Abstract
Bacteria coordinate assembly of the cell wall as well as synthesis of cellular components depending on the growth state. The mycobacterial cell wall is dominated by mycolic acids covalently linked to sugars, such as trehalose and arabinose, and is critical for pathogenesis of mycobacteria. Transfer of mycolic acids to sugars is necessary for cell wall biogenesis and is mediated by mycolyltransferases, which have been previously identified as three antigen 85 (Ag85) complex proteins. However, the regulation mechanism which links cell wall biogenesis and the growth state has not been elucidated. Here we found that a histone-like protein has a dual concentration-dependent regulatory effect on mycolyltransferase functions of the Ag85 complex through direct binding to both the Ag85 complex and the substrate, trehalose-6-monomycolate, in the cell wall. A histone-like protein-deficient Mycobacterium smegmatis strain has an unusual crenellated cell wall structure and exhibits impaired cessation of glycolipid biosynthesis in the growth-retarded phase. Furthermore, we found that artificial alteration of the amount of the extracellular histone-like protein and the Ag85 complex changes the growth rate of mycobacteria, perhaps due to impaired down-regulation of glycolipid biosynthesis. Our results demonstrate novel regulation of cell wall assembly which has an impact on bacterial growth.
Bacteria organize biogenesis of the cell wall as well as synthesis of cellular components depending on the growth state. However, factors linking the growth state and cell wall biogenesis have not been identified.
Mycobacterium tuberculosis is a top killer among bacterial pathogens and is responsible for 2 million deaths annually (6). M. tuberculosis can be quiescent in host cells for a long period of time, growing very slowly or present in a dormant state without multiplication, and it latently infects one-third of the world's human population (22, 43). In 5 to 10% of infected hosts the bacterium reactivates and causes progressive disease during their lifetimes. Most cases of active tuberculosis do not result from the initial infection but instead represent reactivation of previously implanted bacteria (22, 43).
The cell wall is critical for long-term persistence of M. tuberculosis in the hostile environment in the host cells and for progression of tuberculosis (3, 7). Mycobacteria are gram-positive bacilli, but the cell wall structures are different from those of other gram-positive bacteria. Approximately one-half of the cell wall mass is comprised of large (C70 to C90) branched-chain fatty acids called mycolic acids. Mycolic acids are distributed in acid-fast positive bacteria, such as Mycobacterium, Corynebacterium, Rhodococcus, and Nocardia, although mycobacterial mycolic acids are the longest mycolic acids and have the largest side chains (C20 to C24). The cell wall outer layer is composed of extractable glycolipids containing mycolic acids, such as trehalose-6-monomycolate (TMM) and trehalose-6,6′-dimycolate (TDM) (also called cord factor), while the inner layer is composed of mycolic acids covalently linked to the distal portion of the arabinogalactan (AG) moiety (7). TMM- or TDM-derived mycolic acids are transferred to other TMM to synthesize TDM and also to peptidoglycan-linked AG to construct the inner layer of the envelope. Antigen 85 (Ag85) complex proteins (Ag85A, Ag85B, and Ag85C) are mycolyltransferases and catalyze transfer of mycolic acids to free trehalose, TMM, and TDM (4). Ag85 complex proteins are also believed to catalyze the transfer of mycolic acids from TMM or TDM to peptidoglycan-linked AG, because inactivation of Ag85C reduced the level of AG-linked mycolic acids (16).
Regulatory proteins involved in cell wall assembly should localize in the mycobacterial cell wall. We and other groups found that a histone-like DNA-binding protein, which was designated mycobacterial DNA-binding protein 1 (MDP1), laminin-binding protein, histone-like protein (HLP), or HupB, not only localizes in the cytoplasmic space but also occurs externally or is in the mycobacterial cell wall (26, 36, 38). MDP1 is mycobacterium-specific histone-like protein. M. tuberculosis has a single mdp1 gene (Rv2986c, also called hupB) (9, 13), and the mdp1 gene is conserved even in Mycobacterium leprae, which lost many genes during evolution (10). MDP1 likely plays a significant role in DNA functions in mycobacteria, as a transposon-based screen suggested that mdp1 is essential in M. tuberculosis (34). However, the mdp1 gene can be knocked out in Mycobacterium smegmatis, suggesting that another DNA-binding protein may compensate for loss of MDP1 in M. smegmatis. The M. smegmatis MDP1 knockout (KO) strain exhibited growth kinetics similar to those of the wild type in anaerobic culture (19) but was unable to resume growth at 10°C (37), suggesting that MDP1 plays an important role during stress responses in M. smegmatis.
Accumulation of MDP1 in stationary phase or under anaerobic conditions implies that MDP1 is a possible factor that participates in growth-state-dependent regulation of cell wall assembly through binding to sacchariferous components, such as glycolipids, in the cell wall. Based on this hypothesis, here we examined the physiological role of MDP1 in the cell wall.
MATERIALS AND METHODS
Extraction and purification of glycolipids.
M. tuberculosis H37Rv and Mycobacterium bovis bacillus Calmette-Guérin (BCG) were cultivated on Sauton medium at 37°C. M. smegmatis was cultured in Luria-Bertani (LB) medium at 37°C. Bacteria were autoclaved for 10 min, disrupted ultrasonically, and then suspended in chloroform-methanol (4:1, 3:1, or 2:1, vol/vol) to extract lipids. The chloroform layer was collected and dried. TDM was first partially purified by precipitation with acetone, chloroform-methanol (2:1, vol/vol), and tetrahydrofuran-methanol (1:2, vol/vol), followed by passage through a column of silica gel (Wakogel C-200; Wako Pure Chemical, Osaka, Japan) with chloroform-methanol (4:1, vol/vol). The purity of TDM was demonstrated by a single spot on a thin-layer chromatogram. TMM was separated by preparative thin-layer chromatography (TLC) on a silica gel plate (Uniplate; 20 by 20 cm; 250 mm; Analtech, Inc., Newark, DE) using a chloroform-methanol-acetone-acetic acid (80:20:6:1, vol/vol/vol/vol) solvent system. Glycolipids were visualized with a 20% H2SO4 spray, followed by charring at 200°C for analytical purposes or with iodine vapor for a few minutes for preparative purposes. TMM was recovered from the plate immediately after the iodine color had disappeared by passing the plate through a small glass column with the solvent chloroform-methanol (2:1, vol/vol). Finally, TMM was purified until a single spot was obtained by repeating TLC.
Fluorescence microscopy.
MDP1 was purified from BCG by using a method described previously (26). Egg white lysozyme was purchased from Wako. Bovine histone H1 was purchased from Roche Diagnostics. Proteins were labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) by using a fluorescein protein labeling kit (Roche Diagnostics) according to the manufacturer's instructions. BCG was grown in Middlebrook 7H9-ADC medium at 37°C until the optical density at 600 nm (OD600) was 1.5; then it was collected by centrifugation and washed three times with phosphate-buffered saline (PBS) with 10% fetal bovine serum (FBS). FLUOS-labeled proteins, such as MDP1 (50 μg), egg white lysozyme (38 μg), and bovine histone H1 (50 μg), were added to the BCG suspension and incubated at 37°C for 30 min. BCG was washed three times with PBS with 10% FBS and then mounted on a microscope slide and viewed with a confocal scanning laser microscope (LSM510; Carl Zeiss).
