Abstract
HIV Tat, a transactivator of viral transcription, represses transcription of major histocompatibility (MHC) class I genes. Repression depends exclusively on the C-terminal domain of Tat, although the mechanism of this repression has not been known. We now show that repression results from the interaction of Tat with the TAFII250 component of the general transcription factor, TFIID. The C-terminal domain of Tat binds to a site on TAFII250 that overlaps the histone acetyl transferase domain, inhibiting TAFII250 histone acetyl transferase activity. Furthermore, promoters repressed by Tat, including the MHC class I promoter, are dependent on TAFII250 whereas those that are not repressed by Tat, such as SV40 and MuLV promoters, are independent of functional TAFII250. Thus, Tat repression of MHC class I transcription would be one mechanism by which HIV avoids immune surveillance.
HIV-1 infection of cells triggers de novo synthesis of viral gene products and causes altered expression of a variety of cellular genes. These effects are mediated by the HIV-1 encoded protein Tat, which transactivates the viral long terminal repeat (LTR) and various cellular genes (1–7). Tat is also a repressor of some cellular genes (8–10). In particular, it was reported that Tat represses in vivo transcription of major histocompatibility (MHC) class I genes, whose products play a pivotal role in immune surveillance against viral infection (11, 12). Indeed, HIV infection reduces cell surface expression of class I molecules (ref. 13 and unpublished observations).
Two forms of Tat are generated through alternative splicing (6, 14). One form, encoded by a one-exon transcript, is 72 amino acids in length whereas the other, encoded by a two-exon transcript, has an additional C-terminal domain and varies in length between 86 and 101 amino acids, depending on the viral isolate. Both Tat variants transactivate the LTR efficiently, but only the two-exon derived Tat is capable of repressing MHC class I gene transcription (11). Indeed, the second-exon encoded peptide of Tat is both necessary and sufficient for repression: N-terminal domains in the Tat protein that are required for transactivation are not required for repression of MHC class I transcription in vivo (15). Thus, Tat is a bifunctional protein, with distinct domains that mediate repression of the MHC class I promoter and transactivation of the viral LTR.
The mechanisms of Tat-mediated activation and repression are not fully understood. Tat transactivation of the viral LTR depends on both recruitment of the TATAA binding protein (TBP) and interaction with the viral TAR sequence (6, 14, 16). Tat binds to TBP through residues contained within the first exon; mutation of these residues eliminates transactivation (17). In addition, Tat interacts with a variety of cellular factors (17–20), some of which contribute to transactivation.
Significantly less is understood about the mechanism of Tat repression of cellular gene expression. Although the presence of HIV Tat in vivo reduces MHC class I promoter activity in a variety of cell types (12), it is not known whether this repression results from a direct effect of Tat on the class I promoter or from an indirect effect through its activation of other genes. It is known that Tat targets the MHC class I basal promoter for repression but that it does not bind to DNA directly (12). Furthermore, repression is observed only in the presence of Tat’s second exon peptide and does not require the TAR sequence to be associated with the target promoter (11). Together, these observations suggest that the mechanism of Tat-mediated repression is distinct from that of transactivation.
The studies reported here were designed to elucidate the mechanism of Tat repression and to identify any cellular factors with which Tat may interact in repressing MHC class I transcription. We report that Tat interacts with TAFII250, a component of the general transcription factor, TFIID, resulting in repression of MHC class I transcription in vitro. Tat binds to the histone acetyl transferase (HAT) domain of TAFII250, inhibiting its activity. Further, we find that there is a correlation between promoter dependence on TAFII250 and susceptibility to Tat-mediated repression. These observations provide a possible mechanism for Tat repression through its binding to TAFII250.
MATERIALS AND METHODS
Cell Lines and Plasmids.
The human HeLa cell line, the Syrian hamster cell line BHK-21, and the derived cell line tsBN462 were grown as described (12, 21, 22). The reporter constructs containing the GAL4 binding sites with the TATA element from the AdMLP, the Inr element from the TdT, or both, as well as the priming oligonucleotide, were reported (23). The MHC class I promoter construct, 313CAT, consists of 313 bp of 5′ flanking sequences derived from the swine class I gene PD1 ligated to the CAT reporter gene (11). The two-exon Tat proviral construct, pNL-ΔΔ, which encodes only Tat86, and the control proviral construct, pNL-AO, which does not encode any Tat, were as described (11).
