Abstract
Work with pathogens like Vibrio cholerae has shown major differences between genes expressed in bacteria grown in vitro and in vivo. To explore this subject for commensals, we investigated the transcription of the Lactobacillus johnsonii NCC533 genome during in vitro and in vivo growth using the microarray technology. During broth growth, 537, 626, and 277 of the 1,756 tested genes were expressed during exponential phase, “adaptation” (early stationary phase), and stationary phase, respectively. One hundred one, 150, and 33 genes, respectively, were specifically transcribed in these three phases. To explore the in vivo transcription program, we fed L. johnsonii containing a resistance plasmid to antibiotic-treated mice. After a 2-day washout phase, we determined the viable-cell counts of lactobacilli that were in the lumina and associated with the mucosae of different gut segments. While the cell counts showed a rather uniform distribution along the gut, we observed marked differences with respect to the expression of the Lactobacillus genome. The largest number of transcribed genes was in the stomach (n = 786); the next-largest numbers occurred in the cecum (n = 391) and the jejunum (n = 296), while only 26 Lactobacillus genes were transcribed in the colon. In vitro and in vivo transcription programs overlapped only partially. One hundred ninety-one of the transcripts from the lactobacilli in the stomach were not detected during in vitro growth; 202 and 213 genes, respectively, were transcribed under all in vitro and in vivo conditions; but the core transcriptome for all growth conditions comprised only 103 genes. Forty-four percent of the NCC533 genes were not detectably transcribed under any of the investigated conditions. Nontranscribed genes were clustered on the genome and enriched in the variable-genome part. Our data revealed not only major differences between in vitro- and in vivo-expressed genes in a Lactobacillus gut commensal organism but also marked changes in the expression of genes along the digestive tract.
The gastrointestinal tracts of animals harbor microbial communities that impress by sheer cell number and species diversity (13). It was calculated that members of the gut microbiota outnumber human body cells by a factor of 10, and recent sequencing approaches estimated an excess of 500 prokaryotic phylotypes for the human gut microbiota (22). Lactobacilli are common inhabitants of this ecosystem and are numerically more prominent than commensal Escherichia coli (8). In domesticated animals like pigs (27) and chickens (2) and in laboratory animals like mice and rats (28), lactobacilli even represent the predominant bacteria in the proximal parts of the gastrointestinal tract (3). Due to their important role in food and feed fermentation, food microbiologists have intensively investigated lactobacilli (39). Nearly 100 species of lactobacilli have been described (11, 36), and several genomes from lactobacilli have been sequenced (1, 9, 17, 18, 30, 42). A wealth of published reports documents the metabolic characteristics of lactobacilli during in vitro growth and in food fermentation (17). Much less is known about the metabolic states of lactobacilli in the intestine (43). Two recent papers document the gut-specific gene expression of lactobacilli using in vivo expression technology. Walter et al. (44) identified three Lactobacillus reuteri genes that were specifically induced in mice with reconstituted Lactobacillus-free microflora: a xylose isomerase and a peptide methionine sulfoxide reductase, which were expressed in the ceca of the mice, and a hypothetical gene expressed in the forestomach. With a modified resolvase-based in vivo expression technology, Bron et al. (6) identified 72 Lactobacillus plantarum genes whose expression was specifically induced during passage in the gastrointestinal tract. The induced genes included sugar transporters and genes involved in the acquisition and synthesis of amino acids, nucleotides and cofactors, stress proteins, and extracellular proteins. Reverse transcription-PCR confirmed the transcription of a copper-transporting ATPase, an alcohol dehydrogenase, and three cell surface proteins, all of which belonged to the group of in vivo-inducible genes (24). In contrast, whole-genome proteome and transcriptome analyses have not yet been published for lactobacilli in vivo. This lack of data on gut expression of lactobacilli and notably also of commensal E. coli is unfortunate since both have demonstrated in clinical trials beneficial effects against rotavirus diarrhea and ulcerative colitis, respectively (20, 38). There is currently a great interest in probiotic, i.e., health-promoting, bacteria (5). To provide a background for the understanding of probiotic actions, we decided to investigate the expression pattern of Lactobacillus johnsonii in the different gut segments of laboratory mice. We compared this in vivo expression pattern with that obtained during broth growth of the same strain. L. johnsonii showed distinct expression patterns in the different gut segments that were unlike those observed during the different growth phases of the same strain in the broth culture.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. johnsonii NCC533 was obtained from the Nestlé culture collection (NCC) and grown in MRS (http://www.bd.com/ds/productCenter/288130.asp; see the Difco/BBL manual in Related Documents) with 2% glucose as the carbon source, to which filter-sterilized cysteine was added after being autoclaved (final concentration, 0.05% [wt/vol]) as the redox buffer. Cultures were incubated at 37°C under anaerobic conditions using the AnaeroGen system (Oxoid, Basingstoke, United Kingdom) and anaerobic jars.
Growth properties.
For gene expression profiling experiments, cells were grown in a Sixfors fermentor system, composed of four individual 500-ml vessels (Infors, Bottmingen, Switzerland), as described elsewhere (11a). Growth curves were performed at least in triplicate using the four separate fermentation vessels (fermentors 1 to 4), which were inoculated at 0.4% (vol/vol) with four individual overnight cultures in order to reach a starting optical density at 600 nm (OD600) of 0.05. Samples were taken at regular intervals from the four vessels to measure the OD600 and determine the number of CFU/ml until 36 h of fermentation. Aliquots of 15 ml for the early (time points 1 and 2 [T1 and T2, respectively]) and mid-exponential (T3 and T4) phases and 10 ml for the adaptation (T5), mid-stationary (T6), and late stationary (T7) phases were centrifuged for 5 min at 10,000 × g and 4°C. Cell pellets were snap-frozen in liquid nitrogen and stored at −80°C until further use.
Microscopy.
The mouse gut and food pellets were fixed in 2.5% glutaraldehyde in 0.1 M carbonate buffer, pH 7, containing ruthenium red. The material was embedded in Technovit 7100 resin and cut into 2-μm-thick slices by cryosectioning.
Preparation of the bacterial strains and administration to mice.
In order to monitor Lactobacillus johnsonii NCC533 after its administration to the conventional mice, the strain was electrotransformed with plasmid pDP818 (a pGhost derivative into which an erythromycin resistance cassette was introduced). The strain was grown overnight at 37°C (stationary phase), harvested by centrifugation at 3,000 × g for 10 min, washed with fresh MRS medium, and suspended at the appropriate cell concentration of 1010 CFU/ml. Bacterial suspensions were prepared daily for intragastric gavage.
Animal experiments.