MAb preparation.
Ten micrograms of MDP1 and 10 ng of M. tuberculosis DNA were mixed in PBS, emulsified in Freund's incomplete adjuvant, and injected into BALB/c mice subcutaneously (24). Two weeks later, the mice were boosted in the same way; after an additional 2 weeks, the mice were again boosted by injection of 10 μg of MDP1 in PBS into the tail vein. Three days after the final boost, mice were sacrificed and splenocytes were obtained. A monoclonal antibody (MAb) was prepared essentially as described by Harlow and Lane (15) and was screened by an enzyme-linked immunosorbent assay (ELISA) as described below. After an initial screening, several hybridoma cell lines were cloned by two rounds of limiting dilution. A selected subclone was expanded for freezing and for ascites production in pristane-primed mice. The subclass of the hybridoma subclone was determined with a mouse MAb isotyping kit (Amersham). Immunoglobulins (Ig) were purified from ascites fluid by using an Ampure PA kit (Amersham).
ELISA to detect interactions between MDP1 and TDM, TMM, mycolic acids, or Ag85 complex proteins.
TMM, TDM, and mycolic acid methyl esters (MAMEs) purified from M. tuberculosis Aoyama B were purchased from Nakarai and dissolved in N-hexane at a concentration of 50 μg/ml. Glycolipids were immobilized on a 96-well ELISA plate (Sumitomo, Osaka, Japan) by adding 100 μl of glycolipid solution and dried. Ag85A, Ag85B, and Ag85C derived from M. tuberculosis H37Rv and bovine serum albumin (BSA) were immobilized on the 96-well ELISA plate (Sumitomo, Osaka, Japan) by incubation of serial twofold dilutions of the protein solutions in sodium bicarbonate buffer (pH 9.6) at 4°C overnight. MDP1 at a concentration of 1 μg/ml in PBS containing 0.05% Tween 20 (PBS-T) was added to wells and incubated for 1 h at 37°C. The wells were washed with PBS-T four times, and then MAb 3A was added. After incubation at 37°C for 1 h, the wells were washed, peroxidase-conjugated anti-mouse antibody (Dako) diluted 1:2,000 was added, and the plate was incubated for 1 h. After the wells were washed, the level of MDP1 binding was detected by color development with o-phenylenediamine dihydrochloride (Wako, Tokyo, Japan) and measuring the OD492.
Preparation of subcellular fractions.
Subcellular fractions were prepared by a method described previously (1, 27). Briefly, after BCG and M. smegmatis were cultured in Sauton medium and LB medium, respectively, bacteria were collected by centrifugation at 10,000 × g and suspended in ice-cold PBS. The bacteria were then disrupted with a Bioruptor UCD-200T sonicator (Toso), and each suspension was centrifuged at 3,000 × g for 5 min to remove unbroken bacteria. The supernatant was used as the total cellular fraction. The total cellular fraction was fractionated further to obtain a cell wall fraction and a non-cell-wall fraction using the following procedure. The total cellular fraction was centrifuged at 10,000 × g for 10 min. The pellet was rinsed with cold PBS and centrifuged again at 10,000 × g for 10 min. The supernatants were designated the non-cell-wall fraction. The cell wall-containing pellet was suspended again in ice-cold PBS, and then Percoll (Amersham Biosciences) was added to a concentration of 60% and mixed by vortexing. Next, the cell wall-containing fraction was centrifuged at 27,000 × g for 1 h to separate the cell walls from the unbroken cells completely. The cell wall band was collected and washed twice with PBS, and it was designated the cell wall fraction.
Immunoprecipitation assay.
Cell walls derived from BCG as described above were precleaned by using 25 μg of mouse IgG (Chemicon International Temecula) or rabbit Ig in 100 μl of protein G-coupled Sepharose (Amersham Pharmacia Biotech), incubated at 4°C for 2 h, and centrifuged. The supernatants were colleted and incubated with 250 μg of anti-MDP1 MAb 3A, control mouse IgG, anti-Ag85 Ig, or control rabbit Ig at 4°C for 16 h, and then 300 μl of protein G-coated Sepharose was added and the preparation was incubated for 5 h at 4°C. Then the beads were washed with PBS-T three times. After washing, bead-bound proteins were eluted by boiling the preparations in 40 μl of 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (0.125 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% mercaptoethanol). The samples were then fractionated on a 12.5% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The membrane was then blocked for 30 min at room temperature by incubating it in PBS containing 5% skim milk. Then the membrane was probed with anti-MDP1 MAb 3A or anit-Ag85 antibodies overnight at 4°C. After probing, the membrane was washed four times with PBS-T. Next, the membrane was incubated for 4 h at room temperature with peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (Dakopatts A/S, Denmark) diluted 1:10,000. The membrane was then washed as described above, and immunoreactive bands were visualized by using an ECL Western blot detection reagent (Amersham Bioscience, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
Bead-bound glycolipids were eluted with chloroform-methanol (3:1, vol/vol). Thirty microliters of the chloroform layer was spotted on a TLC plate (HPTLC plate; 10 by 10 cm; Silica Gel 60; Merck, Darmstadt, Germany) and developed with the chloroform-methanol-acetone-acetic acid (80:20:6:1, vol/vol/vol/vol) solvent system. The TLC plate was exposed overnight to an immunoprecipitation plate (BAS-MS2025 or BAS-SR2025; Fujifilm, Japan) and visualized with the BAS system (BAS-5000). The radioactivity of separated spots was quantified by using the BAS system's software.
Analysis of mycolyltransferase activity.
The mycolyltransferase catalytic reaction was analyzed by using the method developed by Belisle et al. (4). Twenty-five micrograms of TMM purified from M. tuberculosis H37Rv was immobilized in each glass vial, and then PBS containing 4 μl of a 1-mg/ml dithiothreitol solution and 0.5 μCi of [14C]trehalose (American Radiolabeled Chemicals Inc.) were added. Two hundred micrograms of culture filtrate or 20 μg of Ag85 complex protein and various amounts of MDP1 or BSA were mixed to obtain a total volume of 200 μl and incubated for 30 min at 37°C. Glycolipids were eluted with chloroform-methanol (2:1, vol/vol), 10 μl of the chloroform layer was spotted on a TLC plate (HPTLC plate; 10 by 10 cm; Silica Gel 60; Merck, Darmstadt, Germany), and the plate was developed with the chloroform-methanol-acetone-acetic acid (80:20:6:1, vol/vol/vol/vol) solvent system. TLC plates were analyzed by using the BAS system described above.
Construction of HLP/MDP1 complemented strain.