The Gal4-Tat67–101 vector was constructed by cloning the HindIII-SalI fragment of pSV2Tat into the SmaI/SalI sites of the pAS1-CyH2 yeast expression vector. The resulting Gal-Tat fusion protein expresses the 30 carboxy-terminal amino acids of Tat101 derived from SF2 strain of HIV. PAS1-CYH2 was a kind gift from S. Elledge and W. Harper (Baylor Univ.) (24). The Gal4 activation domain-mouse spleen cell cDNA fusion library was generated as described (25). The pCMVHAXhTAFII250 plasmid was a kind gift from R. Tjian (Univ. of California, Berkeley). The glutathione S-transferase (GST)-Tat101 plasmid was a kind gift from T. Jeang (National Institutes of Health). The K41T mutation of Tat101 was as described (15). GST-Tat67–101 was made by PCR amplification, inserting 5′ BamHI and 3′ EcoRI sites, from a Tat101 template, and insertion into the pGEX2T vector.
Transfections.
HeLa cells (8 × 105) were transfected by the calcium phosphate technique, as described (12), with 5 μg of Gal4 promoter constructs, 1–2 μg of the GalSp1 or GalVP16 expression vectors, and 10 μg of pNL-A0 or pNL-ΔΔ Tat expression vectors. RNA was prepared from transfected cells by using STAT-60 (Tel-Test, Friendswood, TX) according to the manufacturers’ directions. tsBN462 cells and control BHK hamster cells were transfected by calcium phosphate. After transfection, cells were left at 32°C for 24 hr and then either were shifted to 39°C (restrictive temp) or were left at 32°C (permissive temperature) and were harvested after 16 additional hr. DNA concentrations used were 5 μg of the class I promoter constructs pSV2CAT or pSV3CAT; 200 ng RSV luciferase was used as a transfection efficiency control. CAT assays were as described (12) and were normalized to luciferase activity.
Yeast Two-Hybrid Screening.
Saccharomyces cerevisiae strain Y190 was transformed sequentially with the pAS-CHT2-Tat67–101 bait vector and a mouse spleen cDNA library (25) in the GAL4 activation domain vector according to the protocols described for the Matchmaker yeast two-hybrid system (CLONTECH). Preliminary experiments demonstrated that the GAL4-Tat67–101 fusion did not activate in the absence of the GAL4 activation domain in the yeast two-hybrid assay (data not shown). Approximately 3 × 107 cDNA clones were transformed into Y190 cells carrying the GAL4-Tat101 construct and were plated on selection medium lacking Trp, Leu, His and 50 mM 3-aminotriazole. After ≈1 week at 30°C, four clones expressing His3 and β-galactosidase activity were identified. Plasmid DNA from positive clones was recovered by standard method (CLONTECH) and were sequenced on the Applied Biosystems automated Sequencer. DNA sequence analysis and homology searches were by the algorithm of Altschul et al. (26).
Production of TAFII250 and GST Pull Downs.
The HincII fragment of TAFII 250 cDNA cloned into pcDNA3 (1 μg/50 μl reaction) was translated in vitro in the TnT Coupled Reticulocyte Lysate System (Promega) from the T7 polymerase promoter with 35S methionine (Amersham). GST-agarose beads (Pierce) were prewashed in 15 ml of cold BB (20 mM Hepes, pH 7.9/100 mM KCl/12.5 mM MgCl2/0.1 mM EDTA/0.1 mM DTT/0.2% Nonidet P-40/17% glycerol) with 0.5 mg/ml of BSA, were spun at 1500 rpm, and were resuspended in 1 ml BB without BSA. For pull-downs, 5 μg of GST fusion protein was combined with 10 μl reaction mix of 35S-TAFII250 fragment and 30 μl prewashed GST-agarose beads (50% slurry); the final volume was adjusted to 200 μl. The reaction was incubated for 2 hr at 4°C. Beads were washed twice with Wash Buffer (50 mM Tris⋅Cl, pH 7.9/150 mM NaCl/0.2% Nonidet P-40), and samples were resolved on reducing SDS/PAGE gels and were quantified by PhosphorImager (Molecular Dynamics).
