Abstract
Bacteriophage GA-1 infects Bacillus sp. strain G1R and has a linear double-stranded DNA genome with a terminal protein covalently linked to its 5′ ends. GA-1 protein p6 is very abundant in infected cells and binds DNA with no sequence specificity. We show here that it binds in vivo to the whole viral genome, as detected by cross-linking, chromatin immunoprecipitation, and real-time PCR analyses, and has the characteristics of a histone-like protein. Binding to DNA of GA-1 protein p6 shows little supercoiling dependency, in contrast to the ortholog protein of the evolutionary related Bacillus subtilis phage φ29. This feature is a property of the protein rather than the DNA or the cellular background, since φ29 protein p6 shows supercoiling-dependent binding to GA-1 DNA in Bacillus sp. strain G1R. GA-1 DNA replication is impaired in the presence of the gyrase inhibitors novobiocin and nalidixic acid, which indicates that, although noncovalently closed, the viral genome is topologically constrained in vivo. GA-1 protein p6 is also able to bind φ29 DNA in B. subtilis cells; however, as expected, the binding is less supercoiling dependent than the one observed with the φ29 protein p6. In addition, the nucleoprotein complex formed is not functional, since it is not able to transcomplement the DNA replication deficiency of a φ29 sus6 mutant. Furthermore, we took advantage of φ29 protein p6 binding to GA-1 DNA to find that the viral DNA ejection mechanism seems to take place, as in the case of φ29, with a right to left polarity in a two-step, push-pull process.
A large variety of phages that infect bacteria of the genus Bacillus have been characterized. Particular attention has been given to the so-called φ29 family of lytic phages that infect different Bacillus species. They all possess a linear double-stranded DNA genome of about 20 kb, with a terminal protein covalently linked to the 5′ ends. The terminal protein has a dual function in priming the initiation of phage DNA replication at both genome ends (reviewed in reference 43) and in the hydrolytic degradation of the host cell wall during the ejection process (31). On the basis of serological properties, DNA physical maps, peptide maps, and partial or complete DNA sequences, the φ29-like genus of bacteriophages has been classified into three groups (36, 52, 53). The first group includes phages φ29, PZA, φ15, and BS32; the second group includes B103, Nf, and M2Y; and the third group contains GA-1 as its sole member. Phage φ29 has been extensively characterized, being one of the best-studied bacteriophages of gram-positive bacteria. Its mechanism of DNA replication and its regulation of transcription have been reviewed previously (39, 43). Within this family of phages, GA-1 is the one most distantly related to φ29. Its genome is 1.8 kb larger than that of φ29, although they are similarly organized: the early genes are clustered in two operons located at each end of the genome, while the late genes are located in a single operon positioned at the central part of the viral DNA. As in φ29 and B103, the late GA-1 operon is flanked on its left side by an early operon that contains genes required for DNA replication and for transcriptional regulation (genes 6 through 2). These genes are expressed from the early tandemly organized promoters A2b and A2c (22). The right region of the GA-1 genome contains open reading frames (ORFs) whose deduced protein sequences are homologous to those of the φ29 early genes 17 and 16.7, which are involved in DNA replication (12, 30) and genome internalization (3, 20). These genes are expressed from the early promoter C2. However, both ends of the GA-1 genome contain a number of sequences and ORFs that have no counterparts in φ29 or in any of the other related phages characterized (29). The putative proteins encoded by these ORFs could be involved in as-yet-unknown aspects of the phage development, and/or they may have a role in interaction with the infected host. In fact, φ29 infects Bacillus subtilis, while GA-1 infects the poorly characterized Bacillus sp. strain G1R and is unable to infect intact B. subtilis cells (4). The sequence analysis of G1R 16S rRNA showed that Bacillus sp. strain G1R is more than 99% identical to that of Bacillus pumilus, an evolutionary distance that is small enough to consider them two strains of the same species (21).
B. subtilis phage φ29 protein p6 is a small and abundant protein (about 700,000 copies per cell [2]) required for in vivo viral DNA replication (10, 44) and has been shown to activate in vitro the initiation step (6, 35). It is also involved in transcription control, repressing early promoter C2 at the DNA right end (9, 50) and, together with the viral protein p4, repressing early promoters A2b and A2c and activating late promoter A3 (15). In vitro, both the stimulation of the initiation of DNA replication and the control of transcription require the formation of a protein p6-DNA nucleoprotein complex, in which the DNA adopts a right-handed toroidal, and therefore positive, superhelical conformation, wrapping around a multimeric protein core (48). Protein p6 forms multimeric right-handed helical filaments (1) similar to those described for Escherichia coli dnaA (16) or eukaryotic origin recognition complex (11). Wrapping of replication origins around such filaments has been proposed to be a general mechanism to promote DNA unwinding and subsequent assembly of replication forks (33). φ29 protein p6 restraining of positive supercoiling implies that DNA binding is inversely proportional to the degree of negative supercoiling, as shown in vivo and in vitro (19). In addition, in vivo p6 binding to viral DNA increases dramatically upon addition of novobiocin, which inhibits bacterial gyrase, producing a decrease in negative supercoiling (34). Furthermore, novobiocin strongly impairs viral DNA replication. These results demonstrated that φ29 linear DNA, although it has a terminal protein covalently linked to the ends and therefore is not covalently closed, is topologically constrained (18). Protein p6 binds in vivo to most, if not all, of the φ29 DNA (19), so its functions in replication and transcription could be functional outcomes of a more global role as a histone-like protein, which participates in organization and compaction of the viral genome. Such pleiotropic effects are normal properties of this kind of protein and have been described for the four major histone-like proteins of E. coli: HU, IHF, Fis, and H-NS (13, 14, 23, 28, 42).
Although bacteriophage GA-1 protein p6 exhibits similar functional properties than that of φ29, that is, activation of initiation of DNA replication (17) and repression of early transcription from promoter C2 (21), and these properties are the result of the formation of a multimeric complex with the DNA, the structure of this complex differs substantially from that of φ29, as deduced from the different periodicity of DNA binding. As in φ29, the GA-1 protein p6 binds to DNA as a dimer; however, GA-1 protein p6 dimers bind to DNA with a periodicity of 22 bp (17), instead the 24-bp periodicity reported for φ29 (37). The binding of a single φ29 p6 dimer to a covalently closed DNA has been estimated to increase the linking number, and the path of the DNA in the φ29 p6 complex has been described as a right-handed toroidal supercoil with a pitch of 63 bp (46). The different binding periodicity of the GA-1 protein p6 suggests also the formation of a right-handed DNA complex, but with a pitch larger than that of φ29. In fact, it has been shown that GA-1 protein p6 induces lower positive supercoiling on plasmid DNA in comparison to φ29 p6 (17). Thus, the different properties found in vitro between the GA-1 and φ29 proteins prompted us to study the in vivo GA-1 protein p6 binding to DNA and, in particular, how the different structure of the nucleoprotein complex affects the supercoiling binding dependency.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
The bacterial strains used were Bacillus sp. strain G1R (4) and B. subtilis 110NA (trpC2 spo0A3 su−) (32). The plasmids used were the pUB110 derivatives pPR55w6, pPR55ow6 (7), and pPR54 (45). Plasmid pPR55w6 constitutively expresses bacteriophage φ29 protein p6 from the phage λ PR promoter; plasmid pPR55ow6 has gene 6 in its opposite orientation. Plasmid pPR54 has an extra cassette that encodes a temperature-sensitive repressor of the λ PR promoter and was used to clone GA-1 gene 6. Infections were performed at multiplicity of 10 with either of the following phages: GA-1 wild-type (4), φ29 sus6(626), a replication-null mutant (38), or φ29 sus14(1242), a delayed lysis mutant with an otherwise wild-type phenotype (26). The genomes of these mutant φ29 phages contain a suppressible nonsense mutation in the gene indicated.
