Abstract
Nontypeable Haemophilus influenzae (NTHI) causes chronic infections that feature the formation of biofilm communities. NTHI variants within biofilms have on their surfaces lipooligosaccharides containing sialic acid (NeuAc) and phosphorylcholine (PCho). Our work showed that NeuAc promotes biofilm formation, but we observed no defect in the initial stages of biofilm formation for mutants lacking PCho. In this study, we asked if alterations in NTHI PCho content affect later stages of biofilm maturation. Biofilm communities were compared for NTHI 2019 and isogenic mutants that either lacked PCho (NTHI 2019 licD) or were constitutively locked in the PCho-positive phase (NTHI 2019 licON). Transformants expressing green fluorescent protein were cultured in continuous-flow biofilms and analyzed by confocal laser scanning microscopy. COMSTAT was used to quantify different biofilm parameters. PCho expression correlated significantly with increased biofilm thickness, surface coverage, and total biomass, as well as with a decrease in biofilm roughness. Comparable results were obtained by scanning electron microscopy. Analysis of thin sections of biofilms by transmission electron microscopy revealed shedding of outer membrane vesicles by NTHI bacteria within biofilms and staining of matrix material with ruthenium red in biofilms formed by NTHI 2019 licON. The biofilms of all three strains were comparable in viability, the presence of extracellular DNA, and the presence of sialylated moieties on or between bacteria. In vivo infection studies using the chinchilla model of otitis media showed a direct correlation between PCho expression and biofilm formation within the middle-ear chamber and an inverse relationship between PCho and persistence in the planktonic phase in middle-ear effusions. Collectively, these data show that PCho correlates with, and may promote, the maturation of NTHI biofilms. Further, this structure may be disadvantageous in the planktonic phase.
Nontypeable Haemophilus influenzae (NTHI) is a commensal of the human upper airways that can cause localized opportunistic airway infections when mucociliary defenses are compromised (24). NTHI is a leading cause of otitis media with effusion (2), acute otitis media (1), chronic sinusitis (20), and pulmonary infections associated with chronic obstructive pulmonary disease (31). For many of these infections, NTHI bacteria persist within dense biofilm communities that are thought to provide resistance to host clearance (12, 25, 29). NTHI biofilms contain variants expressing lipooligosaccharides (LOS) that contain phosphorylcholine (PCho) and sialic acid (NeuAc) (10, 19, 36, 41). Mutants lacking these structures are unable to establish persistent biofilms in vivo (14, 19, 36). The addition of PCho to LOS occurs in a phase-variable manner, and thus, NTHI populations contain discrete subpopulations of PCho-positive (PCho+) and PCho-negative (PCho−) variants (40). PCho contributes to NTHI colonization and persistence by promoting bacterial adherence to host cells (9, 32, 33), entry into airway epithelial cells (32, 35), resistance to killing by some host-derived antimicrobial peptides (22), and a reduction in the potency of the bacterial LOS (41). We have previously demonstrated, by using static and continuous-flow biofilm systems, that PCho is not required for biofilm formation in vitro (41). However, our more recent work indicates that variants lacking PCho have a defect in the establishment of stable biofilm communities in vivo (14). Of note, in this prior study we also observed that PCho− bacteria were able to persist in the planktonic phase within the chinchilla middle ear.
Therefore, in the present study we asked whether PCho promotes the maturation of NTHI biofilms. Biofilms formed by NTHI 2019 and by isogenic mutants in this background differing in PCho expression (a PCho− licD mutant and a constitutively in-phase PCho+ licON mutant) were compared by confocal laser scanning microscopy (CLSM). Vertical Z-series images were analyzed using the COMSTAT software package to give numeric scores for five different parameters of biofilm structure (maximum thickness, average thickness, biomass, roughness, and surface/volume ratio). These data were then compared by statistical analysis. Biofilms formed by the various NTHI strains were further compared using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). The results of these studies showed a positive correlation of PCho expression with biofilm density, surface coverage, and thickness. For the constitutively PCho+ strain NTHI 2019 licON, while somewhat decreased levels of outer membrane vesicles were observed in biofilms compared to those for strains NTHI 2019 and NTHI 2019 licD, increased amounts of electron-dense extracellular material were observed by SEM, and the presence of a ruthenium red-staining fibrous matrix material that was not observed for the other two strains was observed in ultrathin sections by TEM. Infection studies were also performed in which bacterial persistence and the formation of biofilms by the three strains within the chinchilla middle ear were compared. While PCho promoted biofilm formation in vivo, the opposite result was obtained from analysis of the planktonic bacterial populations within middle-ear fluids, i.e., the PCho+ strain NTHI 2019 licON had significantly lower CFU counts in the planktonic phase than the other two strains. The results clearly demonstrate that biofilm structure and density increase in accordance with PCho content both in vitro and in vivo, and they suggest that the contribution of PCho to bacterial persistence may be specific to bacteria within biofilm communities.
MATERIALS AND METHODS
Bacteria.
