presence of competitive substrates is nearly 1,000-fold weaker (10 to 50 μM). The two different CoA binding modes observed in CoA-PPAT costructures (Fig. 2) (10) may explain the dramatic difference between the two measurements. Perhaps in the absence of PPAT substrates, CoA binds to all monomers in a “tight” mode in which the adenine moiety occupies the novel pocket (see lower monomer in Fig. 2D). The presence of PPAT substrates (i.e., bound to one or more monomers) may shift the protein dynamics such that the “weak” pocket (see upper monomer in Fig. 2D) predominates, resulting in modest kinetic inhibition constants for CoA.
The inhibition constant for CoA in the presence of substrates (10 to 50 μM) is reasonable considering intracellular CoA concentrations are estimated to be 30 to 120 μM (12). PPAT binds acetyl-CoA with ∼10-fold lower affinity than CoA (Table 4) and only weakly inhibits PPAT in steady-state enzyme assays, even at 500 μM acetyl-CoA. The typical concentrations of acetylated-CoA species (20 to 320 μM) in cells grown on various carbon sources (23) are therefore unlikely to inhibit PPAT. The weak potency of acetyl-CoA as a PPAT inhibitor suggests that free CoA, but not CoA-thioesters, is more likely to regulate PPAT activity (Fig. 9A and B).
FIG. 9.
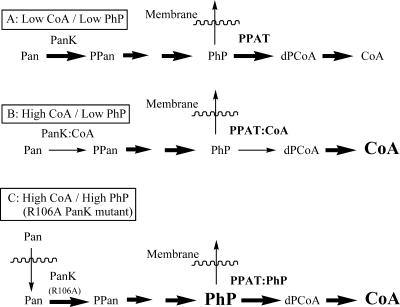
Putative model for the roles of CoA and PPAT in the regulation of CoA biosynthesis. Abbreviations: Pan, pantothenate; PanK, pantothenate kinase; PPan, phosphopantothenate. The thickness of an arrow is representative of the net flux through that step. Higher concentrations of small molecules are represented by a larger font size. (A) Under conditions of low CoA and low PhP concentrations, flux through PanK and PPAT is uninhibited. (B) When CoA levels rise and PhP levels are low, both enzymes are inhibited by CoA binding. (C) When CoA levels and PhP levels are high (such as with the R106A PanK mutant described in the text), PhP displaces CoA and flux through PPAT follows.
Regulation of PPAT by CoA is similar to that of pantothenate kinase (PanK), the first enzyme in CoA biosynthesis. Steady-state kinetic studies of PanK are consistent with an ordered ternary complex mechanism with ATP binding first (20). Similar to PPAT, CoA binds free PanK (Kd of 6 μM) and is competitive with ATP (apparent Ki of 24 μM). Both the dissociation constant and the kinetic Ki value are well below the Km for ATP (ca. 130 to 300 μM). Thus, CoA is an effective PanK inhibitor at physiological concentrations. The importance of PanK regulation of CoA biosynthesis has been shown in studies using pantothenate auxotrophs. When these strains are fed radiolabeled pantothenate, intracellular CoA levels plateau regardless of how much exogenous pantothenate is present. However, when the endogenous E. coli PanK is replaced by a mutant (R106A) unable to bind CoA, the endogenous levels of CoA and PhP increase dramatically in proportion to the amount of exogenous pantothenate supplied (17).
This observation from the R106A PanK mutant strain appears to argue against a regulatory role for PPAT, since high levels of CoA would be expected to inhibit PPAT and block excess CoA biosynthesis. However, the combination of several factors may explain the absence of PPAT-mediated pathway regulation in the mutant cell line. When PhP levels rise dramatically, the low PhP Km (5 μM) and the modest CoA kinetic inhibition constant (10 to 50 μM) likely result in ineffectual inhibition of PPAT by CoA (Fig. 9C). Thus, regulation of PPAT by CoA is only physiologically relevant when CoA levels are high and PhP levels are low. Under these conditions, inhibition of PPAT by CoA may serve as a “back-up” for regulation by PanK. Any flux through CoA-inhibited PanK (leading to PhP) could be diverted through a combination of PhP efflux and PPAT inhibition by CoA. Furthermore, the high affinity of CoA for PPAT in the absence of PhP may help maintain PPAT in an inhibited state, provided PhP levels remain low. One advantage of this mechanism is that CoA-inhibited PPAT can rapidly “reactivate” if PhP levels rise due to increased flux through PanK or through recycling of CoA to PhP and 3′,5′-ADP (22).