HA-tagged full length TAFII250 was prepared from recombinant baculovirus-infected High5 cells (Invitrogen) by one cycle of freeze/thaw in Buffer B (20 mM Tris⋅Cl, pH8.0/5 mM MgCl2/10% glycerol/0.1% Nonidet P-40) supplemented with 420 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 20 μg/ml aprotinin, 5 μg/ml leupeptin, and 10 μg/ml pepstatin. For pull-down experiments, 5 μg of the GST fusion proteins were incubated with 100 μl of High5 extracts expressing HA-hTAFII250 and 30 μl glutathione Sepharose 4B beads (Pharmacia; 1:1 slurry) at 4°C for 60 min. Beads then were washed and resuspended in 2× SDS/PAGE gel loading buffer, and eluates were analyzed as described below for HeLa nuclear extracts.
For analysis of HeLa cell nuclear extracts, 250 μg of nuclear extract were incubated overnight at 4°C with equal amounts of GST, GST-Tat101, or GSTTatK41T bound to agarose beads. Proteins were eluted in sample buffer at 95°C, were subjected to SDS gel electrophoresis, and were transferred to nitrocellulose membranes. After blocking with 5% dried milk in PBS, the blot was incubated with 10 μg of anti-TAFII250 antibody (Santa Cruz Biotechnology) in 5 ml of Blotto (5% dried milk, TBS-T) (Santa Cruz Biotechnology) for 1 hr, was washed twice in TBS-T, and was incubated for 2 hr with goat anti-mouse horseradish peroxidase conjugated anti-mouse IgG (Santa Cruz Biotechnology) at 1:2000 dilution in TBS. The filter was washed three times with TBS-T and was developed with enhanced chemiluminescence reagents.
In Vitro Transcription and Primer Extension Assay.
The in vitro transcription reaction containing 2 μg of 313CAT, 6 mM MgCl2, 0.8 mM dNTPs, and 30 units of HeLa nuclear extract (Promega) in 20 mM Hepes (pH7.9), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and 20% glycerol in a total of 25 μl was incubated at 20°C for 60 min. Where indicated, eluates of GST, GST-Tat101, or GST-TAT67–101, containing between 0.25 and 0.75 μg, were added, keeping the total reaction volume constant. GST-mTAFII250 derived from the mTAFII250 HincII DNA fragment was added at concentrations up to 1.5 μg.
Primer extension reactions were used to monitor both transfections and in vitro transcription, as follows. RNA (10 μg from transfected cells or all of in vitro transcription reaction) was resuspended in H20 and was reprecipitated with 10 ng of 32P-labeled extension oligonucleotide primer. Pellets were resuspended in 10 μl of 1× buffer B plus DTT (50 mM Tris⋅Cl, pH 8.3/75 mM KCl/3 mM MgCl2/10 mM DTT) and were hybridized 90 min at the hybridization temperature; hybridization temperature for the TK oligonucleotide (sequence: GGGGTACGAAGCCATACGCG) was 62°C and for the CAT oligonucleotide (sequence: GGTGGTATATCCAGTGATTTTTTTCTCCAT) was 60°C. Then, 40 μl of reaction mix (1× buffer B plus DTT with 0.5 mM dNTPs) and 200 units Superscript II reverse transcriptase (GIBCO/BRL) were added. The samples were incubated at 42°C for 60 min, were precipitated and resuspended in formamide loading dye, were heated at 75°C for 3 min, and were resolved on an 8% denaturing acrylamide gel.
HAT Assay.
The HAT assays were performed and processed as described in ref. 27 as a modification of Brownell and Allis (28) with 0.5–1.0 μg histones H3/H4 or 1 μg HeLa nuclear histone octamer and varying amount of GST, GST-Snap23, GST-TAT101, or GST-TAT67–101. Alternatively, HAT assays with histone H3 and increasing amounts of GST or GST-TAT101 were resolved on 18% SDS/PAGE gels, were processed, and were quantified by phosphorimaging.