Based on the mdp1/hlp nucleotide sequences, two oligonucleotide primers, forward primer 5′-GGGAAGCTTATTCGCCGCCCACCTAGT-3′ and reverse primer 5′-TAACGCACCAACGCGAAA-3′, were purchased from Sigma Genosys. A PCR was carried out by targeting 10 ng chromosomal DNA of M. smegmatis MC2155 with an automated thermal sequencer (Perkin Elmer). The samples were first denatured by heating them at 94°C for 5 min; then they were subjected to 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 3 min and were finally incubated for 5 min at 72°C. An amplified 0.9-kb DNA fragment, which contained the promoter and structural gene of HLP/MDP1, was cloned into pGEM-T (Promega) utilizing a ligation kit (version 1, Takara), sequenced, excised by digestion with HindIII and NotI, and ligated to the same site of pMV306-Hyg, a one-copy integrated vector for the phage attachment site. pMV306-Hyg was provided by H. I. Boshoff (National Institutes of Health, Bethesda, MD). The resulting plasmid was introduced into an M. smegmatis HLP/MDP1 KO strain by a standard electroporation procedure (17, 39), and then a hygromycin-resistant colony was selected. Complementation was confirmed by protein expression with Western blotting using anti-MDP1 MAb 3A (data not shown).
SEM analysis.
Five-milliliter portions of culture aliquots were concentrated by centrifugation (3,000 × g) before suspension in fresh Middlebrook 7H9-ADC medium. The OD600 was adjusted to 0.1, and then the preparations were placed on poly-l-lysine-coated Thermanox coverslips in 24-well tissue culture plates. The bacteria were allowed to settle for 30 min before gentle decanting and addition of 1 ml of a solution containing 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) with 0.2 M sucrose. The samples were incubated at 4°C overnight before treatment with 2% OsO4 in 0.1 M cacodylate buffer for 2 h at room temperature. A series of sequential ethanol dehydration steps were performed (50, 70, 95, and 100% ethanol, 10 min each) before samples were dried under CO2 using a critical point drier (HCP-2; Hitachi). Samples were Pt-Pd sputter coated (E-1030; Hitachi) and imaged with an Hitachi S-4700 scanning electron microscope (SEM). The SEM analysis was performed in triplicate using three independently grown cultures.
Chase of glycolipid synthesis.
M. smegmatis strains, including the wild type, the MDP1 KO mutant, and the complemented strain, were precultured at 37°C in LB broth (Sigma) containing 1-mm-diameter glass beads. First, bacterial clumps were disrupted with the beads by vortexing, and then the OD600 was adjusted to 0.1. Then 5 μl of each bacterial suspension was added to 5 ml of fresh medium containing glass beads. [1-14C]acetic acid (sodium salt; 37 MBq/ml; PerkinElmer Life & Analytical Sciences, Massachusetts) was added at a final concentration of 1 μCi/ml on day 3 or7. Each bacterial medium was incubated for 16 h and diluted to obtain 6.0 × 107CFU/ml. Two milliliters of this bacterial suspension was collected by centrifugation and washed with pure water three times. After the supernatants were discarded, we added 1 ml of chloroform-methanol (3:1, vol/vol) and sonicated the preparation for 30 min. Thirty microliters of the chloroform layer was spotted on a TLC plate. The radioactivity of the separated spots was quantified by using the BAS system software as described above.
MALDI-TOF mass spectrometry analysis.
A matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis was carried out by using an Ultraflex mass spectrometer (Bruker Daltonics) in the reflectron mode. Samples were dissolved in chloroform-methanol (2: 1, vol/vol) at a concentration of 1 mg/ml and applied to the sample plate as droplets. 2,5-Dihydroxybenzoic acid was used as the matrix. The accelerating voltage was 20 kV.
Construction of BCG-Luc and assessment of its growth.
Linker DNAs including the Shine-Dalgarno sequence (AGCTTAGTACTGGATCCGAGGACCTGCC and GATCGGCAGGTCCTCGGATCCAGTACTA) were synthesized by Sigma Genosys. pGEM-Luc (Promega) was digested with both BamHI and HindIII, and annealed linker DNA was inserted by ligation utilizing a ligation kit (version 1; Takara). The construct was then digested with HindIII and StuI, and the gene fragment containing the Shine-Dalgarno sequence and the luciferase gene was inserted into pSO246 (25), which had been digested with BamHI, blunt ended with T4 DNA polymerase, and digested with HindIII. The resulting plasmid was designated pSO-Luc. pMV261 (39) was digested with KpnI and HindIII, and the Hsp60 promoter region was inserted into the same site of pSO-Luc. The final construct was introduced into BCG by electroporation by the method described previously (25), and the kanamycin-resistant BCG-Luc strain was obtained. BCG-Luc was grown at 37°C in Middlebrook 7H9-ADC medium until mid-log phase and collected by centrifugation. Bacteria were suspended in RPMI 1640 (Sigma) containing 10% FBS with or without MDP1 or Ag85 complex proteins in a 96-well tissue culture plate (Becton Dickinson) and incubated at 37°C. Luciferase activity was determined at each time point as described previously (12).
Statistical analyses.
Data were analyzed by using a Power Macintosh G5 and StatView 5.0 (SAS Institute Inc.) and were expressed as means ± standard deviations. Data that appeared to be statistically significantly different were compared by using an analysis of variance for comparing the means of multiple groups and were considered significantly different if the P value was less than 0.05.
RESULTS
MDP1 binds externally to the cell wall of BCG.
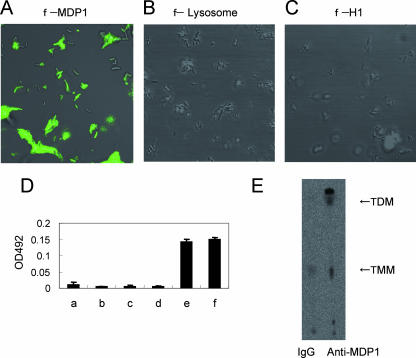
MDP1 acts as an adhesin on the envelope through interaction with glycosaminoglycans on the host cell surface (1, 38). Because MDP1 is retained in the cell wall, we hypothesized that MDP1 is tightly bound to some unknown targets in the cell wall. In order to assess this possibility, first we examined the binding of MDP1 to the cell wall. We incubated FLUOS-labeled MDP1 with BCG and investigated the interaction by fluorescence microscopy. The results revealed that MDP1 (pI 12.4; molecular mass, 21 kDa) bound to the surface of BCG (Fig. 1A). In contrast, other FLUOS-labeled basic proteins, such as egg white lysozyme (pI 9.2; molecular mass, 16 kDa) (Fig. 1B) and bovine histone H1 (pI 11.5; molecular mass, 21 kDa) (Fig. 1C), did not interact with BCG. Bovine serum albumin (pI 5.4) did not interact either (data not shown). Thus, MDP1 can specifically bind externally to the cell wall.
FIG. 1.
MDP1 binds to the cell wall of BCG. (A to C) BCG was incubated with FLUOS-labeled MDP1 (A), egg white lysozyme (B), and bovine histone H1 (C). Bacteria were viewed with a confocal scanning laser microscope. (D) ELISA to detect MDP1-lipid or -glycolipid interactions. Mycolic acids (bar b, alpha-MAME; bar c, methoxy-MAME; bar d, keto-MAME) and glycolipids (bar e, TMM; bar f, TDM) were immobilized on an ELISA plate and reacted with MDP1, and the level of binding was detected with anti-MDP1 MAb 3A. Bar a shows the results for a negative control without immobilization of either lipids or glycolipids. OD492, optical density at 492 nm. (E) Immunoprecipitation assay to detect physiological interaction of MDP1 and glycolipid. Cell wall derived from BCG was incubated with anti-MDP1 MAb 3A or control mouse IgG in the presence of protein G-coated Sepharose. Samples were spotted on a TLC plate and developed with the chloroform-methanol-acetone-acetic acid (80:20:6:1, vol/vol/vol/vol) solvent system.
