The 4th ASM Conference on Biofilms (Quebec City, Quebec, Canada, 25 to 29 March 2007) maintained the format so valued at past conferences (keynote talks, invited speakers, talks selected from abstracts, evening specialty sessions, and a hands-on workshop) while increasing scientific diversity by delegating the selection of invited speakers and of talks from abstracts to session chairs appointed by the organizing committee. In particular, the committee targeted the underrepresentation of research on medically relevant biofilms other than those of Pseudomonas aeruginosa and sought to increase the visibility of clinical aspects of biofilm-based disease research; the fruits of these efforts will become apparent in the descriptions of the sessions that follow. Also, an effort was made to increase participation by non-U.S. investigators. At the 3rd Biofilms Conference, also held in Canada, 34% of attendees came from outside the United States. At the Quebec City meeting, 53% were non-U.S. delegates who hailed from the United Kingdom/Europe (22%), Canada (11%), Scandinavia (8%), Asia (4%), Australia/New Zealand (2%), and India, Central and South America, Israel, and Africa (6%). The number of attendees (ca. 600) and the number of submitted abstracts (ca. 400) did not change from the previous meeting, which suggests that, after three meetings with steady growth, the target population is now well represented at the meeting.
While the major questions and areas of investigation have not changed significantly since the earlier meetings, new perspectives are emerging. First is the recognition that Pseudomonas aeruginosa, while a superb model organism for numerous reasons, is only one of many bacteria important to theoretical as well as applied aspects of biofilm research; other model organisms (e.g., Vibrio spp., Bacillus spp., oral bacteria, and staphylococci) are now being investigated as natural and fitting alternatives to P. aeruginosa. Second, while flow cell work with monocultures grown in laboratory media will continue to provide critical baseline data and continue to be especially useful in examining theoretical questions, it is becoming clear that biofilm behavior and physiology need to be studied in a manner that reflects the natural environment, whether within the human lung or on a soil particle. Experimental systems have therefore become more sophisticated and employ environmentally relevant substrata and media; accordingly, in vivo sampling and experimentation are becoming more common. Last, moving to the fore is the recognition that the vast majority of bacteria, including many of those involved in human disease, must associate with other genera of bacteria as part of their daily existence: multispecies communities are becoming a targeted research area.
This review summarizes the individual sessions through their platform talks, many of which highlighted work presented in greater detail as poster presentations. We hope these descriptions (and the accompanying references, which give a flavor of the topics) will convey the overall high scientific quality not only of the invited talks but also of the talks selected from submitted abstracts. The consensus of the attendees was that this meeting elevated the already high reputation of this conference series. We hope this review will convey some of that feeling to those who have yet to attend the conference.
KEYNOTE TALK 1
E. Peter Greenberg (University of Washington) opened the meeting by speaking on the sociomicrobiology of biofilms: the application to biofilm microbiology of E. O. Wilson's insect society-derived sociobiological concepts. In biofilms, bacteria live in close proximity, the populations are heterogeneous in activity, and individuals exhibit distinct behaviors, all of which can be studied from an ethological perspective. Biofilms offer a unique opportunity to examine the influence of environmental manipulation on gene expression and heritability (45). Greenberg focused on the role of iron in biofilm formation. Iron is frequently a limiting nutrient in the human body as well as in the natural environment, and motility and biofilm initiation in Pseudomonas aeruginosa are dependent on iron (4). The human immune effector lactoferrin sequesters iron; in the presence of lactoferrin (i.e., at low levels of available iron), P. aeruginosa maintains twitching motility, refuses to become sessile, and is thus dramatically attenuated in biofilm formation (58). Production of lactoferrin is seen as a way for the host immune system to slow the rate of biofilm formation so that the microbial intruders can be dealt with before reaching a clinically relevant population density. Greenberg continued this theme by showing that the iron chelator EDTA can disperse a P. aeruginosa biofilm (3), and that citrate as a carbon source can result in a flat biofilm because of citrate's iron chelation capability. Lastly he introduced the use of the iron surrogate gallium as a biofilm inhibitor, a subject explored in greater detail by University of Washington colleagues (28).
DEVELOPMENT AND PHYSIOLOGY
The opening session was chaired by George O'Toole (Dartmouth College) and Paula Watnick (Tufts—New England Medical Center). The chairs put together a set of talks that illustrated not only the diversity but also the commonality in the mechanisms of biofilm development displayed by seven different biofilm-forming species. Roberto Kolter (Harvard University) presented evidence that differentiation within Bacillus subtilis biofilm populations gives rise to six specialist cell types. Each specialist has a defined function, such as matrix building, propagation, or swimming. Time lapse imaging of fluorescent reporter constructs implied that the cells exhibit only one phenotype at a time, although some cells could switch between phenotypes. Kolter suggested that, depending on the local conditions, a certain probability exists that any one cell would switch. Also discussed in the session were various cell-signaling pathways and environmental triggers that result in the modification of biofilm extracellular polymeric substances (EPS) in response to nutrient conditions. These include the release of DNA to the matrix in response to iron starvation in P. aeruginosa, presented by Liang Yang (Danish Technical University, Lyngby, Denmark) (74), and the production of LapA adhesion protein in response to phosphate limitation via a cyclic di-GMP (c-di-GMP) signaling pathway in Pseudomonas fluorescens, presented by Russell Monds (Dartmouth College) (42). Co-chair Watnick showed that Vibrio cholerae knockout mutants in the sugar phosphotransferase system can form a biofilm when grown on glucose but not when grown on mannose. Glucose excess results in the synthesis of EPS rather than of glycogen, whereas under glucose limitation, glycogen is formed and EPS are degraded, thereby releasing the biofilm from the substratum. Carol Kumamoto (Tufts University) used a simple but elegant agar system to show that the switch to an invasive, filamentous phenotype via mitogen-activated-protein kinase sensing in Candida albicans is a response to surface contact, not to oxygen concentration (35). Daniel Verhamme (University of Dundee) presented evidence that the transcription factor DegU regulates more biofilm properties in B. subtilis than previously thought, ranging from attachment and social behavior to colony architecture. Karin Sauer (Binghamton University) identified two phosphorylated proteins, Bfl1 and Bfl2, which appear to regulate the early and late biofilm maturation process; this observation strengthens the hypothesis that biofilm formation by P. aeruginosa occurs by a staged developmental process. Using green fluorescent protein (GFP) reporter fusions and confocal microscopy, Olivier Brun (University of Minnesota) identified biofilm-specific promoters in Mycobacterium marinum; a putative attachment promoter is expressed at the base of the biofilm. Yves Brun (Indiana University) showed how attachment forces could be accurately measured through micromanipulation: the bending of a micropipette in response to a controlled pulling via an attached bacterial cell (66). Using this technique it was shown that Caulobacter crescentus first attached by the flagella and then rotated until pilus tethers stopped the rotation long enough for the organism to create a holdfast.
BIOFILM-HUMAN INTERACTIONS I
In the first of two sessions designed to emphasize the relationships of bacteria to the human host, session chairs were Friedrich Götz (Tübingen University) and Ute Römling (Karolinska Institute). Götz described the role of polysaccharide intercellular adhesin (PIA) in biofilm formation by Staphylococcus epidermidis and Staphylococcus aureus (17). Mutants that do not make PIA are defective in adherence and biofilm formation, and these mutants are much less virulent than the parent strains in animal models of infection. Homologous polysaccharides are found in several other bacteria; thus, this molecular architecture is conserved across bacterial phylogeny. Biofilms of S. aureus and S. epidermidis have transcriptome (50, 51) and proteome profiles that differ from those of planktonic cells with respect to murein and PIA synthesis as well as the physiology of ammonia and acid production. Phage release differs also (49). Holgar Rohde (University of Hamburg) spoke on non-PIA-mediated biofilm formation in S. epidermidis. He discussed the role of embp (extracellular matrix binding protein) in PIA-negative biofilm formation. This newly discovered protein was identified and shown to be crucial for biofilm formation in clinically important PIA-negative S. epidermidis isolates through transposon mutagenesis and subsequent screening for biofilm-negative mutants in a PIA-negative background strain. Fragments of recombinant embp induce cell aggregation and biofilm formation in embp-negative S. epidermidis strains, and the fragments also influence binding to host cells as well as to host components such as fibronectin, findings that underscore the role of the protein in pathogenesis. The protein is broadly distributed in S. epidermidis strains and thus may be central to the colonization of host tissues.