Medium, enzymes, drugs, and reactives.
Bacteria were grown in Luria-Bertani (LB) medium supplemented with 5 mM MgSO4 and 1 μg of phleomycin (Cayla S.A.R.L.)/ml. Micrococcal nuclease was from Amersham Pharmacia Biotech and proteinase K from Boehringer Mannheim. Protein A-Sepharose CL-4B, lysozyme, RNase A, chloramphenicol, and novobiocin were from Sigma-Aldrich. Formaldehyde at 37% was from Calbiochem.
DNAs and oligonucleotides.
Proteinase K-digested GA-1 and φ29 DNA was obtained as described previously (24). The sequences of the oligonucleotides (Isogen) used for PCR amplification of GA-1 DNA regions (coordinates are shown in parentheses) are as follows: region G1 (1 to 300), 5′-AAATAGAGTCCACCCTTCCTCCCCT and 5′-GGCGCAATGAGAAACACCACAGCT; region G2 (6462 to 6823), 5′-GCAAATTTAGCCGTTGTTGTTGGG and 5′-AGTTGTGCGTTGAACTCTTCTAAACTC; region G3 (10695 to 11001), 5′-TTCTGCTCCTACTTATCAAAAGTC and 5′-ATCTGTGTTAAACTTTTTTACCTGC; region G4 (16302 to 16567), 5′-GCTACTGATTTAGAGCTACTTAAG and 5′-GGTATTTTAATAGTTTCTCCTGCG; and region G5 (20817 to 21129), 5′-CATGACATGCGTCAAGATATATCC and 5′-AAATAGATTCCCCATGAACAAGCG. The sequences of the oligonucleotides used for amplification of φ29 DNA regions are as follows: region φ1 (1 to 259), 5′-AAAGTAAGCCCCCACCCTCACATG and 5′-GCCCACATACTTTGTTGATTGG; region φ2 (4895 to 5257), 5′-GATTTCTCTCTGCATCATTTTTGC and 5′-CAAAATATCTTCGTGTTCTTCTGG; region φ3 (7255 to 7528), 5′-GAAGTAGATGATATTAAGGACGCC and 5′-CTGACAGAAGACCAAGCACATCGG; region φ4 (9507 to 9820), 5′-CTGACAACATCGGAAATTACAGCG and 5′-TTGTTGTAAACGTCTCTCTGACCC; region φ5 (11567 to 11778), 5′-GGATTCTCAATGACGGGTTAGA and 5′-CACATACACAGGAAAACCAGACTC; and region φ6 (18998 to 19285), 5′-AAATAGATTTTCTTTCTTGGCTAC and 5′-AAAGTAGGGTACAGCGACAACATAC. A DNA plasmid region present in plasmids pPR55w6, pPR55ow6, and pPR54, named region P1 (2578 to 2873), was amplified with the oligonucleotides 5′-GGGCACAAATCGCATCGTGGAACG and 5′-TCTTGGTCGTCAGACTGATGGGCC.
Cloning and expression of bacteriophage GA-1 gene 6 and Bacillus transformation.
The DNA fragment corresponding to GA-1 gene 6, including its own ribosomal binding site, was obtained by PCR amplification from the viral genome using two primers, primer 1 (5′-TGTTTCCTAGCGCTGCAGTTTAAAATTATA) and primer 2 (5′-GTATATCATAAGGGCTGCAGATTAACAAAC), both containing a PstI site (underlined). The amplified fragment, 373 bp in length, was digested with PstI, purified from agarose gel by using the QIAquick gel extraction kit (QIAGEN) and inserted into the PstI site of the expression vector pPR54. The ligation mixture was used to transform competent B. subtilis YB886 cells (41, 51). Transformants were selected for phleomycin resistance (1 μg/ml). Recombinant plasmid was checked by PstI restriction analysis and DNA sequencing and used to transform competent B. subtilis 110NA cells. B. subtilis 110NA harboring the recombinant plasmid were grown in LB medium at 30°C up to 108 cells/ml and temperature shifted up to 40°C during 15 min. Cells were then centrifuged and resuspended in LB medium at 30°C before infection. B. subtilis 110NA cells transformed by plasmids pPR55w6 and pPR55ow6 were treated in the same way to ascertain the same infection conditions. Competent Bacillus sp. strain G1R cells were also transformed with plasmids pPR55ow6 or pPR55w6 as described previously (41, 51).
Cross-linking, immunoprecipitation, and DNA amplification.
Bacteria were grown at 30°C up to 108 cells/ml and infected according to the conditions described above. Chloramphenicol (34 μg/ml) and novobiocin (500 μg/ml) were added at the indicated times. Novobiocin, a gyrase inhibitor, was used to decrease negative supercoiling. Cross-linking and chromatin immunoprecipitation (X-ChIP) was carried out essentially as described previously (20). Briefly, cells were cross-linked with 1% formaldehyde for 5 min and lysed, and DNA was sheared by sonication using a 150Watt Ultrasonic Disintegrator (MSE, UK). Fragments with an average size of about 750 bp were obtained from 2-ml samples after three pulses of 10 s at 15-μm amplitude, separated by at least 20-s intervals. To avoid heating, samples were kept on ice. Cell debris was eliminated by centrifugation. A 1/20 portion of each sample was kept for total DNA analysis (T sample), and the remainder was split for immunoprecipitation overnight at 4°C with either polyclonal antibodies anti-p6 (αp6 sample) or preimmune serum (pi sample) (20 μl each) and processed as described previously (20). All samples were incubated overnight at 65°C with shaking to reverse cross-links, and DNA was purified by phenol and chloroform extraction, ethanol precipitated, and finally resuspended in water. Analysis of DNA samples was performed by real-time PCR in a LightCycler instrument using a Light Cycler-FastStart DNA master SYBR Green I Hot-start reaction mix (Roche). Amplification conditions included a preheating step of 10 min at 95°C to activate the Taq polymerase, followed by 30 cycles comprising a denaturation step of 15 s at 95°C for all regions. Hybridization or elongation temperature conditions used to amplify GA-1, φ29, and plasmid DNA regions (coordinates are shown in parentheses) were as follows: G1 (1 to 300) and G5 (20817 to 21129), 10 s at 57°C and 40 s at 72°C; G3 (10695 to 11001) and G4 (16302 to 16567), 10 s at 60°C and 40 s at 72°C; G2 (6462 to 6823), 10 s at 59°C and 40 s at 72°C; φ1 (1 to 259) and φ5 (11567 to 11778), 5 s at 53°C and 15 s at 72°C; φ2 (4895 to 5257), φ3 (7255 to 7528), and φ4 (9507 to 9820), 10 s at 51°C and 30 s at 72°C; φ6 (18988 to 19285), 5 s at 50°C and 40 s at 72°C; and region P1 (2578 to 2873) from plasmids pPR55ow6, pPR55w6, and pPR54, 10 s at 50°C and 15 s at 72°C. Finally, a melting analysis was performed by continuous fluorescence measurement from 65 to 95°C to ensure that a single product was amplified. Protein p6 binding values were expressed as the immunoprecipitation coefficient (IC): [(αp6 − pi)/T] × 106, where T stands for total DNA and where αp6 and pi represent DNA immunoprecipitated with serum against p6 and preimmune serum, respectively. Primer hybridization to bacterial DNA was previously ruled out in a control with uninfected cells. The IC values shown in the present study correspond to one representative experiment of at least two, which among themselves differed less than 10%.