Complete lists of primers and of bacterial strains and plasmids (with descriptions and references) used in this study are provided in Tables 1 and 2, respectively. All NTHI strains were cultivated in brain heart infusion (BHI) medium (Difco) supplemented with hemin (ICN Biochemicals) and NAD (Sigma); this medium is referred to below as supplemented BHI (sBHI). NTHI 2019 is a well-characterized strain originally isolated from a sputum sample of a patient with chronic bronchitis (3). NTHI 2019 licD is a previously described PCho− mutant (32). NTHI 2019 licON is a mutant harboring an in-frame deletion of the CAAT repeat region in licA. For the construction of this strain, a ∼2.5-kb DNA fragment containing licA was amplified from NTHI genomic DNA using primers P31 and P32 and was cloned into pCRBlunt (Invitrogen) according to the manufacturer's instructions. This amplicon was excised using EcoRI and cloned into EcoRI-digested pUC19 to generate pUClic1A. An in-frame deletion of the CAAT repeat region was generated by amplifying the plasmid by PCR using primers P34 and P35, followed by ligation with T4 DNA ligase and transformation into Escherichia coli. This amplification results in the insertion of a nonrepetitive DNA fragment (5′-CAGAGTATCAAC-3′) that codes for the QSIN amino acid sequence found in the LicA protein, without the possibility of slipped-strand mispairing resulting in phase variation. The resulting plasmid, pUClicAR3, was digested with BglII and ligated to a gentamicin resistance cassette excised from pGmr with BamHI to yield a selectable marker within an intergenic region upstream of the licA transcriptional start site (pUClicAR3Gm). Deletion of the CAAT repeats was verified by PCR using primers IP1 and IP2, followed by nucleotide sequence analysis. Plasmid pUClicAR3Gm was linearized by digestion with NdeI and introduced into NTHI 2019 by transformation to generate NTHI 2019 licON. For the transformation of NTHI 2019, we used a modification of a colony transformation method used for Neisseria spp. (16). NTHI 2019 bacteria were harvested from an overnight plate culture, patched onto the surface of an sBHI plate, and then incubated at 37°C under 5% CO2 for 4 h, after which 5 μg of NdeI-linearized pUClicAR3Gm plasmid DNA was spotted onto the bacterial patch. After 6 h of additional incubation at 37°C under 5% CO2, the NTHI bacteria were harvested with a cotton swab, suspended in sBHI broth, and incubated for 3 h at 37°C with gentle shaking (75 rpm). NTHI bacteria were pelleted, diluted, plated onto sBHI agar containing 5% Fildes' reagent (Difco) and 5 μg/ml gentamicin (Sigma), and incubated for 48 to 72 h at 37°C. Transformants were screened by PCR using primers IP1 and IP2 as described above. Transformants with the expected genomic deletion were screened for the presence of the PCho epitope on bacterial colonies or purified LOS by using monoclonal antibody HAS or TEPC-15 as described previously (32), and the expected phenotype of increased PCho content in LOS from the licON mutant strain was observed (see Fig. 1).
TABLE 1.
Primers used in this study
| Primers | Target | Sequence |
|---|---|---|
| P31 | licA | 5′-GGTTTTTAGATGGTTTTGCGCCGAGC-3′ |
| P32 | 5′-GCATAATCGCCAAAATTCTCGCCGTG-3′ | |
| P34 | licA (ΔCAAT) | 5′-CATAGCATTTTTGTATTCATTTTTATA-3′ |
| P35 | 5′-ATCAACCAGAGATCAACCAAATTGTAGGATTTGTTAAAACTTGCTAC-3′ | |
| IP1 | ΔCAAT | 5′-CCGAGCTTGGGTAAAAAAGTG-3′ |
| IP2 | 5′-GAAAAAGGAATGGAATGCTGATG-3′ | |
| gfp1 | gfp | 5′-CTCTCTCCCGGGCATATGAGTAAAGGA-3′ |
| gfp2 | 5′-ACACACGGCGCCCAACGGTGGTATATCCAG-3′ | |
| PhelP1 | hel promoter | 5′-CCCGGGATTATGTTCCTATTCATTCAGGGT-3′ |
| PhelP2 | 5′-GATATCCATAATTGCACCTTAGTTTAAATTTTG-3′ | |
| P27 | Region flanking gfp | 5′-GTGCCGATCAACGTCTCATTTTC-3′ |
| P28 | 5′-AGTCGGCGGTGGTGGCTTAG-3′ |
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| NTHI 2019 | Parental NTHI strain | 3 |
| NTHi 2019 licD | PCho transferase mutant | 32 |
| NTHI 2019 licON | Locked-on PCho+ phase | This study |
| Plasmids | ||
| pUClic1A | licA in pUC19 vector | This study |
| pUClicAR3 | lic1 phase locked on (licON); ΔCAAT | This study |
| pUClicAR3Gm | licON linked to gentamicin resistance cassette | This study |
| pBBR1-GFP | Promoterless gfp vector | 26 |
| pCRgfp | gfp in pCR2.1 | This study |
| pCRPhel | hel promoter in pCR2.1 | This study |
| pPhelgfp | hel-gfp | This study |
| pIRA-Cm | Vector for genomic insertion in H. influenzae | 17 |
| pIRAgfp | hel-gfp in pIRA-Cm | This study |
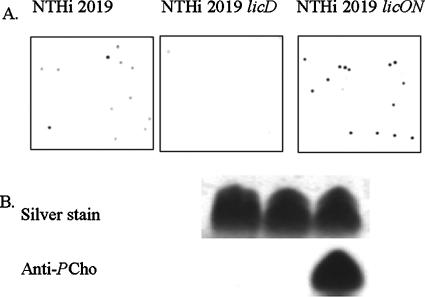
FIG. 1.