Our data also address the half-the-sites reactivity observed in the PPAT crystal structures in the literature. We found all of the published E. coli PPAT structures shown in Fig. 2 to be reproducible (data not shown). However, ITC data and solution ligand binding data at pH 8.0 show that ATP, PhP, CoA, and dPCoA all bind with a stoichiometry of one ligand per PPAT monomer. The PhP and dPCoA complexes are stable enough to isolate and test in single-turnover experiments. Both are catalytically competent since stoichiometric conversion to product is observed.
Binding of CoA to PPAT (pH 8.0) as measured by ITC occurs in two phases. The first phase accounts for only a small fraction of the total binding and yet shows dramatically different thermodynamic parameters relative to the second phase. It is interesting to speculate that the two phases represent the two different binding modes observed in the X-ray crystal structure of PPAT:CoA (10). Nevertheless, when the stoichiometries of the two phases are summed, one molecule of CoA binds to each PPAT monomer.
The differences between the reported crystal structure stoichiometries and the solution stoichiometries may be explained by the large difference in pH between the crystallization conditions (pH 5.0) and the assay and ligand-binding conditions (pH 8.0). Across this pH range, both the protein and the phosphodiester-containing ligands undergo a dramatic change in protonation state (S. Poole, unpublished data). The observation that overexpressed PPAT copurifies with 0.5 equivalents of CoA but is capable of binding one equivalent at pH 8.0 suggests that even a fairly small change in pH (E. coli intracellular pH is maintained at ∼7.6 [19]) may alter the ligand-binding properties of PPAT. The proposed distribution between the “tight” and “weak” CoA binding modes (see above) may also be influenced by pH. Additional kinetic and ligand experiments are required to better understand this phenomenon, since it may suggest another mechanism of PPAT regulation.
Acknowledgments
We thank Jia Liu, Pfizer, Inc., Ann Arbor, MI, for the JL4 genomic DNA; Zuoyu Xu, Pfizer, Inc., Groton, CT, for the S. aureus inorganic pyrophosphatase overexpression clone; and Salwa Poole for measurement of the PhP, dPCoA, and CoA pKa values. We also acknowledge Verna Frasca (Microcal, Inc.) for advice for fitting the CoA ITC binding data and Jack Kirsch (University of California, Berkeley), Zhigang Wang (Pfizer, Inc.), Tod Holler (Pfizer, Inc.), and Barbara Miller (Pfizer, Inc.) for helpful discussions during preparation of the manuscript.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Aghajanian, S., and D. M. Worrall. 2002. Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA synthase). Biochem. J. 365:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J., J. M. Sauder, J. M. Adams, S. Antonysamy, K. Bain, M. G. Bergseid, S. G. Buchanan, M. D. Buchanan, Y. Batiyenko, J. A. Christopher, S. Emtage, A. Eroshkina, I. Feil, E. B. Furlong, K. S. Gajiwala, X. Gao, D. He, J. Hendle, A. Huber, K. Hoda, P. Kearins, C. Kissinger, B. Laubert, H. A. Lewis, J. Lin, K. Loomis, D. Lorimer, G. Louie, M. Maletic, C. D. Marsh, I. Miller, J. Molinari, H. J. Muller-Dieckmann, J. M. Newman, B. W. Noland, B. Pagarigan, F. Park, T. S. Peat, K. W. Post, S. Radojicic, A. Ramos, R. Romero, M. E. Rutter, W. E. Sanderson, K. D. Schwinn, J. Tresser, J. Winhoven, T. A. Wright, L. Wu, J. Xu, and T. J. Harris. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60:787-796. [DOI] [PubMed] [Google Scholar]
- 3.Begley, T. P., C. Kinsland, and E. Strauss. 2001. The biosynthesis of coenzyme A in bacteria. Vitamins Hormones 61:157-171. [DOI] [PubMed] [Google Scholar]
- 4.Daugherty, M., B. Polanuyer, M. Farrell, M. Scholle, A. Lykidis, V. de Crecy-Lagard, and A. Osterman. 2002. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 277:21431-21439. [DOI] [PubMed] [Google Scholar]