Western blot analysis.
Cells were grown at 30°C up to a density of 108 cells/ml and temperature shifted up to 40°C during 15 min. At the time indicated, 1.5-ml aliquots were transferred to ice-cold tubes, concentrated 7.5-fold in loading buffer (60 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate, 5% β-mercaptoethanol, 30% glycerol), and disrupted by sonication. Samples were subjected to sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis, and proteins were transferred by using a Mini-Trans-Blot apparatus (Bio-Rad) at 100 mA and 4°C for 60 min. Immobilon-P membranes (Millipore) were probed with 1:2,000-diluted φ29 αp6 or GA-1 αp6 polyclonal antibodies for 70 min. The membranes were then washed twice and incubated with 1:4,000-diluted anti-rabbit horseradish peroxidase-conjugated antibodies for another 70 min, and the immune complexes were detected by using ECL detection reagents (Amersham). Films were scanned, and bands were subjected to densitometry using ImageQuant software.
DNA replication.
Cells were grown and infected as described earlier. At the times indicated, 1-ml aliquots were collected, and cells were washed and lysed as previously described (7). Samples were treated with proteinase K (50 μg/ml), and DNA was extracted with phenol and chloroform, ethanol precipitated, and resuspended in water. The amount of DNA from the left terminal sequence either from φ29 (φ1) or from GA-1 (G1) was quantified by real-time PCR, using the amplification protocol described above.
Fluorescence measurements.
Fluorescence measurements were performed essentially as described previously (18) in a Variant Cary Eclipse spectrofluorometer and monitored in a 2-mm path length cell at 15°C. The tryptophan residue of φ29 protein p6 was excited at a wavelength of 290 nm, and fluorescence was measured at 350 nm.
RESULTS
Bacteriophage GA-1 protein p6 binding to viral DNA in Bacillus sp. strain G1R.
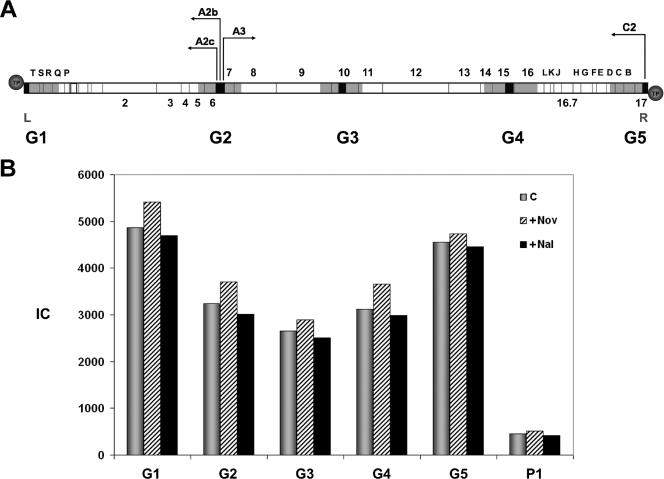
To determine whether p6 binds GA-1 DNA in vivo, Bacillus sp. strain G1R cells were infected with phage GA-1, and DNA binding was measured by X-ChIP. Cells were cross-linked with formaldehyde, and DNA was sonicated and immunoprecipitated with αp6 antibodies. Selected sequences, 300 bp on average, were amplified and quantified by real-time PCR, and protein p6 binding was expressed as IC, as described in Materials and Methods. Figure 1A indicates the location of the five GA-1 DNA regions analyzed, named G1 to G5, scattered along the viral genome. Regions G1 and G5 correspond to the left and right GA-1 DNA ends, respectively, that contain the replication origins; G5 also comprises the early promoter C2. Region G2 includes the central transcriptional control region, with the early promoters A2b and A2c and the late promoter A3. Regions G3 and G4 correspond to the late genes 10 and 15, respectively. Protein p6 binding was measured 15 min postinfection, where the p6/DNA ratio is higher than at later times (results not shown). Figure 1B shows the protein p6 binding to the five GA-1 DNA regions in GA-1-infected Bacillus sp. strain G1R cells. Protein p6 binds to all of the five regions studied, which comprise ∼25% of the total GA-1 DNA (see Fig. 1A); thus, it is likely that protein p6 binds most, if not all, of the viral genome. The highest binding affinity corresponds to regions G1 and G5 located at both DNA ends. As a control, binding to a region, named P1, from plasmid pPR55ow6, a pUB110 derivative (45), was lower than that to any of the GA-1 DNA regions analyzed.
FIG. 1.
(A) Genetic and transcriptional map of the phage GA-1 linear DNA, showing the regions used to measure protein p6 binding by X-ChIP. Since DNA is sheared by sonication to an average size of ∼750 bp, each region would then comprise all of the overlapping DNA fragments that contain the amplified sequence (black boxes). According to the average size of the amplified sequences (∼300 bp) and that of the sonicated DNA (∼750 bp), every DNA region analyzed has ∼1,200 bp (gray boxes). This value would be only ∼750 bp for regions G1 and G5, since the amplified sequences are located at the ends of the linear DNA. The arrows point in the direction of transcription: the early promoters C2, A2b, and A2c transcribe leftward, and the late promoter A3 transcribes rightward. Genes that are conserved in comparison with phage φ29 are indicated with numbers. ORFs located at both genome ends that may encode several proteins, counterparts of which are not present in the genome of φ29, are indicated with letters. Circles represent the terminal protein attached to the 5′ DNA ends. L and R indicate the left and right ends of the GA-1 genome, respectively. (B) Protein p6 binding to GA-1 and plasmid DNA in the absence or presence of novobiocin and nalidixic acid. Bacillus sp. strain G1R cells harboring plasmid pPR55ow6 were grown, infected with GA-1 and, after 15 min, the untreated aliquot (C) was cross-linked and processed as described in Materials and Methods; the other two aliquots were treated with 34 μg of chloramphenicol/ml together with 500 μg of novobiocin/ml (+Nov) or nalidixic acid (+Nal) and cross-linked 10 min later. Protein p6 binding is expressed as the IC (see Materials and Methods). The IC values for each region are as follows (C, +Nov, +Nal): G1 = 4,864, 5,419, and 4,700; G2 = 3,242, 3,703, and 3,002; G3 = 2,659, 2,898, and 2,515; G4 = 3,123, 3,659, and 2,993; G5 = 4,553, 4,732, and 4,453; and P1 = 450, 511, and 420.