Confirmation of PCho phenotypes of NTHI 2019, NTHI 2019 licD, and NTHI 2019 licON. Immunoblot analyses were performed using the anti-PCho monoclonal antibody HAS on colony lifts (A) or purified LOS separated by Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (B).
For the biofilm studies, a constitutively expressed gfp construct was generated and introduced into a noncoding region of the NTHI genome. The gfp open reading frame was amplified from pBBR1-GFP (26) using primers gfp1 and gfp2 (Table 1) and then cloned into pCR2.1 (Invitrogen) to generate plasmid pCRgfp. Because prior work shows that the promoter for hel, which encodes the P4 lipoprotein phosphomonoesterase (orf NTHI0816) (28), is constitutively expressed in H. influenzae (42), this promoter was chosen to drive the expression of gfp. The hel promoter (phel) was amplified from NTHI genomic DNA via PCR using primers phelP1 and phelP2 (Table 1) and then cloned into pCR2.1 to create plasmid pCRPhel. The gfp open reading frame was excised from pCRgfp using SmaI and EcoRV and then ligated into EcoRV-digested pCRPhel to generate pPhelgfp. The orientation of the insert was confirmed by restriction mapping using EcoRI and EcoRV. The sequence for the gfp construct with the hel promoter was excised from pPhelgfp using SmaI and EcoRV and then ligated into an internal SfoI site within pIRA-CM (17). The resulting plasmid (pIRA-HELgfp) was linearized by digestion with NotI and used to transform NTHI as described above. Transformants were screened by fluorescent microscopy, and the presence of the construct within the IRA intergenic region was confirmed by PCR using primers P27 and P28 (Table 1).
Culture of NTHI biofilms.
Bacteria were harvested from overnight sBHI agar plate cultures, inoculated into sBHI broth, and cultured with gentle shaking (150 rpm) to mid-logarithmic phase. Equal inocula of ∼107 bacteria were used to seed biofilm chambers (Stovall, Greensboro, NC), and the bacterial densities of the inocula were confirmed by plate counts. The bacteria were incubated for 2 h at 37°C to allow initial surface attachment, and then a continuous flow of sBHI medium (5 ml/min) was established. At the times indicated above, the biofilms were analyzed by CLSM using a Zeiss 510 inverted microscope. For each experiment, three to five independent comparisons of the three strains were made, all of which gave consistent results. Viability staining of unfixed biofilms was performed using the BacLight Live/Dead kit (Molecular Probes) as indicated.
COMSTAT analyses.
Z-series images were collected from CLSM analyses performed on biofilms at 24, 48, and 72 h postinoculation. For each experiment, 8 to 14 image stacks, each representing a different field of view, were compiled. The analyses of the different strains were repeated in four independent experiments. In each case, the images were obtained at consistent positions (∼5 mm) relative to the inlet. The Z-slice images were exported as tagged-image format files into the MATLAB (version 5.1) package and analyzed in the Image Processing Toolbox as described previously (13).
SEM.
Biofilm samples were fixed within the flow cells for 60 min with 2.5% glutaraldehyde in phosphate-buffered saline (PBS) and were then rinsed twice (for 10 min each time) with PBS prior to dehydration, fixation, and critical point drying for SEM analysis according to standard procedures, as we have described earlier (36). After removal of the flow cell, the biofilm samples were mounted onto stubs and sputter coated with palladium prior to being viewed with a Philips SEM-515 scanning electron microscope.
TEM.
For TEM analyses, biofilm samples were fixed for 60 min in 2.5% glutaraldehyde-PBS, rinsed for 10 min in PBS, and stained with 1% osmium tetraoxide-PBS for 30 min. After another 10-min rinse in PBS, the samples were dehydrated in a graded ethanol series and embedded in Spurr's resin. Ultrathin sections (thickness, ∼90 nm) were cut using a diamond knife. For ruthenium red staining, samples were first fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for 15 min and then stained in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.05% ruthenium red for 30 min at 41°C. After three washes in the same solution, the samples were postfixed for 2 h in 1% osmium tetraoxide containing 0.05% ruthenium red. The samples were then dehydrated, embedded, and sectioned as described above. All sections were placed on grids and viewed with a Philips 400 transmission electron microscope.
Chinchilla infection studies.
Bacterial persistence and the formation of biofilms within the chinchilla middle-ear chamber were compared essentially as described in our prior work (14). Animals were housed for 7 days after delivery to allow for full recovery from shipping-related stress. The animals (3/group) were anesthetized by gas anesthesia and infected via transbullar injection with ∼100 CFU of either NTHI 2019, NTHI 2019 licD, or NTHI 2019 licON. Bacterial inoculum counts were confirmed by plate counting. At 7 and 14 days postinfection, animals were euthanized and the middle-ear chambers aseptically opened. The number of infected ears with macroscopically visible biofilm communities or effusion within the middle-ear chamber was recorded, and the biofilms were photographed as described previously (14). Fluids within the middle ear were recovered, and bacterial counts within the middle-ear fluids were determined by plate counting. For animals with no visible fluid within the middle-ear chamber, we performed lavage using 0.2 ml sterile PBS followed by plate counting.
Statistical analyses.
