Abstract
We have characterized open reading frame RSP0072, which is located within the flgG operon in Rhodobacter sphaeroides. The amino acid sequence analysis of this gene product showed the presence of a soluble lytic transglycosylase domain. The deletion of the N-terminal region (90 amino acids) of the product of RSP0072 yields a leaky nonmotile phenotype, as determined by swarm assays in soft agar. Electron micrographs revealed the lack of flagella in mutant cells. The purified wild-type protein showed lytic activity on extracts of Micrococcus luteus. In contrast, no lytic activity was observed when the residues E57 or E83 were replaced by alanine. Affinity blotting suggests that the protein encoded by RSP0072 interacts with the flagellar rod-scaffolding protein FlgJ, which lacks the muramidase domain present in FlgJ of many bacteria. We propose that the product of RSP0072 is a flagellar muramidase that is exported to the periplasm via the Sec pathway, where it interacts with FlgJ to open a gap in the peptidoglycan layer for the subsequent penetration of the nascent flagellar structure.
Bacteria swim by means of flagella, which are locomotive chemiosmotic nanomachines that are widely distributed among this kingdom. This structure utilizes the energy from the electrochemical gradient to generate torque and propel the cell through its environment. The bacterial flagellum consists of at least three substructures: a basal body, a hook, and a filament. The biogenesis of this organelle is a tightly regulated process and requires the expression of more than 50 genes in a strict hierarchical manner (1, 8, 31). It extends from the cytoplasm to the cell exterior and assembly proceeds outwardly in an orchestrated manner from proximal structures to more distal ones. It uses the Sec pathway for the integral membrane components and a few other components. At a certain point in the assembly process the peptidoglycan layer must be penetrated by the rod, which is apparently thicker (ca. 11 nm) than the peptidoglycan mesh diameter (ca. 4 to 8 nm) (11, 21, 49, 50). Although the chemical composition of the peptidoglycan polymer might vary between different bacteria, the basic architecture is conserved. Fein (15) was the first to propose the necessity for the localized breakdown of peptidoglycan in order to incorporate the flagellum into the cell wall (12, 26). It has been shown that defects in FlgJ in Salmonella enterica serovar Typhimurium arrest the flagellar structure at the MS ring level (27). Within the C-terminal region, Salmonella FlgJ shows homology to the catalytic domains of various muramidases, which suggests a role in the degradation of the rigid peptidoglycan layer. It has also been shown that in Salmonella enterica FlgJ is exported into the periplasm via the flagellar export system and also that it displays peptidoglycan-hydrolyzing activity, which is essential for the formation of the flagellum (32). Recently, Viollier and Shapiro (54) reported that PleA is a lytic transglycosylase that is required for the assembly of pili and the flagella of Caulobacter crescentus.
However, muramidase activity is not absolutely required. Mutations in catalytically relevant residues result in poor flagellation and swarming ability, perhaps because the rod has a low probability of penetrating gaps generated during peptidoglycan biogenesis (38) and also perhaps due to the elastic properties and conformational freedom of the peptide cross-link that can be stretched up to fourfold (28).
The rod is composed by four proteins: FlgB, FlgC, FlgF, and FlgG (23). In addition to these structural components, several more proteins are required for rod assembly (27). Among these, FlgJ protein from S. enterica has been postulated to be a dual-function protein: the N-terminal half could function as a scaffold or cap essential for rod assembly, and the C-terminal half may function as a muramidase that degrades the peptidoglycan layer to facilitate rod penetration (20).