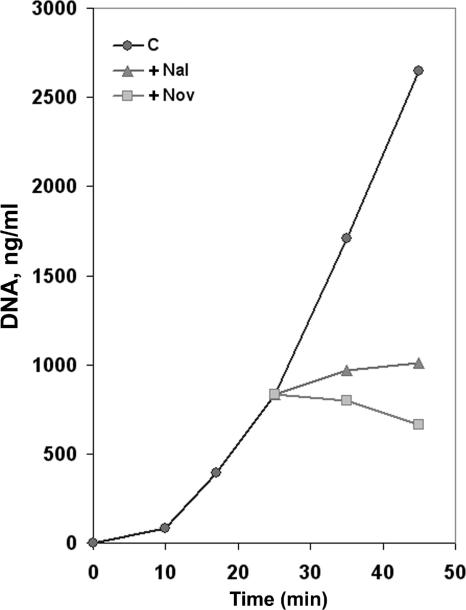
Although phage GA-1 DNA has a terminal protein, and therefore it is not covalently closed, it could be topologically constrained, as has been described for phage T4 (49) or φ29 (18). To test this possibility, protein p6 binding was also measured in the presence of novobiocin, a gyrase inhibitor that generates a loss of DNA negative supercoiling. As a result, if GA-1 DNA were topologically constrained, we would expect an increase of p6 binding. As a control, we used nalidixic acid, which also inhibits gyrase but has no topological effects as it inhibits the topoisomerase nicking-closing activity. As also shown in Fig. 1B, binding to all five GA-1 DNA regions was not significantly increased in the presence of novobiocin. Similarly, the presence of nalidixic acid produced essentially no change in the IC values. This result could be explained if GA-1 DNA were not topologically restricted, if novobiocin and nalidixic acid were not inhibiting gyrase under these conditions, or both. To analyze these possibilities, we studied the effect of both drugs on GA-1 DNA replication. Since the origins of DNA replication in GA-1 DNA are located at the ends of the linear genome, progression of the two replication forks would generate positive supercoiling ahead, due to DNA unwinding that would greatly impair replication if DNA were not allowed to rotate freely. Thus, in a topologically restricted DNA, gyrase would be required for efficient replication. We added novobiocin or nalidixic acid 25 min postinfection, once phage GA-1 DNA replication had started, and measured DNA synthesis. Samples were taken 35 and 45 min postinfection and, using real-time PCR, we quantified accurately the amount of DNA from the left GA-1 DNA terminus (L; shown in black in Fig. 1A). Figure 2 shows that both inhibitors, especially novobiocin, produced a significant impairment in DNA replication, indicating not only that novobiocin and nalidixic acid are functional but that the phage GA-1 genome is topologically constrained in vivo and therefore cellular gyrase activity is required for viral DNA replication. Hence, the results shown in Fig. 1B indicate that GA-1 protein p6 binding to the GA-1 genome or to plasmid DNA is not supercoiling sensitive under the experimental conditions used, in contrast to the results observed in φ29 (18).
FIG. 2.
Effect of novobiocin and nalidixic acid on GA-1 DNA replication. Bacillus sp. strain G1R cells were infected with phage GA-1, and 25 min later the culture was divided into three aliquots and further grown in the presence of novobiocin (+Nov) or nalidixic acid (+Nal) (500 μg of each/ml) or with no addition (C). Aliquots were obtained at the indicated times after infection, and the DNA was purified by phenol extraction and ethanol precipitation. The amount of GA-1 DNA was calculated by real-time PCR of the left terminal sequence (L; see Fig. 1A). The data are expressed as nanograms of full-length GA-1 DNA per milliliter of culture.
Bacteriophage φ29 protein p6 binding to GA-1 DNA in Bacillus sp. strain G1R.
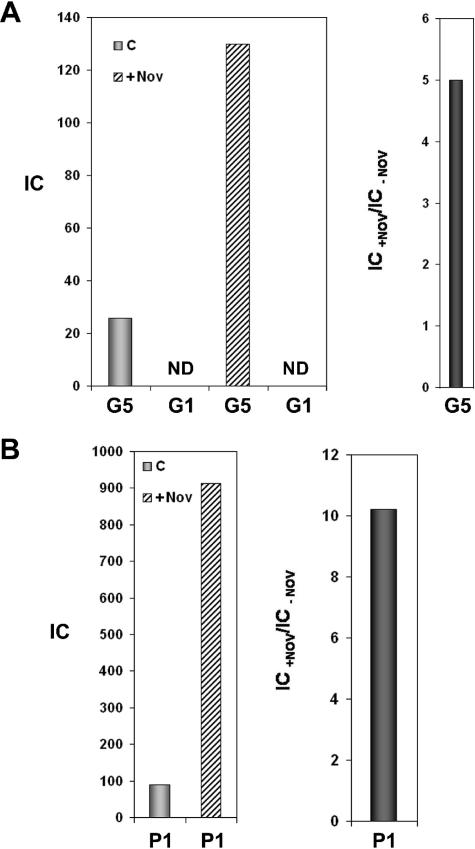
The lack of sensitivity to novobiocin of GA-1 protein p6 binding to GA-1 DNA in Bacillus sp. strain G1R could be due to an intrinsic property of the protein or of the DNA or may even depend on the intracellular background. To discriminate between these possibilities, we expressed φ29 protein p6 in Bacillus sp. strain G1R and analyzed the effect of novobiocin on the protein binding to GA-1 and to plasmid DNA. For this purpose, we transformed G1R cells with plasmid pPR55w6 containing φ29 gene 6. These cells were grown and chloramphenicol was added before GA-1 infection to prevent GA-1 protein p6 synthesis, along with novobiocin, and formaldehyde cross-linking was performed 20 min postinfection. We measured φ29 p6 binding to region G5 and G1 located at the right and left termini, respectively, of GA-1 DNA (see Fig. 1A). Figure 3A shows that under these conditions, novobiocin stimulated fivefold φ29 p6 binding to region G5, in contrast to the result obtained with the GA-1 protein p6 (see Fig. 1B). φ29 protein p6 binding to plasmid DNA was also supercoiling dependent (Fig. 3B) since a 10-fold increase in binding to region P1 from plasmid pPR55w6 was obtained in the presence of novobiocin, again in contrast to the result obtained with the GA-1 protein (see Fig. 1B). These results indicate that the different dependency on supercoiling found for GA-1 and φ29 p6 proteins is a feature that depends on the protein rather than on the DNA or the intracellular background. The failure to detect binding to region G1 (see Fig. 3A) does not seem to be due to a particularly low affinity of φ29 protein p6 for this region, since the affinities of φ29 protein p6 for both GA-1 genome ends in vitro, as measured by fluorescence quenching, were almost identical (with effective binding constant values of about 1.2 × 106 μM−1; result not shown). The absence of in vivo binding to the left terminal region would rather reflect the lack of internalization of the left terminus of the GA-1 genome, strongly suggesting that, as previously described for phage φ29 (20), the ejection mechanism of phage GA-1 is a two-step process in which the entrance of the left (second) part of the viral genome requires the synthesis of proteins encoded by the right genome end and therefore is prevented in the presence of chloramphenicol.
FIG. 3.
φ29 protein p6 binding to GA-1 right terminus (G5) and plasmid region P1 in Bacillus sp. strain G1R. Cells harboring plasmid pPR55w6 were grown, and the culture was treated with 34 μg of chloramphenicol/ml and infected with phage GA-1. The culture was divided into two aliquots and, after 20 min, one aliquot was cross-linked (C), and the other was treated with 500 μg of novobiocin/ml (+Nov) and cross-linked 10 min later. φ29 protein p6 binding is expressed as the IC. (A) IC values for regions G1 and G5 (GA-1 left and right genome ends, respectively) are as follows: G1, not detected (ND); G5, 25.8 (C) and 130 (+Nov). (B) IC values for plasmid region P1 are as follows: 89.4 (C) and 911.9 (+Nov). The binding increase in the presence of novobiocin, expressed as the IC+Nov/IC−Nov ratio, is shown in the right panel.
Bacteriophage GA-1 protein p6 binding to the φ29 right DNA end in B. subtilis 110NA cells.