Paul Stoodley (Center for Genomic Sciences, Pittsburgh, PA) presented work on in vivo imaging of bacterial biofilms in infections. A combination of fluorescence in situ hybridization (FISH), antibodies, and general stains was used to show biofilms associated with infected sutures and an infected arthroplastic remodeling (62), as well as in mucosal epithelia from upper respiratory tract infections (adenoids and middle ear mucosae) in clinical specimens (Fig. 1) and the chinchilla model of otitis media (18). Several of these cases were culture negative even though symptoms of bacterial infection were presented. Despite the success in demonstrating biofilms, Stoodley cautioned that biofilm imaging as a routine clinical diagnosis practice is a long way from acceptance. Ed Swords (Wake Forest University) expanded on biofilm formation in the chinchilla model of otitis media by describing the relationship between phosphorylcholine (PCho) modification of lipooligosaccharides in nontypeable Haemophilus influenzae (72) and the infection. A mutant that does not add PCho formed biofilms of lower density than did the parent strain and also caused increased inflammation levels in the animal. Thus, it seems that PCho modification results in tolerance of the infection by the animal (reduced initial inflammation) as well as in stable growth and maturation of the biofilm.
FIG. 1.
Pediatric adenoid-associated biofilm that meets the four criteria suggested by Parsek and Singh (45a) for the determination of a biofilm: (i) the pathogenic bacteria are surface associated or adherent to a substratum; (ii) direct examination reveals bacteria in clusters and encased in a matrix of bacterial or host constituents; (iii) the infection is localized; (iv) the infection is resistant to antibiotic therapy. The first, second, and third criteria were determined by microscopic examination. The fourth criterion was fulfilled by empirical evidence: unsuccessful resolution with antibiotic treatment. The fresh specimen was stained with the nucleic acid stain Syto 9 (red), which stained bacterial cocci (black arrows) as well as host nuclei (white arrows). Less defined nuclei are deeper in the tissue. The biofilm EPS was stained with lectins (green), and the surface of the adenoid (blue) was imaged using reflected confocal microscopy. Scale symbol (black), 10 μm in each dimension. Image courtesy of Laura Nistico, Paul Stoodley, and Luanne Hall-Stoodley (Center for Genomic Sciences, Pittsburgh, PA).
Several talks focused on P. aeruginosa and cystic fibrosis, both from the laboratory perspective and through examination in vivo. The development of a tissue culture model system for the study of interactions between P. aeruginosa and cystic fibrosis-affected human airway epithelial cells was described by Greg Anderson (Dartmouth College). Addition of the bacteria to confluent eukaryotic-cell cultures caused detachment of the eukaryotic cells within a few hours. Detachment could be greatly reduced by the presence of tobramycin in the medium; although the number of bacteria was reduced, the system retained some bacteria despite the antibiotic addition. The toxicity of nitrite and its derivatives (e.g., nitric oxide) to anaerobically grown P. aeruginosa biofilms was discussed by Dan Hassett (University of Cincinnati). He showed that mucA mutants (the mucoid strains that predominate in cystic fibrosis lung infections) are extremely sensitive to NO at a pH of 6.5, the approximate pH of lung airway mucus according to his measurements in lungs recently removed from transplant patients; however, he found limited NO toxicity at a pH of 7 or above. He further related the toxicity to inefficient nitrite reductase and nitric oxide reductase activities in mucA mutants, and he suggested that nitrite or NO could be useful as a therapeutic agent for cystic fibrosis patients (76) if the bacteria in situ are growing anaerobically. Janus Haagensen (Danish Technical University) used FISH to show the presence of P. aeruginosa cell aggregates within the sputa of cystic fibrosis patients who had coinfections with mucoid and nonmucoid strains; however, these aggregates were absent from the sputa of patients infected solely with nonmucoid strains. Further, aggregates were shown to contain other bacteria in addition to P. aeruginosa; antibiotic treatment of the patients resulted in reductions in mucoidy and aggregation. Niels Høiby (University of Copenhagen) presented new ideas on P. aeruginosa infection that emphasized the focal nature of the infection and the importance of understanding the role of oxygen tension in the lung in bacterial growth and subsequent tissue destruction. On the basis of autopsy and sputum specimens, he described how the infection compartmentalizes in the lung. In the respiratory zone, where oxygen tension is high relative to that in the bronchi, no thick mucous layer is present but mucoid P. aeruginosa colonies are found surrounded by polymorphonuclear leukocytes (PMN); inflammation and tissue destruction occur in this zone, and Høiby believes this is where the infection has its true genesis. These mucoid colonies, together with destroyed lung tissue, are then transported by coughing to the bronchi (conductive zone), where they become embedded in the naturally occurring thick mucous layer. The mucous layer creates steep oxygen gradients; single P. aeruginosa cells, as well as mucoid colonies, can be found in the mucus.
Co-chair Römling spoke on second-messenger c-di-GMP signaling and phenotype switching (25, 57). Low levels of c-di-GMP encourage motility (twitching and swimming), whereas high levels encourage a sessile phenotype through production of exopolymers and extracellular appendages. GGDEF/EAL domain proteins have phosphodiesterase as well as guanylate cyclase activity and thus regulate c-di-GMP levels; these proteins are widespread in bacteria, which emphasizes the universality of this regulatory mechanism. In Salmonella enterica serovar Typhimurium, which has 20 GGDEF/EAL proteins, the “rdar” (red, dry, and rough) phenotype is controlled by c-di-GMP levels. This phenotype is characterized by a rough colony morphology, red colonies, and clumpy, adherent cells. Expression of the transcriptional regulator cgsD, which is controlled by environmental signals (such as temperature) as well as by the c-di-GMP level, results in production of the rdar phenotype through synthesis of curli as well as by indirect activation of cellulose exopolymer synthesis; csgD knockouts have a smooth colony morphology and are nonadherent and nonclumping. AdrA is one of four GGDEF/EAL proteins that influence the transcription of cgsD. These c-di-GMP “networks” have been shown to be crucial to the development of a biofilm phenotype in several bacteria. Paul Rainey's talk, discussed later in this paper, explored c-di-GMP networks in an evolutionary context.
BIOFILMS IN NATURAL ENVIRONMENTS
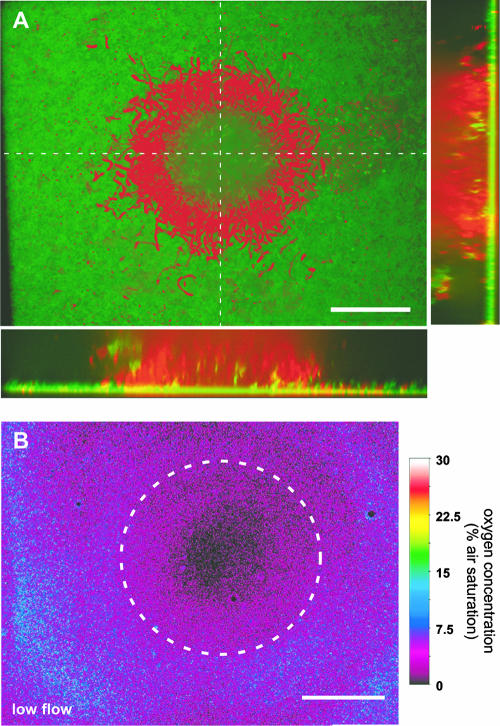
Michael Kühl (University of Copenhagen) and Steven Lindow (University of California—Berkeley) were the chairs of a session designed to examine the structure and function of biofilms in terrestrial and marine ecosystems, including bacteria as commensals or pathogens of plant or animal hosts. Kühl spoke on spatiotemporally resolved techniques for imaging chemical species, particularly oxygen, within microbial mats and corals. Much of this work employs films that contain a photochemically active sensor species (21), which reports the concentration of the soluble target species (Fig. 2). Lifetime images are produced that describe how a chemical parameter changes in response to changes in the environment. Examples include changes in photosynthetic O2 production as a function of the light level (33) or changes in oxygen concentration as a function of the flow rate in a bacterial biofilm (34).