All experiments employing mice were performed using protocols approved by the ethical committee of the Canton de Vaud. Conventional C3H/HeJ mice (Nestlé Research Centre, Lausanne, Switzerland) with an average age of 8 weeks were placed in Macrolon cages (five mice per cage) and housed in a room with a cycle of 12 h of light and 12 h of darkness and at a temperature of 22°C as previously described (11a). The normal intestinal flora (mainly Lactobacillus flora [data not shown]) was decreased by the addition of erythromycin (at 10 μg·ml−1) in the drinking water of mice. Water intake was controlled as is normally done (data not shown). Then, L. johnsonii NCC533 (Emr) was administrated by intragastric gavage at 109 CFU to a group of five animals for three successive days. Forty-eight hours after the last gavage, mice were euthanized using 3% isoflurane. Whole tissues corresponding to the stomach, duodenum, jejunum, ileum, cecum, and colon were dissected from each mouse, washed with phosphate-buffered saline, scraped, and then flash-frozen in liquid nitrogen.
RNA isolation.
Cell pellets from the incubation vessels or frozen tissue scraping samples from mice were crushed and suspended in equal volumes of Tris-EDTA buffer (pH 8) and phenol (pH 4.2), and total RNA was extracted by the Macaloid method described by Kuipers et al. (21). The cells were disrupted at maximum speed using a Mini-BeadBeater-8 apparatus (BioSpec Products, Bartlesville, OK) at 4°C for 1 min for three cycles with a resting period of 1 min on ice between each cycle. RNA was purified by phenol-chloroform extraction followed by ethanol precipitation. Pellets were resuspended in nucleotide-free water, and 100 μg of total RNA was treated with 200 units of DNase I (Ambion, Huntingdon, United Kingdom) for 2 h at 37°C to eliminate contaminating DNA. The RNeasy mini-kit (QIAGEN, Basel, Switzerland) that was used for further purification includes an additional on-column DNase digestion step. RNA concentrations were determined spectrophotometrically. RNA of L. johnsonii from tissue scraping samples was extracted from 100 μg of a mouse bacterium RNA mixture using a MICROBEnrich kit (Ambion, Huntingdon, United Kingdom) and amplified using a MessageAmp II-Bacteria prokaryotic-RNA kit (Ambion, Huntingdon, United Kingdom). RNA integrity for all samples was tested using the Agilent RNA 6000 Nano assay (Agilent, Waldbronn, Germany).
Microarray design.
DNA-based arrays were produced by Eurogentec S.A. (Liege, Belgium). Oligonucleotide primers were designed to amplify segments, ranging in size from 127 to 800 bp, based on the open reading frames (ORFs) identified from the L. johnsonii NCC533 genome sequence (30). In total, 1,857 ORF-specific amplicons were generated and spotted in duplicate on glass slides, thus covering approximately 96% (corresponding to 1,756 ORFs) of the L. johnsonii NCC533 genome.
cDNA synthesis, array hybridization, and analysis.
Total RNA extracted from in vivo or in vitro samples (T1, T2, T3, T4, T5, T6, or T7) was hybridized onto DNA arrays together with RNA extracted from mid-exponential-phase cells (T4). For each hybridization, 2 μg of total RNA was labeled using the 3DNA Array 350RP Genisphere kit (Genisphere Inc., Hatfield, PA), by following the protocol provided by the supplier. Luciferase control mRNA (10 ng) (Promega, Zurich, Switzerland) was mixed with total RNA before being labeled to balance the two channels during scanning. After the hybridization procedure, array slides were scanned with a Scanarray 4000 machine (Packard Biochip Technologies, Billerica, MA). Data were extracted from the scanned images using the software Imagene 5.6 (BioDiscovery, El Segundo, CA). Spot signal intensities of each channel were corrected by subtracting the corresponding local background values. Spots displaying low intensity (i.e., less than threefold the local background standard deviation) were considered empty. For all technical replicates, the control for fluorescence labeling by dye swapping was done. As the method is based on qualitative detection, negative and positive controls were used to confirm the absence of cross- and/or nonspecific hybridization.
In vitro, the absolute expression pattern was determined from at least three independent biological samples. A gene was called “expressed” when it was detected in all three experiments. Many published microarray analyses express the in vitro transcription profile quantitatively by reference to mid-exponential broth growth. In the present study, we called a gene expressed when its signal exceeded by threefold the background standard deviation. This procedure allows a comparison of the different growth phases, but it yields only qualitative data. To avoid genes expressed by the autochthones microflora, spots displaying detectable signal with total RNA from erythromycin-treated mice without gavage of L. johnsonii NCC533 were excluded from the in vivo analysis (fewer than 20 spots representing mainly ribosomal protein were detected).
In vivo, a gene was called “expressed” when it was detected in two biological experiments. Three biological replicates were analyzed for gene expression in the cecum and the colon, and two biological replicates (and two technical replicates) were analyzed for the stomach and the jejunum.
Microarray data accession number.
The microarray data were deposited in the GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE9236.
RESULTS
In vitro growth.
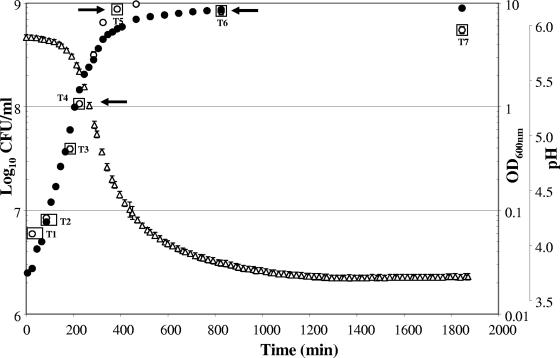
L. johnsonii NCC533 was grown in MRS broth supplemented with 2% glucose as the carbon source and 0.05% cysteine as the redox buffer. Under these conditions, an overnight culture showed rapid growth without a lag phase and achieved a high OD600 corresponding to a cell titer of 109 CFU/ml (Fig. 1). The culture maintained a high titer of 5.5 × 108 CFU of viable cells/ml over an extended period in stationary phase. Under comparable growth conditions, dairy lactobacilli like Lactobacillus delbrueckii showed a lag phase and slower growth kinetics but the same final OD600 (29).
FIG. 1.
Growth curve of L. johnsonii NCC533 in MRS broth. Growth was monitored over time by measuring OD600 (filled circles), viable colony counts on MRS-cysteine agar (open circles), and drops in pH due to lactic acid production (triangles). Data points are arithmetic means with standard errors of the means (bars) from four independent experiments. The time points (T1 to T7) investigated for microarray expression analysis are boxed.
In vitro transcription profiles.