MDP1 binds to TMM and TDM but not to free mycolic acids.
Mycolic acids form an ordered structure in the envelope and are believed to be some of the outermost covalently linked elements (3, 20). M. tuberculosis produces three kinds of mycolic acids, the alpha-, methoxy-, and keto-mycolates. Three types of MAMEs (alpha-, methoxy-, and keto-mycolates), as well as TMM and TDM derived from M. tuberculosis strain Aoyama B, were immobilized on an ELISA plate and reacted with MDP1, and the interaction was detected with anti-MDP1 MAb 3A (subclass IgG1, light chain κ). The affinity of MAb 3A for MDP1 was 1.39 e−9 M as determined by surface plasmon resonance analysis with a Biacore biosensor (Biacore) (data not shown). This MAb binds to neither TMM nor TDM (data not shown). The results showed that MDP1 bound to TMM and TDM but not to any type of MAME (Fig. 1D). This indicates that MDP1 specifically recognizes the covalent linkage of mycolic acids to the 6-hydroxyl group of trehalose, because MDP1 does not bind to any mycolic acid (Fig. 1D) or free trehalose (1).
Physiological interaction between MDP1 and glycolipid in the mycobacterial cell wall.
Next we examined whether MDP1 actually associated with TMM and TDM in the mycobacterial envelope by using an immunoprecipitation assay. A cell wall fraction derived from BCG by the method described previously (27) was incubated with MAb 3A or control mouse IgG. Then MDP1-bound glycolipids were precipitated with protein G-coupled beads, extracted with an organic solvent (chloroform-methanol, 3:1 [vol/vol]), and analyzed by TLC. Both TMM and TDM were precipitated by MAb 3A but not by control IgG (Fig. 1E), showing that MDP1 associated with TDM and TMM in the cell wall. The data also showed that MDP1 bound to other unknown lipids that migrated above TDM, as well as below TMM (Fig. 1E).
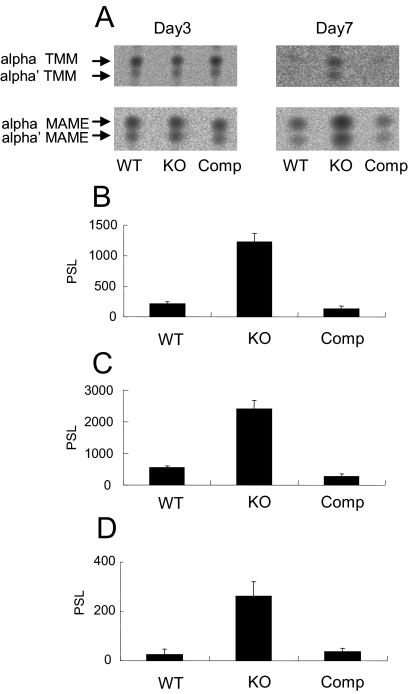
MDP1 regulates transfer of mycolic acids by Ag85 complex proteins in vitro.
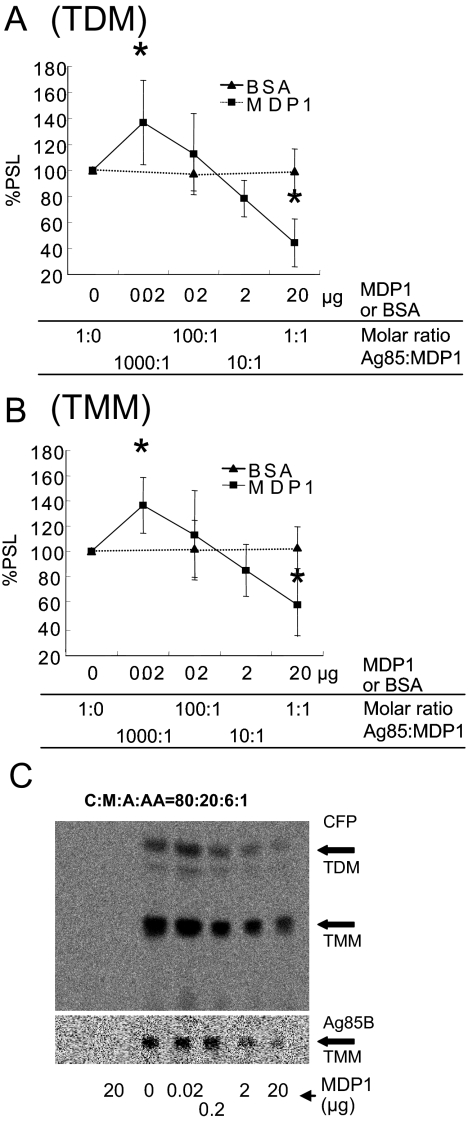
We next examined the effect of MDP1 on transfer of mycolic acids by Ag85 complex proteins, because MDP1 can bind TMM and TDM, which are substrates of Ag85 complex proteins. We purified Ag85 complex proteins, as well as Ag85A, Ag85B, and Ag85C individually, from M. tuberculosis H37Rv by the method described previously (28). Purified Ag85 complex proteins transferred mycolic acids to 14C-labeled trehalose as described previously (4). A molar amount of MDP1 equivalent to that of the Ag85 complex reduced synthesis of TDM and TMM by 44.4% ± 18.4% and 57.4% ± 25.2%, respectively (Fig. 2A and 2B). In contrast, a molar ratio of MDP1 to Ag85 proteins of 1/1,000 enhanced synthesis of TDM and TMM by 36.8% ± 32.4% and 36.4% ± 22.1%, respectively (Fig. 2A and 2B). The same results were obtained when total culture filtrates were used (Fig. 2C, upper chromatogram) or when purified Ag85B was used (Fig. 2C, lower chromatogram). These results demonstrate that MDP1 possesses an activity to control the function of mycolyltransferases.
FIG. 2.
MDP1 regulates mycolyltransferase activity in vitro. (A and B) Radioactivity of 14C-labeled TDM (A) and TMM (B) synthesized by Ag85 complex proteins in vitro was quantified by using the BAS system software. Radioactivity was expressed as a percentage of the value for the sample without MDP1, which was defined as 100%. Similar experiments were performed utilizing BSA instead of MDP1, and the results were compared. PSL, photo-stimulated luminescence. Asterisk, P < 0.05 for a comparison with control BSA (0 or 20 μg) (as determined by analysis of variance). (C) Culture filtrate (CFP) (upper chromatogram) or Ag85B (lower chromatogram) was added to each vial and incubated for 30 min at 37°C in the presence of various doses of MDP1 and [14C]trehalose, and then glycolipids were eluted with chloroform-methanol (2:1, vol/vol). Biosynthesis of glycolipids was analyzed by using the BAS system after TLC plates were developed with the chloroform-methanol-acetone-acetic acid (C:M:A:AA) (80:20:6:1, vol/vol/vol/vol) solvent system. The lane on the left contained a negative control without culture filtrate and Ag85B.