We have previously reported that the FlgJ protein of Rhodobacter sphaeroides lacks the C-terminal muramidase domain and that mutations in this protein yield a Fla− phenotype (16). The absence of the muramidase domain in this protein suggests that another polypeptide must accomplish this function. More recently, Pallen et al. have also reported the absence of a muramidase domain in FlgJ of another set of bacteria (40). In the present study we describe the characterization of the protein encoded by the gene RSP0072 that belongs to the flagellar flgG operon and contains a putative muramidase domain.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The bacterial strains, plasmids, and oligonucleotides used in the present study are described in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristics or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM103 | hsdR4 Δ(lac-pro) F′ traD36 proAB lacIqZΔΜ15 | 3 |
| M15[pREP4] | Lac− Ara− Gal− Mtl− F′ RecA+ Uvr+ Lon+ | QIAGEN |
| BL21DE3pLysS | F′ ompT hsdSB(rB−mB−)gal dcm(DE3)pLysS | Novagen |
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7; Tpr Smr | 45 |
| R. sphaeroides | ||
| WS8-N | Wild type; spontaneous Nalr | 47 |
| SltF1 | WS8 sltFΔ1::aadA derivative; Fla− Spcr | This study |
| Plasmids | ||
| pQE30 | Expression vector; Apr N-terminal His6 tag | QIAGEN |
| pQE60 | Expression vector; Apr C-terminal His6 tag | QIAGEN |
| pRSJ | flgJ cloned into the NcoI/BglII sites of pQE60 sites | This study |
| pRS0072 | RSP0072 cloned into NcoI/BamHI sites of pQE30 sites | This study |
| pRS0072E57A | RSP0072 E57A cloned into pQE30 | This study |
| pRS0072E83A | RSP0072 E83A cloned into pQE30 | This study |
| pJQ200mp18 | Sucide vector used for gene replacement | 42 |
| pTZ19R | Cloning vector, pUC derivative, Apr | Pharmacia |
| pRK415 | pRK404 derivative; used for expression in R. sphaeroides | 24 |
| pWM5 | pUC derivative carrying the omega-Spcr cassette | 34 |
| pBG313 | 3.8-kb BamHI fragment carrying RSP0072 from WS8-N | |
| Cloned into pTZ19R | 16 | |
| pRKsltF | 1.4-kb PstI/PstI fragment from pBG313 subcloned into pRK415 carrying sltF (RSP0072); Tcr | This study |
| Oligonucleotides | ||
| F1 | 5′-CTGATCTAGACCCTCCGGCCCCGGCCACGGTG-3′ | This study |
| R2 | 5′-CCGGAATTCGACGAGCGCGAACCTCATGCC-3′ | This study |
| F3 | 5′-CGGGAATTCCTCAACTGGCGCTGGCATGCG-3′ | This study |
| R4 | 5′-CTGATCTAGAGGGGTCACGCCCGAGAGACAG-3′ | This study |
| SPC1 | 5′-CCTGGATATCGGGCAGATCCGTGC-3′ | This study |
| SPC2 | 5′-TCATGATATCTCTCCCAATTTGTG-3′ | This study |
| orf72F | 5′-CATGGAGCTCGCGGACGAGGGCTGCGAGACG-3′ | This study |
| orf72R | 5′-CCCGAAGCTTTCACGGTTGCATTGCGAGCAG-3′ | This study |
| orf72F57A | 5′-GGCGATTGCCCGCGTGGCGTCGGGCCGGGGCGGGC-3′ | This study |
| orf72R57R | 5′-GCCCGCCCCGGCCCGACGCCACGCGGGCAATCGCC-3′ | This study |
| orf72F83A | 5′-CTTCGAGACCCGGGCCGCGGCGGTGCGCATGCTC-3′ | This study |
| orf72R83A | 5′-GAGCATGCGCACCGCCGCGGCCCGGGTCTCGAAG-3′ | This study |
| JF | 5′-CATGCCATGGATCTGAAGCTTCAGTCC-3′ | This study |
| JR | 5′-GGAAGATCTCGACTTGCCGTCCCTGACGAGAG-3′ | This study |
Spcr, spectinomycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim; Apr, ampicillin resistance; Smr, streptomycin resistance; Nalr, nalidixic acid resistance.
Media and growth conditions.
R. sphaeroides cell cultures were grown in liquid Sistrom's culture medium (46) at 30°C under constant illumination in completely filled screw-cap tubes. Motility plates were prepared using 0.25% agar and propionate 0.1 mM in M9 minimal culture medium. When required, the following antibiotics were added at the indicated concentrations nalidixic acid, 20 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 1 μg/ml. Strains of Escherichia coli were grown in Luria-Bertani medium (3). When needed, antibiotics were added at the following concentrations: spectinomycin, 50 μg/ml; gentamicin, 30 μg/ml; and ampicillin, 200 μg/ml. Standard molecular biology techniques were used for the isolation and purification of chromosomal DNA from R. sphaeroides WS8-N (3). Plasmid DNA and PCR fragments were purified with the QIAprep Spin and QIAquick PCR kits, respectively (QIAGEN GmbH).
Isolation of a mutant in RSP0072.
