Abstract
The CIII protein encoded by the temperate coliphage lambda acts as an inhibitor of the ubiquitous Escherichia coli metalloprotease HflB (FtsH). This inhibition results in the stabilization of transcription factor λCII, thereby helping the phage to lysogenize the host bacterium. λCIII, a small (54-residue) protein of unknown structure, also protects σ32, another specific substrate of HflB. In order to understand the details of the inhibitory mechanism of CIII, we cloned and expressed the protein with an N-terminal six-histidine tag. We also synthesized and studied a 28-amino-acid peptide, CIIIC, encompassing the central 14 to 41 residues of CIII that exhibited antiproteolytic activity. Our studies show that CIII exists as a dimer under native conditions, aided by an intersubunit disulfide bond, which is dispensable for dimerization. Unlike CIII, CIIIC resists digestion by HflB. While CIII binds to HflB, it does not bind to CII. On the basis of these results, we discuss various mechanisms for the antiproteolytic activity of CIII.
An important event in the life cycle of bacteriophage lambda is the choice between its two alternate modes of development, viz., lytic and lysogenic (14, 39). This choice involves the participation of proteins from the virus, as well as from its bacterial host, Escherichia coli. The small 54-residue phage protein λCIII plays an important role in this process by stabilizing λCII, the transcription factor essential for the establishment of lysogeny (24). In the absence of CIII, CII is rapidly degraded by the ATP-dependent host metalloprotease HflB (FtsH) (36, 37), driving λ toward the lytic cycle. On the other hand, overexpression of λCIII promotes the lysogenic mode of phage development (1).
The E. coli σ32 protein, another substrate of HflB (22, 38), is also protected by CIII, generating a heat shock response in the cell (6). Thus, the primary function of CIII appears to be inhibition of the protease activity of HflB, although another function, viz., acting as a molecular chaperone, has also been proposed (33). There has been just one recent report on the molecular mechanism of this antiproteolytic function of CIII (31), where it has been shown that CIII prevents the association between HflB and CII. Interestingly, CIII is itself an unstable protein and a substrate for HflB (23, 30), suggesting competitive inhibition as a possible mechanism for its action. An important development in the understanding of CIII action was the observation that the 24-residue central region of CIII (amino acids 14 to 37) is both necessary and sufficient for its activity (32). This domain is highly homologous among CIII proteins from λ, P22, and HK022 and is likely to form an amphipathic helix. Mutations in this region lead to loss of CIII activity, while those outside this region have little effect (32). Nevertheless, how this putative helical portion of CIII protects CII or σ32 from HflB remains to be understood.
To unravel the mechanism of CIII action, it is important to know the structure of CIII, as well as to understand its possible interaction with CII and HflB. We therefore overexpressed and purified CIII with an N-terminal hexahistidine tag. This protein showed inhibition of CII digestion by HflB in vitro and allowed us to attempt a structural characterization of the protein. Further, we synthesized a 28-residue peptide, CIIIC, encompassing residues 14 to 41 of CIII and partially characterized it. Study of this region of CIII at the peptide level provided additional information. We found that (i) CIIIC is a helical homodimer that protects CII from HflB; (ii) full-length CIII is also dimeric, stabilized by an intersubunit S-S linkage that is dispensable for dimer formation; and (iii) unlike CIII, CIIIC is not digested by HflB. We also found a direct interaction between HflB and CIII, while there was no such interaction between CII and CIII. On the basis of these results, we suggest that inhibition of HflB by CIII involves a direct CIII-HflB interaction, leading to occlusion of the substrate.
MATERIALS AND METHODS
Materials.
Chelating Sepharose-FF, deoxynucleoside triphosphates, and ATP were from Amersham Biosciences, Sweden. Primers were custom synthesized and obtained from MWG Biotech, Germany. Rink amide resin, N-α-9-fluorenylmethoxy carbonyl (N-α-Fmoc), and the necessary side chain-protected amino acids were purchased from Novabiochem (San Diego, CA). Coupling reagents 1-hydroxy-benzotriazole and N,N′-diisopropylethylamine and 2,4,6-trinitrobenzenesulfonic acid, used for checking the coupling reaction, were purchased from Sigma.
Expression of genes and purification of proteins.
The cIII gene from the lambda genome was PCR amplified and cloned between the NdeI and BamHI sites of the vector pET15b (Novagen Inc.). The resultant plasmid, pAB905, was used for expression of the recombinant His6-CIII protein. For purification, E. coli strain BL21(DE3) harboring this plasmid was grown in Terrific broth (35) at 37°C until the absorbance at 590 nm reached 0.6, followed by induction with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at the same temperature. The cells were harvested, washed with 0.9% (wt/vol) NaCl solution, and broken by sonication in buffer A (20 mM Tris-HCl [pH 8.0], 200 mM NaCl) containing 8 M urea and 10 mM imidazole. The lysate was centrifuged (at 14,500 × g for 30 min), and the supernatant was loaded onto a 5-ml chelating Sepharose column charged with Ni2+ and preequilibrated with the same buffer. The column was washed with 5 volumes of buffer A containing 6 M urea and 100 mM imidazole, followed by elution of the protein in buffer A containing 6 M urea and 300 mM imidazole. The eluent was extensively dialyzed against buffer R (20 mM Tris-HCl [pH 8.0], 200 mM NaCl) for renaturation. This renatured protein was used for all subsequent experiments. The concentration of this protein was determined by absorbance measurements by using ɛ280 = 15,470 M−1 cm−1 (20). Typically, 2 to 3 mg of pure protein was obtained from a 1-liter culture. His6-CII was prepared in an identical manner by cloning a 297-bp NdeI-BglII fragment from the lambda (cI857Sam7) genome containing the cII gene into the pET15b vector to obtain plasmid pAB412. E. coli BL21(DE3) cells harboring this plasmid were grown and harvested as described above, washed with 0.9% NaCl, and lysed by sonication in buffer C (20 mM Tris-HCl, 0.5 M NaCl, 5 mM MgCl2, 10 mM imidazole, 5% [vol/vol] glycerol, 0.5 mM phenylmethylsulfonyl fluoride [pH 8.0]). The resultant solution was centrifuged as described above and mixed with Talon resin equilibrated in the same buffer. The beads were sequentially washed in buffer C, buffer C2 (20 mM Tris-HCl, 0.5 M NaCl, 10 mM imidazole [pH 8.0]), and buffer C3 (20 mM Tris-HCl, 0.5 M NaCl, 50 mM imidazole [pH 8.0]), followed by elution in 20 mM Tris-HCl-0.5 M NaCl-0.5 M imidazole (pH 8.0) as described in reference 15.