The statistical significances of differences observed in the COMSTAT and infection data were determined by unpaired t tests with Welch's correction for data sets with unequal variances, using GraphPad Prism, version 4.00 (GraphPad Software, San Diego, CA).
RESULTS
Construction and confirmation of NTHI 2019 licON.
In order to generate a constitutively locked-on PCho+ mutant strain, we used PCR to construct a licA allele from which the (CAAT)n repeat region had been deleted (see Materials and Methods). The phenotype of a mutant strain harboring this allele was confirmed by colony immunoblotting and Western blot analysis of purified LOS (Fig. 1). On the colony blot, the parental strain, NTHI 2019, had differing levels of reactivity with anti-PCho antibody HAS (Fig. 1A), as has been reported previously (32). As expected, we observed no reactivity for strain NTHI 2019 licD, which is known to lack all PCho+ LOS forms (32). For strain NTHI 2019 licON, we observed not only that all of the colonies were reactive with the antibody (and thus were PCho+) but also that all had high levels of reactivity (Fig. 1A). In Western blot analyses of purified LOS, we observed minimal reactivity for LOS from the parental strain (Fig. 1B). In contrast, there was a high degree of reactivity with LOS purified from NTHI 2019 licON. Thus, not only does the licON mutant express a greater proportion of PCho+ variants within the population, but PCho within the LOS glycoforms has greater accessibility for antibody binding.
Comparison of biofilm structure and density by CLSM and COMSTAT.
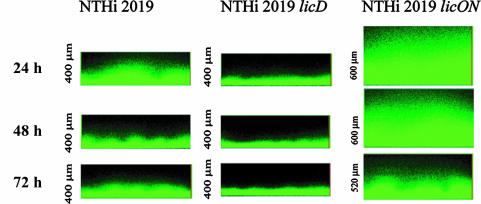
NTHI 2019, NTHI 2019 licD, and NTHI 2019 licON strains with constitutively expressed chromosomal gfp fusions were generated (see Materials and Methods). These strains were used to test the contribution of PCho to biofilm structure, as measured by CLSM. As seen in reconstructed vertical cross sections from stacked Z-series (Fig. 2), the biofilms formed by NTHI 2019 and NTHI 2019 licON were notably denser and thicker than those formed by NTHI 2019 licD at 24 h, 48 h, and 72 h postinoculation. Notably, the thicknesses of the biofilms formed by all three strains were decreased at 72 h postinoculation, a time that may correspond with the beginning of biofilm dispersal. There were no significant differences in the viability of NTHI bacteria within the biofilms of the three strains as measured by qualitative viability staining with the BacLight Live/Dead kit (Molecular Probes) (data not shown).
FIG. 2.
Vertical Z-series images of biofilms formed by NTHI 2019, NTHI 2019 licD, and NTHI 2019 licON. Each bacterial strain harbored a constitutively expressed gfp construct as described in Materials and Methods. The measurement at the left of each image is the height of the Z-stack.
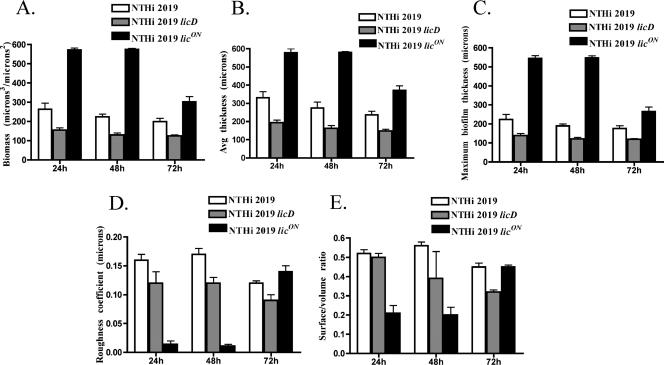
Quantitative analysis of biofilm structure by COMSTAT.
To quantify the results of the CLSM analyses, the data were processed using the COMSTAT software package (13). Numerical scores for the total biomass, average thickness, maximum thickness, roughness, and surface/volume coverage of the biofilms were generated (Fig. 3). Biofilms formed by NTHI 2019 licON had significantly higher biomass, average thickness, and maximum thickness than those formed by the parental strain, NTHI 2019, at all time points (Table 3). Conversely, for biofilms formed by strain NTHI 2019 licD, lacking PCho, all of these parameters were significantly decreased compared to those for the parental strain. The roughness coefficient, reflecting diversity in thickness and biomass through a biofilm, was significantly lower for strain NTHI 2019 licON than for the parental strain at the 24-h and 48-h time points but was not significantly different at the 72-h time point, possibly reflecting decreased biofilm density at some points in the biofilm structure in accordance with biofilm dispersal. The roughness of the NTHI 2019 licD biofilm was not statistically different from that of the parental strain at 24 h (P = 0.06) but was significantly less at 48 h and 72 h.
FIG. 3.
Analysis of biofilm properties by COMSTAT. Measures of total biomass (A), average thickness (B), maximum thickness (C), roughness (D), and surface/volume ratio (E) for all strains and time points are shown.
TABLE 3.