In order to obtain a mutant in RSP0072 a segment of 600 bp located upstream of RSP0072 was amplified by PCR using the oligonucleotides F1 and R2. The region downstream was amplified by using oligonucleotides F3 and R4 with chromosomal DNA from R. sphaeroides WS8-N as a template. From pWM5 plasmid, we amplified the aadA gene (which confers spectinomycin resistance), using oligonucleotides SPC1 and SPC2 (34). The resistance cassette was cloned in the joining point of the two PCR products. This construction was subcloned into suicide vector pJQ200mp18 (42), which was introduced to R. sphaeroides by diparental conjugation with E. coli S17-1 (10). The deletion of the soluble lytic transglycosylase (Slt) domain reported here runs from V15 to L105. This is in contrast to a previously reported mutation of this gene (16), which carries an aadA insertion at the ClaI site corresponding to I122.
Site-directed mutagenesis of RSP0072.
Replacement of the residues E57 and E83 by alanine was carried out according to the QuikChange mutagenesis method (Stratagene), and the presence of the desired mutations was confirmed by sequencing. These mutant proteins were overproduced and purified as described below.
Motility assays.
A 5-μl sample of a stationary-phase culture was placed on the surface of swarm plates (3) and incubated aerobically in the dark at 30°C. Swarming ability was recorded as the ability of bacteria to move away from the inoculation point after 24 to 36 h of incubation.
Electron microscopy.
Cultures were grown for 16 h under phototrophic conditions. The cells were washed and treated with 2% uranyl acetate for 10 min and observed at 80 kV with a JEM-1200EXII JEOL electron microscope.
Overexpression and purification of FlgJ and the product of RSP0072.
The flgJ and RSP0072 genes were amplified from chromosomal DNA of R. sphaeroides WS8-N. The oligonucleotides used were orf72F and orf72R for RSP0072 and JF and JR for flgJ. The PCR products were cloned into overexpression vectors pQE30 and pQE60, respectively. E. coli cells (strain M15 pREP4) were grown at 37°C to an optical density at 600 nm (OD600) of 0.6, and at this point, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM, and cell growth was allowed to proceed for 6 to 7 h at 25°C. To overproduce FlgJ, we used the E. coli BL21(DE3)pLysS strain; in this case, the culture was grown to an OD600 of 1.5, and then the cells were harvested.
For the purification of FlgJ and the product of RSP0072, the cells were sonicated, and the cell debris was removed by centrifugation. The supernatant from the overexpressed product of RSP0072 was incubated with nickel-Sepharose beads for 1 to 2 h and washed several times with 50 mM imidazole. In the case of FlgJ the protein was obtained from inclusion bodies that were resuspended in 5 M guanidine-HCl, and the solubilized protein was treated as mentioned above for RSP0072 except that the nickel-Sepharose beads were washed with 10 mM imidazole. Finally, the proteins were eluted with imidazole 200 mM. After purification, FlgJ was renatured by dialysis against a buffer containing 50 mM sodium phosphate (pH 6.5). Protein determination was carried out by the method described by Lowry et al. (30), and the purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (29). The structural integrity of FlgJ was verified by circular dichroism (AVIV model 202-01). Antibodies to the protein encoded by RSP0072 were raised in rabbits according to standard procedures (18), and anti-FliC antibodies were obtained as reported previously (16).
FliC secretion assays.
R. sphaeroides was grown photoheterotrophically for 16 h (OD600 = 2.0) and harvested by centrifugation at 16,000 × g for 15 min. The supernatant was centrifuged and filtered (pore size, 0.22 μm; Millipore Corp., Bedford, MA); the flowthrough was treated with chloroform-methanol as described previously (55) and resuspended in 20 μl of sample buffer. The cell pellet was resuspended in 200 μl of SDS sample buffer. The protein concentration was determined according to the method of Lowry (30). After SDS-PAGE, the proteins were transferred to nitrocellulose membranes and incubated with a 1:10,000 solution of anti-FliC polyclonal antiserum and developed by using an ECL immunoblotting detection kit (Amersham International, Little Chalfont, United Kingdom).
Muramidase activity assays.
To determine the muramidase activity, a zimodot or lisoplate technique was used. Petri dishes filled with soft 1% agar containing lyophilized Micrococcus luteus cells used as a substrate at a concentration of 50 mg/ml in phosphate buffer (50 mM NaH2PO4 [pH 6.5] as described in reference 4). The lisoplates were inoculated with 7 μg of protein and incubated for 12 h at 37°C.