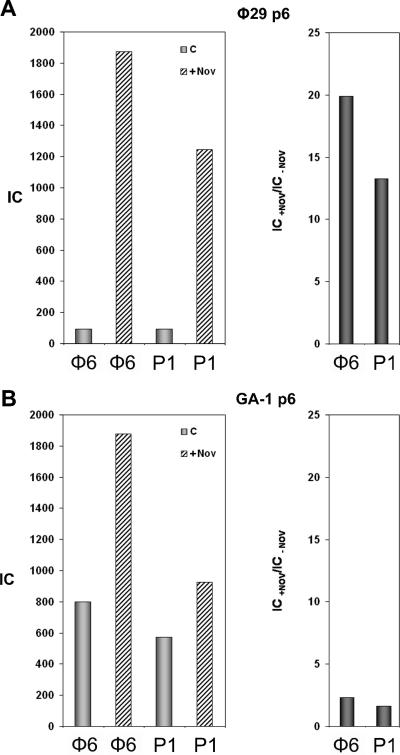
To further study the properties of GA-1 protein p6, we measured its binding to DNA in B. subtilis 110NA cells and performed a direct comparison with the φ29 ortholog. To accomplish this, we cloned the GA-1 gene 6 into the pPR54 plasmid, which allows the temperature-dependent protein expression driven under the control of phage λ PR promoter (45) and transformed B. subtilis 110NA cells (see Materials and Methods). Thus, two different strains were available: one producing GA-1 protein p6 and the other producing φ29 protein p6. To compare the properties of both proteins, induction conditions were found in which the amount of GA-1 protein p6 synthesized was equivalent to the amount of φ29 protein p6 constitutively expressed from plasmid pPR55w6, as determined by Western blot analysis (results not shown). Both strains were infected with φ29 sus14(1242) in the presence of chloramphenicol to prevent exogenous p6 expression. It should be noticed that under these conditions only the ∼65% right part of φ29 DNA penetrates in the cell, since viral proteins are required for complete ejection (20). Thus, we measured the binding of both p6 proteins to the φ29 DNA right terminus (φ6, see Fig. 5A), that penetrates in the first (push) step of ejection, and to the plasmid region P1. Cross-linking was performed 40 min postinfection to maximize DNA internalization. Figure 4A (left panel) shows the binding of the φ29 protein p6 in the absence or presence of novobiocin. The presence of novobiocin increased binding to φ29 and plasmid DNA regions 13.5- and 19.9-fold, respectively (Fig. 4A, right panel). In contrast, the GA-1 protein p6 results show only a small increase (Fig. 4B, right panel). These results, obtained with φ29 DNA and B. subtilis, confirm those found with GA-1 DNA and Bacillus sp. strain G1R and further indicate that GA-1 protein p6 binding to DNA has a very low supercoiling dependency, in contrast to the φ29 ortholog.
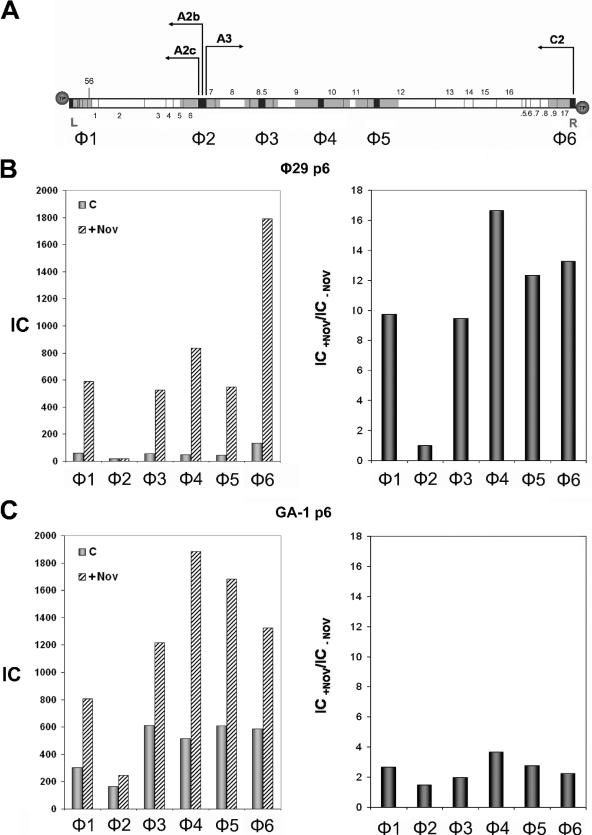
FIG. 5.
(A) Genetic and transcriptional map of phage φ29 linear DNA. The sequences selected for PCR amplification are shown in black and the regions analyzed for protein p6 binding, φ1-φ6, are shown in gray, as explained in Fig. 1A. The position of genes 1 to 17 and 56 is indicated. .5 to .9 stands for 16.5 to 16.9, respectively. Circles represent the terminal protein attached to the 5′ DNA ends. L and R indicate the left and right ends of the φ29 genome, respectively. The arrows point the direction of transcription: the early promoters C2, A2b, and A2c leftward and the late promoter A3 rightward. (B) φ29 protein p6 binding to φ29 DNA. B. subtilis 110NA cells producing φ29 protein p6 were infected with φ29 sus6(626) and, 20 min later, an aliquot (c) was cross-linked as described in Materials and Methods; another aliquot (+Nov) was treated with 34 μg of chloramphenicol/ml together with 500 μg of novobiocin/ml and cross-linked 40 min later. Protein p6 binding for each region is shown in the left panel. The IC values (c and +Nov) are as follows: φ1, 60.5 and 590; φ2, 18.5 and 18.6; φ3, 55.8 and 527.7; φ4, 50.1 and 834.4; φ5, 44.4 and 548.5; φ6, 134.9 and 1791; and P1, 205.9 and 767.5. Binding increase for each φ29 DNA region in the presence of novobiocin, expressed as IC+Nov/IC−Nov ratio, is shown in the right panel. (C) GA-1 protein p6 binding to φ29 DNA. B. subtilis 110NA cells producing GA-1 protein p6 were infected with φ29 sus6(626) and aliquots (c) and (+Nov) were obtained as described above. Protein p6 binding for each region is shown in the left panel. The IC values (c and +Nov) are the following: φ1, 303.7 and 805.9; φ2, 162.9 and 245.3; φ3, 613.4 and 1,216; φ4, 515.7 and 1,885.5; φ5, 607.3 and 1,681.8; φ6, 585.4 and 1,325; and P1, 586.3 and 975. The binding increase for each φ29 DNA region in the presence of novobiocin, expressed as the IC+Nov/IC−Nov ratio, is shown in the right panel.
FIG. 4.
(A) φ29 protein p6 binding to the φ29 right terminus (φ6) and plasmid region P1. B. subtilis 110NA cells producing φ29 protein p6 were grown and treated with 34 μg of chloramphenicol/ml together with or without 500 μg of novobiocin/ml. Cells were infected with φ29 sus14(1242) and cross-linked 40 min later. IC values for each DNA region in the absence or presence of novobiocin are shown in the left panel. The binding increase in the presence of novobiocin, expressed as the IC+Nov/IC−Nov ratio, is shown in the right panel. (B) GA-1 protein p6 binding to the φ29 right terminus (φ6) and plasmid region P1. GA-1 protein p6 synthesis was induced in B. subtilis 110NA cells as described in Materials and Methods. Cells processed as before were infected with φ29 sus14(1242) and cross-linked 40 min later. IC values in the absence or presence of novobiocin are shown in the left panel. The binding increase in the presence of novobiocin, expressed as the IC+Nov/IC−Nov ratio, is shown in the right panel.
Bacteriophage GA-1 protein p6 binding to the φ29 genome in B. subtilis 110NA cells.