RESULTS
HIV Tat Binds To TAFII250.
Because Tat targets the basal class I promoter but does not bind to DNA directly (12), we surmised that repression of transcription results from the interaction of Tat’s C-terminal domain with cellular factors. To identify such factors, a yeast two-hybrid screen was performed by using a fusion of the C-terminal peptide of Tat and the GAL4 DNA binding domain (GAL4-Tat67–101). The C-terminal domain of Tat was used as bait to avoid isolating any of the cellular factors (including TBP, RNA polymerase II, and TAFII55) known to bind the N-terminal transactivation domain of Tat (17–19). To further increase the possibility of only isolating factors involved in repression, a mouse spleen cDNA library was screened. In mouse cells, Tat represses class I transcription but does not transactivate the HIV LTR (15), so Tat-interacting cellular factors involved in repression should be identified preferentially in a mouse cDNA library.
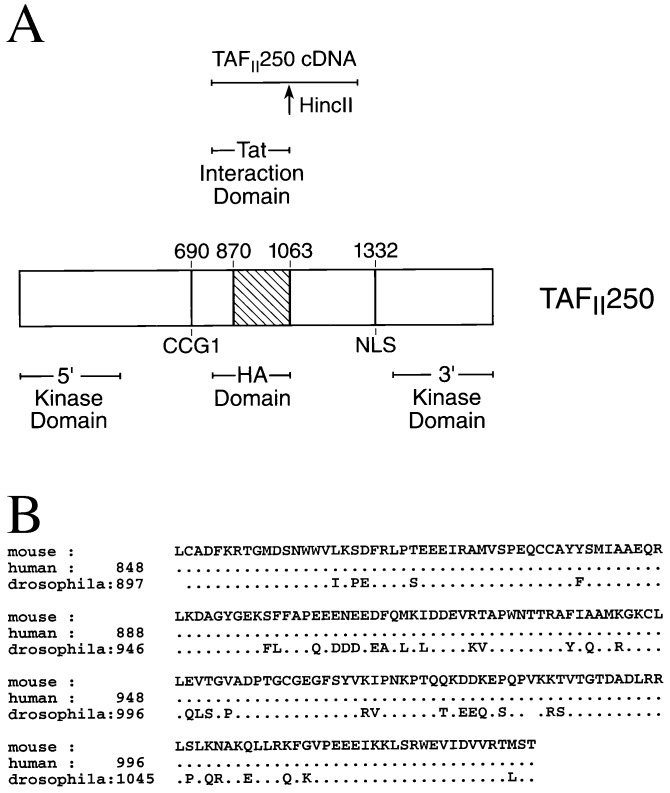
Three clones were isolated that depended on the presence of the GAL4-Tat67–101 fusion protein to generate β-galactosidase activity; neither the Gal4 DNA binding domain vector (pAS-CHY2) alone nor an unrelated Gal4-syntaxin5 construct yielded prototrophic, β-galactosidase+ colonies in conjunction with the cDNA clones (data not shown). Two of the clones contained a 1,299-bp insert with a 433-aa ORF. The DNA sequence was homologous to the TAFII250 genes of various species: 91% to human, 94% to hamster, and 69% to drosophila [data not shown; the sequence has been deposited in the GenBank database (accession no. AF022178)]. The encoded peptide is 99% homologous to human TAFII250 and 61% homologous to drosophila TAFII230, leading to the conclusion that it is a fragment of mouse TAFII250 (mTAFII250) (Fig. 1) and that it specifically interacts with the second-exon Tat peptide 67–101 in vivo in yeast.
Figure 1.
Isolation of TAFII250 by yeast two-hybrid screening by using a second exon Tat fragment. (A) Map of the entire human TAFII250 protein indicating the regions containing the CCG1 mutation and the 5′ and 3′ kinase domains as well as the histone acetylase (HA) domain. The region homologous to the cloned mouse TAFII250 segment that interacts with the second exon of Tat is noted. (B) Homology of the translated mouse TAFII250 HincII fragment with the human and drosophila TAFII250 protein sequences.