Affinity blotting.
The proteins SltF (the product of RSP0072), FlgJ, and CheY6 were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, Richmond, CA). Membranes were blocked overnight at room temperature in Tris-buffered saline containing 0.1% (vol/vol) Tween 20 (TTBS)-5% nonfat milk powder. For immunoblotting, these membranes were probed with specific anti-SltF polyclonal antiserum at a 1:5,000 dilution in TTBS solution containing 0.1% nonfat milk powder. Detection was performed by using an ECL immunoblotting detection kit (Amersham).
For affinity blotting experiments, nitrocellulose membranes were incubated with purified SltF (1.8 μg/ml) for 1 h at room temperature, washed with TTBS, and probed with anti-SltF polyclonal antiserum at a 1:5,000 dilution. Detection was performed as described above for immunoblots.
Immunoprecipitation.
Sepharose CL-4B coupled to protein A (20 μl) (Sigma Chemicals) was incubated with 4 μg of anti-SltF gamma globulins in 1 ml of 50 mM phosphate buffer (pH 6.5) for 12 h at 4°C; the tube was then centrifuged, and the supernatant was discarded. To evaluate the interaction of SltF with FlgJ, 0.07 μM concentrations of each protein were incubated for 30 min at 4°C before the addition of the specific anti-SltF gamma globulins attached to protein A Sepharose. The mixture was then incubated for 30 min at 4°C and washed five times with 1 ml of phosphate buffer. The resulting pellet was suspended in 26 μl of sample buffer and boiled for 10 min. The samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes treated as described above and developed using anti-His6 antibodies at a 1:10,000 dilution (Pierce Chemicals).
Gene context.
We analyzed the genomic context of genes similar to RSP0072 by using GeConT (9).
RESULTS
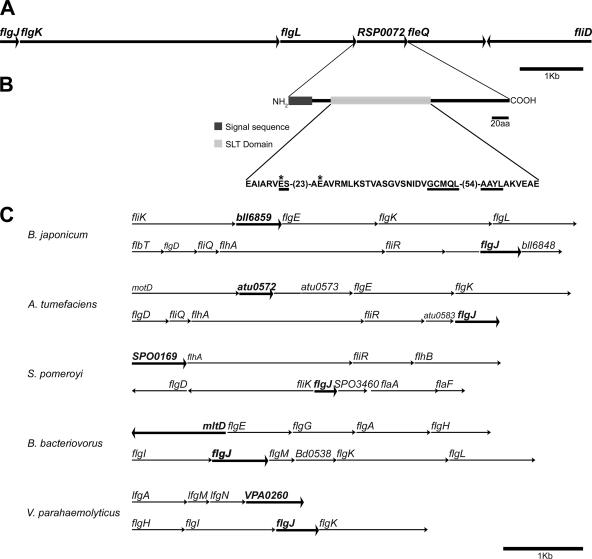
We have previously reported (16) the presence of an open reading frame downstream of flgL that codes for a protein of 265 amino acid residues (Fig. 1) that did not give an apparent phenotype on swarm plates after it was interrupted. This gene has been annotated in the genome of R. sphaeroides as RSP0072 (http://genome.ornl.gov/microbial/rsph/). Further analysis of the sequence with PSI-BLAST (2) revealed that the protein encoded by RSP0072 is similar to one type of muramidases also known as a soluble lytic transglycosylase (Slt). We determined that this protein shows a sec-type export sequence (39) that was detected by using SignalP-3 (5). Using the program GeConT, we identified genes similar to RSP0072, as well as their genomic context (9) (Fig. 1C). It should be noted that the five examples shown correspond to putative Slt proteins from the group of alphaproteobacteria. In all cases, the genes similar to RSP0072 are flanked by flagellar genes. Therefore, we tentatively named this putative soluble flagellar lytic transglycosylase encoded by RSP0072 as SltF.
FIG. 1.
(A) Genetic context of flgJ and RSP0072 in R. sphaeroides. (B) Representation of the protein coded by RSP0072 that contains an export signal sequence and an Slt domain. Also shown is the amino acid sequence of the Slt domain with underlined conserved signatures and marked with asterisks the two catalytically relevant glutamic residues that were exchanged in the present study for alanine. (C) Genetic context of genes coding for FlgJ that lack the muramidase domain. Also shown (in boldface) are genes that code for putative Slt proteins in the same microorganisms.