The phage GA-1 protein p6 binding studies to φ29 DNA described above were restricted to the right DNA end, since they were performed in the presence of chloramphenicol. We further extended these studies measuring the binding to the whole φ29 DNA. As before, we made use of the X-ChIP technique in φ29 sus6(626)-infected B. subtilis 110NA cells producing either GA-1 or φ29 protein p6. However, since the φ29 p6-producing cells support the growth of φ29 sus6(626) phage (7), cross-linking was performed 20 min postinfection, just before the onset of viral DNA replication takes place. Thus, the results obtained with GA-1 and φ29 proteins could be compared under similar protein/DNA ratios. Binding, both in the absence and in the presence of novobiocin, was measured to the φ29 DNA regions shown in Fig. 5A. The φ29 protein p6 binding pattern to regions φ1 to φ6, shown in Fig. 5B, left panel, is in agreement with the one previously described (18, 19) despite the fact that, under these conditions, 20 min after infection, viral DNA ejection is not completed, since synthesis of p17 and p16.7 proteins from promoter C2, required for the second (pull) step of ejection, is strongly repressed in vivo in protein p6-producing cells (3). This is reflected by the IC value for region φ1 lower than that for region φ6, despite the fact that the in vivo intrinsic affinity of φ29 protein p6 for region φ1 is about twofold higher than that for region φ6 (18). Also, as described, φ29 p6 binding values for each region are highly increased in the presence of novobiocin (18), except for region φ2 (Fig. 5B, right panel). On the other hand, although GA-1 protein p6 is able to bind to all φ29 DNA regions analyzed (Fig. 5C, left panel), it generated a binding pattern different from the one obtained with φ29 protein p6 (Fig. 5B, left panel) and also different from the one obtained with GA-1 DNA in Bacillus sp. strain G1R, where the highest binding corresponds to both viral genome ends (see Fig. 1B). The DNA-binding increase in the presence of novobiocin is shown in Fig. 5C, right panel, where the IC ratios plotted for each φ29 DNA region show much lower values than in the case of the φ29 p6, the highest one being only 3.7-fold for the φ4 region. These results clearly indicate that the two viral proteins have a very different binding behavior with respect to DNA topology. Furthermore, although not as much as for GA-1 DNA in G1R cells (see Fig. 1B), GA-1 protein p6 binding to φ29 DNA displays a lower supercoiling dependency than the φ29 counterpart.
Bacteriophage GA-1 protein p6 complex with φ29 DNA is not functional.
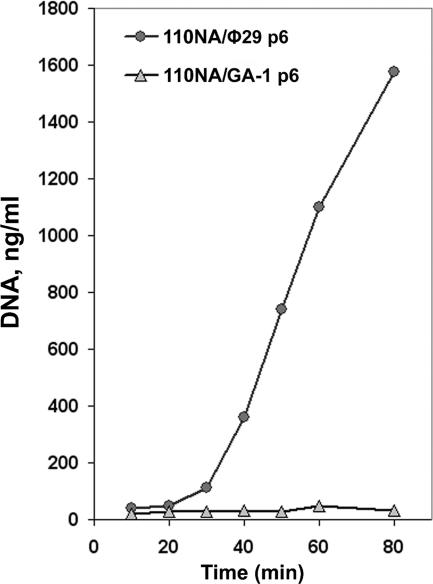
Since GA-1 protein p6 was able to bind to φ29 DNA, and the formation of the nucleoprotein complex was a requirement for φ29 DNA replication, we studied the functionality of the GA-1 p6-φ29 DNA complex in a transcomplementation assay. For this, GA-1 and φ29 protein p6-producing B. subtilis 110NA cells were infected with φ29 sus6(626), and viral DNA replication was measured by real-time PCR. While, as previously described (7), φ29 sus6(626) DNA can replicate in the φ29 p6-producing cells, the GA-1 protein p6-producing cells fail to support DNA replication (Fig. 6). Therefore, we conclude that the nucleoprotein complex formed by GA-1 protein p6 with φ29 DNA has no biological activity. If GA-1 protein p6 is not forming a functional nucleoprotein complex with φ29 DNA, either it is not repressing in vivo the φ29 C2 promoter, or repression is weaker than the one observed with the φ29 protein. Therefore, the φ29 ejection process in GA-1 p6-producing cells should be little or not delayed. In agreement with this, the difference between the IC values obtained, either in the absence or in the presence of novobiocin, for regions φ1 and φ6 (Fig. 5C, left panel) is lower than the one obtained for the same regions in φ29 p6-producing cells (Fig. 5B, left panel).
FIG. 6.
Transcomplementation of φ29 DNA synthesis in B. subtilis 110NA cells producing φ29 or GA-1 protein p6. B. subtilis 110NA cells producing φ29 or GA-1 protein p6 were infected with φ29 sus6(626), aliquots were taken at the indicated times after infection, and the DNA was purified by phenol extraction and ethanol precipitation. The amount of φ29 DNA was measured by real-time PCR of the left terminal sequence (L). The data are expressed as nanograms of full-length φ29 DNA per milliliter of culture.
DISCUSSION
Bacteriophage GA-1 protein p6 activates the initiation of GA-1 DNA replication (17) and represses transcription from the right early C2 promoter (21). Both biological functions require the formation of a nucleoprotein complex with the DNA; therefore, we studied the in vivo DNA- binding properties of GA-1 protein p6 by means of X-ChIP coupled to real-time PCR. GA-1 protein p6 binding was detected to all of the GA-1 DNA regions analyzed, suggesting that GA-1 p6 has a role in the organization of the viral chromosome, in the manner of a histone-like protein, as previously described for phage φ29 protein p6 (19). The binding of p6 was highest at both genome ends (regions G1 and G5), where the origins of replication are located, which may be related to the requirement for protein p6 in GA-1 DNA replication (17).
Previous results showed that GA-1 protein p6 induced lower positive supercoiling on plasmid DNA than the φ29 protein (17), suggesting that DNA binding to negative supercoiled DNA could be less restricted than with φ29 p6. Indeed, in contrast to the results obtained with the φ29 protein, GA-1 p6 binding to GA-1 DNA was not substantially increased in the presence of novobiocin, that is, under these conditions the GA-1 protein p6 binding was not significantly affected by the DNA topology. This finding was unexpected since it has been proposed for φ29 protein p6 that supercoiling is the main factor that rules protein p6 binding specificity in vivo (18, 19). Thus, we tested the effect of novobiocin and nalidixic acid by measuring viral DNA replication. Both drugs arrested replication, indicating that gyrase is required for GA-1 DNA replication and that the GA-1 genome is topologically constrained, most probably due to membrane attachment through the terminal proteins, as it has been previously described for phage φ29 (18). To discriminate among protein, DNA, or intracellular environment as factors that could be responsible for the low supercoiling dependency of GA-1 p6 binding to the viral genome in Bacillus G1R cells, we constructed φ29 p6-producing Bacillus sp. strain G1R cells that were infected with phage GA-1, and we measured φ29 p6 binding to DNA. The binding was found to increase 5- to 10-fold in the presence of novobiocin. Thus, the low supercoiling dependency binding to DNA seems to be an intrinsic protein property. Since infection with GA-1 was performed in the presence of chloramphenicol, no viral proteins could be synthesized. In these conditions, protein p6 binds to the right (G5 region) but not to the left DNA end (G1 region). This strongly suggests that the GA-1 DNA ejection mechanism is a two-step process in which the second step requires viral proteins to pull inside the cell the DNA left end (20).