For in vitro binding interaction studies, CII (without a histidine tag) was obtained by overexpressing recombinant plasmid pAB305 and purification by two steps of ion-exchange chromatography as described earlier (16). HflB was overexpressed as a glutathione S-transferase (GST) fusion protein from plasmid pAD101 containing the hflB gene cloned in vector pGEX4T and was purified with a glutathione-Sepharose column (GSTrap FF; Amersham Biosciences, Sweden) as described by Shotland et al. (37).
Peptide synthesis and characterization.
The 28-mer peptide CIIIC, encompassing the central region of CIII (residues 14 to 41), was synthesized by the solid-phase synthesis method by using Fmoc chemistry (4, 5) with a Biolynx 4175 peptide synthesizer. Amino acids were added as either Fmoc-Xaa-OH or Fmoc-Xaa-OpfP esters. The coupling of each amino acid was confirmed by a 2,4,6-trinitrobenzenesulfonic acid test. After the synthesis was over, the peptide was cleaved from the resin and the side chain-protecting group was completely removed by incubation in reagent K (trifluoroacetic acid-phenol-thioanisole-ethane dithiol at 94:2:2:2, vol:vol:vol:wt) (29). The resulting peptide was finally purified by reverse-phase high-performance liquid chromatography on a C18 (Hypersil) column with 0 to 80% acetonitrile gradient containing 0.1% trifluoroacetic acid. The molecular mass of the peptide was confirmed by electrospray ionization mass spectrometry.
CD spectroscopy.
Far-UV circular-dichroism (CD) spectra were recorded at 25°C in a JASCO J600 spectropolarimeter with a cuvette with a 1-mm path length both for refolded His6-CIII (10 μM) and for the peptide CIIIC (40 μM). The buffer used was 20 mM Tris-HCl (pH 8.0) containing 200 mM NaCl. The spectra were deconvoluted with the software CDNN (2, 9) for estimating the secondary-structure components.
Measurement of inhibition of proteolysis by HflB.
The activity of CIII (or CIIIC) was measured by its ability to inhibit the HflB-mediated proteolysis of λCII (37). Reaction mixtures (40 μl) were prepared by using 0.4 nmol of CII in buffer P (50 mM Tris-acetate [pH 7.2], 100 mM NaCl, 5 mM MgCl2, 25 μM Zn-acetate, 1.4 mM β-mercaptoethanol [β-ME]) containing 5 mM ATP. CIII (5 nmol) or CIIIC (3 nmol) was also added. Proteolysis was carried out by adding 40 pmol of HflB, followed by incubation at 37°C for specified time intervals. Reactions were stopped by the addition of 5 mM EDTA and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, followed by heating in a boiling water bath for 5 min. The amount of CII that remained after proteolysis was analyzed by subjecting the samples to 17.5% SDS-PAGE, followed by regular Coomassie blue staining and quantitation in a gel document system (Bio-Rad Gel Doc 1000).
CIII was also subjected to proteolysis by HflB in the presence of CIIIC. The experiment was performed in two ways. (i) HflB (40 pmol) was added to a mixture of CIII (2 nmol) and CIIIC (3 nmol) as described above, or (ii) HflB was preincubated with CIIIC before adding CIII and starting the proteolytic reaction.
Proteolysis of CIII and CIIIC by HflB.
Both CIII and CIIIC were also individually subjected to proteolysis by HflB under the same conditions as described above. In these experiments, 2 nmol of protein or peptide was used. Following proteolysis, samples were analyzed by 17.5% SDS-PAGE as described above.
Tryptic cleavage of CIII.
The His6-CIII protein was also digested by trypsin (Sigma) in buffer R, with 10 μg of protein and 0.1 μg of trypsin at room temperature (25 ± 2°C) for specified time intervals. The reaction was stopped by 1 mM phenylmethylsulfonyl fluoride and SDS-PAGE loading buffer and by heating in boiling water. The proteolytic fragments were analyzed by subjecting the samples to 18% SDS-PAGE and visualized by Coomassie blue staining.
Determination of the accessibility of the cysteine residue of CIII.
The accessibility of the single cysteine residue (C13) present in CIII was measured by treating 900 μl of protein solution in 100 mM sodium phosphate buffer (pH 8.0) with 100 μl of 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) at 25°C. The final concentrations of protein and DTNB were 12 μM and 300 μM, respectively. The number of free cysteine residue was determined from the change in absorbance at 412 nm as 2-nitro-5-thiobenzoate molecules released in the reaction, using 13,600 M−1 cm−1 as the ɛ of 2-nitro-5-thiobenzoate at 412 nm (19).
Size exclusion chromatography.
Size exclusion chromatography experiments were performed in an AKTA system (Amersham Biosciences, Sweden) with a Superdex 75 HR 10/30 column with buffer R. A 200-μg portion of the sample was injected at a time. The standard molecular mass marker proteins used were obtained from Amersham Biosciences. The Rf value was calculated with the formula Rf = (Ve − V0)/(VT − V0), where Ve is the elution volume, V0 is the void volume, and VT is the total volume of the column.
In vitro binding assays.
Overexpressed GST-HflB was immobilized on glutathione-Sepharose beads (Amersham Bioscience) at 4°C for 1 h in phosphate-buffered saline with 0.5% NP-40. The beads were then washed with buffer P and each of three 100-μl aliquots of bound GST-HflB (20 μg) was mixed with 25 μg of His6-CII, 25 μg of His6-CIII, and 25 μg of CIIIC separately. The final volume was made up to 500 μl, and the mixture was incubated on a rotating machine at 4°C for 4 h. The beads were then washed thrice with buffer P and resuspended in SDS gel loading buffer for elution of proteins from the beads. The eluted proteins were separated by 17.5% SDS-PAGE and transferred to nitrocellulose membrane, immunoblotted by anti-His or anti-CIII polyclonal antibody, and detected by Lumi-Light Western blotting substrate (Roche). As a negative control, the same experiment was also performed with GST protein replacing GST-HflB.