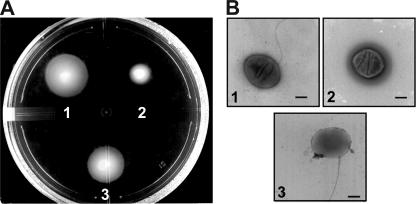
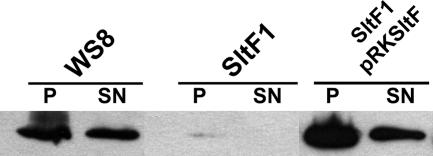
We noticed that in the previously reported mutant (16), the Slt domain was left basically intact. Therefore, we isolated a new mutant strain in RSP0072 with a deletion of the Slt domain (the details of these mutants are in Materials and Methods). Figure 2A shows that swimming in such a mutant is significantly impaired compared to the wild-type WS8-N strain. When the mutant was complemented with the wild-type gene, motility was recovered to levels similar to those of wild-type cells (Fig. 2A). This mutant was analyzed with the electron microscope and compared to WS8-N cells and also to the complemented mutant (Fig. 2B). It should be noted that the deletion of the Slt motif renders cells that lack the flagellar filament, which explains the leaky motile phenotype observed in swarm plates. The absence of filament was further confirmed by analyzing the presence of flagellin, either attached to the surface of the bacterium or secreted to the medium. Specific polyclonal anti-flagellin antibodies were used to determine by immunoblotting the presence of this protein in either a low-speed pellet (cell bodies) or supernatant. Figure 3 shows that mutant cells do not produce or export flagellin, in contrast to what was observed with wild-type WS8-N and complemented cells.
FIG. 2.
Characterization of SltF1 mutant. (A) Swimming assay in 0.25% soft agar. Spots: 1, wild-type strain WS8-N; 2, mutant SltF1 strain (sltFΔ1::aadA); 3, SltF1 strain complemented with the wild-type RSP0072 gene. (B) Electron micrographs of cells from the strains shown in the swimming assay (same order as in panel A). Bar, 500 nm.
FIG. 3.
Immunoblotting of wild-type, SltF1 mutant, and SltF1/pRKsltF strains with a polyclonal anti-FliC antibody. Lanes: P, pellet; SN, supernatant. Fractions were obtained as indicated in Materials and Methods.
SltF contains a conserved glutamic residue (E57) that has been reported to be responsible for catalysis in all of the Slt proteins reported previously (51; data not shown). From the alignment of these proteins, we also detected a region that is present in VirB but is absent in many Slts.
We purified SltF to determine catalytic activity. In a first attempt, we amplified by PCR the complete region encoding SltF, and this product was cloned into pQE30. However, a few minutes after induction, the optical density of the culture decreased, presumably due to cell lysis (data not shown). We then cloned a PCR product that lacked the sequence encoding the predicted signal peptide of SltF. In this case the protein was successfully induced and then purified, and its activity was tested in agar plates containing a cell extract from M. luteus. As shown in Fig. 4, both SltF and lysozyme produce a lysis area. Two mutant versions of SltF in which glutamic acid residues E57 or E83 were changed by site-directed mutagenesis to alanine were tested in this assay. Figure 4 shows that these two mutant proteins lack enzymatic activity.
FIG. 4.
Activity of wild-type SltF and mutant proteins. Agar plates containing M. luteus as a substrate were used to test the activity of the pure proteins SltF (spot 1), SltF E57A (spot 2), SltF E83A (spot 3), egg white lysozyme (EWL) (spot 4). The clear zone is due to enzymatic activity degrading peptidoglycan from “M. lysodeikticus.” Pure protein (7 μg) was deposited, and the lisoplate was incubated 12 h at 37°C.
The putative sec-dependent export sequence found in the N-terminal region of SltF suggests that this protein is not exported through the flagellar type III secretion system (35). How then does this lytic transglycosylase find its target in order to allow growth of the flagellar structure at a precise location? One possibility could be that, once it is exported to the periplasmic space, SltF is guided by FlgJ, which is exported to the periplasm through the flagellar export system.