Statistical analysis of COMSTAT dataa
| Time point and strain | Avg thickness (μm) | P | Maximum thickness (μm) | P | Biomass (μm3/μm2) | P |
|---|---|---|---|---|---|---|
| 24 h | ||||||
| NTHI 2019 | 263 ± 32 | 331 ± 33 | 223 ± 27 | |||
| NTHI 2019 licD | 155 ± 12 | 0.012 | 193 ± 14 | 0.004 | 138 ± 11 | 0.017 |
| NTHI 2019 licON | 572 ± 9 | <0.001 | 578 ± 6 | 0.001 | 544 ± 15 | <0.001 |
| 48 h | ||||||
| NTHI 2019 | 224 ± 14 | 274 ± 33 | 189 ± 11 | |||
| NTHI 2019 licD | 130 ± 10 | <0.001 | 162 ± 16 | 0.012 | 121 ± 8 | 0.001 |
| NTHI 2019 licON | 575 ± 5 | <0.001 | 580 ± 4 | <0.001 | 547 ± 11 | <0.001 |
| 72 h | ||||||
| NTHI 2019 | 199 ± 17 | 236 ± 20 | 176 ± 14 | |||
| NTHI 2019 licD | 125 ± 5 | 0.003 | 148 ± 9 | 0.002 | 119 ± 4 | 0.003 |
| NTHI 2019 licON | 302 ± 27 | 0.004 | 371 ± 25 | 0.004 | 265 ± 24 | 0.005 |
P values refer to comparison with NTHI 2019.
The surface/volume ratios were significantly less for strain NTHI 2019 licON than for the parental strain at 24 h and 48 h, reflecting the greater density and overall surface coverage of these biofilms prior to dispersal. Overall, these data support the conclusion that PCho promotes biofilm maturation.
Comparison of biofilms by electron microscopy.
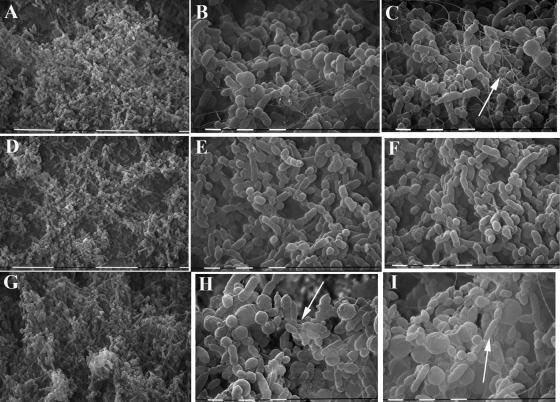
We also performed qualitative SEM analyses on mature biofilms formed by each strain. The results were consistent with the CLSM/COMSTAT data in that the biofilms differed in thickness and apparent density in accordance with PCho content (Fig. 4). NTHI 2019 formed biofilms that covered the majority of the slide surface, with visible 3-dimensional projections (i.e., “towers”) arising from the chamber surface (Fig. 4A). A fibrous matrix material was visible at higher levels of magnification (Fig. 4B and C). In contrast, biofilms formed by NTHI 2019 licD had a lesser degree of substratum coverage (note the open patches in Fig. 4D) and a general lack of visible matrix material (Fig. 4E and F). NTHI 2019 licON produced visibly denser biofilms with more-pronounced 3-dimensional projections (Fig. 4G). Examination at higher magnifications revealed increased amounts of electron-dense extracellular material being released from the bacterial surfaces (Fig. 4H and I), along with visible fibrous matrix material like that observed for the parental strain.
FIG. 4.
Analysis of biofilms by SEM. Images were obtained from biofilms formed by NTHI 2019 (A to C), NTHI 2019 licD (D to F), or NTHI 2019 licON (G to I) in a continuous-flow biofilm system at 72 h postinfection. Arrows indicate fibrous matrix material (B and C) or blebs (H and I).
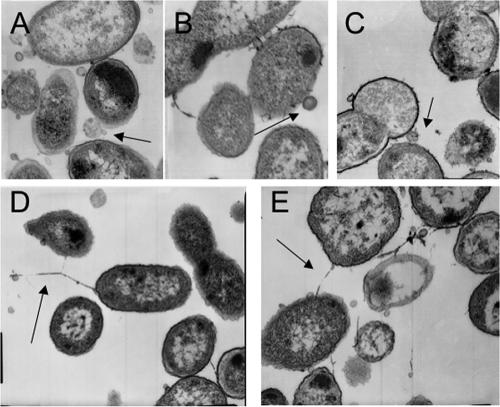
The biofilms were further examined by TEM. As shown in Fig. 5, outer membrane vesicles were observed in the thin sections of all three strains. On the sections stained with ruthenium red, we observed a fibrous matrix material in biofilms formed by NTHI 2019 licON (Fig. 5E and F). No such staining was observed for strain NTHI 2019 or NTHI 2019 licD. Immunogold staining using an anti-PCho antibody showed that this fibrous material did not contain PCho+ LOS forms, which, as expected, were found on the cell surfaces of strains NTHI 2019 and NTHI 2019 licON but not of strain NTHI 2019 licD (data not shown).
FIG. 5.
Analysis of biofilms by TEM. Images show thin sections of biofilms formed by NTHI 2019 (A), NTHI 2019 licD (B), and NTHI 2019 licON (C), as well as ruthenium red-stained sections of an NTHI 2019 licON biofilm (D and E). Arrows indicate outer membrane vesicles (A, B, and C) or matrix (D and E).
Biofilm formation in vivo.