- 5.Eom, S. J., H. J. Ahn, H. W. Kim, S. H. Baek, and S. W. Suh. 2003. Crystallization and preliminary X-ray crystallographic studies of phosphopantetheine adenylyltransferase from Helicobacter pylori. Acta Crystallogr. D Biol. Crystallogr. 59:561-562. [DOI] [PubMed] [Google Scholar]
- 6.Freiberg, C., B. Wieland, F. Spaltmann, K. Ehlert, H. Brotz, and H. Labischinski. 2001. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 3:483-489. [PubMed] [Google Scholar]
- 7.Geerlof, A., A. Lewendon, and W. V. Shaw. 1999. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J. Biol. Chem. 274:27105-27111. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izard, T. 2002. The crystal structures of phosphopantetheine adenylyltransferase with bound substrates reveal the enzyme's catalytic mechanism. J. Mol. Biol. 315:487-495. [DOI] [PubMed] [Google Scholar]
- 10.Izard, T. 2003. A novel adenylate binding site confers phosphopantetheine adenylyltransferase interactions with coenzyme A. J. Bacteriol. 185:4074-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izard, T., and A. Geerlof. 1999. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 18:2021-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackowski, S. 1996. Biosynthesis of pantothenic acid and coenzyme A, p. 687-694. In C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 13.Kang, J. Y., H. H. Lee, H. J. Yoon, H. S. Kim, and S. W. Suh. 2006. Overexpression, crystallization, and preliminary X-ray crystallographic analysis of phosphopantetheine adenylyltransferase from Enterococcus faecalis. Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 62:1131-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. R., J. T. Herberg, M. Tomilo, M. C. McCroskey, and B. J. Feilmeier. 2007. Streptococcus pneumoniae gyrase ATPase: development and validation of an assay for inhibitor discovery and characterization. Anal. Biochem. 365:132-143. [DOI] [PubMed] [Google Scholar]
- 15.Morris, V. K., and T. Izard. 2004. Substrate-induced asymmetry and channel closure revealed by the apoenzyme structure of Mycobacterium tuberculosis phosphopantetheine adenylyltransferase. Protein Sci. 13:2547-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagase, O. 1967. Investigations on pantothenic acid and its related compounds. IV. Chemical studies. 3. Syntheses of d-pantetheine 4′-phosphate and N-d-pantothenoyl-l-cysteine 4′-phosphate. Chem. Pharm. Bull. 15:648-654. [DOI] [PubMed] [Google Scholar]
- 17.Rock, C. O., H. W. Park, and S. Jackowski. 2003. Role of feedback regulation of pantothenate kinase (CoaA) in control of coenzyme A levels in Escherichia coli. J. Bacteriol. 185:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segel, I. H. 1975. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley & Sons, Inc., New York, NY.
- 19.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song, W. J., and S. Jackowski. 1994. Kinetics and regulation of pantothenate kinase from Escherichia coli. J. Biol. Chem. 269:27051-27058. [PubMed] [Google Scholar]
- 21.Takahashi, H., E. Inagaki, Y. Fujimoto, C. Kuroishi, Y. N. Nakamura, F. Arisaka, K. Yutani, S. Kuramitsu, C. Shigeyuki, et al. 2004. Structure and implications for the thermal stability of phosphopantetheine adenylyltransferase from Thermus thermophilus. Acta Crystallogr. D Biol. Crystallogr. D60:97-104. [DOI] [PubMed] [Google Scholar]
- 22.Vallari, D. S., and S. Jackowski. 1988. Biosynthesis and degradation both contribute to the regulation of coenzyme A content in Escherichia coli. J. Bacteriol. 170:3961-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallari, D. S., S. Jackowski, and C. O. Rock. 1987. Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 262:2468-2471. [PubMed] [Google Scholar]
- 24.Webb, M. R. 1992. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA 89:4884-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, L., N. M. Allanson, S. P. Thomson, J. K. Maclean, J. J. Barker, W. U. Primrose, P. D. Tyler, and A. Lewendon. 2003. Inhibitors of phosphopantetheine adenylyltransferase. Eur. J. Medicinal Chem. 38:345-349. [DOI] [PubMed] [Google Scholar]