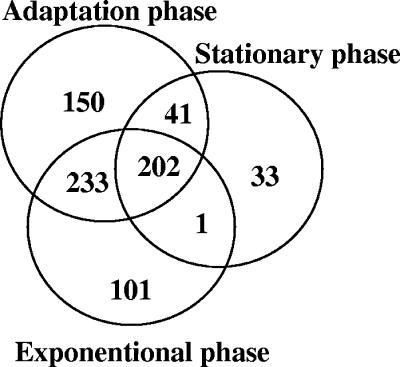
For mRNA isolation L. johnsonii NCC533 cells were harvested from the broth culture at several time points after inoculation (Fig. 1). In a total of 27 hybridization experiments, the first four time points yielded transcription profiles that differed in less than 5% of their expressed genes. Genes were scored as expressed when their signal was detected in all hybridization experiments (with two technical replicates per slide), with mRNA preparations being obtained from three independent growth experiments. Overall, 761 genes were expressed in vitro (41% of all NCC533 genes on the microarray). Specifically, 537 (71% of all in vitro-transcribed genes), 626 (82%), and 277 (36%) genes were transcribed in the exponential phase (225 min), in the “adaptation” period from exponential to stationary growth (385 min), and in the early stationary phase (825 min), respectively (Fig. 2). A comparable transcription profile was observed at the 825- and 1,820-min time points (early and late stationary phases). The overall intensities of the hybridization signals in the stationary phase diminished with respect to those of the preceding adaptation and exponential phases. During the exponential and adaptation phases, the ribosomal proteins and the glycolytic enzymes (and their feeder pathway proteins) represented the most abundantly expressed mRNAs; within the 50 genes showing the highest transcript intensities, 27 and 8 genes belonged to these two categories in the exponential phase and 21 and 13 genes belonged to these two categories in the adaptation phase, respectively. The remaining transcripts represented a variety of functions; only two were represented with two genes (ATP synthase and peptidoglycan synthesis genes in exponential phase). Within the 50 most abundantly transcribed genes of the stationary phase, we observed a clearly distinct transcription pattern with eight ABC transporters, five chaperons, four transcriptional regulators, and three NADH oxidoreductases, in addition to nine ribosomal proteins and seven glycolytic enzymes.
FIG. 2.
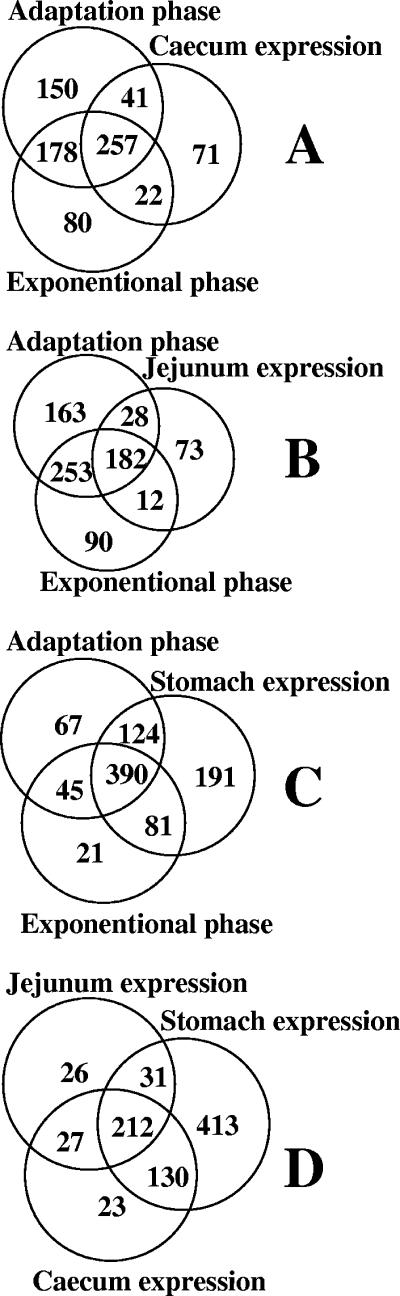
Numbers of L. johnsonii NCC533 genes expressed in vitro as assessed by microarray hybridization. The Venn diagram displays the numbers of genes expressed during the time points T4 (225 min, exponential growth phase), T5 (385-min adaptation growth phase), and T6 (825 min, early stationary phase) of the in vitro growth indicated by arrows in Fig. 1.
Analysis of the in vitro transcription profile.
The 537 transcripts of the exponential phase corresponded to a transcription of 31% of the ORFs from the NCC533 genome. One hundred one ORFs were specifically transcribed in the “exponential” phase; i.e., they were not detected in the “adaptation” or “stationary” phase (Fig. 2). This group of genes was specific for DNA replication, cell wall synthesis, glycosyltransferases, and exporter proteins. Striking, but expected, was the prominence of mRNA for cell division proteins (FtsH, -W, -Q, and -Y; division protein A; and chromosome partition protein) and enzymes involved in lipopolysaccharide synthesis (LPS gycosyltransferases from two genetic loci and flippase).
Likewise, 150 of the 626 ORFs transcribed in the adaptation phase were specific to this growth phase (Fig. 2). This group of genes contains transcriptional regulators, hydrolases, and transporter proteins. According to the expression pattern, amino acids, oligopeptides, proteins, and also sugar alcohols and other sugar derivatives are transported into the cell, where they experience further hydrolytic degradation. In both the exponential and the adaptation phase, we dealt with metabolically active cells, as was also suggested by the pH drop during the adaptation phase.
Finally, 33 of the 277 ORFs transcribed in the early stationary phase were restricted to this time point. This list contained three gene clusters: a trehalose phosphotransferase system (PTS), an ABC transporter, and a copper-exporting ATPase. Notably, these genes comprise only a single stress protein and no transcriptional repressor. Apparently, NCC533 is not locked in a repressed state during the stationary phase, as was also suggested by the qualitatively similar transcription profiles in early- and late-stationary-phase cells (273 and 241 genes for T6 and T7, respectively; 238 genes were shared between both phases) and the quickly resumed growth of the stationary cells upon transfer into a fresh medium (Fig. 1). In fact, an overnight culture showed after only 5 min of incubation in a fresh medium a substantial part of the transcripts that characterizes the expression profile of the exponential phase (data not shown). Apparently, during the stationary phase, the NCC533 strain capitalizes on maintaining an energy metabolism and does not prepare for a longer starvation period.
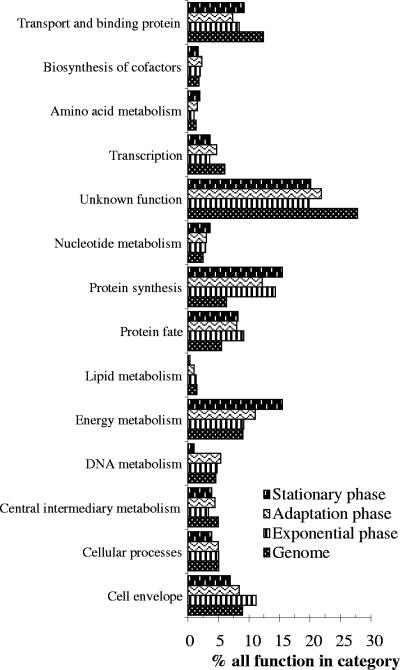
In all, 761 ORFs were expressed during the broth growth of NCC533, corresponding to 43% of all ORFs. The functional classification of NCC533 genes present on the array is presented in Fig. 3 and is compared to the functional classification of the in vitro transcripts. In a comparison with the functional categories in the genome, the transcripts from all in vitro phases were enriched in the functions of “protein fate” (protein secretion and trafficking) (9, 8, and 8% of the transcribed genes in exponential phase, during adaptation, and in stationary phase, respectively, belonged to this category compared to 6% of the genes in the genome) and “protein synthesis” (where 14, 12, and 16% of the transcribed genes belonged to this category, compared to a 6% representation of these genes in the genome). In contrast, transcribed genes were underrepresented in the category “unknown functions” (20, 22, and 20% of the transcripts compared to 28% of the genes in the genome had unknown functions). The exponential phase was enriched in genes with functions in the cell envelope category (11% of transcripts versus 9% of genes in the genome). In the adaptation phase, we observed an increased transcription of genes involved in energy metabolism (11% of transcripts versus 9% of the genome). The stationary-phase transcripts exhibited less DNA metabolism (1%) and less lipid metabolism (1%) but more energy metabolism (16%) functions than transcripts in the preceding phases.