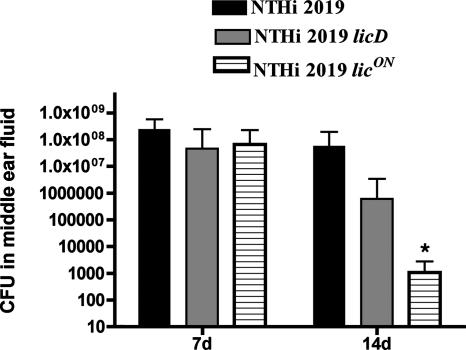
The chinchilla has been established as a valid infection model system for testing biofilm formation in vivo (14, 18, 19, 27). We infected animals with comparable infectious doses (∼102 CFU) of NTHI 2019, NTHI 2019 licD, or NTHI 2019 licON. The animals were monitored for signs of otitis disease, and groups of 3 animals/strain/time point were euthanized at 7 and 14 days postinfection. The middle-ear chambers were aseptically opened posteuthanasia and examined for the presence of macroscopically visible biofilms and effusion fluids. The results of the infection studies are summarized in Table 4 and Fig. 6 and 7. Biofilms and effusions were observed in animals infected with any of the three strains at 7 days postinfection (Fig. 6; Table 4). However, at 14 days postinfection, we observed a markedly greater proportion of biofilm-positive ears in the group infected with NTHI 2019 licON (Fig. 6E and F) than in the groups infected with the other two strains (Fig. 6), and none of the animals infected with NTHI 2019 licD showed visible biofilms (Table 4). Also of note, while the numbers of bacteria in planktonic populations of NTHI 2019 and NTHI 2019 licD were comparable, we observed a high planktonic bacterial load (∼105 CFU) only for one ear infected with NTHI 2019 licON; the remaining ears of animals infected with this strain had planktonic populations of <103 CFU (Fig. 7).
TABLE 4.
Results of chinchilla infection experiments
| Strain | Inoculum (CFU) | No. of positive ears/total no. of ears
|
|||
|---|---|---|---|---|---|
| Biofilm
|
Effusion
|
||||
| 7 days | 14 days | 7 days | 14 days | ||
| NTHI 2019 | 310 | 5/6 | 2/6 | 5/6 | 3/6 |
| NTHI 2019 licD | 220 | 6/6 | 0/6 | 6/6 | 2/6 |
| NTHI 2019 licON | 180 | 3/6 | 4/6 | 3/6 | 4/6 |
FIG. 6.
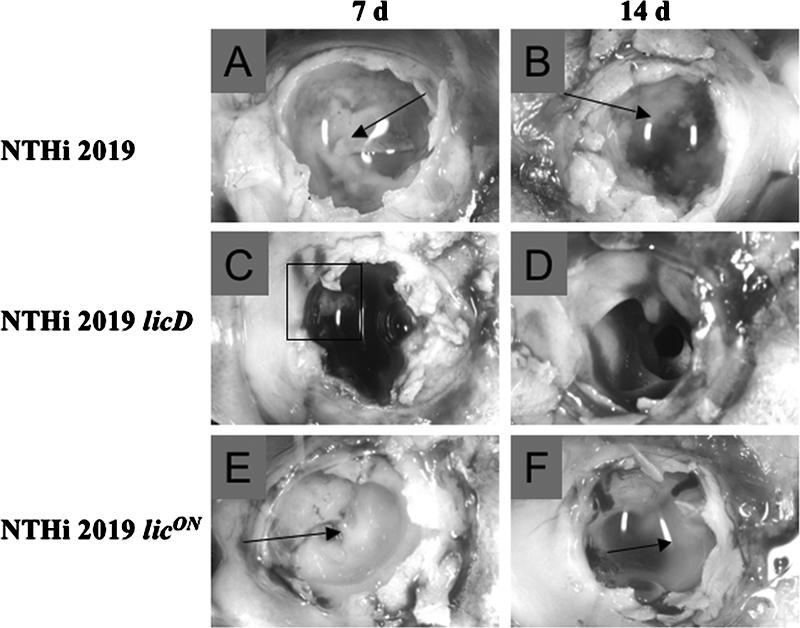
PCho promotes biofilm density and persistence in vivo. Images show macroscopically visible biofilms formed by NTHI 2019 (A and B), NTHI 2019 licD (C and D), and NTHI 2019 licON (E and F). d, days. Arrows and boxed region indicate visible biofilms.
FIG. 7.
Bacterial survival in the planktonic phase in vivo. Bars represent mean CFU counts obtained from effusion or lavage fluids from ears infected with NTHI 2019, NTHI 2019 licD, or NTHI 2019 licON. Error bars, standard deviations. d, days. Asterisk indicates a statistically significant difference from the count for the parental strain.
DISCUSSION
Like many mucosal pathogens, H. influenzae is able to modulate the expression of surface moieties by phase variation. Factors that undergo phase variation are commonly thought to confer some fitness advantage on the bacterial population only during a discrete phase of the infectious cycle, outside of which they may confer no benefit or even be detrimental (34, 37). PCho, for example, is known to confer a number of advantages on NTHI (22, 32, 35, 38, 39) while also (in at least some structural settings) targeting bacteria for the binding of C-reactive protein or antibody and subsequent clearance (6-8, 21). Consistent with this idea, Humphries and High showed that constitutive expression of the lic1 locus (and thus, presumably, expression of PCho) resulted in attenuation of encapsulated H. influenzae in an infection model of invasive disease (15).