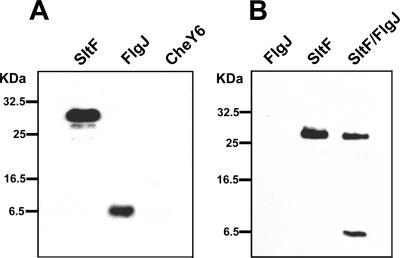
To explore possible interactions between FlgJ and SltF, we used affinity blotting and coimmunoprecipitation. These methods have been used as direct proof of interaction between different flagellum specific proteins such as the switch proteins FliM, FliN (52), FliI, FliC, and FlgE (44), as well as protein kinase C and its substrate (43). An unrelated protein was included and was used as a control that allowed us to distinguish between specific and nonspecific interactions in the case of affinity blotting. FlgJ, SltF, and CheY6 were subjected to SDS-PAGE and transferred to nitrocellulose membranes, which were incubated with purified SltF for 1 h at room temperature, and then probed with anti-SltF antibodies. Figure 5A shows that anti-SltF antibodies reveal SltF interacting with FlgJ but not with CheY6. To further confirm the specific interaction between SltF and FlgJ, we used protein A-Sepharose beads carrying anti-SltF gamma globulins. The result shows that protein A-Sepharose beads bind SltF as well as SltF-FlgJ complex but do not retain FlgJ (Fig. 5B).
FIG. 5.
Affinity blot and coimmunoprecipitation. (A) An affinity blot was performed as described in Materials and Methods. In this case, pure protein (2 μg) was subjected to SDS-17.5% PAGE, transferred to a nitrocellulose membrane, incubated with SltF, and probed with anti-SltF antibodies. (B) Coimmunoprecipitation assay showing specific interaction between SltF and FlgJ. Protein A-Sepharose beads to which specific anti-SltF gamma globulins were bound were incubated with FlgJ, SltF, or both proteins previously mixed. The retained proteins were subjected to SDS-PAGE and transferred to nitrocellulose membranes developed with anti-His6 histidine commercial antibodies (see Materials and Methods).
DISCUSSION
The flagellar morphogenetic pathway has been extensively studied in several bacterial species. In its early stages, the assembly process relies on a specific muramidase that locally digests the peptidoglycan layer to allow penetration of the nascent rod (31). In R. sphaeroides the open reading frame RSP0072, which we propose after the present study be named SltF, is located within a flagellar region that comprises 35 kb in chromosome I of this photosynthetic bacterium. It is flanked by flgL and fleQ that code for HAP3 (16) and a flagellum-specific enhancer-binding protein (41). The genetic context strongly suggests that SltF is the flagellar muramidase. In the present study we analyzed the relevance of this gene product in the biogenesis of the flagellum by means of several experimental approaches. Our results show that the deletion of the N-terminal region of SltF renders a nonmotile phenotype in swarm assays after an incubation period of 24 h. Nevertheless, if the incubation time is extended to 5 to 6 days the swarming spot becomes apparent (data not shown). A possible explanation for this could be the redundancy of the lytic transglycosylases present in many bacteria or to the elastic properties of the peptidoglycan layer or both. It has been shown in E. coli that mutations in either one of seven lytic transglycosylases do not show an observable phenotype (19). The authors of that study suggested that amidases, lytic transglycosylases, and endopeptidases are involved in cell separation after cell division. Similarly, the genome of R. sphaeroides shows at least four putative lytic transglycosylases (http://genome.ornl.gov/microbial/rsph/), which could partially complement the function lost after the deletion of the N-terminal region of SltF. An alternative explanation for the recovery of the swimming behavior is the high exchange rate of the peptidoglycan layer that has been estimated to reach levels of 50 to 60% in one generation (17, 53). Therefore, the possibility exists that eventually gaps and flagellar biogenesis coincide, allowing the nascent structure to penetrate this barrier, as has been suggested by Hirano et al. (20). We have consistently observed that the wild-type phenotype is recovered with the reintroduction of sltF, suggesting that the protein is necessary for efficient morphogenesis. The absence of a flagellar structure as well as the lack of flagellin (FliC) in either the cytoplasm or the extracellular medium strongly suggests that the structure is arrested at the MS ring level and that FlgM, which is the anti-sigma factor, is present in the cytoplasm repressing the expression of the class IV genes, among which is fliC (41).