FIG. 3.
Functional attributions of the in vitro-expressed genes of L. johnsonii NCC533 to the main TIGR-CMR (Comprehensive Microbial Resource) role classes. We based the analysis on 1,756 ORFs, excluding 110 ORFs belonging to the mobile-DNA category. For each category, the lowest bar represents the percentage of genes in that category as detected in the sequenced genome of NCC533. The three bars on top of it indicate the percentages of the genes transcribed during the T4, T5, and T6 growth periods (exponential, adaptation, and stationary phases, from bottom to top [Fig. 1]) which fall into this category. “% all function in category” refers to the percentage of transcribed genes belonging to the indicated functional category relative to all transcribed genes. “Genome” refers to the percentage of genes belonging to the indicated functional category relative to all genes in the genome.
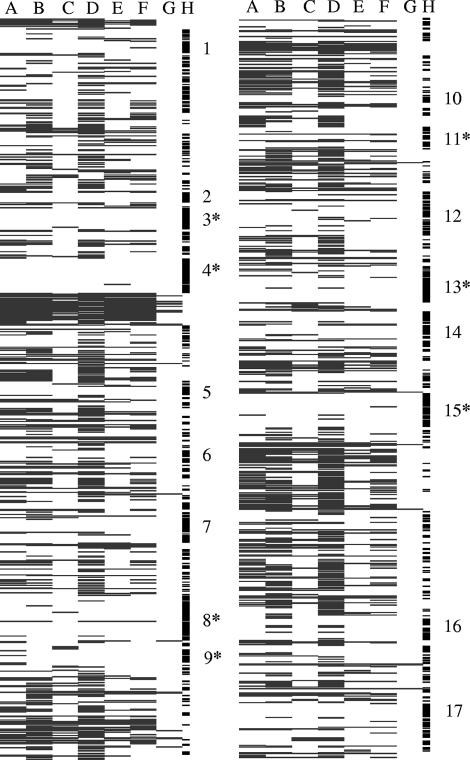
A total of 995 genes, corresponding to 57% of all ORFs, were not detectably transcribed under any in vitro growth condition. These “nontranscribed” genes were not randomly distributed over the NCC533 genome but belonged to about 20 gene clusters (Fig. 4). Three large clusters of nontranscribed genes correspond to mobile DNA elements (two prophages), but most nontranscribed genome regions were much smaller, like, for example, an arsenate resistance cassette. Nontranscribed genes were enriched in the variable-genome part determined in previous DNA-DNA microarray experiments (4).
FIG. 4.
Projection of the L. johnsonii transcripts observed during in vitro and in vivo growth on the genome map of L. johnsonii. The circular L. johnsonii NCC533 genome was split into two halves after ORF LJ0910. The genes are ordered from top to bottom (from LJ0001 to LJ0910 for the left columns and from LJ0911 to LJ1857 for the right columns) according to their positions in the NCC533 genome. Each row corresponds to an amplicon on the array, and the row is marked in black if the amplicon was transcribed under the specified growth conditions and left white if no transcripts were detected in our microarray system. The columns represent the growth conditions. Columns A to C show the expressed genes during the in vitro growth, with column A representing the exponential phase, column B the adaptation phase, and column C the stationary phase. Columns D to G show the expressed genes during in vivo growth of NCC533 in the specified segments of the mouse gut; column D specifies the transcribed amplicons in the stomach, column E those in the jejunum, column F those in the cecum, and column G those in the colon. The small vertical bars in column H represent the gene clusters that were not detectably transcribed during either in vitro or in vivo growth. These clusters contain, according to their gene annotations, the following genes: multidrug resistance and transcriptional regulators (cluster 1), maltose transport genes (2), stress protein and DNA repair genes (3), Lj965 prophage genes (4), transcriptional regulator and succinate metabolism cluster genes (5), PTS system and β-galactosidase cluster genes (6), ABC and PTS transporter genes (7), ABC and PTS system mannose-specific genes (8), bacteriocin cluster genes (9), ComE (competence operon) genes (10), exopolysaccharide operon genes (11), ABC transporter and transcriptional regulator genes (12), acetyltransferase cluster and regulator genes (13), cadmium/manganese and ABC transporter genes (14), Lj928 prophage genes (15), PTS system genes (16), and cobalt transport genes (17). Asterisks represent genes from NCC533 that were not detected by microarray hybridization using genomic DNA from other L. johnsonii strains (4).
Gut distribution of NCC533.
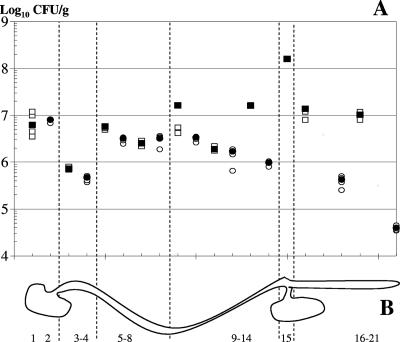
Before exploring the in vivo transcription profile of NCC533 during gut passage, we investigated the segmental distribution of NCC533 in the murine gut. To increase the ratio of NCC533 organisms to autochthonous lactobacilli, we transformed the strain with an antibiotic resistance plasmid and then fed it (at 109 CFU) to five conventional, antibiotic-treated mice over 3 days. Two days after the last feeding, the mice were killed and the distribution of NCC533 genes along the gut was investigated. Similar in situ titers of lactobacilli were determined on MRS plates containing or lacking the applied antibiotic, suggesting that under the used experimental conditions, the intestinal lactobacilli were mainly the orally added NCC533 cells. In the luminal parts of the different gut segments, titers ranged from 8 × 105 to 2 × 108 CFU/g (wet weight) (Fig. 5A). The luminal titers fluctuated between 2 × 106 and 2 × 107 CFU/g throughout the majority of the gut segments; peak titers were found in the cecum and trough titers in the duodenum. Scraping of the flushed gut mucosae revealed lactobacilli associated with the gut surface. In the upper gut segments, the mucosa-associated Lactobacillus cell counts were similar to those observed in the lumen, while in the lower gut segments, the mucosa-associated bacteria showed a titer at least 10-fold lower than that of the luminal bacteria (Fig. 5A).