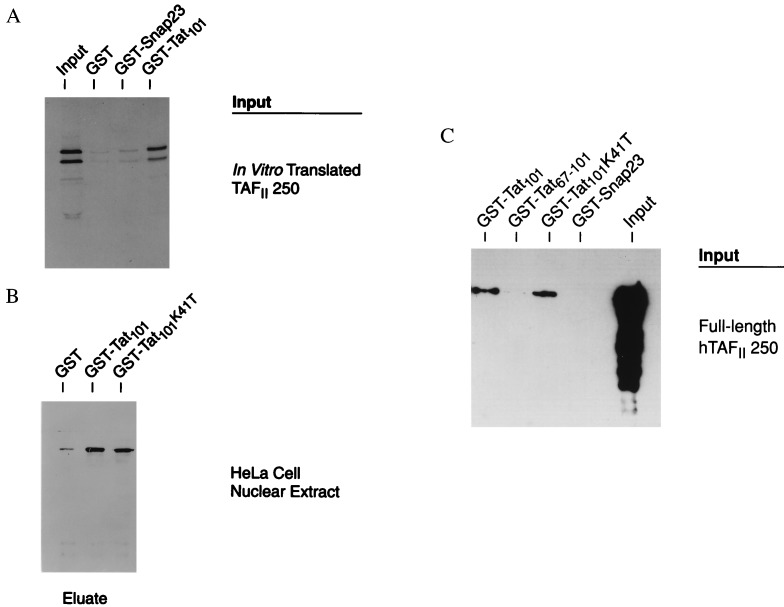
To determine whether this interaction also occurs in vitro, GST pull-down assays were performed by using GST-Tat101 and either (i) in vitro-translated fragment of mouse TAFII250, (ii) full length recombinant human TAFII250 (hTAFII250), or (iii) native hTAFII250 from nuclear extracts. Tat101 binds efficiently to the mTAFII250 fragment: GST-Tat101 bound to the in vitro-translated fragment of mTAFII250 significantly better than did either GST alone or an irrelevant fusion protein, GST-SNAP23 (Fig. 2A; Table 1). GST-Tat101 also bound native hTAFII250 from HeLa nuclear extracts significantly above the background levels of GST alone (Fig. 2B). Tat is known to bind TBP, another component of the TFIID complex (17). To eliminate the possibility that binding of Tat to hTAFII250 occurred indirectly through TBP, a Tat101 derivative, Tat101K41T, that no longer binds TBP was tested. Whereas the wild-type GST-Tat101 bound TBP, the mutant GST-Tat101K41T did not (data not shown). However, the mutant GST-Tat101K41T still bound TFIID from HeLa nuclear extracts (Fig. 2B), suggesting an interaction with TAFII250. Both GST-Tat101 and GST-Tat101K41T also bound efficiently to recombinant full-length human TAFII250 (Fig. 2C). Finally, binding of recombinant hTAFII250 by the second exon fragment of Tat, GST-Tat67–101, was detectable and reproducible, although relatively inefficient (Fig. 2C). Taken together, these results demonstrate a specific interaction between Tat and TAFII250.
Figure 2.
HIV Tat Binds TAFII250 in vitro. (A) In vitro-translated 35S-labeled mTAFII250 HincII fragment was incubated with recombinant GST, GST-SNAP23, or GST-Tat101, was captured on glutathione-agarose beads, and was analyzed in SDS/PAGE. Shown is a representative autoradiogram. Results of multiple experiments are summarized in Table 2. (B) Western blot of hTAFII250 precipitated from HeLa nuclear extract by GST-Tat101 or GST-Tat101K41T but not by GST alone. Recombinant GST, GST-Tat101, or GST-Tat101K41T were added to excess HeLa nuclear extract and were recovered on glutathione-agarose beads; hTAFII250 was detected with antibody after SDS/PAGE. (C) Western blot of recombinant full length hTAFII250 precipitated by GST-Tat101, GST-Tat101K41T, or GST-Tat67–101, but not the irrelevant GST-SNAP23 fusion protein. Bound hTAFII250 was detected as in B.
Table 1.