We recently showed that NTHI mutants lacking PCho caused increased early inflammation in the chinchilla model and had a defect in the establishment of stable biofilm communities (14). Surprisingly, these mutants were able to survive as well as the parental strain in the planktonic phase (middle-ear fluids), suggesting that the observed biofilm defect was not solely a consequence of failure of NTHI to survive within the middle-ear chamber. We therefore revisited the question of whether PCho affects NTHI biofilm structure by comparing biofilm formation in vitro and in vivo by isogenic strains differing in PCho expression. Our data clearly show that the expression of PCho is positively correlated with the maturation of NTHI biofilms in vitro as well as in vivo. As we reported previously, an NTHI mutant lacking PCho had an in vivo biofilm defect (Fig. 5) yet survived as well as the parent in the planktonic phase (Fig. 6). However, a phase-locked NTHI mutant overexpressing PCho had a significant planktonic survival defect (Fig. 6) while producing a greater proportion of biofilm-positive ears in vivo (Fig. 5; Table 4). Our interpretation of these data is that expression of PCho is necessary for full biofilm maturation, while it is not required for survival outside of the biofilm mode of growth. It is possible that PCho expression increases the susceptibility of NTHI to killing by complement or other factors within the middle-ear chamber. In a related observation, we previously found no PCho-related difference in the killing of NTHI by chinchilla beta-defensin or serum (14); however, it remains possible that the levels of C-reactive protein or other factors within the middle ear in vivo following vascular perfusion were above the thresholds we tested.
Our results with regard to outer membrane vesicles and matrix material are also noteworthy. Vesicles are a known constituent of biofilm matrices (30) and may be a conduit for the delivery of quorum-signaling molecules (23). Therefore, it seems counterintuitive that we would observe an increase in biofilm formation by the licON mutant, while the number of vesicles observed within biofilms was somewhat diminished for this strain (Fig. 4). It may be that the increased amount of ruthenium red-staining matrix material observed in the NTHI 2019 licON biofilms underlies the increased biofilm density for this strain. Because we did not observe immunogold labeling of this matrix material with an anti-PCho antibody, the current data do not support the conclusion that PCho+ LOS forms are directly forming the biofilm matrix. Other matrix components associated with NTHI biofilm development include NeuAc-LOS or, possibly, other sialylated moieties (10, 36) and extracellular DNA (18). However, we observed comparable levels of NeuAc and extracellular DNA within colonies and biofilms formed by NTHI 2019, NTHI 2019 licD, and NTHI 2019 licON (data not shown). Thus, the exact composition of the matrix material observed in the NTHI 2019 licON biofilms remains an open question.
Biofilms are thought to promote bacterial persistence by allowing bacteria to resist clearance (4, 5, 11). For NTHI, it has recently become clear that there are bacterial subpopulations that do not form biofilms and yet have the capacity to persist within a planktonic community (14, 19). Beyond and apart from their resistance to clearance, biofilm communities are also associated with chronic infections that evoke a low (albeit continual) level of inflammation, as opposed to acute infections, which are associated with high levels of inflammation. Our recent work showed that adaptations occurring during the biofilm mode of growth (including PCho) are central to the maintenance of a chronic infection and appear to diminish the level of inflammation (14, 41). It is tempting to speculate that populations of NTHI and other bacteria undergo a programmed shift during the formation and maturation of biofilms that leads to the establishment and maintenance of a chronic infection. Understanding how bacteria in these communities, and in planktonic communities, resist clearance will be key to better understanding, and eventual eradication, of chronic airway infections such as otitis media. Because these infections are among the most common and costly public health problems worldwide, this is an important topic for additional study.
Acknowledgments
We acknowledge helpful discussions and suggestions from our colleagues in the WFUHS Biofilm Group. Ken Grant, Bilinda Dawson, and Paula Means provided expert assistance in the microscopy work, which was performed in the WFUHS MicroMed core facility. Luyan Ma and Gina Parise made helpful suggestions for the continuous-flow biofilm systems and microscopy. Gayle Foster and Vadim Ciobanu provided excellent technical assistance.
This work was supported by NIH grants AI054425 and DC007444 (to W.E.S.). S.W.-B. was supported by an individual predoctoral fellowship from NIH (AI61830).