FIG. 5.
Viable counts of L. johnsonii cells along the mouse gastrointestinal tract. (A) Numbers of live bacteria expressed as log10 numbers of CFU/g (wet weight) in the lumina (squares) or associated with the mucosae (circles) of antibiotic-treated conventional mice 48 h after the last forced-feeding with strain NCC533 (containing an antibiotic resistance plasmid) in the specified gut segments. The filled symbols give the median value for each group of five mice, and the open symbols give the individual values. If fewer than five symbols are seen, they are overlaid by the median. (B) Schematic representation of the murine gastrointestinal tract. 1, forestomach lumen; 2, forestomach (the corpus mucosa of which was scraped); 3-4, lumen and mucosa of duodenum; 5-8, proximal lumen, mucosa, distal lumen, and mucosa of jejunum; 9-14, ileum (which was sectioned into the proximal ileum, middle and distal parts, first lumen, and mucosa); 15, cecum (only the lumen is represented, since there is no cecal mucosa that can be scraped off); 16-21, colon (which was sectioned into a proximal and a distal part, and then from each half, the lumen was squeezed out [fractions 16 and 19], the tube was washed [fractions 17 and 20], bacteria were not counted, and the mucosa was scraped off [fractions 18 and 21] [there are thus four colon samples]).
The bacterial-titer distribution is likely to represent a temporary steady-state situation and not a washout phase. When a similar amount of E. coli K-12 cells was given to control mice, the E. coli titer in the lumen of the distal colon was at the limit of detection (5 × 104 CFU/g), and not at the 107 CFU/g found for NCC533, 24 h after the last forced feeding (11a).
In vivo transcription.
We selected four anatomical sites for bacterial mRNA isolation: the stomach, the jejunum, the cecum, and the colon (see Fig. 5B for an anatomical orientation). Due to the higher cell count, we first targeted luminal bacteria for mRNA isolation. We obtained good-quality prokaryotic mRNA for the cecum and the colon, while only small amounts of low-quality bacterial RNA was recovered from the lumina of the stomach and the jejunum. With some effort, we obtained sufficient amounts of RNA from mucosa-associated bacteria for the stomach and the jejunum to conduct four microarray hybridization experiments. We determined the presence of 786, 296, 391, and 26 transcribed NCC533 genes for the stomach, jejunum, cecum, and colon, respectively. At first glance, it is difficult to believe that only 26 genes are expressed in viable cells found at high titers in the colon. The low number of genes expressed from colon-derived L. johnsonii was not an artifact of low viable-bacterial-cell recovery, nor was the RNA visibly degraded upon biochemical analysis. For the in vivo arrays, we depend on enrichment of bacterial mRNA followed by linear amplification of the isolated bacterial RNA using commercially available kits. To test whether these procedures worked well, we investigated the mRNA expression of another bacterium, Bifidobacterium longum, with the same technique. B. longum contains a number of genes in its genome comparable to the number in L. johnsonii (32), but this commensal shows a completely different gut distribution and gene expression pattern, transcribing 146, 426, and 917 genes in the small intestine, cecum, and colon, respectively (11a). We observed also a good correlation when the levels of expression of selected genes in the colon were compared by real-time reverse transcription-PCR and our microarray system (11a). These data exclude a kit-related bias, which systematically underestimates the transcription of commensal bacteria in the colon.
We thus observed not only substantial differences in gene expression between the three phases of in vitro growth but also even greater differences in expression between the various in vivo growth locations. In view of this great variability, a comparison across the two subsets of expression data is only partially insightful and will therefore be treated independently. This separate treatment is furthermore justified by the very different nutritional conditions encountered during in vitro and in vivo growth.
Comparison of in vitro and in vivo growth conditions.
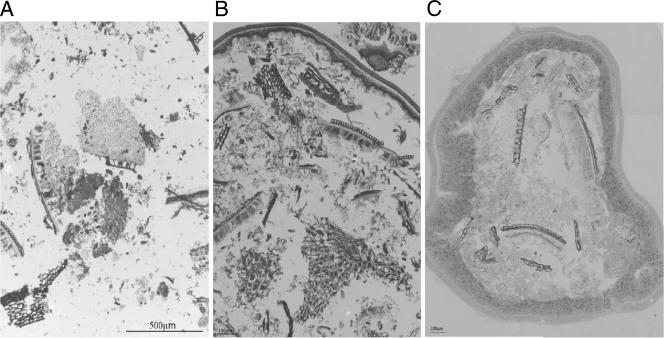
Our supplemented MRS medium contained peptone, beef extract, yeast extract, dextrose, and glucose, which support the growth of all lactobacilli from oral, fecal, and dairy sources. The chow of mice consisted of 69% (wt/wt) cereals, 20% soybean meal and yeast, and 6% fish meal, constituting carbohydrate, protein, and lipid contents of 52, 21, and 5%, respectively. L. johnsonii showed a rapid titer increase to 108 CFU/ml on a broth consisting of crushed chow (data not shown). Microscopic analysis of the chow (Fig. 6A) revealed the aleuron layer from wheat and maize, which stained intensively because of its cytoplasmic-protein content, overlaid by the cell walls of the cross cell layer. Below the aleuron layer is the endosperm, containing starch granules. Soybean material was easily recognized by the elongated protein bodies of their cotyledons. The darkest spots were identified as yeast cells. When the stomachs of mice were sectioned 5 h after the last feeding, no substantial protein digestion was observed; however, the starch in the endosperm was hydrolyzed (Fig. 6B). Finally, in the colons of the same mice, we identified fragments of wheat, maize, and soybeans with relatively intact cell wall structures, while the aleuron layer of the cereals appeared empty and no protein bodies were stained in the soybean cotyledons (Fig. 6C). These microscopic data suggest carbohydrate digestion in the proximal mouse gut and protein digestion in the distal mouse gut segments.
FIG. 6.
Microscopic comparison of the mouse chow before digestion (A) and as food content of the stomach (B) and the colon (C) of a mouse reared on this food. The pictures are 2-μm-thick cryosections through the food pellets and the specified gut cross sections. The mounting medium was 1% toluidine blue in 1% sodium borax and 40% glycerol.
During in vitro growth, the pH of the MRS medium decreased from 5.9 to 3.7 (Fig. 1), while low pH values were encountered in vivo only in the stomach (pH 3.5 in the proximal part and pH 2.2 in the pyloric part of the murine stomach [14]). In vitro growth occurred in a jar under anaerobic conditions, while in vivo growth occurred under relatively aerobic conditions in the stomach. Oxygen pressure fell along the small intestine to become anaerobic only in the distal parts of the gut (34).
Analysis of the in vivo transcription profile.