TAFII250 binds to GST-TAT but not to GST or GST-Snap23
| GST fusion | TAFII250 Relative binding |
|---|---|
| GST alone | 1.0 |
| GST-Snap23 | 0.9 ± 1.3 |
| GST-TAT101 | 22.1 ± 3.7 |
In vitro translated 35S-labeled TAFII250 HincII fragment was incubated with the various GST fusion proteins, as described in Materials and Methods. Binding of the TAFII250 was quantitated relative to the GST control. The results represent the average of four independent experiments.
HIV Tat Represses MHC Class I Promoter Transcription in Vitro.
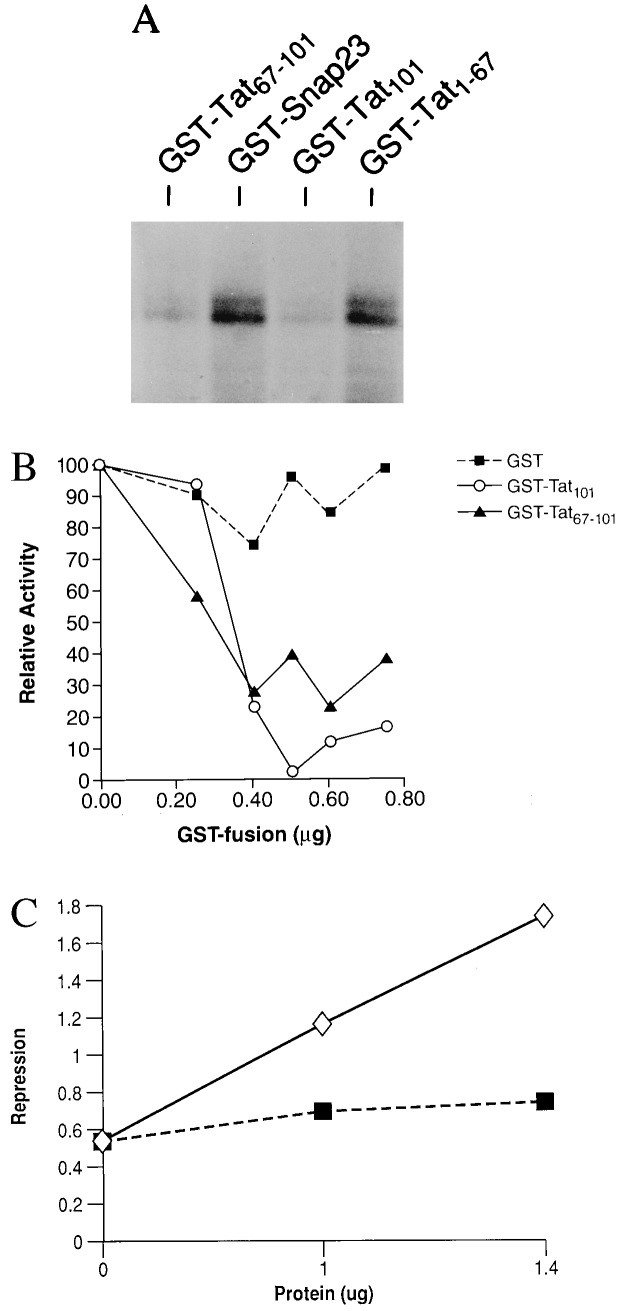
The finding that Tat binds to TAFII250 leads to the prediction that Tat repression results from a direct effect at the class I basal promoter. To test this, Tat’s effect on in vitro transcription was assessed. A class I promoter construct extending from the transcription initiation site to 313 bp upstream directs the in vitro synthesis of a correctly initiated transcript; transcription is α-amanitin-sensitive (data not shown). In vitro transcription was inhibited markedly by the addition of a GST-Tat101 fusion protein but not an irrelevant fusion protein, GST-SNAP23 (Fig. 3A; Table 2). The extent of repression increased with increasing concentrations of Tat protein (Fig. 3B), achieving a magnitude comparable to that observed in vivo (12). Repression does not depend on the ability of Tat to bind to TBP because the mutant Tat101K41T, which does not bind TBP, is as effective in repressing class I transcription as native Tat101 (Table 2). Finally, repression is specific for the class I promoter: Tat does not repress in vitro transcription of either the HIV LTR or an unrelated promoter, the MuLV LTR (Table 2). Thus, Tat directly represses class I promoter activity.