For CII-CIII interactions, purified His6-CIII was immobilized at 4°C for 1 h on Ni2+-nitrilotriacetic acid (NTA) beads in buffer A containing 10 mM imidazole and washed with buffer P. A 50-μl volume of His6-CIII-bound beads (25 μg) was mixed with 25 μg of CII (without the histidine tag) and incubated and detected as described before.
Coimmunoprecipitation.
Purified CII (10 μg) and GST-HflB (10 μg) were mixed with purified His6-CIII (10 μg) separately in buffer I (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) on a rotating machine for 3 h at 4°C. The proteins were then incubated with anti-His antibody along with protein A-agarose beads for 12 h. Immunoprecipitates were isolated by centrifugation, washed thrice with buffer I, and resuspended in SDS sample buffer. Proteins were separated by 17.5% SDS-PAGE; transferred to nitrocellulose membrane; separately immunoblotted by anti-CII, anti-GST, and anti-His antibodies; and detected as described above.
Complementation by tagged proteins.
The in vivo activities of tagged CII, CIII, and HflB were tested by complementation assays. For His6-CII and His6-CIII, strain BL21(DE3) containing pET15b-derived plasmid pAB412 or pAB905 was used. These host cells were subjected to infection by phage λcII68 (CII−) or λcIII67 (CIII−), respectively. The bacterial cultures were grown in Luria-Bertani medium (35) with 0.4% maltose and 100 μg/ml ampicillin at 37°C. The phages were adsorbed on ice for 30 min at a multiplicity of 0.1. Complementation by the histidine-tagged proteins was judged by examining the morphology of the resulting plaques (turbid or clear).
For testing of GST-tagged HflB, E. coli strain AK525, which has very low expression of HflB (27), was used. It was infected with the chloramphenicol-resistant wild-type phage λc+Cam105 (21), and the frequency of lysogenization was measured as the ratio of chloramphenicol-resistant clones to the number of infected cells at 37°C. A 10-μg/ml concentration of chloramphenicol was used in the plates. The lysogenization frequency was also measured for AK525 containing plasmid pAD101 expressing GST-HflB. Addition of IPTG was not necessary for the plasmid-containing BL21(DE3) cells because of leaky expression from the T7 promoters on the plasmids. For AK525, 1 mM IPTG was used for induction.
RESULTS
CIII and CIIIC are largely helical.
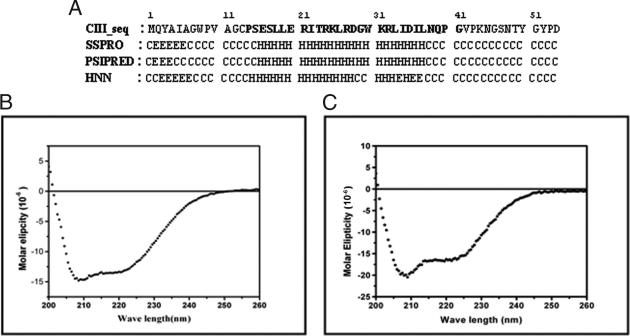
λCIII was prepared as a recombinant protein, His6-CIII (henceforth referred to as CIII), with an N-terminal tag, MGSSHHHHHHSSGLVPRGSH. It was purified under denaturing conditions and refolded by single-step dialysis as described above, yielding a preparation that was ≥95% pure as judged by SDS-PAGE and Coomassie blue staining. From the amino acid sequence, a large portion (∼50%) of CIII is predicted to be unstructured; the structured region is confined to the central part of the molecule and is largely helical (Fig. 1A). This helical region makes up the core of the molecule, which was synthesized as the peptide CIIIC. The calculated values for secondary structural elements predicted from the sequence of CIII and as analyzed from the far-UV CD spectra of CIII and CIIIC (Fig. 1B and C) are shown in Table 1. The CD spectrum of CIII was also recorded under reducing conditions in the presence of β-ME. No change from the spectrum obtained under nonreducing conditions (Fig. 1B) was observed (data not shown).
FIG. 1.
Predicted secondary structure and CD spectra of CIII. (A) Secondary structure of CIII predicted by the various algorithms indicated. H, helix; E, beta strands; C, coil. The sequence of CIIIC, the central core region of CIII, is shown in bold. (B) Far-UV CD spectrum of CIII. (C) Far-UV CD spectrum of CIIIC.
TABLE 1.
Secondary structure of λCIII
| Secondary structure | % as predicted from sequencea | % as obtained from far-UV CD datab |
|---|---|---|
| Helix | 33-40 | 30.1 (53.6) |
| Beta strands | 6-15 | 18.9 (9.6) |
| Beta turns | 0 | 17.4 (13.9) |
| Random coil | 50-54 | 35.0 (21.3) |
Shown is the range of values predicted by the following programs available at the ExPASy website (www.expasy.ch): SSPRO (http://www.igb.uci.edu), PSIPRED (11, 34), and HNN (http://npsa-pbil.ibcp.fr).
Inhibition of HflB-mediated proteolysis by CIII and by CIIIC.