The major colon transcripts from L. johnsonii were detected under all growth conditions and encoded ribosomal proteins, stress proteins (DnaK, ClpE, and a multidrug exporter), and a few enzymes. The colon transcription did not correspond to that of the in vitro stationary phase.
The expression pattern of L. johnsonii in the cecum documents a metabolically active cell, in contrast to the transcriptional silence in the colon (Fig. 7A). Most of the cecum-expressed genes belonged to transcripts active during both the exponential and the adaptation phase of the in vitro growth. These genes included several sugar PTS importers. The L. johnsonii genes transcribed in the cecum are mainly a subgroup of the genes also transcribed in the stomach (Fig. 7D). Only 16 genes transcribed in the cecum were not transcribed in other gut segments or in vitro. A galactosamine PTS transporter was the only operon in this group of cecum-specific genes.
FIG. 7.
Venn diagrams comparing the L. johnsonii expression patterns obtained under various growth conditions. (A to C) Comparisons of the NCC533 genes expressed in the cecum (A), jejunum (B), and stomach (C) with those expressed in the exponential and adaptation phases during the in vitro growth of NCC533. (D) Distribution of the genes according to their expression in the three specified gut segments.
L. johnsonii isolated from the jejunum showed transcripts for 296 genes. Most of them were also transcribed during both the exponential and adaptation phases of the in vitro growth (Fig. 7B). We identified 59 genes transcribed in the jejunum but not in vitro; this figure decreased to 26 genes when genes transcribed in other gut segments were subtracted. The jejunum-specific genes included four transcriptional regulators and seven transporters, including three PTS transporters annotated with fructose, glucose, and cellobiose specificity.
With respect to expression level, no clear dominance of genes could be detected (transcripts from ribosomal proteins were excluded from the analysis because they cross-hybridized with other lactobacillus species in the murine gut). In the stomach, transporters, cell wall synthesis, nucleotide metabolism, and transcriptional regulator genes represented, respectively, 7, 5, 5, and 3 transcripts within the 50 most highly transcribed genes. Within the 50 most often expressed NCC533 genes in the jejunum, we detected 6 genes encoding sugar-digesting enzymes, 3 transporter genes, and 22 hypothetical genes. In the cecum, transporter proteins (n = 12) figured most prominently in the 50 most highly transcribed genes.
The largest number of L. johnsonii genes was transcribed in the stomach. Most genes expressed during in vitro growth were also transcribed in L. johnsonii recovered from the stomach. The stomach expression profile was not closer to the adaptation than to the exponential phase of in vitro growth (Fig. 7C). In fact, the stomach contained additional transcripts not expressed in vitro, such that the stomach profile defines a growth state sui generis. More specifically, 191 of the 785 stomach transcripts were not expressed in vitro (Fig. 7C), and 119 were expressed neither in vitro nor in any other gut segments. Neither the categorization of the transcripts into functional classes nor the visual screening of the gut segment-specific transcripts revealed a clear-cut metabolic differentiation of the NCC533 strain when it was recovered from the stomach, jejunum, and cecum (Fig. 8). However, there is nevertheless some relevant information in the transcription pattern even if we have difficulties in reading the patterns. For example, when NCC533's transcription in the stomach was analyzed, we observed the expression of several putative multidrug transport proteins, a cation efflux protein, and, specifically, a copper-transporting ATPase; the corresponding genes were not transcribed in vitro. These genes were previously also identified in Lactobacillus plantarum when in vivo expression technology in mice (6) or PCR technology (24) was used. Several PTS system genes of both Lactobacillus species, sometimes with the same substrate specificity (e.g., galactosamine), and DNA polymerase III were specifically transcribed in the gut (6).
FIG. 8.
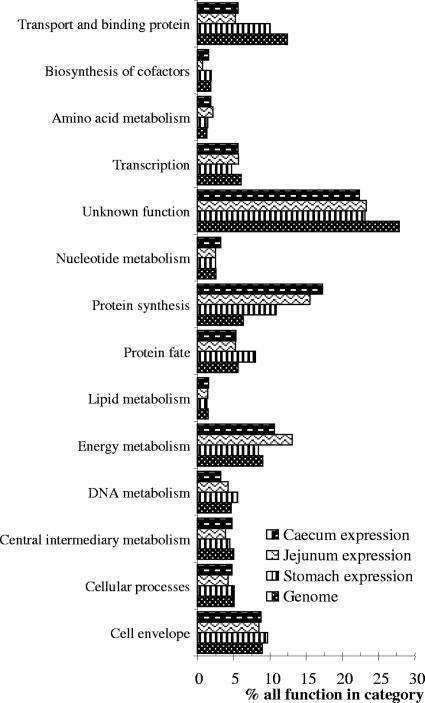
Functional attribution of the in vivo-expressed genes of L. johnsonii NCC533 into the main TIGR-CMR role classes. For each category, the lowest bar represents the percentage of genes in that category as detected in the sequenced genome of NCC533. The three bars on top of it display the percentages of the genes which fall into this category that are transcribed in the stomach, jejunum, and cecum, respectively, from bottom to top. See the legend to Fig. 3 for “% all function in category” and “Genome.”
Finally, we analyzed the functional attributions of the genes transcribed in the different gut locations with respect to the functional categories in the genome (Fig. 8). Fewer of the transcripts from all in vivo phases had unknown functions (23, 23, and 22% of the genes that were expressed in the stomach, jejunum, and cecum, respectively, had unknown functions, compared with 28% of genes in the whole genome). In vivo transcripts were enriched for functions of protein synthesis (10, 16, and 17% of the transcribed genes in the stomach, jejunum, and cecum, respectively, were enriched for protein synthesis, compared with 6% of the genes in the genome). Genes expressed in the stomach were enriched for the function of protein fate (8% of transcribed versus 6% of all genes). In the jejunum and cecum, we observed an increased transcription of genes involved in energy metabolism (13 and 11%, respectively, versus 9% of the all genes) and a decreased transcription of genes from the transport and binding protein categories (5 and 6%, respectively, versus 12% of all genes) (Fig. 8).
Core transcriptome.
We observed that 202 (Fig. 2) and 212 (Fig. 7) genes were expressed under all in vitro and all in vivo conditions, respectively. A total of 103 genes were transcribed under both in vitro and in vivo conditions. They represented a type of “core transcriptome” encoding mainly ribosomal proteins and translation factors, ATP synthase chains, DNA/RNA polymerases, and key carbohydrate catabolic enzymes.