The protein sequence of SltF clearly shows the reported motifs that identify the family of proteins known as Slts (6, 13, 25). These enzymes differ from lysozymes in that they induce a molecular rearrangement during lysis of the peptidoglycan layer that converts the glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine into an internal 1→6 anhydro N-acetylmuramyl (hemiacetal) bond (22). Furthermore, these enzymes require a single catalytic glutamic acid residue to carry out its function. In contrast, lysozymes require an aspartic and a glutamic acid residue for catalysis (48). Some examples of this are the hen egg white lysozyme (33) and lysozyme from T4 bacteriophage (14). In the present study we show that SltF contains two glutamic acid residues (E57 and E83) that, when mutated to alanine, rendered the enzyme inactive. From the sequence alignment (data not shown), we deduced that E57 might be the catalytic residue. However, further studies are needed to determine the role of E83 in catalysis, i.e., whether it is structural or functional.
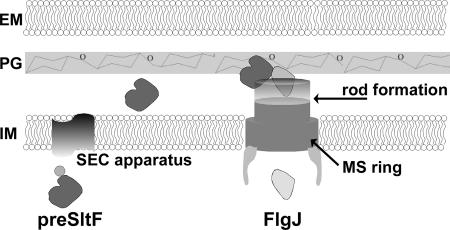
We also addressed the question of the possible interaction between FlgJ and SltF that would help explain the mechanism by which this lytic transglycosylase (SltF) would be directed to the site where the flagellar structure penetrates the cell wall (see Fig. 6). According to this model, SltF would be exported by the general secretion pathway to the periplasm, where it would interact with FlgJ and perform the localized degradation of the cell wall, this would allow the growing rod to cross the peptidoglycan wall. There are examples of lytic transglycosylases involved in various secretion systems such as VirB1 of the type IV secretion system from Agrobacterium tumefaciens (36) or IpgF in the type III secretion system from Shigella sonnei (56).
FIG. 6.
Model showing the possible mode of action of SltF during flagellar biogenesis. The external membrane (EM), peptidoglycan cell wall (PG), and inner membrane (IM) are indicated.
Further questions remain to clarify how the catalytic activity of the lytic transglycosylase that has been exported to the periplasmic space is regulated in order to avoid random generation of gaps in the peptidoglycan layer. Regarding this issue, preliminary data show that the interaction between FlgJ and SltF does not modify the activity of the muramidase (F. J. de la Mora et al., unpublished results).
Recently, Nambu et al. (37) found that various alphaproteobacteria, including R. sphaeroides, possess proteins homologous to FlgJ that lack the lytic transglycosylase domain, whereas beta- and gammaproteobacteria possess FlgJ that contains the N-terminal domain fused to the lytic C-terminal moiety. It should be noted that this catalytic domain does not show muramidase enzymatic activity (21). These proteins behave more like amidases similar to the T7 lysozyme that breaks bonds linking muramic acid and l-alanine (7, 38, 40).
Acknowledgments
We thank Sebastian Poggio for helpful discussions, Aurora Osorio for helpful technical assistance, and the IFC Molecular Biology Unit for sequencing facilities as well as the Microscopy Unit for the electron micrographs.