MDP1 binds to Ag85 complex proteins.
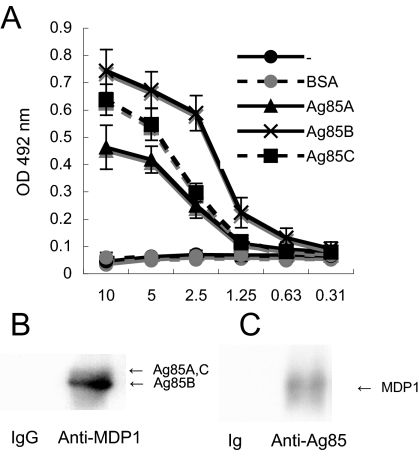
Because MDP1 controlled transfer of mycolic acids in the presence of excess amounts of substrate (TMM) (Fig. 2), we considered the possibility that MDP1 interacts not only with TMM but also with Ag85 complex proteins, which we examined next. Ag85A, Ag85B, or Ag85C was immobilized on ELISA plates by serial concentration and then incubated with MDP1. The level of binding of MDP1 to Ag85 complex proteins was detected by anti-MDP1 MAb. The results showed that the interaction between MDP1 and each Ag85 complex protein produced standard binding curves (Fig. 3A). BSA did not bind to MDP1. These results suggested that MDP1 binds to all Ag85 proteins.
FIG. 3.
MDP1 binds to Ag85 complex proteins. (A) ELISA to detect the MDP1-Ag85 complex protein interaction. BSA, Ag85A, Ag85B, and Ag85C were immobilized on an ELISA plate by incubating various concentrations of proteins and reacted with MDP1, and the binding was detected with anti-MDP1 MAb. −, results without immobilized proteins. OD 492 nm, optical density at 492 nm. (B and C) Immunoprecipitation assay to detect a physiological interaction between MDP1 and Ag85 complex proteins. Cell walls derived from BCG were incubated with (B) anti-MDP1 MAb 3A (Anti-MDP1) or control mouse IgG (IgG) or (C) with anti-Ag85 Ig (Anti-Ag85) or control rabbit Ig (Ig) in the presence of protein G-coated Sepharose. Samples for Western blotting were probed with rabbit anti-Ag85 Ig (B) or anti-MDP1 MAb (C).
Ag85 complex proteins bind to fibronectin by the motif conserved in the complex, which is comprised of 11 amino acid residues (29). We examined if this region also participated in the MDP1-Ag85 complex protein interaction. However, neither human fibronectin nor synthetic peptide corresponding to the region from position 98 to position 108 of Ag85B, which inhibits an Ag85 complex-fibronectin interaction, inhibited an MDP1-Ag85 complex protein interaction (data not shown). Thus, Ag85 complex proteins associate with MDP1 through a region other than the fibronectin-binding site.
We next examined whether MDP1 interacted with Ag85 complex proteins in the mycobacterial cell wall. A cell wall fraction derived from BCG was immunoprecipitated with MAb 3A or control mouse IgG and separated by SDS-PAGE. One gel was stained with Coomassie brilliant blue R250, and another gel was used for Western blot analysis. Although we observed only faint bands corresponding to Ag85 complex proteins (Ag85A and Ag85C at 32 kDa and Ag85B at 30 kDa) in the stained gel, in the Western blot analysis we observed that the bands reacted with anti-Ag85 complex protein Ig in the precipitates when anti-MDP1 MAb was used but not when control IgG was used (Fig. 3B). Similar experiments were performed with anti-Ag85 Ig. Anti-Ag85 Ig, but not control Ig, precipitated MDP1 from the cell wall fraction of BCG (Fig. 3C). Taken together, these results suggest that the Ag85 complex proteins associate with MDP1 in the cell wall.
Subcellular localization of MDP1 in the course of culture.
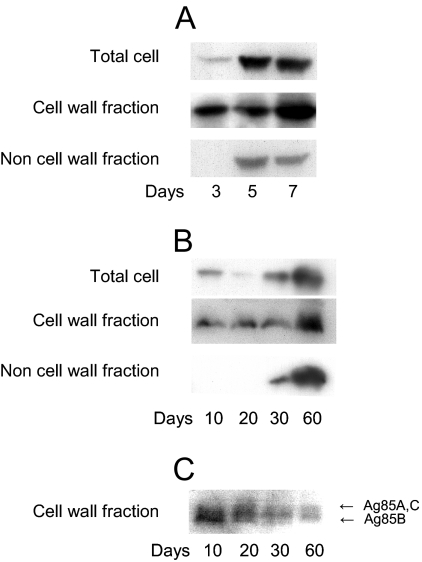
We analyzed the MDP1 content of the cell wall during the course of culture. We cultured M. smegmatis in LB medium for 3, 5, and 7 days, and each subcellular fraction was purified. In this experiment, bacteria grew to stationary phase by day 4 (Fig. 4A). The MDP1 content in each fraction was analyzed by Western blotting. The results showed that MDP1 accumulated in both cell wall and other cellular fractions (membrane, ribosome, and cytoplasmic fractions) at the stationary growth phase (Fig. 5A).
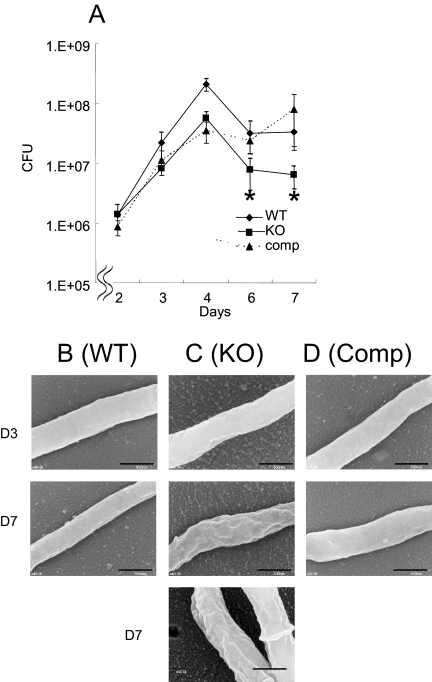
FIG. 4.
Effect of MDP1 deficiency on growth kinetics and cell surface morphology. (A) Wild-type M. smegmatis mc2155, MDP1 KO, and complemented strains were cultured in LB broth, and bacterial numbers at various time points were determined and expressed as CFU after serially diluted samples on LB agar were harvested. Asterisk, P < 0.05 for a comparison with the wild-type strain (as determined by analysis of variance). The results are the results of one representative experiment of seven experiments in which similar results were obtained. (B to D) Visualization of cell surface structure of the wild-type (WT) (B), MDP1 KO (KO) (C), and complemented (Comp) (D) strains by SEM. Bars = 0.5 μm. Bacteria in both exponential (day 3 [D3]) and stationary (day 7 [D7]) phases were analyzed.