DISCUSSION
Microarray-based expression analysis allows a bird's eye view of the metabolic state of a bacterial cell under different culture conditions. The difficulty of isolating enough mRNA from the investigated bacterial strain in its natural environment has prevented a wider application of this technique. With respect to gut bacteria, which coexist with hundreds of other bacterial species, some researchers used mono-colonized axenic mice (12, 35). This approach yields larger amounts of specific bacterial mRNA, but the situation is artificial since the investigated bacterium is no longer under competition pressure from other bacteria, which is a fundamental characteristic for the gut microbiota (23). In the present work, we opted for a combination of force-feeding (to introduce the study bacterium) and antibiotic treatment (to decrease the competition by the endogenous microbiota without eliminating it). However, there is not only a quantity problem but also quality problems: in the upper parts of the gut, only mucosa-associated, and not luminal, bacteria yielded mRNA suitable for microarray analysis. We suspect that the large amount of enzymes secreted into the upper intestine had a negative impact on mRNA extraction. As mucosa-associated bacteria were in limited supply, we could conduct only hybridization experiments with a few biological samples from the upper gut, while we could conduct more experiments with the luminal material from the lower gut (colon, cecum). Despite that caveat, it is clear that L. johnsonii is transcriptionally very active in the stomach and exhibited here a very broad expression pattern exceeding that of in vitro growth qualitatively, but not quantitatively. This conclusion is quite plausible since lactobacilli are the dominant microbiota in the murine stomach (26, 41). This observation is explained by the peculiar physiological compartmentalization of the murine stomach, where one part develops only moderate acidity, thus allowing a permanent colonization with acid-resistant lactobacilli (31).
A relatively homogeneous distribution of lactobacilli was found along the length of the intestine (26). This observation is compatible with one of two interpretations: lactobacilli are active in all gut segments or they are seeded from their colonization site in the stomach into the more distal parts of the intestine, where they just transit and survive. In a microarray analysis, L. johnsonii showed an expression pattern in the stomach distinct from that in the jejunum and cecum, while it showed a dramatically down-regulated transcription in the colon. L. johnsonii organisms recovered from the gut do not represent the orally applied bacteria in transit but a temporary steady-state population. This interpretation is backed by the longer gut residence time of L. johnsonii than that of E. coli or L. plantarum (24). Apparently, L. johnsonii expresses its genome differently when located in different gut segments. A straightforward interpretation is complicated by the facts that data from the lower gut segments (cecum and colon) were derived from luminal bacteria while those from the upper segments (stomach to ileum) came from mucosa-associated bacteria. However, the active transcription observed in L. johnsonii isolated from the lumen of the cecum excludes the possibility that luminal gut bacteria are necessarily down-regulated for gene expression. The low-level colon transcription is not a technical artifact since Bifidobacterium longum investigated in our lab with the same technique showed the highest transcription levels specifically in the colon (11a). In addition, the few colon-specific transcripts were highly expressed.
We do not know enough about the ecology of L. johnsonii to interpret its transcriptional down-regulation in the colon with respect to its survival strategy. L. johnsonii has been isolated from humans, the crops of chicken, the intestines of mice and pigs, and the stomachs of rats (7, 15, 16, 28, 30). Since mice are coprophageous, murine intestinal lactobacilli might simply rely on a direct fecal-oral transmission route. Lactobacilli must therefore ensure their survival in the feces for only a day and for the rapid up-regulation of the fecal bacteria when they meet again the permissive growth conditions of the stomach. In fact, 1-day-old fecal samples from mice yielded rapidly growing colonies on agar plates. Interestingly, in contrast to other gut bacteria like Bifidobacterium longum (18a), L. johnsonii does not lose cell viability in the stationary phase of in vitro growth and overnight L. johnsonii cultures changed within less than 10 min their transcription pattern when transferred into a fresh medium, thus excluding the possibility of a longer-lasting repressed state.
Our data showed that, during in vitro growth, early, middle, and late exponential phases are characterized by a unique “exponential” transcription pattern. In contrast, the early, middle, and late stationary phases cover at least two distinct transcription patterns (“adaptation” and “stationary”). The “adaptation” phase showed a higher number of transcribed genes than the exponential phase (626 versus 537). Even if these bacteria are no longer growing, the cells are transcriptionally and metabolically very active, as was demonstrated by the sharp pH decline in the growth medium observed in the transition period between the exponential and stationary phases. When we analyzed the functional annotations of the transcribed genes, we diagnosed a shift from cell division gene expression in the exponential phase to more carbohydrate metabolism gene transcription in the “adaptation” phase. Later on in the growth cycle, L. johnsonii changes its transcription pattern again: fewer genes are transcribed, and also the overall level of transcription decreases such that the term stationary phase is justified. This transcription level was then maintained over at least a day, although with less signal intensity.
As determined by our analysis, less than half of the L. johnsonii genome is transcribed in vitro. There are certainly genes that are transcribed below the sensitivity threshold of our microarray system. However, two observations argue against a serious underestimation of the in vitro transcription in our study. Nontranscribed genes were clustered on the genome, and mobile-DNA and variable-genome segments were overrepresented in this category. Apparently, only part of the genetic program of L. johnsonii is activated during broth growth. This conclusion was supported by the substantial number of gut-expressed genes that were silent during the broth growth of L. johnsonii. Numerous genes might be carried in bacterial genomes that manifest their selective value only under peculiar in vivo growth conditions. As the murine stomach is a potential ecological niche for L. johnsonii, its differential gene expression at this location might be of physiological relevance. It could mean that to stay in this anatomical site, L. johnsonii must activate more genes than are necessary for broth growth. Notably, the in vivo expression of drug and cation transporters, which were not transcribed during in vitro growth, suggests stress reactions of L. johnsonii, as was observed with L. plantarum isolated from the murine gut. Our study accounts for only 982 transcribed genes (56% of the genome) when all in vitro and in vivo transcripts are combined. As the ecology of L. johnsonii has not been characterized in detail, we cannot even speculate under what conditions the remaining genes are expressed. Notably, 33% of the nontranscribed genes belonged to the unknown-function category. For bacterial genomes, in silico ORF prediction commonly shows up to 30% of genes without database matches. Molecular microbiologists will have to study their bacteria under both in vitro and in vivo conditions; the latter should include various combinations of specified conditions. However, the possibilities that strain-specific ORFs that are not transcribed under standard in vitro and in vivo growth conditions do not play a role in the physiology of the cell and represent a type of genetic noise cannot be excluded. Moreover, intraspecies comparisons frequently reveal gene differences between strains of 15%; this is also the case for L. johnsonii (4).
Distinct in vivo and in vitro expression profiles were also published for other gut bacteria. Campylobacter jejuni investigated in an ileal loop showed transcriptional adaptation to an oxygen-limited, nutrient-poor, and hyper-osmotic environment (37). The most striking case is that of Vibrio cholerae. Transcriptional profiling of V. cholerae from stool samples of patients revealed a unique hyperinfectious state defined by a high expression of genes required for nutrient acquisition and motility and a down-regulation of chemotactic signaling compared with that in in vitro-propagated V. cholerae (25). The growth of V. cholerae in a rabbit ileal loop suggested that nutrient limitation and anaerobiosis are major stresses experienced by V. cholerae during in vivo growth (45).