This study was supported by Consejo Nacional de Ciencia y Tecnología grants 47172/A-1 and P42600-Q.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 4.Becktel, W. J., and W. A. Baase. 1985. A lysoplate assay for Escherichia coli cell wall-active enzymes. Anal. Biochem. 150:258-263. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, N. T., and A. J. Clarke. 2001. Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 52:78-84. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, X., X. Zhang, J. W. Pflugrath, and F. W. Studier. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 91:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciria, R., C. Abreu-Goodger, E. Morett, and E. Merino. 2004. GeConT: gene context analysis. Bioinformatics 20:2307-2308. [DOI] [PubMed] [Google Scholar]
- 10.Davis, J., T. J. Donohue, and S. Kaplan. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demchick, P., and A. L. Koch. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkstra, B. W., and A. M. Thunnissen. 1994. ‘Holy’ proteins. II: the soluble lytic transglycosylase. Curr. Opin. Struct. Biol. 4:810-813. [DOI] [PubMed] [Google Scholar]
- 14.Fastrez, J. 1996. Phage lysozymes. EXS. 75:35-64. [DOI] [PubMed] [Google Scholar]
- 15.Fein, J. E. 1979. Possible involvement of bacterial autolytic enzymes in flagellar morphogenesis. J. Bacteriol. 137:933-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Pedrajo, B., J. de la Mora, T. Ballado, L. Camarena, and G. Dreyfus. 2002. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta 1579:55-63. [DOI] [PubMed] [Google Scholar]
- 17.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 19.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano, T., T. Minamino, and R. M. Macnab. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359-369. [DOI] [PubMed] [Google Scholar]
- 21.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Höltje, J. V., D. Mirelman, N. Sharon, and U. Schwarz. 1975. Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 124:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homma, M., K. Kutsukake, M. Hasebe, T. Iino, and R. M. Macnab. 1990. FlgB, FlgC, FlgF, and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211:465-477. [DOI] [PubMed] [Google Scholar]
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Koonin, E. V., and K. E. Rudd. 1994. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem. Sci. 19:106-107. [DOI] [PubMed] [Google Scholar]
- 26.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of gram-negative bacteria. Cell Mol. Life Sci. 60:2371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 28.Labischinsky, H., G. Barnickel, and D. Naumann. 1983. The state of order of bacterial peptidoglycan, p. 49-54. In R. Hackenbeck, J. V. Höltje, and H. Labischinsky (ed.), The target of penicillin. Walter Gruyter, Berlin, Germany.
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 31.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 32.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207-217. [DOI] [PubMed] [Google Scholar]
- 33.Malcolm, B. A., S. Rosenberg, M. J. Corey, J. S. Allen, A. de Baetselier, and J. F. Kirsch. 1989. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc. Natl. Acad. Sci. USA 86:133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 35.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA 93:7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nambu, T., Y. Inagaki, and K. Kutsukake. 2006. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet. Syst. 81:381-389. [DOI] [PubMed] [Google Scholar]
- 38.Nambu, T., T. Minamino, R. M. Macnab, and K. Kutsukake. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181:1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver, D. 1985. Protein secretion in Escherichia coli. Annu. Rev. Microbiol. 39:615-648. [DOI] [PubMed] [Google Scholar]
- 40.Pallen, M. J., C. W. Penn, and R. R. Chaudhuri. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13:143-149. [DOI] [PubMed] [Google Scholar]
- 41.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol. Microbiol. 58:969-983. [DOI] [PubMed] [Google Scholar]
- 42.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 43.Robles-Flores, M., and J. A. García-Sáinz. 1994. Immunological crossreactivity of G-protein beta subunit and receptors for activated C-kinase. Biochem. Mol. Biol. Int. 34:465-473. [PubMed] [Google Scholar]
- 44.Silva-Herzog, E., and G. Dreyfus. 1999. Interaction of FliI, a component of the flagellar export apparatus, with flagellin and hook protein. Biochim. Biophys. Acta 1431:374-383. [DOI] [PubMed] [Google Scholar]
- 45.Simon, R., Priefer, U., and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 46.Sistrom, W. R. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 28:607-616. [DOI] [PubMed] [Google Scholar]
- 47.Sockett, R. E., J. C. A. Foster, and J. P. Armitage. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473-479. [Google Scholar]
- 48.Strynadka, N. C., and M. N. James. 1996. Lysozyme: a model enzyme in protein crystallography. EXS 75:185-222. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, H., K. Yonekura, K. Murata, T. Hirai, K. Oosawa, and K. Namba. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J. Struct. Biol. 124:104-114. [DOI] [PubMed] [Google Scholar]
- 50.Tampakaki, A. P., V. E. Fadouloglou, A. D. Gazi, N. J. Panopoulos, and M. Kokkinidis. 2004. Conserved features of type III secretion. Cell Microbiol. 6:805-816. [DOI] [PubMed] [Google Scholar]
- 51.Thunnissen, A. M., A. J. Dijkstra, K. H. Kalk, H. J. Rozeboom, H. Engel, W. Keck, and B. W. Dijkstra. 1994. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature 367:750-753. [DOI] [PubMed] [Google Scholar]
- 52.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN, and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 53.Uehara, T., K. Suefuji, N. Valbuena, B. Meehan, M. Donegan, and J. T. Park. 2005. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viollier, P. H., and L. Shapiro. 2003. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol. Microbiol. 49:331-345. [DOI] [PubMed] [Google Scholar]
- 55.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 56.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. E. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]