FIG. 5.
Subcellular localization of MDP1 and Ag85 in the course of culture. (A to C) Total cellular protein (Total cell), the cell wall fraction (Cell wall fraction), and the residual material after isolation of the cell wall fraction (Non cell wall fraction) were obtained from M. smegmatis (A) and BCG (B and C) at each time point indicated. The samples were analyzed by Western blotting with anti-MDP1 MAb 3A (A and B) or anti-Ag85B Ig (C). The data are representative of three to five experiments.
We next carried out a similar experiment with BCG. Total cellular proteins, the cell wall fraction, and other cellular fractions were obtained from BCG after growth for 10, 20, 30, and 60 days on Sauton medium. A Western blot analysis showed that the cellular content of MDP1 increased in both the cell wall and other cellular fractions (Fig. 5B), while the levels of Ag85 complex proteins in the cell wall decreased with time (Fig. 5C).
Role of MDP1 in glycolipid biosynthesis.
MDP1 is presumed to be essential in slow growers, such as M. tuberculosis (34). However, the mdp1/hlp gene can be knocked out in M. smegmatis (19). In order to determine the physiological role of MDP1 in assembly of the cell wall, we employed an M. smegmatis MDP1/HLP KO strain constructed by the group of Thomas Dick (19). We additionally generated an MDP1-complemented strain by insertion of a single copy of the M. smegmatis MDP1 gene into the genome of the MDP1/HLP KO strain.
We first analyzed the growth kinetics of the wild type, the MDP1 KO strain, and the complemented strain in LB medium. In this experiment, all strains reached stationary phase on day 4, but the bacterial density of the MDP1 KO strain was lower than that of the wild-type strain during the stationary growth phase (Fig. 4A). This phenotype was almost completely reversed by complementation. Next, we analyzed bacterial surface morphology by SEM. All strains produced similar smooth structures at exponential phase (Fig. 4B, C, and D, upper panels), and both the wild-type and complemented strains were normal with a smooth shape even in the stationary growth phase (Fig. 4B and D, middle panels). However, at the same time point, the MDP1 KO strain displayed a crenellated structure (Fig. 4C, middle and lower panels), implying that MDP1 influences cell envelope structure during stationary phase in M. smegmatis.
In order to analyze the effect of MDP1 disruption on glycolipid biosynthesis, we chased synthesis of mycolates and TMM by adding 14C-labeled acetic acid to cultures of the wild-type, MDP1 KO, and complemented strains. Incorporation of radioactivity into TMM and MAMEs was analyzed by using the BAS system after fractionation by high-performance TLC (Fig. 6). TLC analysis revealed two TMM spots (Fig. 6A). M. smegmatis produces three types of mycolic acids, alpha, alpha′, and epoxy mycolates. To determine which types of mycolates were present in each TMM spot, we determined the molecular mass of each spot by MALDI-TOF mass spectrometry. The major peak of the upper TMM spot showed a pseudomolecular ion [M+Na]+ at m/z 1556, which was identified as alpha-C83 TMM, while the major peak of the lower spot showed a pseudomolecular ion [M+Na]+ at m/z 1306, which was presumed to be alpha′-C65 TMM (data not shown) (14). Thus, high-performance TLC analysis can separate alpha TMM and alpha′ TMM.
FIG. 6.
MDP1 mediates cessation of glycolipid biosynthesis at the stationary growth phase. (A) Lipid synthesis was measured by addition of [14C]acetate to a bacterial culture. The TMM and MAMEs from 108 bacteria, including the M. smegmatis parent strain (wild type [WT]), the MDP1 KO strain (KO), and the complemented strain (Comp), were extracted on days 3 and 7 and fractionated by TLC. The radioactivities of synthesized TMM and MAMEs were visualized by using the BAS system. (B to D) Radioactivities of TMM (B), alpha-MAME (C), and alpha′-MAME (D) extracted from a bacterial culture after 7 days were quantified by using the BAS system software and compared for the wild-type, MDP1 KO, and complemented strains. The values are means ± standard deviations of five experiments. PSL, photo-stimulated luminescence.
The three strains synthesized similar amounts of TMM and MAMEs during exponential growth (day 3). However, during stationary phase (day 7), in both the wild-type and complemented strains mycolate synthesis was reduced strongly (day 7). By contrast, the MDP1-deficent strain continued production of both mycolates and TMM, synthesizing 9.9-fold-higher amounts of TMM (Fig. 6B), 4.3-fold-higher amounts of alpha-MAME (Fig. 6C), and 5.9-fold-higher amounts of alpha′-MAME (Fig. 6D) than the wild type. This phenotype was completely reversed by complementation, indicating that a lack of MDP1 impaired the down-regulation of biosynthesis of mycolates and TMM in stationary phase.
Altering the growth rate with Ag85 complex proteins and MDP1.
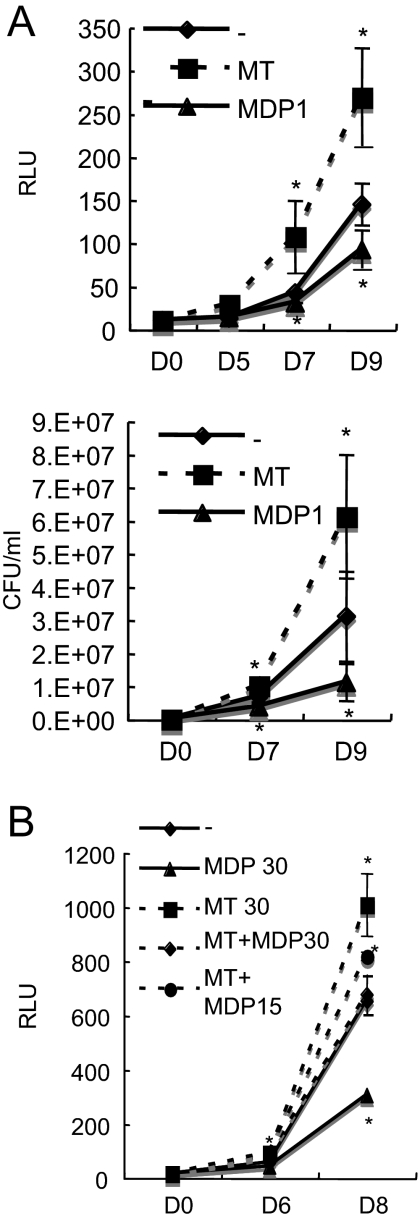
Because cell wall biogenesis is a biological event involved in multiplication of bacteria, we next examined whether regulation of the transfer of mycolic acids by MDP1 influences mycobacterial growth. We assessed the effects of exogenously added MDP1 in culture media on this growth. We generated a luciferase-producing BCG strain (BCG-Luc) to estimate the growth of BCG. The activity of luciferase paralleled the CFU assay results up to 104 CFU/ml (data not shown). We cultured BCG-Luc in the presence or absence of MDP1 or Ag85 complex proteins. Both the luciferase-based assay and determination of the CFU showed that exogenously added MDP1 suppressed growth of BCG, while Ag85 complex proteins enhanced growth (Fig. 7A). We next added serial doses of MDP1 to the culture of BCG-Luc in the presence of Ag85 complex proteins. We found that a low dose of MDP1 further boosted Ag85 complex-induced growth enhancement, while a high dose of MDP1 suppressed Ag85-dependent growth enhancement (Fig. 7B).