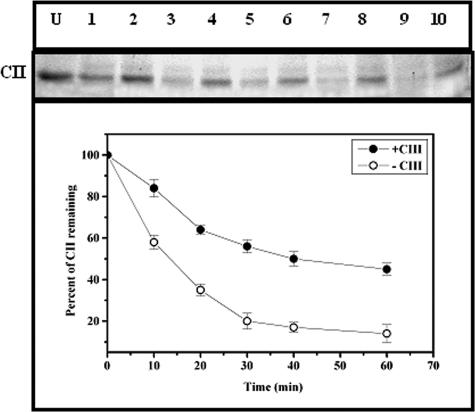
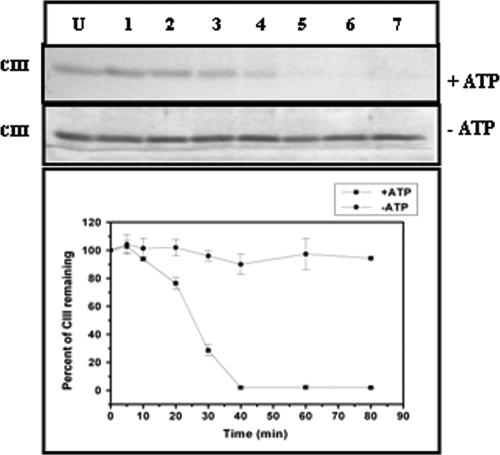
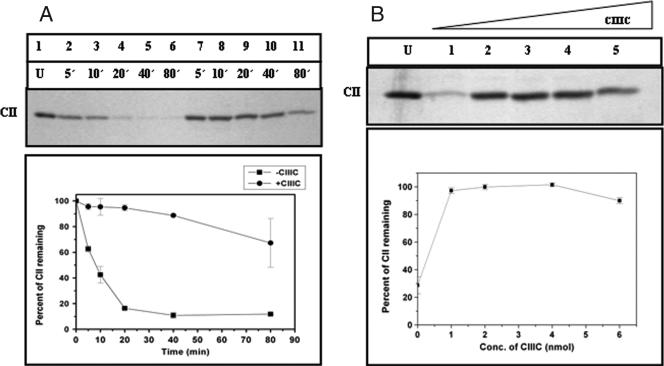
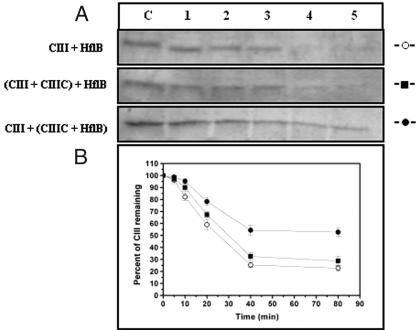
The antiproteolytic activity of CIII was tested by an experiment in which the degradation of CII by GST-HflB was carried out at 37°C for various time intervals in the presence or absence of ATP. The amount of CII remaining was checked by 17.5% SDS-PAGE. The inhibitory effect of CIII was judged by including a molar excess of CIII (HflB-CII-CIII at 1:10:125) in the reaction mixture before the addition of HflB. Partial protection of CII was observed when CIII was present (Fig. 2). Since CIII itself is a substrate of HflB (22, 29), proteolysis of CIII by HflB was also examined in the absence or presence of ATP (Fig. 3). In accordance with an earlier report (23), CIII was found to be rapidly proteolyzed by HflB, but only in the presence of ATP. However, compared to the proteolysis of CII, the proteolysis of CIII was initially slow, as evident from the initial slopes of the proteolysis curves in Fig. 2 and 3. To examine the inhibitory effect of CIIIC, the proteolysis of CII was also carried out in the presence of CIIIC (Fig. 4A, HflB-CII-CIIIC at 1:10:75). The results of Fig. 2 and 4A show that both CIII and CIIIC inhibit the proteolysis of CII by HflB; apparently, CIIIC protects CII better than CIII.
FIG. 2.
Inhibition of proteolysis of CII from HflB by CIII. The top panel shows the CII band on a 17.5% SDS-PAGE, and the bottom panel represents the amount of CII remaining at various times of digestion, obtained by densitometry of the gel. Lane U, undigested CII; lanes 1, 3, 5, 7, and 9, CII (0.4 nmol) treated with HflB (0.04 nmol) for 10, 20, 30, 40, and 60 min; lanes 2, 4, 6, 8, and 10, CII treated with HflB for 10, 20, 30, 40, and 60 min in the presence of CIII (5 nmol). Each data point shown is the average of three identical experiments.
FIG. 3.
Proteolysis of CIII by HflB. The top panels show the band corresponding to CIII on a 17.5% SDS-PAGE, in the presence or absence of ATP (5 mM). A densitometric scan of the above gels showing the amount of CIII (2 nmol) remaining after treatment with HflB (0.04 nmol) is shown in the bottom panel. Lane U, undigested CIII; lanes 1 to 7, CIII treated with HflB for 5, 10, 20, 30, 40, 60, and 80 min.
FIG. 4.
Inhibition of proteolysis of CII from HflB by CIIIC. (A) The top part of the panel shows the CII band on a 17.5% SDS-PAGE, and the bottom part of the panel represents the amount of CII remaining at various times of digestion, obtained by densitometry of the gel. Lane U, undigested control; lanes 2 to 6, digestion of CII (0.4 nmol) by HflB (0.04 nmol) for the indicated times (5 to 80 min); lanes 7 to 11, digestion of CII in the presence of CIIIC (3 nmol). Each data point is the mean value of three identical experiments. (B) The top part of the panel shows the CII band on a 17.5% SDS-PAGE, and the bottom part of the panel represents the amount of CII (0.4 nmol) remaining after digestion by HflB (0.04 nmol) for 60 min in the presence of different concentrations of CIIIC. Lane U, undigested control; lanes 1 to 5, 0 to 6 nmol of CIIIC.
When CII was digested by HflB in the presence of increasing concentrations of CIIIC, it was found that near-total protection occurred at 1 nmol of CIIIC (corresponding to HflB-CII-CIIIC at 1:10:25) and there was no further inhibition of CII proteolysis at higher CIIIC concentrations (Fig. 4B).
Inhibition of CIII proteolysis by CIIIC.
The ability of CIIIC to inhibit the proteolysis of CIII by HflB was also tested. When HflB was added to a mixture of CIII and CIIIC, CIII was digested as CIII alone, with little effect of CIIIC. However, when CIII was subjected to proteolysis by HflB preincubated with CIIIC, partial protection of CIII was observed (Fig. 5).
FIG. 5.
Inhibition of proteolysis of CIII by CIIIC. (A) CIII band on a 17.5% SDS-PAGE. Two nanomoles of CIII, alone (top part of the panel) or in the presence of 3 nmol of CIIIC (middle part of the panel), was digested by HflB (40 pmol). CIII was also digested by HflB preincubated with CIIIC (bottom part of the panel). Lane U, undigested control; lanes 1 to 5, treated with HflB for 5, 10, 20, 40, or 80 min. (B) Amounts of CIII remaining after various times of digestion, obtained by densitometry of the gels. Symbols: ○, CIII alone; ▪, HflB added to the CIII-CIIIC mixture; •, HflB preincubated with CIIIC prior to the addition of CIII.