FIG. 2.
Combined microscopic imaging of oxygen concentration and bacterial colonization within a biofilm reprinted from reference 34 with permission. (A) Projection and vertical slices through a confocal image stack of a biofilm structure consisting of GFP-marked Pseudomonas putida (green) and heterotrophic bacteria labeled with Syto 60 (red) imaged through an O2 coverslip-sensor mounted on top of a flow chamber. (B) Corresponding O2 distribution at the base of the biofilm. The central cell cluster is outlined by the dashed curve. Scale bars, 40 μm.
Two talks on Bahamian marine stromatolithic mats highlighted interesting properties of this system. Chris Dupraz (University of Lausanne) presented data on extracellular polysaccharide inhibition of biogenic calcium carbonate formation (16). When mineralization does occur, changes in the local pH result in the formation of differently shaped carbonate crystals, thus providing a snapshot of the pH at that site in the biofilm. Alan Decho (University of South Carolina) discussed layering and microbial activity in the same biofilms. Microautoradiographic data together with FISH studies show that in this system, in sharp contrast to the spatial organization in other mat systems, active sulfate reduction occurs at the very surface of the biofilm (6). Decho suggested that this may be set up through homoserine lactone signaling.
Plant-associated biofilms were introduced by Clay Fuqua (Indiana University), who related inorganic phosphate (Pi) deprivation to enhanced biofilm formation in Agrobacterium tumefaciens. Pi is frequently limiting in the environment; the phosphate response regulator protein PhoB is activated during phosphate starvation, and in vitro, phosphate-starved A. tumifaciens biofilms have an elevated biomass relative to that of normal biofilms. When phoB is induced, the cells bind to substrata using their poles, and they also show wheat germ agglutinin binding at their poles. A transposon mutant selected for the lack of polar binding is not recognized by wheat germ agglutinin, and the mutated gene bears similarity to a gene in the holdfast biosynthesis pathway of Caulobacter crescentus. In A. tumefaciens, low Pi levels may be a cue for the production of a specialized polymer and thus for biofilm initiation. Co-chair Steve Lindow continued the theme of plant pathogen biofilms in his talk on Xylella fastidiosa (43). This bacterium infects plant xylem vessels, which then become blocked; the densely packed biofilms in blocked vessels have restricted access to nutrients and thus have very low activity, as assessed by propidium iodide staining. Solitary bacteria in unblocked vessels can move from vessel to vessel. Bacterial biomass and the level of the diffusible signal factor DSF (the product of rpfF) are low early in infection, and movement from vessel to vessel (biofilm metastasis) is encouraged. Late in infection, biomass and signal levels increase and cue polymer production, with resultant vessel blockage. Lindow proposed to treat infection by introducing signal-hyperproducing strains to push early infections toward the late-stage phenotype, thereby sacrificing a few xylem vessels but concomitantly inhibiting the spread of the infection and saving the plant. Susanne von Bodman (University of Connecticut) also spoke on a plant xylem pathogen: Pantoea stewartii (31). Cell density-dependent synthesis of EPS is required for virulent biofilm formation; mutants with mutations in the esaI and esaR quorum-sensing loci form less-dense biofilms and do not disperse effectively in planta. The bacterium was shown to specifically colonize annular rings and spiral wall thickenings within the xylem.
Black band disease in corals was discussed by Laurie Richardson (Florida International University). Comparison of the bacterial community from diseased coral with that from healthy coral (54) has shown that the migration of certain bacteria, in particular that of Beggiatoa spp., produces sharp gradients of sulfide and oxygen that kill the coral. Further, toxin-producing cyanobacteria have been identified from the community and may be responsible for regional differences in the disease. Dhana Rao (University of the South Pacific, Fiji) spoke on the interaction between Pseudoalteromonas tunicata and Roseobacter gallaeciensis, two surface colonizers of the marine alga Ulva australis (47). P. tunicata can outcompete other natural community members when grown as a biofilm in natural seawater, but R. gallaeciensis outcompetes P. tunicata under the same circumstances. Likewise, P. tunicata cannot invade an extant biofilm composed of other species, but R. gallaeciensis can invade and eventually comes to dominate such a biofilm. Interestingly, P. tunicata is unable to persist in sterile-filtered seawater.
Two talks on biofilms in terrestrial systems highlighted the importance of water availability and of local chemistry. Water potential is critical to soil biofilm development, and Larry Halverson (Iowa State University) showed that production of the extracellular polysaccharide alginate is upregulated in Pseudomonas putida biofilms under matric water stress but not under osmotic stress. Biofilms with reduced alginate levels are less dense and flatter. Alginate seems to protect against desiccation by maintaining a hydrated environment for the cells (11, 12, 70). Ryan Hunter (University of Guelph) showed that in an in vitro P. aeruginosa biofilm, precipitated iron is heterogeneously distributed. This may be correlated with the local pH, and the bacterium alters its lipopolysaccharide chemistry in response to pH (22, 23). Less precipitate was found associated with metabolically active cells than with carbonyl cyanide m-chlorophenylhydrazone (CCCP)-inhibited cells; protons transported to the cell surface by respiration appear to compete with metal cations for reactive cell surface sites.
CROSS-KINGDOM INTERACTIONS IN BIOFILMS
Leo Eberl (University of Zurich) chaired a session on soluble signals and interactions of bacteria with eukaryotes. Carsten Matz (Helmholtz Center for Infection Research, Braunschweig, Germany) spoke on protozoan grazing effects on biofilms (38, 39). Grazing by amoebae induces changes in bacterial gene regulation, e.g., rhamnolipid production is increased. Bacterial defense mechanisms against grazing include microcolony formation (protective shielding) and the production of soluble inhibitors of amoebal growth.
Soluble signaling molecules were the theme of several talks, and in a second example of cross-kingdom inhibition, Bastiaan Krom (University Medical Center, Groningen, The Netherlands) showed that homoserine lactone from P. aeruginosa inhibits hyphal formation in Candida albicans. Further, breakdown products of homoserine lactone inhibit germ tube formation. C12 and C14 homoserine lactones had the greatest effect. In an example of beneficial cross-kingdom signaling, Miguel Cámara (University of Nottingham) discussed how homoserine lactone signaling molecules produced by biofilms influence the attachment of Ulva linza (marine alga) zoospores (63). Using lactones synthesized by Vibrio anguillarum as model compounds, he showed that spore attachment increases with increasing lactone concentration. Using natural bacterial isolates, three patterns of increasing zoospore attachment were seen as bacterial numbers increased: a linear relationship, an exponential relationship, and a bell-shaped curve. These results suggest a role in nature for this signaling mechanism.
Two talks addressed cross-kingdom interactions in pathogenesis. Costi Sifri (University of Virginia Health System) discussed the nematode Caenorhabditis elegans as a model organism for bacterial infection (56). Virulence is measured as the reduction in the mean lifetime of the worm; S. aureus colonizes the alimentary tract, and known virulence factors such as PIA have been demonstrated to apply to the worm. Coagulase-negative staphylococci have also been shown to be virulent in this model, and PIA-negative mutants do not kill as effectively as the parent strain. Thomas Rasmussen (Chr. Hansen A/S, Hørnsholm, Denmark) talked about quorum sensing in Pseudomonas aeruginosa biofilms and how it affects interactions between the biofilm and PMN. Wild-type P. aeruginosa biofilm supernatants lyse PMN, but supernatants of quorum-sensing mutant biofilms do not. Microarray analysis of wild-type biofilms exposed to PMN showed upregulation of PQS (Pseudomonas quinolone signal), phenazine, phospholipase, elastase, and rhamnolipid synthesis genes, whereas quorum-sensing mutant biofilms upregulated catalase. These results suggest that wild-type biofilms mount an aggressive attack on PMN and that mutant biofilms respond to the PMN oxidative burst by shielding themselves with catalase.