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Bluestone, C. D. 2000. Clinical course, complications and sequelae of acute otitis media. Pediatr. Infect. Dis. J. 19:S37-S46. [DOI] [PubMed] [Google Scholar]
- 2.Bluestone, C. D. 1998. Epidemiology and pathogenesis of chronic suppurative otitis media: implications for prevention and treatment. Int. J. Pediatr. Otorhinolaryngol. 42:207-223. [DOI] [PubMed] [Google Scholar]
- 3.Campagnari, A. A., M. R. Gupta, K. C. Dudas, T. F. Murphy, and M. A. Apicella. 1987. Antigenic diversity of lipooligosaccharides of nontypeable Haemophilus influenzae. Infect. Immun. 55:882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erwin, A. L., Y. A. Brewah, D. A. Couchenour, P. R. Barren, S. J. Burke, G. H. Choi, R. Lathigra, M. S. Hanson, and J. N. Weiser. 2000. Role of lipopolysaccharide phase variation in susceptibility of Haemophilus influenzae to bactericidal immunoglobulin M antibodies in rabbit sera. Infect. Immun. 68:2804-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldenberg, H. B., T. L. McCool, and J. N. Weiser. 2004. Cross-reactivity of human immunoglobulin G2 recognizing phosphorylcholine and evidence for protection against major bacterial pathogens of the human respiratory tract. J. Infect. Dis. 190:1254-1263. [DOI] [PubMed] [Google Scholar]
- 8.Gould, J. M., and J. N. Weiser. 2001. Expression of C-reactive protein in the human respiratory tract. Infect. Immun. 69:1747-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould, J. M., and J. N. Weiser. 2002. The inhibitory effect of C-reactive protein on bacterial phosphorylcholine platelet-activating factor receptor-mediated adherence is blocked by surfactant. J. Infect. Dis. 186:361-371. [DOI] [PubMed] [Google Scholar]
- 10.Greiner, L. L., H. Watanabe, N. J. Phillips, J. Shao, A. Morgan, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2004. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72:4249-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 14.Hong, W., K. M. Mason, J. Jurcisek, L. A. Novotny, L. O. Bakaletz, and W. E. Swords. 2007. Phosphorylcholine decreases early inflammation and promotes establishment of stable biofilm communities of nontypeable Haemophilus influenzae in the chinchilla model for otitis media. Infect. Immun. 75:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphries, H., and N. J. High. 2002. The role of licA phase variation in the pathogenesis of invasive disease by Haemophilus influenzae type b. FEMS Immunol. Med. Microbiol. 34:221-230. [DOI] [PubMed] [Google Scholar]
- 16.Janik, A., E. Juni, and G. Heym. 1976. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 4:71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, P. A., N. A. Samuels, N. J. Phillips, R. S. Munson, J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type B strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 18.Jurcisek, J., and L. O. Bakaletz. 23 February 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed]
- 19.Jurcisek, J., L. Greiner, H. Watanabe, A. Zaleski, M. A. Apicella, and L. O. Bakaletz. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect. Immun. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalcioglu, M. T., B. Durmaz, E. Aktas, O. Ozturan, and R. Durmaz. 2003. Bacteriology of chronic maxillary sinusitis and normal maxillary sinuses: using culture and multiplex polymerase chain reaction. Am. J. Rhinol. 17:143-147. [PubMed] [Google Scholar]
- 21.Lysenko, E., J. C. Richards, A. D. Cox, A. Stewart, A. Martin, M. Kapoor, and J. N. Weiser. 2000. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35:234-245. [DOI] [PubMed] [Google Scholar]
- 22.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mashburn-Warren, L. M., and M. Whiteley. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61:839-846. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, T. F., and M. A. Apicella. 1987. Nontypable Haemophilus influenzae: a review of clinical aspects, surface antigens and the human response to infection. Rev. Infect. Dis. 9:1-15. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, T. F., C. Kirkham, S. Sethi, and A. J. Lesse. 2005. Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol. Med. Microbiol. 44:81-89. [DOI] [PubMed] [Google Scholar]
- 26.Ouahrani-Bettache, S., F. Porte, J. Teyssier, J. P. Liautard, and S. Kohler. 1999. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques 26:620-622. [DOI] [PubMed] [Google Scholar]
- 27.Post, J. C. 2001. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111:2083-2094. [DOI] [PubMed] [Google Scholar]
- 28.Reilly, T. J., D. L. Chance, and A. L. Smith. 1999. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J. Bacteriol. 181:6797-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanderson, A. R., J. G. Leid, and D. Hunsaker. 2006. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope 116:1121-1126. [DOI] [PubMed] [Google Scholar]
- 30.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swords, W. E., B. Buscher, K. Ver Steeg, W. Nichols, A. Preston, J. N. Weiser, B. Gibson, and M. A. Apicella. 2000. Nontypeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells by an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 33.Swords, W. E., D. L. Chance, L. A. Cohn, J. Shao, M. A. Apicella, and A. L. Smith. 2002. Acylation of the lipooligosaccharide of Haemophilus influenzae and colonization: an htrB mutation diminishes the colonization of human airway epithelial cells. Infect. Immun. 70:4661-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swords, W. E., P. A. Jones, and M. A. Apicella. 2003. The lipooligosaccharides of Haemophilus influenzae: an interesting assortment of characters. J. Endotoxin Res. 9:131-144. [DOI] [PubMed] [Google Scholar]
- 35.Swords, W. E., M. R. Ketterer, J. Shao, C. A. Campbell, J. N. Weiser, and M. A. Apicella. 2001. Binding of the nontypeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signaling. Cell. Microbiol. 3:525-536. [DOI] [PubMed] [Google Scholar]
- 36.Swords, W. E., M. L. Moore, L. Godzicki, G. Bukofzer, M. J. Mitten, and J. VonCannon. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiser, J. N. 2000. The generation of diversity by Haemophilus influenzae. Trends Microbiol. 8:433-435. [DOI] [PubMed] [Google Scholar]
- 38.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 39.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West-Barnette, S. L., A. Rockel, and W. E. Swords. 2006. Biofilm growth increases phosphorylcholine content and decreases potency of nontypeable Haemophilus influenzae endotoxins. Infect. Immun. 74:1828-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, S. M., and B. J. Akerley. 2005. Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol. Microbiol. 55:724-738. [DOI] [PubMed] [Google Scholar]