CIIIC is not a substrate for HflB.
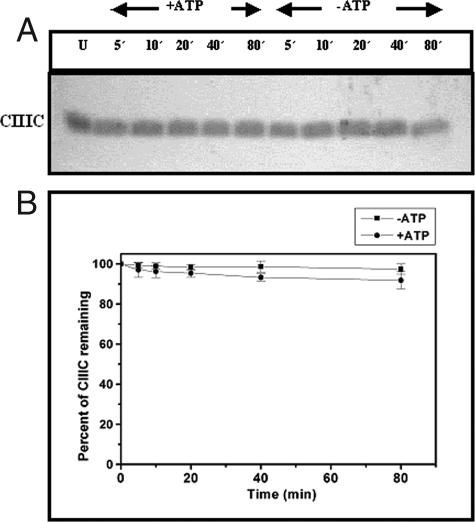
When CIIIC was subjected to proteolysis by HflB, it was found that no cleavage occurred, irrespective of the presence of ATP (Fig. 6). It is interesting that, in this respect, the core region of CIII behaves differently than the full-length protein.
FIG. 6.
Proteolysis of CIIIC by HflB. (A) A 17.5% SDS-PAGE analysis showing the time course of digestion of CIIIC (2 nmol) by HflB (0.04 nmol) in the presence or absence of ATP (5 mM). Times of digestion in minutes are indicated at the top of each lane. An undigested CIIIC control is shown in lane U. (B) Amounts of CIIIC remaining after various times of digestion, obtained by densitometry of the gels.
CIIIC is a dimer with a disulfide bond that is dispensable for dimerization.
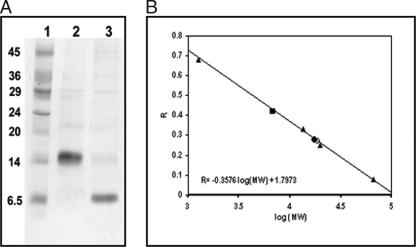
In the absence of the three-dimensional structure of CIII, we sought to examine its structure by subjecting the protein to DTNB treatment. CIII has one cysteine residue (C13) located near the N terminus. Estimation of the number of accessible cysteine residues in CIII yielded a value of 0.01 per monomer, implying that this cysteine was practically inaccessible. In the presence of 7 mM β-ME, 0.75 cysteine residue per monomer of CIII became accessible to DTNB (data not shown), indicating the possible presence of a disulfide bridge linking two monomeric CIII molecules through their sole C13 residues. SDS-PAGE analysis of CIII was carried out to check whether dimeric CIII was indeed held by an S-S bridge. Figure 7A shows that while CIII migrated as a dimer (corresponding to a molecular mass of ∼17 kDa, lane 2), in the presence of β-ME it migrated as a monomer (corresponding to a molecular mass of ∼8 kDa, lane 3). Thus, CIII is a homodimer in which the two monomers are held by a disulfide bond linking the C13 residues.
FIG. 7.
CIII and CIIIC are dimeric. (A) A 17.5% SDS-PAGE analysis of CIII under nonreducing (lane 2) or reducing (lane 3) conditions. A 7 mM concentration of β-ME was used as the reducing agent. Lane 1 shows the molecular weight marker. The values on the left are molecular masses (kilodaltons). (B) Gel filtration analysis of CIII and CIIIC. The curves show the plot of Rf against the log10 molecular mass for chromatography on a Superdex 75 HR 10/30 column previously calibrated with the standard proteins (▴) bovine serum albumin (66 kDa), soybean trypsin inhibitor (20 kDa), RNase A (13.5 kDa), and vitamin B12 (1.25 kDa). The linear fit is also shown along with the equation. This equation was used to calculate the molecular masses of CIII (•) and CIIIC (▪) from the observed Rf values indicated. CIII was also run on this column in the presence of 7 mM β-ME and migrated as shown (○).
Analytical gel filtration chromatography carried out with CIII and with CIIIC (Fig. 7B) showed that CIII eluted as a molecule with a molecular weight of 17,100 (calculated monomeric molecular mass = 8.2 kDa), vindicating the above finding that CIII was dimeric. However, CIIIC also migrated as a dimer, corresponding to 6,780 Da (calculated monomeric molecular mass = 3.3 kDa). It may be noted that there is no cysteine residue in CIIIC (Fig. 1). Therefore, we conclude that additional contacts are present between the two monomers of the CIII dimer, which can retain the dimeric organization even without the disulfide bond. Gel filtration of CIII was also carried out under identical conditions in the presence of β-ME, and the protein eluted as a dimer (Fig. 7B), supporting the above conclusion.
Proteolysis of CIII by trypsin.
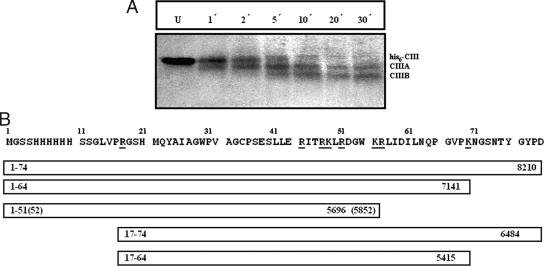
Figure 8 shows the pattern of limited proteolysis of CIII by trypsin. There are two predominant cleavage products, CIIIA and CIIIB (with approximate molecular masses of ∼ 5.5 to 7 kDa). An analysis of the possible tryptic sites shown along the sequence of His6-CIII (Fig. 8B) suggests that in CIII, the susceptible points for tryptic digestion which can give rise to CIIIA or CIIIB are located at either or both termini of the molecule and not near the central region.
FIG. 8.