Some data suggest that the transcription profile of well-investigated bacteria like E. coli can be used to probe the nutritional constraints of an in vivo environment (33). Unfortunately, microarray expression data are lacking for E. coli commensals growing in the gut (10). These data would be especially interesting, since we know that growth in the murine gut selects for a coccoid form of E. coli, while in vitro growth selects again for rod-shaped E. coli, the form known to microbiologists (19). As long as we do not understand the nutrition of such well-studied bacteria as commensal E. coli in the large intestines of mice, we miss key data for a more complete understanding of microbial gut colonization. However, such data are also important for less well characterized bacteria like L. johnsonii, which belongs to the growing list of probiotic bacteria (40). A rational use of health-promoting bacteria must be based on a sound understanding of their interactions with competing gut bacteria and mammalian gut cells.
Finally, a word of caution. Microarray techniques create large data sets, which are not an easy starting point for post hoc hypothesis building. It is easier to test a priori-formulated hypotheses with microarray expression data or to use them after fusion with other data sets for screening purpose. We compared, for example, two L. johnsonii strains differing in their in vivo phenotypes (gut persistence times in mice) using DNA-DNA and intestinal-expression microarrays. We identified three gene loci specific to the long-persistence strain, which were also expressed in the murine gut. Knockout mutants demonstrated that two of the three mutants showed a shortened gut persistence time in mice (11a). Thus, only when combined with other approaches do expression microarrays reveal their analytical power.
Acknowledgments
We thank Catherine Schwartz and Massimo Marchesini for assistance with the animal experiments and Marie-Lise Dillmann and Marietta Weiss for microscopy. We also thank Annick Mercenier for comments and fruitful discussions.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amit-Romach, E., D. Sklan, and Z. Uni. 2004. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 83:1093-1098. [DOI] [PubMed] [Google Scholar]
- 3.Bateup, J. M., M. A. McConnell, H. F. Jenkinson, and G. W. Tannock. 1995. Comparison of Lactobacillus strains with respect to bile salt hydrolase activity, colonization of the gastrointestinal tract, and growth rate of the murine host. Appl. Environ. Microbiol. 61:1147-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, B., R. D. Pridmore, C. Barretto, F. Delmas-Julien, K. Schreiber, F. Arigoni, and H. Brüssow. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 189:1311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergonzelli, G. E., S. Blum, H. Brüssow, and I. Corthésy-Theulaz. 2005. Probiotics as a treatment strategy for gastrointestinal diseases? Digestion 72:57-68. [DOI] [PubMed] [Google Scholar]
- 6.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, P. G., G. D. Casey, G. E. Gardiner, M. Tangney, C. Stanton, R. P. Ross, C. Hill, and G. F. Fitzgerald. 2004. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett. Appl. Microbiol. 39:431-438. [DOI] [PubMed] [Google Scholar]
- 8.Castillo, M., S. M. Martin-Orue, E. G. Manzanilla, I. Badiola, M. Martin, and J. Gasa. 2006. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 114:165-170. [DOI] [PubMed] [Google Scholar]
- 9.Chaillou, S., M. C. Champomier-Verges, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongere, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 10.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dellagio, F., and G. E. Felis. 2005. Taxonomy of lactobacilli and bifidobacteria, p. 25-49. In G. W. Tannock (ed.), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom.
- 11a.Denou, E. 2006. From genes to transcripts to phenotypes: preliminary characterization of commensalism with Lactobacillus, Bifidobacterium and Escherichia coli in the murine gut. Ph.D. thesis. University of Caen, Caen, France.
- 12.Drouault, S., J. Anba, and G. Corthier. 2002. Streptococcus thermophilus is able to produce a beta-galactosidase active during its transit in the digestive tract of germ-free mice. Appl. Environ. Microbiol. 68:938-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egert, M., A. A. de Graaf, H. Smidt, W. M. de Vos, and K. Venema. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14:86-91. [DOI] [PubMed] [Google Scholar]
- 14.Freter, R., E. Stauffer, D. Cleven, L. V. Holdeman, and W. E. Moore. 1983. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect. Immun. 39:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, L. L., K. E. Hagen, G. W. Tannock, D. R. Korver, G. M. Fasenko, and G. E. Allison. 2003. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl. Environ. Microbiol. 69:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan, C. W., J. C. Astaire, M. E. Sanders, B. S. Reddy, and C. L. Kitts. 2001. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 67:1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 18.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Klijn, A. W. P. E. 2005. Physiological and molecular characterisation of stress responses in Bifidobacterium longum NCC 2705. Ph.D. thesis. University College Cork, Cork, Ireland.
- 19.Krogfelt, K. A., L. K. Poulsen, and S. Molin. 1993. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect. Immun. 61:5029-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruis, W., P. Fric, J. Pokrotnieks, M. Lukas, B. Fixa, M. Kascak, M. A. Kamm, J. Weismueller, C. Beglinger, M. Stolte, C. Wolff, and J. Schulze. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 22.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 23.Lievin-Le Moal, V., and A. L. Servin. 2006. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 19:315-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marco, M. L., R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norin, K. E., A. K. Persson, H. Saxerholt, and T. Midtvedt. 1991. Establishment of Lactobacillus and Bifidobacterium species in germfree mice and their influence on some microflora-associated characteristics. Appl. Environ. Microbiol. 57:1850-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen, K., and G. W. Tannock. 1989. Colonization of the porcine gastrointestinal tract by lactobacilli. Appl. Environ. Microbiol. 55:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pena, J. A., S. Y. Li, P. H. Wilson, S. A. Thibodeau, A. J. Szary, and J. Versalovic. 2004. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl. Environ. Microbiol. 70:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petry, S., S. Furlan, M. J. Crepeau, J. Cerning, and M. Desmazeaud. 2000. Factors affecting exocellular polysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus grown in a chemically defined medium. Appl. Environ. Microbiol. 66:3427-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roach, S., D. C. Savage, and G. W. Tannock. 1977. Lactobacilli isolated from the stomach of conventional mice. Appl. Environ. Microbiol. 33:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnenburg, J. L., C. T. Chen, and J. I. Gordon. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 36.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 37.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73:1797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szymanski, H., J. Pejcz, M. Jawien, A. Chmielarczyk, M. Strus, and P. B. Heczko. 2006. Treatment of acute infectious diarrhoea in infants and children with a mixture of three Lactobacillus rhamnosus strains—a randomized, double-blind, placebo-controlled trial. Aliment. Pharmacol. Ther. 23:247-253. [DOI] [PubMed] [Google Scholar]
- 39.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tannock, G. W. 2005. Probiotics and prebiotics. Caister Academic Press, Norfolk, United Kingdom.
- 41.Tannock, G. W., O. Szylit, Y. Duval, and P. Raibaud. 1982. Colonization of tissue surfaces in the gastrointestinal tract of gnotobiotic animals by lactobacillus strains. Can. J. Microbiol. 28:1196-1198. [DOI] [PubMed] [Google Scholar]
- 42.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lacto-bacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaughan, E. E., H. G. Heilig, K. Ben Amor, and W. M. de Vos. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 44.Walter, J., N. C. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]