Figure 3.
Tat specifically inhibits transcription from the MHC class I promoter. (A) In vitro transcription of the MHC class I promoter construct 313CAT was performed in the presence or absence of 0.2 μg of recombinant proteins, GST-Tat101, GST-Tat1–67, GST-Tat67–101, or GST-SNAP23. Transcripts were detected by primer extensions, as detailed in Materials and Methods. Shown is a representative autoradiogram. (B) Titration of the effects of Tat101, Tat67–101 fragment, and GST on in vitro transcription of the class I promoter. Tat 101 and Tat67–101 were both added as GST fusion proteins. The results are plotted relative to the level of transcription in the absence of added protein. The data shown in A and B are from separate experiments. (C) mTAFII250 fragment relieves Tat-mediated repression of transcription. In vitro transcription reactions of 313CAT were performed in the presence and absence of 0.5 μg Tat101 and increasing concentrations of either mTAFII250 fragment (open symbol) or control SNAP23 protein (closed symbol). The magnitude of Tat repression was determined at each concentration of competitor.
Table 2.
HIV-1 TAT101 inhibits in vitro transcription of the MHC class I, but not the HIV LTR or MuLV LTR, promoter
| Relative promoter
activity in vitro
|
|||
|---|---|---|---|
| MHC Class I | HIV LTR | MuLV | |
| A. | |||
| No additions | 1.0 | 1.0 | |
| GST | 0.84 | 0.92 | |
| GST-Tat101 | 0.41 | 2.5 | |
| B. | |||
| GST | – | 1.0 | |
| GST-SNAP23 | 1.0 | – | |
| GST-Tat101 | 0.32 | 1.05 | |
| GST-Tat67-101 | 0.32 | 1.0 | |
| GST-Tat101K41T | 0.50 | ||
| GST-Tat1-67 | 0.91 | 1.05 | |
In vitro transcription reactions with each of the three promoters—class I, HIV LTR, and MuLV—were performed by using the promoters fused to the CAT gene. Recombinant GST fusion proteins (0.5 μg) were added where indicated. A and B quantitate the results of two independent and representative experiments using the class I promoter. Analysis of the HIV and MuLV LTRs were done in parallel with the class I promoter experiments; each has been repeated twice.
The second exon-encoded C-terminal domain of Tat (amino acids 67–101) is responsible for repression in vivo (11, 15). In fact, in transfected HeLa cells, a fusion protein consisting of the isolated 67- to 101-aa C-terminal fragment and the DNA binding domain of GAL4 (GAL4-Tat67–101) repressed by 3- to 10-fold the activity of a class I promoter containing 5 gal4 sites (data not shown). This second exon encoded peptide also is capable of repressing transcription in vitro. Thus, addition to the in vitro transcription reaction of GST-Tat67–101 efficiently repressed class I transcription (Fig. 3 A and B). In contrast, a GST-Tat fusion protein containing only first exon sequences, namely GST-Tat 1–67, had no effect on class I promoter activity in vitro (Fig. 3A; Table 2). Thus, the C terminus of Tat is sufficient to repress class I promoter activity.
The above data suggest that repression of class I transcription results from the interaction between Tat and TAFII250. To determine whether this is the case, we tested the ability of the Tat-binding mTAFII250 fragment (amino acids 848–1034) to relieve Tat repression of class I promoter activity in vitro. As shown in Fig. 3C, addition of increasing amounts of the mTAFII250 fragment reversed Tat-mediated repression. Addition of an irrelevant control protein had no effect. (It is interesting to note that the mTAFII250 fragment, at the highest concentration, modestly activated transcription; this observation is under investigation.) Taken together, these data are most consistent with the interpretation that Tat represses in vitro transcription through a direct interaction with TAFII250.
HIV Tat Inhibits the Histone Acetyl Transferase Activity of TAFII250.