Digestion of CIII by trypsin. (A) CIII was digested by trypsin for 1 to 30 min as indicated and then analyzed by 18% SDS-PAGE. Lane U shows the undigested protein. The positions of CIII and the tryptic fragments CIIIA and CIIIB are indicated. (B) Sequence of CIII with the possible tryptic sites marked (underlined). The 20 N-terminal residues in this sequence make up the histidine tag. Possible cleavage fragments between 5.5 kDa and 7 kDa are shown below as boxes. The calculated molecular mass is shown on the right, and the terminal residue numbers for each fragment are shown on the left.
CIII interacts with HflB but does not interact with CII.
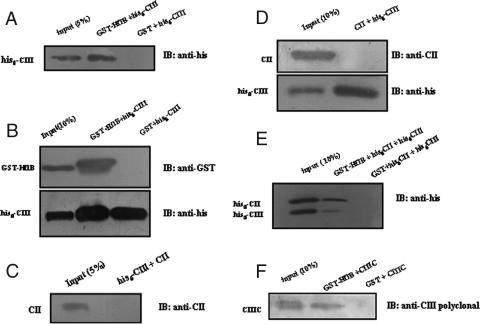
The interaction between GST-HflB and His6-CIII was tested by in vitro binding assay with GST pull down and coimmunoprecipitation (Fig. 9A and B). HflB was found to interact with CIII. A similar study of the interaction between CII and His6-CIII by in vitro Ni-NTA pull-down assay, as well as by coimmunoprecipitation, failed to produce any evidence for CII-CIII interaction (Fig. 9C and D). When an equimolar mixture of His6-CII and His6-CIII (1.9 nmol each) was mixed with GST-HflB (0.2 nmol) and tested by GST pull-down assay, both CII and CIII were found to bind to HflB (Fig. 9E); however, the interaction between CIII and HflB appeared to be weaker than the CII-HflB interaction. CIIIC also showed a direct interaction with HflB (Fig. 9F), which was again weaker compared to the HflB-CIII interaction.
FIG. 9.
In vitro binding of CIII-HflB, CII-CIII, and CIIIC-HflB. Direct interaction between His-CIII and GST-HflB from GST pull down and immunoblotting (IB) with anti-His antibody (A) or immunoprecipitation with anti-His antibody and immunoblotting with the indicated antibodies (B). Absence of interaction between CII and His-CIII from Ni-NTA pull down and immunoblotting with anti-CII antibody (C) or immunoprecipitation with anti-His antibody and immunoblotting as described above (D). (E) Interactions between His-CII and GST-HflB and between His-CIII and GST-HflB when all three were mixed together, as judged by GST pull down and immunoblotting with anti-His antibody. (F) Interaction between CIIIC and GST-HflB obtained from GST pull down, followed by immunoblotting with anti-CIII antibody. For GST pull-down assays, GST-HflB was immobilized on glutathione-Sepharose beads; mixed with CII, His-CII, His-CIII, or CIIIC; and analyzed by 17.5% SDS-PAGE and immunoblotting with the antibodies indicated. For Ni-NTA pull down, His-CIII was immobilized on Ni-NTA beads. Coimmunoprecipitation was carried out by precipitation of His-CIII by anti-His antibody and protein A-agarose and analyzed by SDS-PAGE and immunoblotting as described above. Band positions are shown to the left of each gel, and the antibody used for immunoblotting is shown to the right.
In separate control experiments, the tagged proteins GST-HflB, His6-CII, and His6-CIII were found to complement an hflB mutant host or λcII and λcIII mutants (Table 2), respectively. The effect of GST-tagged HflB on λ lysogeny was as follows. In E. coli AK525 (which has a low level of hflB expression), no protein is expressed from a plasmid, and the frequency of lysogenization (defined as the number of Cmr colonies divided by the number of infective centers times 100) was 9.1 ± 0.9 (mean ± standard deviation). In strain AK525/pAD101, GST-HflB is expressed from plasmid pAD101 and the frequency of lysogenization was 0.9 ± 0.2. These values are from six identical experiments. Thus, all of the tagged proteins used were active in vivo.
TABLE 2.
Effect of histidine-tagged proteins on λ plaque morphologya
| Strain | Protein expressed from plasmid | λ c+ | λ cII68 | λ cIII67 |
|---|---|---|---|---|
| BL21(DE3) | None | T | C | C |
| BL21(DE3)/pAB305 | CII | T | T | ND |
| BL21(DE3)/pAB412 | His6-CII | T | T | ND |
| BL21(DE3)/pAB905 | His6-CIII | T | ND | T |
T, turbid plaque; C, clear plaque; ND, not done.
DISCUSSION
Phage-encoded protein CIII was identified as a factor that stabilized the transcription activator CII in lambdoid bacteriophages, promoting the lysogenic pathway (1, 24). CIII also stabilizes the heat shock-specific subunit σ32 of RNA polymerase, inducing a heat shock response in cells (6). Both λCII and E. coli σ32 are degraded by the ATP-dependent integral membrane metalloprotease HflB (FtsH) (8, 12, 23, 25, 37). It has been suggested that λCIII acts as an antiprotease (13, 24, 32). It is believed that CIII inhibits the proteolytic activity of HflB against CII or σ32 (23). On the basis of structure prediction, the existence of an amphipathic helix was proposed for λCII, HK022 CII, and σ32, and it was suggested that an amphipathic helix that was conserved among CIII proteins from lambda, P22, or HK022 was responsible for the action of CIII as a competitive protease inhibitor (32). However, it was subsequently found that the point of HflB attack on λCII was not the putative amphipathic helix but its C-terminal tail (30), which was shown to be flexible (17, 18).
The HflB inhibitor λCIII is also its substrate (23). However, little is known about the structure of CIII. Recently, the inhibitory function of CIII has been attributed to disruption of the CII-HflB interaction by CIII, which forms dimers and higher oligomers and binds to HflB (31). In the present work, we have found that CIII forms dimers, with a disulfide bond linking the two monomers. The largely helical central core of this molecule lacks any cysteine residue. Nevertheless, the peptide CIIIC (containing the entire core region of CIII, residues 14 to 41) is also dimeric, emphasizing the existence of strong noncovalent interactions between the central helical core in the two subunits forming the CIII dimer. Moreover, limited proteolysis by trypsin revealed susceptible regions at the terminal ends of CIII. Thus, it is possible that HflB cleaves CIII too from one end. Indeed, HflB is known to recognize disordered regions in proteins (3), apart from its specific substrates such as σ32 (23), SecY (27), or YccA (28). HflB is a hexameric molecule comprising two rings, and the protease domain consists of helices forming a flat hexagon covered by AAA domains (26). It has been suggested that the degradation mechanism requires entry of the substrate through a narrow cleft, accompanied by ATP hydrolysis, unfolding of the substrate, and its translocation to the active site (7, 10). We found that CIIIC, which lacks the terminal tails, was resistant to HflB and also exhibited better antiprotease activity than CIII. Therefore, it is very likely that for both CIII and CII, disordered terminal regions act as targets for HflB proteolysis.