To further study GA-1 protein p6 binding to DNA, B. subtilis cells producing GA-1 protein p6 were infected with φ29, and the binding was measured to six different regions of φ29 DNA. Under this cellular background, GA-1 protein p6 was able to bind all of the φ29 DNA regions analyzed. The particularly low affinity of φ29 p6 protein in vivo and in vitro for the transcriptional control region φ2 was shown to be due to the presence of an intrinsic curvature (18) located at the promoter A2b (5, 40). GA-1 protein p6 lowest affinity was also detected for region φ2, indicating that the rigidity of this DNA sequence may also impair GA-1 p6 binding. However, in Bacillus sp. strain G1R, the lowest GA-1 p6 affinity was not detected for the GA-1 transcriptional control region, G2, but for the internal region G3, located in late gene 10. These differences could indicate that the intrinsic curvature found in φ29 is not present in the analogous GA-1 transcriptional control region, since no impairment in p6 binding is detected in comparison with other GA-1 DNA regions. Thus, in phage GA-1, the switch from early to late transcription mechanism would have differences in comparison with phage φ29, in agreement with previous results (22). Although GA-1 protein p6 binds in vivo to φ29 DNA, the nucleoprotein complex formed does not have the proper structure to complement in trans the DNA replication deficiency of φ29 sus6(626). This lack of biological activity of the GA-1 p6-φ29 DNA nucleoprotein complex may not prevent the in vivo transcription from the right φ29 DNA early promoter, and thus ejection of φ29 DNA in GA-1 protein p6-producing cells would not be so delayed as in the case of φ29 p6-producing cells.
The low supercoiling dependency of the GA-1 protein p6 observed with GA-1 DNA is in contrast to the described high supercoiling dependency of the φ29 protein p6 binding to φ29 DNA (18). This is reflected by the fact that when the φ29 DNA negative supercoiling was decreased in the presence of novobiocin, the binding of the φ29 protein increased more than that of the GA-1 protein. This lower dependency on supercoiling of GA-1 p6 with respect to φ29 p6 could be explained by the different structure of the nucleocomplex formed by each protein. In the case of φ29, p6 monomers bind DNA every 12 bp (48), while GA-1 p6 monomers bind to DNA every 11 bp (17). Thus, for φ29 p6 the surface-related DNA helical repeat, hc, is 12, and the protein forms a right-handed superhelix, restraining positive supercoiling in a covalently closed DNA, implying that the binding increases as negative supercoiling is decreased. On the other hand, the GA-1 protein p6 hc is lower (11 bp) than the φ29 protein p6 hc (12 pb) (47) but, in any case, higher than the absolute DNA helical repeat (hc = 10.5 bp), so the supercoiling restrained per unit length would be lower and the DNA affinity of GA-1 protein p6 should be less supercoiling dependent. Nevertheless, these arguments would only apply provided other parameters that define DNA topology, such as twist, remain constant.
It has been proposed that the φ29 protein p6 preferential binding to viral DNA in vivo is mainly based on its lower negative superhelicity (18, 19). Since phage GA-1 protein p6 displays a much lower dependency on supercoiling, other factors may be involved in the viral DNA-binding specificity. For instance, architectural proteins present in the nucleoid may prevent protein p6 binding to bacterial DNA when the viral infection takes place. It should be also noted that φ29 DNA replication is membrane associated (8, 25), and when protein p6 is synthesized around the periphery of the cell, where ribosomes have been shown to localize (27), it could predominantly bind to the adjacent viral DNA, decreasing the opportunities to interact with the nucleoid. In addition, GA-1 protein p6-DNA interaction may be regulated by other viral proteins such as those encoded by the different ORFs located at both GA-1 genome ends that are not present in phage φ29.
The present comparative study of φ29 and GA-1 protein p6 in DNA binding has shown that, whereas the main characteristics of these proteins are conserved, interesting differences are also found. These differences will be important tools in further unraveling the molecular mechanism by which these proteins act and in understanding in more detail the evolutionary adaptation of these proteins with respect to their function for their specific phage.
Acknowledgments
We are grateful to A. Bravo for providing plasmid pPR54 and B. subtilis strains 110NA/pPR55w6 and 110NA/pPR55ow6.
This study was supported by research grant BFU2005-00733 from the Spanish Ministry of Education and Science to M.S. and by an Institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” M.A. was a postdoctoral fellow of the Ministry of Education and Science.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Abril, A. M., S. Marco, J. L. Carrascosa, M. Salas, and J. M. Hermoso. 1999. Oligomeric structures of the phage φ29 histone-like protein p6. J. Mol. Biol. 292:581-588. [DOI] [PubMed] [Google Scholar]
- 2.Abril, A. M., M. Salas, J. M. Andreu, J. M. Hermoso, and G. Rivas. 1997. Phage φ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry 36:11901-11908. [DOI] [PubMed] [Google Scholar]
- 3.Alcorlo, M., V. González-Huici, J. M. Hermoso, W. J. Meijer, and M. Salas. 2007. The phage φ29 membrane protein p16.7, involved in DNA replication, is required for efficient ejection of the viral genome. J. Bacteriol. 189:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arwert, F., and G. Venema. 1974. Protease-sensitive transfection of Bacillus subtilis with bacteriophage GA-1 DNA: a probable case of heterologous transfection. J. Virol. 13:584-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthelemy, I., and M. Salas. 1989. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J. Mol. Biol. 208:225-232. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, L., J. Gutiérrez, J. M. Lázaro, A. Bernad, and M. Salas. 1986. Replication of phage φ29 DNA in vitro: role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 14:4923-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo, A., J. M. Hermoso, and M. Salas. 1994. A genetic approach to the identification of functional amino acids in protein p6 of Bacillus subtilis phage φ29. Mol. Gen. Genet. 245:529-536. [DOI] [PubMed] [Google Scholar]
- 8.Bravo, A., G. Serrano-Heras, and M. Salas. 2005. Compartmentalization of prokaryotic DNA replication. FEMS Microbiol. Rev. 29:25-47. [DOI] [PubMed] [Google Scholar]
- 9.Camacho, A., and M. Salas. 2001. Repression of bacteriophage φ29 early promoter C2 by viral protein p6 is due to impairment of closed complex. J. Biol. Chem. 276:28927-28932. [DOI] [PubMed] [Google Scholar]
- 10.Carrascosa, J. L., A. Camacho, F. Moreno, F. Jiménez, R. P. Mellado, E. Viñuela, and M. Salas. 1976. Bacillus subtilis phage φ29; characterization of gene products and functions. Eur. J. Biochem. 66:229-241. [DOI] [PubMed] [Google Scholar]
- 11.Clarey, M. G., J. P. Erzberger, P. Grob, A. E. Leschziner, J. M. Berger, E. Nogales, and M. Botchan. 2006. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 13:684-690. [DOI] [PubMed] [Google Scholar]
- 12.Crucitti, P., J. M. Lázaro, V. Beneš, and M. Salas. 1998. Bacteriophage φ29 early protein p17 is conditionally required for the first rounds of viral DNA replication. Gene 223:135-142. [DOI] [PubMed] [Google Scholar]
- 13.Dorman, C. J., and P. Deighan. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179-184. [DOI] [PubMed] [Google Scholar]