FIG. 7.
Growth control of BCG by exogenously added MDP1 and Ag85 complex proteins. BCG-Luc was grown in RPMI 1640 containing 10% FBS in the presence of various doses of MDP1, Ag85 complex, or a mixture of MDP1 and the Ag85 complex in a 96-well tissue culture plate. The total culture volume was 200 μl in each well. Representative results of three independent experiments are presented. (A) Luciferase activity (expressed in relative luciferase units [RLU]) was measured on days 0, 5, 7, and 9 (upper graph), and CFU were counted on days 0, 7, and 9 (lower graph). MT, 30 μg/well Ag85 complex; MDP1, 30 μg/well MDP1; −, BCG-Luc alone. The values are the means ± standard deviations of three experiments. (B) Luciferase activity was measured on days 0, 6, and 8. MT 30, 30 μg/well Ag85 complex; MDP 30, 30 μg/well MDP1; MT+MDP30, 30 μg/well Ag85 complex plus 30 μg/well MDP1; MT+MDP15, 30 μg/well Ag85 complex plus 15 μg/well MDP1; −, BCG-Luc alone. The values are the means ± standard deviations of three experiments. Asterisk, P < 0.05 for a comparison with the no-protein control (as determined by analysis of variance).
DISCUSSION
In this study, we analyzed the role of the mycobacterial histone-like protein MDP1 in the cell wall. We found that MDP1 plays an important role in tuning cell wall assembly by controlling transfer of mycolic acids to sugars by Ag85 complex proteins.
We showed that the cellular content of MDP1 was increased in advanced cultures of both M. smegmatis and BCG (Fig. 5). Previously, we and other groups showed that MDP1/HLP was accumulated in growth-retarded phases, including dormant bacilli of M. tuberculosis and M. smegmatis (19, 26, 37), by one-dimensional SDS-PAGE analysis. MDP1 is resistant to analysis by two-dimensional gel electrophoresis because of its strong positive charge (pI of BCG, 12.4; pI of M. tuberculosis, 12.45). However, in spite of the extensive analysis of gene expression profiles with DNA microarrays, increased expression of mdp1 mRNA in a stationary or anaerobic culture has not been observed (5, 18, 30, 35, 42). These results imply that accumulation of MDP1 in growth-retarded phases is largely due to posttranscriptional regulation. Pethe et al. found that lysines of the C-terminal region of laminin-binding protein/MDP1 are methylated by an unknown enzyme, which is present at a high level in the cell wall and confers resistance to proteolysis (31). Thus, posttranslational modification stabilizes MDP1 and might be involved in accumulation of MDP1 during a growth-retarded phase. In addition, methylation of basic charged amino acids may help the association of MDP1 with glycolipids by negating the charge of the protein. Posttranslational modifications, resembling eukaryotic histones, may control the cellular function and stability of MDP1. This issue should be clarified by additional study. By contrast, Ag85 complex proteins were localized in the cell wall during culture, but the contents gradually decreased with time in BCG (Fig. 5C) and M. smegmatis cultures (data not shown). It is likely that mycobacteria regulate transfer of mycolic acids by altering amounts of both MDP1 and Ag85 complex proteins in the cell wall.
The density of the MDP1 KO strain was lower in stationary phase (Fig. 4A). Lee et al. reported that an HLP/MDP1 KO strain displayed the same growth kinetics in Dubos Tween-albumin broth (19). We confirmed that an M. smegmatis MDP1 KO strain exhibited similar levels of cell density and survival at stationary phase when it was cultured in Dubos Tween-albumin broth (data not shown). Furthermore, alteration of the surface structure at stationary phase was not revealed by SEM analysis of the surface of the MDP1 KO strain cultured in Dubos Tween-albumin broth (data not shown).
TMM-derived mycolic acids are a major source of TDM and AG-linked mycolic acids (40, 41). Thus, the amount of TMM is an important factor for determining the level of cell wall assembly. The MDP1 KO strain exhibited continued synthesis of TMM other than MAMEs (Fig. 6) and TDM (data not shown) at the stationary growth phase. TMM could be synthesized from TMM and TDM by Ag85 complex proteins, once the substrates (TMM and TDM) were synthesized. However, the primary enzyme that catalyzes synthesis of TMM is still not known. Takayama et al. proposed that TMM could be synthesized in the cytoplasm (40). However, recently, Tropis et al. demonstrated that TMM is synthesized outside the plasma membrane but not inside the cytoplasm (41). The Ag85 complex is the most abundant protein secreted by M. tuberculosis (around 10 to 30%) and is a possible candidate enzyme for transfer of mycolic acids to trehalose to synthesize the TMM precursor (β-keto-acyl trehalose), as deduced from the structure of Ag85C (33). MDP1 suppresses Ag85 complex-dependent secondary synthesis of TMM (Fig. 2B), but we cannot eliminate the possibility that MDP1 also inhibits the primary synthesis of the TMM precursor whenever it is catalyzed by Ag85 complex proteins or undetermined enzymes.
An activity controlling glycolipid biosynthesis prompted us to examine whether exogenously added MDP1 and Ag85 complex proteins influence bacterial growth. We showed that growth of BCG could be altered by a combination of Ag85 complex proteins and MDP1 (Fig. 7). The MDP1 content in the total proteins of the cell wall increased at the stationary growth phase, while that of the Ag85 complex proteins decreased (Fig. 3). It can be speculated that a change in the ratio of MDP1 to Ag85 complex proteins in the cell wall is involved in determining the growth rate.
Inhibition of specific cellular metabolism causes cell death, as many antibiotics kill bacteria. However, expression of MDP1 does not kill mycobacteria; instead, it just suppresses growth (23). This is probably due to suppression of whole cellular metabolism, including macromolecular biosynthesis of DNA, RNA, and proteins (23) and cell wall assembly, as shown in this study. This can be caused by multiple interacting activities of different classes of macromolecules. The mechanism of such multiple binding activities should be resolved by structure-based studies, like those done on bacterial proteins such as SecB (32, 44). Recently, it has been reported that the histone-like protein HN-S mediates silencing of global gene expression in growth-retarded Salmonella (2, 8, 21). In humans, histone H2A seems to be involved in X chromosome inactivation (11). It can be speculated that use of a nonspecific DNA-binding protein to inactivate cellular metabolism is a general strategy. Here we found an alternative role of a histone-like protein in bacterial metabolism. Our data suggest that MDP1-medaited control of glycolipid biosynthesis is involved in the mechanism linking the growth state and cell wall biogenesis. Although we conducted experiments using nonpathogenic or slightly pathogenic mycobacteria, such as M. smegmatis and BCG, these bacteria share the basic structure of the cell wall with pathogenic mycobacteria, including M. tuberculosis. Taken together, our data provide significant information for understanding both the coordination of bacterial growth and virulence.