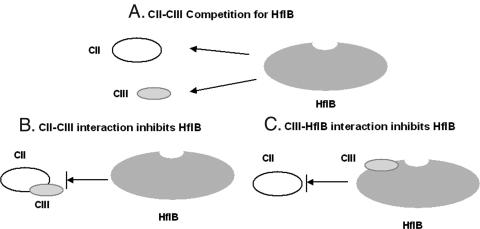
The question that remains to be answered is how does CIII act as an antiprotease, inhibiting HflB? When HflB encounters a mixture of CII and CIII, three distinct scenarios are possible. In model A, HflB competes for CII and CIII, resulting in the partial protection of each in the presence of the other. In model B, CIII forms a complex with CII and inhibits the proteolysis of the latter. In model C, CIII binds to HflB, inhibiting its proteolytic activity. These possibilities are shown in Fig. 10. CIIIC acts as an inhibitor but is itself not a substrate for HflB. Assuming that CIII and CIIIC do not act differently with respect to HflB, the simple competition model (model A) does not explain the antiproteolytic activity of CIIIC. Model B requires an interaction between CII and CIII, which is ruled out by our in vitro binding data obtained by Ni-NTA pull-down assay and coimmunoprecipitation (Fig. 9). For the third possibility (model C), direct binding of CIII to HflB is required. Such a direct CIII-HflB interaction was demonstrated by GST pull down and coimmunoprecipitation (Fig. 9). The fact that σ32 is protected by CIII also favors this possibility. A direct CIII-HflB interaction is in agreement with the antiproteolytic mechanism of CIII proposed by Kobiler et al. (31). However, mechanisms C and A are not mutually exclusive. Thus, CII and CIII may compete for binding to HflB, and the relative binding affinities may play an important role, as discussed below.
FIG. 10.
Alternative models for the protection of CII from HflB by CIII. In model A, CII and CIII compete for HflB. In model B, CIII binds to CII and inhibits the HflB digestion of CII. In model C, CIII binds to HflB near the substrate entry site (shown as a small depression in the HflB cartoon) and inhibits the access of CII to this site.
In the recently published structure of HflB (7), the proposed site for the entry of substrates such as CII is lined with Phe-234 from all of the subunits, forming a hydrophobic pocket. There are a large number of helical regions in the neighborhood of this entry cleft (7). It is possible that CIII binds to HflB near this cleft by aligning its central helical region, inhibiting the entry of other substrates by steric hindrance. Simultaneously, the unstructured terminal regions of a CIII dimer may gain access to the entry channel, initiating CIII proteolysis. On the other hand, proteolysis of CII would require an interaction between the C termini of tetrameric CII, a larger molecule, and the HflB entry channel. This could lead to a difference in how the two molecules are attacked by HflB, which is reflected in the initial modes of their proteolysis; while CII is readily cleaved upon the addition of HflB (Fig. 2), CIII proteolysis is somewhat delayed (Fig. 3 and 5). This is evident upon examination of the initial slope of the cleavage reactions as a function of time. Indeed, a slow in vivo degradation of CIII has been reported (23). This mode of action also explains the binding of CIIIC to HflB, its resistance to proteolysis, and its failure to effectively protect CIII from HflB digestion. Limiting concentrations of CIII do not inhibit the proteolysis of CII by HflB (our unpublished data). CIIIC can partially protect CIII only when CIIIC is allowed to first interact with HflB (Fig. 5). From the data in Fig. 9E and F, it appears that the binding strengths for interaction with HflB follows the order CII > CIII > CIIIC (supported by densitometry data [not shown]). This could account for the inability of CIIIC to protect CIII when the two are added together. Therefore, the protection of CII by CIII would depend on the relative affinities for HflB-CII and HflB-CIII interactions. These affinities would dictate the amounts of CII and CIII present as free molecules or as complexes with HflB in a mixture when all three are present and direct the fate of the cellular substrate of HflB.
Acknowledgments
This work was supported by research fellowships to S. Halder and A. B. Datta from the Council of Scientific and Industrial Research, Government of India.
We thank Mrinal Das, S. R. Majhi, and Shri Asim Banerjee of the Bose Institute for help with the densitometry of gels, with peptide synthesis, and with high-performance liquid chromatography, respectively, and the Indian Institute of Chemical Biology, Kolkata, for mass spectrometry. Various bacterial and viral strains were obtained from Sankar Adhya, NIH; Koreaki Ito, Kyoto, Japan; and Don Court, NIH. Dipak Dutta, CDFD, Hyderabad, and Kaustav Bandyopadhyay of the Bose Institute contributed many useful suggestions, and G. Basu and P. Chakrabarti of the Bose Institute helped clarify several ideas by stimulating discussions.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Altuvia, S., and A. B. Oppenheim. 1986. Translational regulatory signals within the coding region of the bacteriophage λ cIII gene. J. Bacteriol. 167:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 3.Asahara, Y., K. Atsuta, K. Motohashi, H. Taguchi, M. Yohda, and M. Yoshida. 2000. FtsH recognizes proteins with unfolded structure and hydrolyzes the carboxyl side of hydrophobic residues. J. Biochem. (Tokyo) 127:931-937. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, E., and R. C. Sheppard. 1989. Solid phase peptide synthesis: a practical approach. IRL Press at Oxford University Press, Oxford, England.