- 14.Drlica, K., and J. Rouvière-Yaniv. 1987. Histonelike proteins of bacteria. Microbiol. Rev. 51:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elías-Arnanz, M., and M. Salas. 1999. Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of φ29 early-late transcriptional switch. Genes Dev. 13:2502-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erzberger, J. P., M. L. Mott, and J. M. Berger. 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13:676-683. [DOI] [PubMed] [Google Scholar]
- 17.Freire, R., M. Serrano, M. Salas, and J. M. Hermoso. 1996. Activation of replication origins in φ29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J. Biol. Chem. 271:31000-31007. [DOI] [PubMed] [Google Scholar]
- 18.González-Huici, V., M. Alcorlo, M. Salas, and J. M. Hermoso. 2004. Binding of phage φ29 architectural protein p6 to the viral genome: evidence for topological restriction of the phage linear DNA. Nucleic Acids Res. 32:3493-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. Genome wide, supercoiling-dependent, in vivo binding of a viral protein involved in DNA replication and transcriptional control. Nucleic Acids Res. 32:2306-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. The push-pull mechanism of bacteriophage φ29 DNA injection. Mol. Microbiol. 52:529-540. [DOI] [PubMed] [Google Scholar]
- 21.Horcajadas, J. A., W. J. Meijer, F. Rojo, and M. Salas. 2001. Analysis of early promoters of the Bacillus bacteriophage GA-1. J. Bacteriol. 183:6965-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horcajadas, J. A., M. Monsalve, F. Rojo, and M. Salas. 1999. The switch from early to late transcription in phage GA-1: characterization of the regulatory protein p4(G). J. Mol. Biol. 290:917-928. [DOI] [PubMed] [Google Scholar]
- 23.Hwang, D. S., and A. Kornberg. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267:23083-23086. [PubMed] [Google Scholar]
- 24.Inciarte, M. R., J. M. Lázaro, M. Salas, and E. Viñuela. 1976. Physical map of bacteriophage φ29 DNA. Virology 74:314-323. [PubMed] [Google Scholar]
- 25.Ivarie, R. D., and J. J. Pène. 1973. DNA replication in bacteriophage φ29: the requirement of a viral-specfic product for association of φ29 DNA with the cell membrane of Bacillus amyloliquefaciens. Virology 52:351-362. [DOI] [PubMed] [Google Scholar]
- 26.Jiménez, F., A. Camacho, J. De la Torre, E. Viñuela, and M. Salas. 1977. Assembly of Bacillus subtilis phage φ29. 2. Mutants in the cistrons coding for the nonstructural proteins. Eur. J. Biochem. 73:57-72. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, P. J., S. D. Thaker, and J. Errington. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLeod, S. M., and R. C. Johnson. 2001. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 4:152-159. [DOI] [PubMed] [Google Scholar]
- 29.Meijer, W. J., J. A. Horcajadas, and M. Salas. 2001. φ29 family of phages. Microbiol. Mol. Biol. Rev. 65:261-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meijer, W. J., A. Serna-Rico, and M. Salas. 2001. Characterization of the bacteriophage φ29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol. Microbiol. 39:731-746. [DOI] [PubMed] [Google Scholar]
- 31.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, F., A. Camacho, E. Viñuela, and M. Salas. 1974. Suppressor-sensitive mutants and genetic map of Bacillus subtilis bacteriophage φ29. Virology 62:1-16. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell, M., and D. Jeruzalmi. 2006. Helical proteins initiate replication of DNA helices. Nat. Struct. Mol. Biol. 13:665-667. [DOI] [PubMed] [Google Scholar]
- 34.Osburne, M. S., S. M. Zavodny, and G. A. Peterson. 1988. Drug-induced relaxation of supercoiled plasmid DNA in Bacillus subtilis and induction of the SOS response. J. Bacteriol. 170:442-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastrana, R., J. M. Lázaro, L. Blanco, J. A. García, E. Méndez, and M. Salas. 1985. Overproduction and purification of protein p6 of Bacillus subtilis phage φ29: role in the initiation of DNA replication. Nucleic Acids Res. 13:3083-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pečenková, T., and V. Pačes. 1999. Molecular phylogeny of φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by gram-positive bacteria. J. Mol. Evol. 48:197-208. [DOI] [PubMed] [Google Scholar]
- 37.Prieto, I., M. Serrano, J. M. Lázaro, M. Salas, and J. M. Hermoso. 1988. Interaction of the bacteriophage φ29 protein p6 with double-stranded DNA. Proc. Natl. Acad. Sci. USA 85:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reilly, B. E., V. M. Zeece, and D. L. Anderson. 1973. Genetic study of suppressor-sensitive mutants of the Bacillus subtilis bacteriophage φ29. J. Virol. 11:756-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rojo, F., M. Mencía, M. Monsalve, and M. Salas. 1998. Transcription activation and repression by interaction of a regulator with the alpha subunit of RNA polymerase: the model of phage φ29 protein p4. Prog. Nucleic Acids Res. Mol. Biol. 60:29-46. [DOI] [PubMed] [Google Scholar]
- 40.Rojo, F., A. Zaballos, and M. Salas. 1990. Bend induced by the phage φ29 transcriptional activator in the viral late promoter is required for activation. J. Mol. Biol. 211:713-725. [DOI] [PubMed] [Google Scholar]
- 41.Rottländer, E., and T. A. Trautner. 1970. Genetic and transfection studies with Bacillus subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol. Gen. Genet. 108:47-60. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, V. T., J. E. Grimwade, J. E. Camara, E. Crooke, and A. C. Leonard. 2004. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF, and DnaA. Mol. Microbiol. 51:1347-1359. [DOI] [PubMed] [Google Scholar]
- 43.Salas, M. 1999. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. 21:159-171. [DOI] [PubMed] [Google Scholar]
- 44.Schachtele, C. F., B. E. Reilly, C. V. De Sain, M. O. Whittington, and D. L. Anderson. 1973. Selective replication of bacteriophage φ29 deoxyribonucleic acid in 6-(p-hydroxyphenylazo)-uracil-treated Bacillus subtilis. J. Virol. 11:153-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano-Heras, G., M. Salas, and A. Bravo. 2005. A new plasmid vector for regulated gene expression in Bacillus subtilis. Plasmid 54:278-282. [DOI] [PubMed] [Google Scholar]
- 46.Serrano, M., C. Gutiérrez, M. Salas, and J. M. Hermoso. 1993. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage φ29 DNA replication. J. Mol. Biol. 230:248-259. [DOI] [PubMed] [Google Scholar]
- 47.Serrano, M., M. Salas, and J. M. Hermoso. 1993. Multimeric complexes formed by DNA-binding proteins of low sequence specificity. Trends Biochem. Sci. 18:202-206. [DOI] [PubMed] [Google Scholar]
- 48.Serrano, M., M. Salas, and J. M. Hermoso. 1990. A novel nucleoprotein complex at a replication origin. Science 248:1012-1016. [DOI] [PubMed] [Google Scholar]
- 49.Sinden, R. R., and D. E. Pettijohn. 1982. Torsional tension in intracellular bacteriophage T4 DNA. Evidence that a linear DNA duplex can be supercoiled in vivo. J. Mol. Biol. 162:659-677. [DOI] [PubMed] [Google Scholar]
- 50.Whiteley, H. R., W. D. Ramey, G. B. Spiegelman, and R. D. Holder. 1986. Modulation of in vivo and in vitro transcription of bacteriophage φ29 early genes. Virology 155:392-401. [DOI] [PubMed] [Google Scholar]
- 51.Yasbin, R. E., P. I. Fields, and B. J. Andersen. 1980. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene 12:155-159. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa, H., J. H. Elder, and J. Ito. 1986. Comparative studies on the small Bacillus bacteriophages. J. Gen. Appl. Microbiol. 32:39-49. [Google Scholar]
- 53.Yoshikawa, H., K. J. Garvey, and J. Ito. 1985. Nucleotide sequence analysis of DNA replication origins of the small Bacillus bacteriophages: evolutionary relationships. Gene 37:125-130. [DOI] [PubMed] [Google Scholar]