EVOLUTION AND DIVERSITY
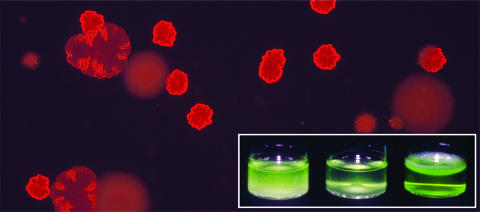
Paul Rainey (Massey University, New Zealand) chaired the session on evolution and diversity and gave the opening talk, presenting biofilm development in Pseudomonas fluorescens as a process of mutation and subsequent selection. Numerous mutations that generate biofilm-forming genotypes were identified; the majority occurred within negative regulators of various diguanylate cyclases. The net effect was to reduce negative regulator function, with concomitant overproduction of c-di-GMP. In turn, enzymes involved in the production of a cellulose-based polymer were activated, causing cells not only to remain attached after cell division but also to generate a mat of polymer-linked cells at the air-liquid interface in broth microcosms. Continued selection of mat-forming genotypes (known “wrinkly spreaders” [Fig. 3]) resulted in the evolution of genotypes that no longer overproduced the polymer but could still grow within the mat. These selfish (cheating) genotypes did not contribute to mat strength or integrity and thus caused the mat to collapse. Molecular analyses of cheaters revealed a range of different mutations whose net effect is to abolish the activity of the previously activated diguanylate cyclases. The process can continue; mat-forming (cooperating) genotypes can once again evolve from the cheating types (5). Thus, diversity and development in biofilms may occur simply by the environment selecting for “standard” mutations, not through a hard-wired developmental cycle particular to the bacterium.
FIG. 3.
Colony variants—including two different wrinkly spreader genotypes—of P. fluorescens that arise during selection in spatially structured microcosms. (Inset) Niches occupied by different variants. Wrinkly spreader genotypes form a biofilm at the air-liquid interface (culture at right). Image courtesy of Paul Rainey (Massey University, New Zealand).
John Roth (University of California—Davis) addressed the question of whether stress (such as could occur during biofilm growth) increases mutation frequency. Problems in defining exactly how a mutation arises and in accurate counting of mutations have left this question unresolved for decades, and studies of mutations in biofilms underscore the uncertainty. Roth presented theoretical arguments against the existence of a “hypermutable” state under any circumstances, as well as an alternative interpretation of the data from the best-accepted example of hypermutation (10) whereby the observations could be due to gene duplication (32). Roth believes that selection is more important than the mutation rate.
Pradeep Singh (University of Washington) described experiments that suggest an increase in the occurrence of genetic/phenotypic variants in biofilm systems (8). Singh showed that these variants are generated by error-prone repair (recA and recBC mutants) of double-stranded breaks in DNA arising from oxidative stress; addition of an antioxidant to the biofilm reduces the occurrence of variants. Once the variants appear, they are selected for and come to dominate the biofilm. Continuing the theme of variants, Alfred Spormann (Stanford University) argued that the stability of Vibrio cholerae biofilms is related to the appearance of small-colony variants (SCV). He described how these biofilms fall apart after the appearance of the SCV and how the SCV can be sorted into four classes based on motility. Characterization of SCV physiology is now under way using 2,000-parameter Biolog plates, and Spormann argued for an increased emphasis on physiology in biofilm experiments.
Jaione Valle (Public University of Navarre, Pamplona, Spain) concluded the session by describing how insertion sequence IS256 mediates phenotypic switching in Staphylococcus aureus (69). Knockouts of transcription factor σB have increased IS256 activity and produced biofilm-negative variants with dramatically higher IS256 copy numbers in the chromosome at a high frequency. Osmotic stress can complement the lack of σB and restore the wild-type phase variation frequency, suggesting that σB is not directly involved. σB appears to act as a regulator of IS256 activity and thereby as a controller of biofilm-negative phenotype occurrence.
KEYNOTE TALK 2
Ivan Matic (University of Paris) continued the discussion of genetic variation in biofilms from the perspective of mutation and repair. Observed mutation rates are typically low because most mutations are deleterious or neutral and because the cell has extensive investments in repair mechanisms. However, populations of natural isolates contain strong mutators that display mutation rates 100-fold higher than those of laboratory strains; these mutators are deficient in repair (15). They can be selected for in gnotobiotic mice by sequential multiple-antibiotic treatments; the surviving cells are mutators. Also present in natural populations are weak mutators whose mutation rates are 4 to 5 times those of laboratory strains. The difference in the mutation rate is a by-product of selection for improved fitness, and mutation rates can be very different in the host environment than in the laboratory.
THE SLIME MATRIX
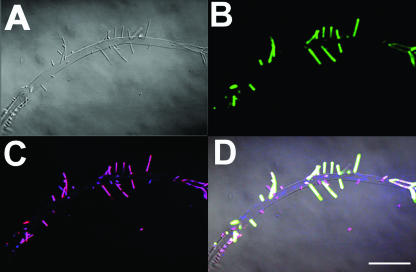
Tony Romeo (Emory University) chaired the session focusing on the regulation, composition, and function of EPS. The characterization of EPS chemistry and function has remained elusive even though EPS is a hallmark that distinguishes biofilm populations from planktonic cultures. Romeo discussed the regulation of poly-β1,6-N-acetyl-d-glucosamine (PGA), which is a common component of EPS in gram-positive as well as gram-negative bacteria. Romeo discussed how the carbon storage regulator (Csr) RNA-binding protein, which represses PGA and biofilm formation, can be sequestered by the noncoding short RNAs csrB and csrC (2). In turn, csrB and csrC can be degraded by csrD. Romeo concluded that this regulatory network was adapted for fine-tuning homeostatic adjustment rather than being an “on-off” system. Matt Parsek (University of Washington) discussed the contribution of the EPS genes pel and psl in P. aeruginosa colony morphology variants isolated from in vitro biofilm systems. Similar colony morphology variants were isolated from cystic fibrosis patients; thus, the in vitro system may reflect certain in vivo conditions. By knocking out c-di-GMP in a sticky mutant, the wild-type morphology was recovered; c-diGMP signaling was important in directing adhesion and cohesion (30). Jean-Marc Ghigo (Pasteur Institute, Paris, France) presented evidence for secretion of valine into EPS. This secretion acts not only as a “carbon dump” but also to provide an antimicrobial effect against invading planktonic bacteria, which have been shown to be more sensitive than biofilm cells to valine. Per Nielsen (Aalborg University, Aalborg, Denmark) introduced evidence for the presence of amyloid protein (originating from fimbriae) as a ubiquitous component of EPS produced by many different species and found in freshwater, brackish-water, and seawater environments. Nielsen used antibodies and FISH (Fig. 4) to demonstrate that this previously overlooked protein is produced by as many as 50% of the species in a given environment. Sarah Schooling (University of Guelph) concluded the session by presenting evidence that membrane vesicles are an important component of P. aeruginosa biofilm EPS (53). DNA from vesicles was estimated to make up at least 0.33% of the extracellular DNA. Vesicle production may play a significant role in EPS function for biofilms. The data presented in this session, the lively discussion at the specialty evening session on EPS chaired by Hans-Curt Flemming and Dan Wozniak, and the final keynote address by Bill Costerton on structured EPS illustrate how EPS is no longer seen as an amorphous carbohydrate material that serves simply to provide a physical matrix for cells but rather as a chemically complex mechanically and structurally adaptive material that has multiple biological functions.