Acknowledgments
We are grateful to Keizou Oka, Department of Bioscience, INCS, Ehime University, for technical assistance with preparation of MAbs and to Thomas Dick, Novartis Institute for Tropical Diseases, for providing the M. smegmatis HLP/MDP1 KO strain. We also thank Todd P. Primm and Charles Scanga for editing the manuscript and Sara Matsumoto for heartfelt encouragement.
This work was supported by grants from the Ministry of Health, Labor and Welfare (Research on Emerging and Re-emerging Infectious Diseases, Health Sciences Research Grants), The Japan Health Sciences Foundation, the Ministry of Education, Culture, Sports, Science and Technology, and The United States-Japan Cooperative Medical Science Program against Tuberculosis and Leprosy.
We have no competing interests.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Aoki, K., S. Matsumoto, Y. Hirayama, T. Wada, Y. Ozeki, M. Niki, P. Domenech, K. Umemori, S. Yamamoto, A. Mineda, M. Matsumoto, and K. Kobayashi. 2004. Extracellular mycobacterial DNA-binding protein 1 participates in Mycobacterium-lung epithelial cell interaction through hyaluronic acid. J. Biol. Chem. 279:39798-39806. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, H., K. Kawamoto, T. Shirahata, and S. Makino. 2004. Changes in Salmonella enterica serovar Oranienburg viability caused by NaCl-induced osmotic stress is related to DNA relaxation by the H-NS protein during host infection. Microb. Pathog. 36:147-151. [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, R. E. Lee, K. Mdluli, A. E. Sampson, B. G. Schroeder, R. A. Slayden, and Y. Yuan. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37:143-179. [DOI] [PubMed] [Google Scholar]
- 4.Belisle, J. T., V. D. Vissa, T. Sievert, K. Takayama, P. J. Brennan, and G. S. Besra. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420-1422. [DOI] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, B. R. 2002. Tuberculosis—the global view. N. Engl. J. Med. 346:1434-1435. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. C., H. Y. Wu, M. Y. Chou, C. H. Huang, and A. Majumder. 2005. LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J. Biol. Chem. 280:15111-15421. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 11.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 12.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita, Y., T. Naka, T. Doi, and I. Yano. 2005. Direct molecular mass determination of trehalose monomycolate from 11 species of mycobacteria by MALDI-TOF mass spectrometry. Microbiology 151:1443-1452. [DOI] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 16.Jackson, M., C. Raynaud, M. A. Laneelle, C. Guilhot, C. Laurent-Winter, D. Ensergueix, B. Gicquel, and M. Daffe. 1999. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31:1573-1587. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, W. R., Jr., M. Tuckman, and B. R. Bloom. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532-535. [DOI] [PubMed] [Google Scholar]
- 18.Karakousis, P. C., T. Yoshimatsu, G. Lamichhane, S. C. Woolwine, E. L. Nuermberger, J. Grosset, and W. R. Bishai. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp. Med. 200:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, B. H., B. Murugasu-Oei, and T. Dick. 1998. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol. Gen. Genet. 260:475-479. [DOI] [PubMed] [Google Scholar]
- 20.Liu, J., C. E. Barry III, G. S. Besra, and H. Nikaido. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545-29551. [DOI] [PubMed] [Google Scholar]
- 21.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nat. Med. 6:1327-1329. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, S., M. Furugen, H. Yukitake, and T. Yamada. 2000. The gene encoding mycobacterial DNA-binding protein 1 (MDP1) transformed rapidly growing bacteria to slowly growing bacteria. FEMS Microbiol. Lett. 182:297-301. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto, S., M. Matsumoto, K. Umemori, Y. Ozeki, M. Furugen, T. Tatsuo, Y. Hirayama, S. Yamamoto, T. Yamada, and K. Kobayashi. 2005. DNA augments antigenicity of mycobacterial DNA-binding protein 1 and confers protection against Mycobacterium tuberculosis infection in mice. J. Immunol. 175:441-449. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, S., M. Tamaki, H. Yukitake, T. Matsuo, M. Naito, H. Teraoka, and T. Yamada. 1996. A stable Escherichia coli-mycobacteria shuttle vector ′pSO246′ in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 135:237-243. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, S., H. Yukitake, M. Furugen, T. Matsuo, T. Mineta, and T. Yamada. 1999. Identification of a novel DNA-binding protein from Mycobacterium bovis bacillus Calmette-Guerin. Microbiol. Immunol. 43:1027-1036. [DOI] [PubMed] [Google Scholar]
- 27.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 28.Naito, M., N. Ohara, S. Matsumoto, and T. Yamada. 1998. Immunological characterization of alpha antigen of Mycobacterium kansasii: B-cell epitope mapping. Scand. J. Immunol. 48:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Naito, M., N. Ohara, S. Matsumoto, and T. Yamada. 1998. The novel fibronectin-binding motif and key residues of mycobacteria. J. Biol. Chem. 273:2905-2909. [DOI] [PubMed] [Google Scholar]
- 30.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pethe, K., P. Bifani, H. Drobecq, C. Sergheraert, A. S. Debrie, C. Locht, and F. D. Menozzi. 2002. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc. Natl. Acad. Sci. USA 99:10759-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall, L. L., and S. J. Hardy. 1995. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem. Sci. 20:65-69. [DOI] [PubMed] [Google Scholar]
- 33.Ronning, D. R., T. Klabunde, G. S. Besra, V. D. Vissa, J. T. Belisle, and J. C. Sacchettini. 2000. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7:141-146. [DOI] [PubMed] [Google Scholar]
- 34.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 35.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimoji, Y., V. Ng, K. Matsumura, V. A. Fischetti, and A. Rambukkana. 1999. A 21-kDa surface protein of Mycobacterium leprae binds peripheral nerve laminin-2 and mediates Schwann cell invasion. Proc. Natl. Acad. Sci. USA 96:9857-9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shires, K., and L. Steyn. 2001. The cold-shock stress response in Mycobacterium smegmatis induces the expression of a histone-like protein. Mol. Microbiol. 39:994-1009. [DOI] [PubMed] [Google Scholar]
- 38.Soares de Lima, C., L. Zulianello, M. A. Marques, H. Kim, M. I. Portugal, S. L. Antunes, F. D. Menozzi, T. H. Ottenhoff, P. J. Brennan, and M. C. Pessolani. 2005. Mapping the laminin-binding and adhesive domain of the cell surface-associated Hlp/LBP protein from Mycobacterium leprae. Microbes Infect. 7:1097-1109. [DOI] [PubMed] [Google Scholar]
- 39.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 40.Takayama, K., C. Wang, and G. S. Besra. 2005. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18:81-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tropis, M., X. Meniche, A. Wolf, H. Gebhardt, S. Strelkov, M. Chami, D. Schomburg, R. Kramer, S. Morbach, and M. Daffe. 2005. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J. Biol. Chem. 280:26573-26585. [DOI] [PubMed] [Google Scholar]
- 42.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, J., and Z. Xu. 2005. The structural view of bacterial translocation-specific chaperone SecB: implications for function. Mol. Microbiol. 58:349-357. [DOI] [PubMed] [Google Scholar]