- 5.Atherton, E., D. L. Clive, and R. C. Sheppard. 1975. Polyamide supports for polypeptide synthesis. J. Am. Chem. Soc. 97:6584-6585. [DOI] [PubMed] [Google Scholar]
- 6.Bahl, H., H. Echols, D. B. Straus, D. Court, R. Crowl, and C. P. Georgopoulos. 1987. Induction of the heat shock response of E. coli through stabilization of σ32 by the phage λ cIII protein. Genes Dev. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 7.Bieniossek, C., T. Schalch, M. Bumann, M. Meister, R. Meier, and U. Baumann. 2006. The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaszczak, A., C. Georgopoulos, and K. Liberek. 1999. On the mechanism of FtsH-dependent degradation of the σ32 transcriptional regulator of Escherichia coli and the role of the DnaK chaperone machine. Mol. Microbiol. 31:157-166. [DOI] [PubMed] [Google Scholar]
- 9.Bohm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 10.Bruckner, R. C., P. L. Gumyuzlu, and R. L. Stein. 2003. Coupled kinetics of ATP and peptide hydrolysis by Escherichia coli FtsH protease. Biochemistry 42:10843-10852. [DOI] [PubMed] [Google Scholar]
- 11.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36-W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmona, M., and V. deLorenzo. 1999. Involvement of the FtsH (HflB) protease in the activity of σ54 promoters. Mol. Microbiol. 31:261-270. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, H. H., P. J. Muhlrad, M. A. Hoyt, and H. Echols. 1988. Cleavage of the cII protein of phage λ by purified HflA protease: control of the switch between lysis and lysogeny. Proc. Natl. Acad. Sci. USA 85:7882-7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Court, D. L., A. B. Oppenheim, and S. L. Adhya. 2007. A new look at bacteriophage λ genetic networks. J. Bacteriol. 189:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta, A. B. 2003. The lysis-lysogeny switch of bacteriophage lambda: a structural and biophysical analysis. Ph.D. dissertation. Jadavpur University, Kolkata, India.
- 16.Datta, A. B., P. Chakrabarti, H. S. Subramanya, and P. Parrack. 2001. Purification and crystallization of CII: an unstable transcription activator from phage λ. Biochem. Biophys. Res. Commun. 288:997-1000. [DOI] [PubMed] [Google Scholar]
- 17.Datta, A. B., S. Roy, and P. Parrack. 2003. Disorder-order transition of λCII promoted by low concentrations of guanidine hydrochloride suggests a stable core and a flexible C-terminus. Eur. J. Biochem. 270:4439-4446. [DOI] [PubMed] [Google Scholar]
- 18.Datta, A. B., S. Roy, and P. Parrack. 2005. Role of C-terminal residues in oligomerization and stability of λCII: implications for lysis-lysogeny decision of the phage. J. Mol. Biol. 345:315-324. [DOI] [PubMed] [Google Scholar]
- 19.Ellman, G. L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:70-77. [DOI] [PubMed] [Google Scholar]
- 20.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Baroch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 21.Herman, C., T. Ogura, T. Tomoyasu, S. Hiraga, Y. Akiyama, K. Ito, R. Thomas, and P. Bouloc. 1993. Cell growth and λ phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc. Natl. Acad. Sci. USA 90:10861-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herman, C., D. Thévenet, R. D'Ari, and P. Bouloc. 1997. The HflB protease of Escherichia coli degrades its inhibitor λcIII. J. Bacteriol. 179:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyt, M. A., D. M. Knight, A. Das, H. I. Miller, and H. Echols. 1982. Control of phage λ development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell 31:565-573. [DOI] [PubMed] [Google Scholar]
- 25.Ito, K., and Y. Akiyama. 2005. Cellular functions, mechanism of action, and regulation of FtsH. Annu. Rev. Microbiol. 59:211-231. [DOI] [PubMed] [Google Scholar]
- 26.Karata, K., C. S. Verma, A. J. Wilkinson, and T. Ogura. 2001. Probing the mechanism of ATP hydrolysis and substrate translocation in the AAA protease FtsH by modelling and mutagenesis. Mol. Microbiol. 39:890-903. [DOI] [PubMed] [Google Scholar]
- 27.Kihara, A., Y. Akiyama, and K. Ito. 1995. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. USA 92:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kihara, A., Y. Akiyama, and K. Ito. 1998. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol. 279:175-188. [DOI] [PubMed] [Google Scholar]
- 29.King, D. S., C. G. Fields, and G. B. Fields. 1990. A cleavage method which minimizes side reaction following F-moc solid phase peptide synthesis. Int. J. Pept. Protein Res. 36:255-266. [DOI] [PubMed] [Google Scholar]
- 30.Kobiler, O., S. Koby, D. Teff, D. Court, and A. B. Oppenheim. 2002. The phage λ CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc. Natl. Acad. Sci. USA 99:14964-14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobiler, O., A. Rokney, and A. B. Oppenheim. 2007. Phage lambda CIII: a protease inhibitor regulating the lysis-lysogeny decision. PloS ONE 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornitzer, D., S. Altuvia, and A. B. Oppenheim. 1991. The activity of CIII regulator of lambdoid bacteriophages resides within a 24-amino-acid protein domain. Proc. Natl. Acad. Sci. USA 88:5217-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latala, B., M. Obuchowski, and G. Wegrzyn. 2001. Bacteriophage λ cIII gene product has an additional function apart from inhibition of cII degradation. Virus Genes 22:127-132. [DOI] [PubMed] [Google Scholar]
- 34.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed, p. A.1-A.2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Shotland, Y., S. Koby, D. Teff, N. Mansur, D. A. Oren, K. Tatematsu, T. Tomoyasu, M. Kessel, B. Bukau, T. Ogura, and A. B. Oppenheim. 1997. Proteolysis of the phage CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol. 24:1303-1310. [DOI] [PubMed] [Google Scholar]
- 37.Shotland, Y., A. Shifrin, T. Ziv, D. Teff, S. Koby, O. Kobiler, and A. B. Oppenheim. 2000. Proteolysis of bacteriophage λ CII by Escherichia coli FtsH (HflB). J. Bacteriol. 182:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemory, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wegrzyn, G., and A. Wegrzyn. 2005. Genetic switches during bacteriophage λ development. Prog. Nucleic Acid Res. Mol. Biol. 79:1-48. [DOI] [PubMed] [Google Scholar]