FIG. 4.
Detection of amyloids in a brackish-water biofilm, with simultaneous labeling of amyloid adhesins by antibodies and identification of bacteria by FISH. (A) Transmitted light image showing small rods attached to an empty sheath, presumably from a cyanobacterium (judging from the morphology and the ability to produce red fluorescent phycoerythrin). (B) Epifluorescence image of the same field of view as that in panel A. Green fluorescence marks antibodies raised against conformational epitopes of amyloid adhesins. (C) Epifluorescence image of the same field of view as that in panel A. Shown are results of FISH using the red fluorescent oligonucleotide probe CFB719, targeting the class Bacteriodetes, and blue fluorescent EUBmix, targeting most members of the domain Bacteria. (D) Overlay of panels A, B, and C. Colocalization of the three fluorescent probes shows as white. Scale bar, 20 μm. Image courtesy of P. Larsen, J. L. Nielsen, D. Otzen, and P. H. Nielsen (Aalborg University, Aalborg, Denmark).
CELL SIGNALING IN BIOFILMS
A function for cell signaling in a biofilm context was first described for P. aeruginosa in 1998 (14). The concept that a sessile bacterial population could coordinate its phenotype and organization through quorum-sensing mechanisms quickly captured the imagination, and it was thought that signaling might be a magic bullet for the control of biofilms. Since then, the field has gained in complexity, because the diversity of signal molecules and mechanisms has grown. Matt Parsek, who was involved in the earliest biofilm signaling work, chaired the session, which included examples of signaling in three different experimental systems. Susanne Häussler (Helmholtz Center for Infection Research, Braunschweig, Germany) discussed PQS in P. aeruginosa. She presented data extending the role of PQS from iron chelation (9) to the release of DNA to the EPS matrix in response to stress. Häussler suggested that PQS is a regulator of coordinated dispersal (movement of bacterial cells from the biofilm to the bulk liquid), because of its promotion of detergent tolerance. Deborah Hogan (Dartmouth College) presented an example of competitive survival of two organisms that involves response to, as well as interference with, each other's signaling systems. P. aeruginosa and C. albicans are commonly isolated in cases of opportunistic infections (71), and in coculture, P. aeruginosa can colonize and kill C. albicans hyphae through the production of virulence factors, such as the fungicide pyocyanin (29), which are regulated by the Las/Rhl signaling system. However, the production by P. aeruginosa of its virulence factor-stimulating signaling molecule OdDHL [N-(3-oxododecanoyl)-l-homoserine lactone] causes C. albicans to switch its morphotype to the colonization-resistant but host cell-invasive yeast form. In addition to this defensive response, C. albicans mounts an offensive against P. aeruginosa through the production of farnesol. Farnesol, a signal for C. albicans to block the formation of hyphae, also depresses PQS signaling and pyocyanin production in P. aeruginosa. In this cross-kingdom interaction, signals coordinate the phenotype of the producing organism while at the same time altering the phenotype of the competitor. Cell signaling in the social bacterium Myxococcus xanthus was discussed by Heidi Kaplan (University of Texas Medical School). She presented evidence for a quorum-sensing system sensitive to membrane integrity. M. xanthus responded to an accumulation of lysed cell wall components, such as peptidoglycan, in the culture by initiating sporulation. Intriguingly, this recognition and response to damage in the population is not unlike the “danger model” of the mammalian immune system response (40), in which recognition of cell damage is the immune activator. It remains to be seen how widespread sensing and response to self-damage is among prokaryotes. Finally, Jan Kreft (University of Bonn) presented modeling evidence that cell-signaling mechanisms in gram-negative bacteria may be adapted to regulate behavior over the small distances between adjacent cells and within small clusters (19). Based on the observation that the diversity of signals in gram-negative species is small (primarily acetyl-homoserine lactones) but that a large number of species respond to these molecules, Kreft hypothesized that signaling in biofilms is local. Signals are directed toward nearest-neighbor, closely related cells (i.e., toward clones), rather than toward spatially distant and therefore, presumably, less closely related cells. Kreft's modeling suggests that current assumptions regarding the distance over which signaling operates may need to be reevaluated.
BIOFILM-HUMAN INTERACTIONS II
The second human microbiology session was chaired by Michael Otto (NIH Rocky Mountain Labs) and David Stickler (University of Cardiff). The first three talks focused on biofilms in the urinary tract. Stickler discussed Proteus mirabilis biofilm formation in urinary catheters and the resultant struvite crystallization (61). Multispecies biofilms attenuated crystallization by modifying the pH; patient urine chemistry and biofilm species composition were important factors in the rate of catheter blockage. Barbara Trautner (Baylor College of Medicine) then presented evidence that biofilm infection of urinary catheters could be reduced by a probiotic approach: seeding the catheter with avirulent urinary tract isolates of Escherichia coli (64, 65). Seeding with these E. coli isolates reduced the incidence of symptomatic infection nearly 20-fold: from 2.7 per 100 catheter days to 0.15 per 100 catheter days. Control of device-related biofilm infections is perhaps best addressed not by attempting to render the surface sterile, which has proved extremely challenging and is possibly futile, but rather by prophylactic inoculations. Trautner is currently designing E. coli strains through genetic modification that will be even more efficient at “competitive exclusion.” Mark Schembri (University of Queensland, Brisbane, Australia) discussed the role of antigen 43 as an important adhesin and virulence factor for uropathogenic E. coli in a mouse model (68). Adhesins such as antigen 43 may be useful targets for modification to create probiotic E. coli strains.
The next two presentations were on a rapidly growing area of interest in medical biofilms: chronic wound infections. Klaus Kirketerp-Møller (Bispebjerg University Hospital, Copenhagen, Denmark) presented evidence from clinical specimens suggesting that the persistence of P. aeruginosa in inflamed chronic wound infections was due to its protected location within microcolonies, where it remained impervious to, and could kill, leukocytes. Garth James (Montana State University) used confocal microscopy and denaturing gradient gel electrophoresis to show the broad diversity of infecting bacteria, even in similar wound types. Importantly, 16S rRNA analysis showed that, in addition to the routinely cultured aerobic and facultatively anaerobic species, a number of uncultured obligate anaerobes were present. James hypothesized that strict anaerobes may inhabit anaerobic microniches created through oxygen consumption by aerobic biofilms.
Session co-chair Michael Otto then gave the first of two talks on the molecular pathogenesis of staphylococcal infections. He showed that spontaneous mutation in the biofilm-repressing agr locus causes S. epidermidis to form different biofilm phenotypes. The difference in architecture between the heterogeneous wild type and the thicker but flatter agr mutant was caused by the phenol-soluble modulin (PSM) (75). GFP reporters of PSM synthesis, together with exogenous PSM addition, suggest that PSM is responsible for controlled detachment, possibly through surfactant properties. Anna Muench (University of Maryland Dental School) used microarrays to identify genes unique to clinical isolates of S. aureus. Differential expression showed that approximately 50% of the genes were unique to clinical strains, that many of these genes were involved in stress response and the production of virulence factors, and that they were upregulated in biofilms. In contrast, only 10% of genes unique to laboratory strains were upregulated in biofilms of laboratory strains. These data indicate that continued laboratory passage of clinical strains may cause a loss of virulence, because no selective pressure exists to maintain virulence genes, and that many of these genes are important in the context of biofilms.
In addition to the infectious disease-related talks, two talks on oral biofilms were given. Rob Palmer (NIDCR, NIH) used immunofluorescence confocal microscopy and enterobacterial repetitive intergenic consensus PCR to investigate phenotypic and genotypic diversity within a Veillonella sp. in natural dental plaque biofilms from human volunteers. In as little as 4 h, changes in coaggregation, antibody reactivity, and enterobacterial repetitive intergenic consensus fingerprinting patterns occurred (44). Palmer concluded that the only way to fully understand the complexity of the time-dependent microdiversity present in dental biofilms was through a polyphasic approach. Mary Ellen Davey (Forsyth Institute, Harvard University) discussed the role of capsule expression and biofilm formation in Porphyromonas gingivalis. Interestingly, Davey found that encapsulated P. gingivalis strains did not form biofilms, while strains lacking a capsule did (13). Further, capsule expression was found to be dependent on high nutrient levels and rapid growth, explaining why encapsulated forms are more commonly observed on culture plates. Bernd Kreikemeyer (University Clinic, Rostock, Germany) discussed the development of a probiotic approach to reduce the incidence of nasopharyngeal infections by group A streptococci (37). In a simple coculture, it was found that Streptococcus salivarius inhibited the development of Streptococcus pyogenes biofilms; however, in more-complex consortia, the effect was not as straightforward, because other nonpathogenic species, including the probiotic E. coli Nissle strain, attenuated the interference effect.
BIOFILMS IN INDUSTRIAL AND ENGINEERED SYSTEMS
Much early pioneering work in biofilm research was conducted in the context of fouling prevention in industrial and marine systems, and also for optimization of bioconversion processes in wastewater treatment and bioremediation. Satoshi Okabe (Hokkaido University, Japan) and Linda Blackall (University of Queensland, Brisbane, Australia) were the chairs for this session on biofilms in engineering applications. Korneel Rabaey (University of Queensland) described the use of microbial electron transfer systems to create microbial fuel cells. He showed how power output could be optimized by varying species composition and growth conditions, while indicating that initial applications may be limited to low-current situations such as sensors and feedback switches (1). Christian Picioreanu (Delft University) and Andrew Kato-Marcus (Arizona State University) each presented mathematical models that could be used to optimize these fuel cells. Picioreanu's model showed how biofilm structure may be an important factor for charge density on the anode, while Kato-Marcus modeled the anode as an electron acceptor, with the biofilm itself as an integral part of the anode. Moshe Herzberg (Yale University) illustrated how molecular, microscopic, and expression techniques could be applied to identify biofilm activity in reverse osmosis systems.
Three talks focused on multispecies biofilms in wastewater treatment. Co-chair Linda Blackall showed how the development of structured biofilm communities in flocs could be used for the simultaneous removal of ammonium, nitrate/nitrite, and phosphorus from wastewater. Electron microscopy suggested that the flocs may have structured EPS, which appeared similar to the honeycomb-like structures introduced later in the meeting by Bill Costerton. Co-chair Satoshi Okabe used a polyphasic approach that employed FISH, microautoradiography, and microelectrodes to characterize the distribution and activity of anaerobic ammonium-oxidizing (anammox) bacteria in a mixed wastewater biofilm (67). These bacteria were distributed throughout the biofilm, but rates of ammonium and nitrite oxidation differed at different locations in the biofilm. Silvia Weber (Technical University of Munich) used similar techniques to investigate granular flocs. A FISH protocol was optimized for use in thick, dense biofilms; this approach may have applications in other biofilm systems such as microbial mats. Maria Giao (University of Minho, Portugal) showed the importance of nutrients and hydrodynamics to Legionella pneumophila biofilm formation. Low carbon levels increased the number of viable but nonculturable bacteria in the biofilm, suggesting that environmental factors should be taken into account when one is culturing from drinking water systems. Carsten Schwermer (Max Planck Institute for Marine Microbiology, Bremen, Germany) presented industrially funded research that employed FISH and microelectrodes to determine the influence of nitrate on a biofilm community from an oilfield seawater injection system that contains sulfate-reducing bacteria (SRB). Nitrate achieved the desired effect of reducing SRB activity and sulfide production and may see widespread use throughout the industry as a more environmentally compatible alternative to conventional biocides used for SRB control. Finally, Chuanwu Xi (University of Michigan) presented work on optical coherence tomography, a high-resolution imaging technique that does not require staining, for real-time monitoring of living biofilms. The instrument offers two advantages over conventional microscopes, especially in process situations: it can be positioned much further from the biofilm, and it can penetrate opaque materials (73).
PREVENTION AND TREATMENT OF BIOFILMS
Michael Givskov (Danish Technical University) and Phil Stewart (Montana State University) were session chairs. Stewart's talk on the visualization of antimicrobial action in biofilms described the use of calcein AM as a viability indicator. When treated with the quaternary amine Barquat, biofilm microcolonies quickly lost calcein fluorescence at the edges but showed time-dependent retention of fluorescence at the center (Fig. 5). Treatment with chlorite caused loss of fluorescence only in a thin band at the edge of the colony. Treatment with nisin led to a rapid and uniform fluorescence loss. This approach allows spatiotemporally resolved measurement of toxicity and suggests that differences in susceptibility to particular agents exist within the biofilm.
FIG. 5.
Visualization of antimicrobial action against a Staphylococcus epidermidis biofilm. Bacteria in a flow cell-grown biofilm were loaded with the green fluorescent dye calcein AM and then exposed to 50 mg liter−1 alkyl dimethyl benzyl ammonium chloride (ADBAC) under continuous flow. Time lapse confocal scanning laser microscopy examination of fluorescence loss shows permeabilization of cells by the biocide. The number in each panel is the time, in minutes, after the introduction of ADBAC into the flow cell. In the first and last panels, biomass was imaged in transmission mode (gray scale). Image courtesy of W. Davison and P. Stewart (Center for Biofilm Engineering, Montana State University).
Thomas Neu (Helmholtz Center for Environmental Research, Magdeburg, Germany) described fluid dynamic gauging, a method of monitoring biofilm stability by which the shear strength of fouling layers can be measured (41). The base layers of the biofilm were found to be highly stable, whereas the upper layers were easily removed. Neu also showed magnetic resonance imaging data that allow real-time, nondestructive visualization of biofilm biomass, as well as of flow velocity and shear stress around the biofilm.
Persister cells, first reported by Spoering and Lewis (59), are defined as a numerically small (<1%), antimicrobial-resistant, slow-growing subpopulation of cells that is maintained in both planktonic and biofilm cultures; these cells were the subject of talks by Peter Gilbert (University of Manchester) and Kim Lewis (Northeastern University). Using high-throughput cell sorting, Gilbert showed that an Escherichia coli population labeled with GFP contained a very small fraction of poorly metabolizing (weakly fluorescent) cells that also demonstrated multidrug resistance. Large volumes of culture were processed to collect sufficient mRNA for microarray analysis. High levels of prophage transcripts and a group of candidate “persister” genes including the putative regulator ykgK were identified. Antisense suppression of pin, ykgK, and ykgM eliminated persister cells, while overexpression increased persisters. These genes may be regulators of the persistence state and thus potential drug targets for recalcitrant biofilm infections. Lewis' main observations on weakly metabolizing cells being persisters (55) are in agreement with those of Gilbert. Lewis also used transcriptional profiling to identify a number of candidate genes, but he identified genes different from those discovered by Gilbert; Lewis' persister genes included the dehydrogenase-encoding gene glpD and the lipid biosynthesis gene plsB (60). Lewis thought that the differences between candidate genes from the two studies may be a result of strain differences. He also reported that Candida albicans, in contrast to bacteria, forms persister cells only in a biofilm; persisters are not part of the population during planktonic culture (36).
Jennifer Kofonow (Northern Arizona University) spoke on the use of biofilm-specific antibodies to detect biofilm infections in a rabbit model. Antigens are being identified by a proteomics approach as present in infected animals but absent in healthy animals; because the targets are eventually to be used in the diagnosis of human biofilm infections, human blood must be screened as well, and one candidate protein has emerged. The development of antimicrobials from marine invertebrates was reported on by Domenico Schillaci (University of Palermo). Cytosol of the sea urchin Paracentrotus lividus has activity against S. epidermidis and S. aureus. A 5-kDa peptide fraction inhibits bacterial adhesion and interferes with bacterial respiration in biofilms.
Co-chair Michael Givskov spoke on quorum-sensing blockers as drugs. Advantages of this approach are (i) that development of resistance to compounds similar to those generated naturally by the bacterium is highly unlikely and (ii) that blockers can be used to modulate antibiotic resistance and to generate synergistic effects with other antimicrobials (48). Givskov suggested that rhamnolipid is a necrotic agent for PMN (7, 24) (Fig. 6) and that reduced rhamnolipid synthesis may partly explain the more rapid clearance of quorum-sensing-defective Pseudomonas aeruginosa than of the wild type in a mouse model of lung infection (20). A promising garlic-based quorum-sensing blocker (46) has been synthesized and tested; it is less active than garlic extract, but it is possible that a mixture of garlic-based compounds can reach or exceed the efficacy of natural garlic extract.
FIG. 6.
Quorum-sensing-controlled killing of PMN. Freshly isolated, Syto 62-stained PMN (red) interact in vitro with a GFP-tagged biofilm (green). (Left) Wild-type bacteria kill the PMN through quorum-sensing-controlled production of rhamnolipid. The PMN are lysed and disappear before they can eliminate the biofilm. (Right) In the presence of the P. aeruginosa LasR/RhlR double-quorum-sensing mutant, the PMN readily traverse the biofilm and phagocytose the virulence-factor-deficient bacteria. Images courtesy of Thomas Bjarnsholt (Danish Technical University).
The topic of predatory/parasitic bacteria as antibiofilm agents was covered by Daniel Kadouri (University of Medicine and Dentistry of New Jersey). Bdellovibrio bacteriovorus can greatly diminish the biofilm biomass of E. coli as well as that of P. fluorescens (26). The less well known exoparasitic bacterium Micavibrio aeruginosavorus is likewise able to reduce the biofilm biomass of laboratory and clinical strains of P. aeruginosa, Klebsiella pneumoniae, and Burkholderia cepacia (27). Closing the session, David Davies (Binghamton University) spoke on a biofilm-dispersing activity found in the organic fraction of P. aeruginosa spent culture medium. The activity reduces P. aeruginosa biofilm thickness by 50%; it also acts against mixed-species biofilms, and it has a synergistic effect with antibiotics whereby a lower antibiotic concentration is required to achieve the same level of killing as that seen in the absence of the dispersing activity.
KEYNOTE TALK 3
The prebanquet address was given by Bill Costerton, the person who, beginning at the University of Calgary and later as the director of Montana State University's Center for Biofilm Engineering, brought biofilms to the forefront of microbiological research and who is now developing a biofilm research center at the University of Southern California. Costerton introduced his ideas on structured biofilm EPS, which he termed “caserna,” named after the temporary stone forts built by soldiers of ancient Rome. These are networks and honeycomb-shaped scaffolds with pore sizes of several micrometers (Fig. 7) which develop in several-day-old cultures of S. epidermidis isolates (52), although they have been seen in other bacterial cultures as well. Based on scanning electron microscopic, freeze-substitution transmission electron microscopic, and confocal and light microscopic evidence, these structures appear to form from the contents (especially ribosomes) of lysing cells that merge with one another. Costerton termed the process “bacterial coalescence” and noted that the repeated structure, which can form macroscopic structures with dimensions of millimeters, likely resulted from a genetically regulated process. As discussed in the “slime matrix” session, little is known about the structural and chemical composition of biofilm EPS; the possibility of higher-order structural organization, such as that described for proteins, is provocative. Casernum production in natural environments and a role for caserna in biofilm function remain to be demonstrated. However, the possibility of structured EPS hints at yet further levels of complexity in biofilm systems.
FIG. 7.
Scanning electron microscopy of a part of a macroscopic casernum formed in liquid culture by the MH strain of S. epidermidis. Note the coalescence of cells to form the parallel planar walls of the basic honeycomb structure. Image courtesy of Christoph Schaudinn and Bill Costerton (University of Southern California).
CODA
In summary, the meeting elevated its reputation for showcasing high-quality, exciting science from a rapidly moving field and for demonstrating the broad applicability of biofilm biology to microbiological research. The organizing committee took steps to attract an audience and a program that were demographically as well as scientifically diverse; this benchmark for the meeting seems to have been heartily embraced, and the committee hopes that this approach will continue.
Acknowledgments
The committee (Niels Hoiby, University of Copenhagen; Søren Molin, Technical University of Denmark; Robert J. Palmer, Jr., NIDCR, NIH; Matthew Parsek, University of Washington; Paul Stoodley, Allegheny-Singer Research Institute) thanks ASM for its continued support and development of this conference series. In particular, we thank ASM Conferences staff members Lisa Nalker and Latonya Nichols for outstanding work in bringing the 4th Conference on Biofilms to fruition.
Robert J. Palmer, Jr., is supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health. Paul Stoodley is supported by Allegheny-Singer Research Institute and Philips Oral Healthcare.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Aelterman, P., K. Rabaey, P. Clauwaert, and W. Verstraete. 2006. Microbial fuel cells for wastewater treatment. Water Sci. Technol. 54:9-15. [DOI] [PubMed] [Google Scholar]
- 2.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10:156-163. [DOI] [PubMed] [Google Scholar]
- 3.Banin, E., K. M. Brady, and E. P. Greenberg. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72:2064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 102:11076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bantinaki, E., R. Kassen, C. G. Knight, Z. Robinson, A. J. Spiers, and P. B. Rainey. 2007. Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics 176:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgartner, L. K., R. P. Reid, C. Dupraz, A. W. Decho, D. H. Buckley, J. R. Spear, K. M. Przekop, and P. T. Visscher. 2006. Sulfate reducing bacteria in microbial mats: changing paradigms, new discoveries. Sediment. Geol. 185:131-145. [Google Scholar]
- 7.Bjarnsholt, T., P. O. Jensen, M. Burmolle, M. Hentzer, J. A. J. Haagensen, H. P. Hougen, H. Calum, K. G. Madsen, C. Moser, S. Molin, N. Hoiby, and M. Givskov. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373-383. [DOI] [PubMed] [Google Scholar]
- 8.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bredenbruch, F., R. Geffers, M. Nimtz, J. Buer, and S. Häussler. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 8:1318-1329. [DOI] [PubMed] [Google Scholar]
- 10.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, W.-S., M. van de Mortel, L. Nielsen, G. N. de Guzman, X. Li, and L. J. Halverson. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189:8290-8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, W. S., and L. J. Halverson. 2003. Reduced water availability influences the dynamics, development, and ultrastructural properties of Pseudomonas putida biofilms. J. Bacteriol. 185:6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 188:5510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 15.Denamur, E., and I. Matic. 2006. Evolution of mutation rates in bacteria. Mol. Microbiol. 60:820-827. [DOI] [PubMed] [Google Scholar]
- 16.Dupraz, C., and P. T. Visscher. 2005. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 13:429-438. [DOI] [PubMed] [Google Scholar]
- 17.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 18.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hense, B. A., C. Kuttler, J. Müller, M. Rothballer, A. Hartmann, and J.-U. Kreft. 2007. Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5:230-239. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann, N., T. B. Rasmussen, P. O. Jensen, C. Stub, M. Hentzer, S. Molin, O. Ciofu, M. Givskov, H. K. Johansen, and N. Hoiby. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 73:2504-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holst, G., O. Kohls, I. Klimant, B. Konig, M. Kuhl, and T. Richter. 1998. A modular luminescence lifetime imaging system for mapping oxygen distribution in biological samples. Sensors Actuators B 51:163-170. [Google Scholar]
- 22.Hunter, R. C., and T. J. Beveridge. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter, R. C., and T. J. Beveridge. 2005. High-resolution visualization of Pseudomonas aeruginosa PAO1 biofilms by freeze-substitution transmission electron microscopy. J. Bacteriol. 187:7619-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, P. O., T. Bjarnsholt, R. Phipps, T. B. Rasmussen, H. Calum, L. Christoffersen, C. Moser, P. Williams, T. Pressler, M. Givskov, and N. Hoiby. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329-1338. [DOI] [PubMed] [Google Scholar]
- 25.Kader, A., R. Simm, U. Gerstel, M. Morr, and U. Römling. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602-616. [DOI] [PubMed] [Google Scholar]
- 26.Kadouri, D., and G. A. O'Toole. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71:4044-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadouri, D., N. C. Venzon, and G. A. O'Toole. 2007. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 73:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko, Y., M. Thoendel, O. Olakanmi, B. E. Britigan, and P. K. Singh. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Investig. 117:877-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerr, J., G. Taylor, A. Rutman, N. Hoiby, P. Cole, and R. Wilson. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52:385-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutsoudis, M. D., D. Tsaltas, T. D. Minogue, and S. B. von Bodman. 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 103:5983-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kugelberg, E., E. Kofoid, A. B. Reams, D. I. Andersson, and J. R. Roth. 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 103:17319-17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kühl, M., G. Holst, A. W. D. Larkum, and P. J. Ralph. Mapping of the oxygen distribution and its dynamics within the endolithic algal community of the massive coral Porites lobata (Dana). J. Phycol., in press. [DOI] [PubMed]
- 34.Kühl, M., L. F. Rickelt, and R. Thar. 2007. Combined imaging of oxygen and bacteria in biofilms. Appl. Environ. Microbiol. 73:6289-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaFleur, M. D., C. A. Kumamoto, and K. Lewis. 2006. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50:3839-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lembke, C., A. Podbielski, C. Hidalgo-Grass, L. Jonas, E. Hanski, and B. Kreikemeyer. 2006. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl. Environ. Microbiol. 72:2864-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 39.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102:16819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matzinger, P. 2002. The danger model: a renewed sense of self. Science 296:301-305. [DOI] [PubMed] [Google Scholar]
- 41.Möhle, R. B., T. Langemann, M. Haesner, W. Augustin, S. Scholl, T. R. Neu, D. C. Hempel, and H. Horn. 9 April 2007. Structure and shear strength of microbial biofilms as determined with confocal laser scanning microscopy and fluid dynamic gauging using a novel rotating disc biofilm reactor. Biotechnol. Bioeng. doi: 10.1002/bit.21448. [DOI] [PubMed]
- 42.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656-679. [DOI] [PubMed] [Google Scholar]
- 43.Newman, K. L., R. P. P. Almeida, A. H. Purcell, and S. E. Lindow. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 101:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188:4117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsek, M. R., and E. P. Greenberg. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13:27-33. [DOI] [PubMed] [Google Scholar]
- 45a.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 46.Persson, T., T. H. Hansen, T. B. Rasmussen, M. E. Skinderso, M. Givskov, and J. Nielsen. 2005. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org. Biomol. Chem. 3:253-262. [DOI] [PubMed] [Google Scholar]
- 47.Rao, D., J. S. Webb, and S. Kjelleberg. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72:5547-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, T. B., and M. Givskov. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology 152:895-904. [DOI] [PubMed] [Google Scholar]
- 49.Resch, A., B. Fehrenbacher, K. Eisele, M. Schaller, and F. Gotz. 2005. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol. Lett. 252:89-96. [DOI] [PubMed] [Google Scholar]
- 50.Resch, A., S. Leicht, M. Saric, L. Pasztor, A. Jakob, F. Gotz, and A. Nordheim. 2006. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6:1867-1877. [DOI] [PubMed] [Google Scholar]
- 51.Resch, A., R. Rosenstein, C. Nerz, and F. Gotz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaudinn, C., P. Stoodley, A. Kainovic, T. O'Keeffe, B. Costerton, D. Robinson, M. Baum, G. D. Ehrlich, and P. Webster. 2007. Bacterial biofilms, other structures seen as mainstream concepts. Microbe 2:231-237. [Google Scholar]
- 53.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekar, R., D. K. Mills, E. R. Remily, J. D. Voss, and L. L. Richardson. 2006. Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl. Environ. Microbiol. 72:5963-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah, D., Z. G. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119-127. [DOI] [PubMed] [Google Scholar]
- 57.Simm, R., A. Lusch, A. Kader, M. Andersson, and U. Römling. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 59.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spoering, A. L., M. Vulic, and K. Lewis. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stickler, D. J., S. M. Jones, G. O. Adusei, M. G. Waters, J. Cloete, S. Mathur, and R. C. L. Feneley. 2006. A clinical assessment of the performance of a sensor to detect crystalline biofilm formation on indwelling bladder catheters. BJU Int. 98:1244-1249. [DOI] [PubMed] [Google Scholar]
- 62.Stoodley, P., S. Kathju, F. Z. Hu, G. Erdos, J. E. Levenson, N. Mehta, B. Dice, S. Johnson, L. Hall-Stoodley, L. Nistico, N. Sotereanos, J. Sewecke, J. C. Post, and G. D. Ehrlich. 2005. Molecular and imaging techniques for bacterial biofilms in joint arthroplasty infections. Clin. Orthop. Relat. Res. 2005:31-40. [DOI] [PubMed] [Google Scholar]
- 63.Tait, K., I. Joint, M. Daykin, D. L. Milton, P. Williams, and M. Camara. 2005. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 7:229-240. [DOI] [PubMed] [Google Scholar]
- 64.Trautner, B. W., R. A. Hull, and R. O. Darouiche. 2003. Escherichia coli 83972 inhibits catheter adherence by a broad spectrum of uropathogens. Urology 61:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trautner, B. W., R. A. Hull, J. I. Thornby, and R. O. Darouiche. 2007. Coating urinary catheters with an avirulent strain of Escherichia coli as a means to establish asymptomatic colonization. Infect. Control Hosp. Epidemiol. 28:92-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsang, P. H., G. L. Li, Y. V. Brun, L. B. Freund, and J. X. Tang. 2006. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. USA 103:5764-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsushima, I., Y. Ogasawara, T. Kindaichi, H. Satoh, and S. Okabe. 2007. Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors. Water Res. 41:1623-1634. [DOI] [PubMed] [Google Scholar]
- 68.Ulett, G. C., J. Valle, C. Beloin, O. Sherlock, J.-M. Ghigo, and M. A. Schembri. 9 April 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed]
- 69.Valle, J., M. Vergara-Irigaray, N. Merino, J. R. Penades, and I. Lasa. 2007. σB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 189:2886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Mortel, M., and L. J. Halverson. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735-750. [DOI] [PubMed] [Google Scholar]
- 71.Wargo, M. J., and D. A. Hogan. 2006. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr. Opin. Microbiol. 9:359-364. [DOI] [PubMed] [Google Scholar]
- 72.West-Barnette, S., A. Rockel, and W. E. Swords. 2006. Biofilm growth increases phosphorylcholine content and decreases potency of nontypeable Haemophilus influenzae endotoxins. Infect. Immun. 74:1828-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xi, C., D. Marks, S. Schlachter, W. Luo, and S. A. Boppart. 2006. High-resolution three-dimensional imaging of biofilm development using optical coherence tomography. J. Biomed. Optics 11:034001. [DOI] [PubMed] [Google Scholar]
- 74.Yang, L., K. B. Barken, M. E. Skindersoe, A. B. Christensen, M. Givskov, and T. Tolker-Nielsen. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318-1328. [DOI] [PubMed] [Google Scholar]
- 75.Yao, Y. F., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191:289-298. [DOI] [PubMed] [Google Scholar]
- 76.Yoon, S. S., R. Coakley, G. W. Lau, S. V. Lymar, B. Gaston, A. C. Karabulut, R. F. Hennigan, S. H. Hwang, G. Buettner, M. J. Schurr, J. E. Mortensen, J. L. Burns, D. Speert, R. C. Boucher, and D. J. Hassett. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J. Clin. Investig. 116:436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]