The fragment of mTAFII250 isolated in the yeast two-hybrid screen and shown to interact with Tat extends from amino acids 848 to 1280. The drosophila homolog of TAFII250, dTAF II230, recently has been found to contain HAT activity, which maps to a region between 885 and 1140 amino acids (27). Initial mapping on mouse TAFII250 of the Tat binding site localized it to a polypeptide, encoded by a HincII DNA fragment, that is 100% homologous to human TAFII250 protein, amino acids 848 to 1034, and 77% homologous to the drosophila TAFII230 (dTAFII230) (Fig. 1B and data not shown). Because the Tat interaction domain of TAFII250 overlaps with the corresponding HAT domain in dTAFII230 (Fig. 1A), we examined the effect of Tat on HAT enzymatic activity. Tat101 protein, added as a GST fusion protein, efficiently inhibited the HAT activity of dTAFII230 (Table 3). Inhibition increased as a function of the concentration of added Tat101 (data not shown). This inhibition is significant and was not observed with control GST protein or with an irrelevant fusion protein, GST-SNAP23 (Table 3). Inhibition of HAT activity depended on the second exon peptide because one exon Tat1–67 did not significantly affect the HAT activity (Table 3B). Tat101 itself was not acetylated by dTAFII230 and thus did not act simply as a competitive sink (data not shown). This inhibition by Tat101 of the dTAFII230 HAT activity suggests that Tat101 also would inhibit hTAFII250 HAT activity. The ability of two-exon, but not one-exon, Tat to inhibit dTAFII230 HAT activity also correlates with the ability of two-exon, but not one-exon, Tat to repress MHC class I promoter activity.
Table 3.
HIV Tat inhibits the HAT of dTAFII230
| Relative HAT activity | P* | |
|---|---|---|
| A. dTAFII230 alone | 1.0 | |
| +GST-Tat101 | 0.35 ± 0.04 | <0.015 |
| +GST | 0.79 ± 0.10 | |
| B. dTAFII230 | ||
| +GST-Snap23 | 1.0 | |
| +GST-Tat101 | 0.6 ± 0.01 | <0.0005 |
| +GST-Tat1-67 | 0.82 ± 0.05 | =0.05 |
HAT activity was determined as described in Materials and Methods. Recombinant GST fusion proteins (0.5 μg) were added where indicated. The data in A are derived from four independent experiments, using two different assays, with a single enzyme preparation. The data from B are derived from three independent filter assays, with 250 ng of two different enzyme preparations.
The two-tailed Student’s t test was used to compare the effect of GST-Tat101 to that of GST alone.
Promoters Susceptible to HIV Tat Repression Depend on TAFII250 for Function.
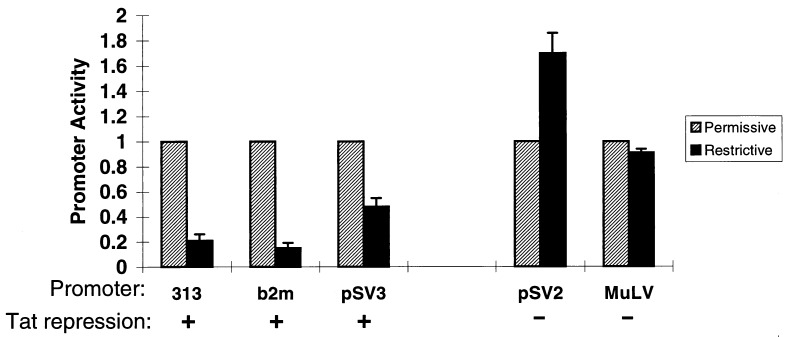
The role of TAFII250 in transcription is not fully understood (27, 29). Analysis of cell lines carrying a temperature-sensitive point mutation of TAFII250 reveals that many, but not all, promoters depend on TAFII250 (21, 22, 30–33). The observed interaction between Tat101 and TAFII250 leads to the prediction that the MHC class I promoter should be among those dependent on TAFII250. To assess the TAFII250 dependence of the class I promoter, we examined its activity in the TAFII250 temperature-sensitive cell line tsBN462. As shown in Fig. 4, class I promoter activity was impaired markedly at the nonpermissive temperature, demonstrating that it depends on TAFII250. Inhibition of class I promoter activity is a consequence of the TAFII250 mutation because transfection of wild-type hTAFII250 into the tsBN462 cells restored promoter activity (data not shown). Similarly, the β2-microglobulin promoter, which is repressed also by Tat (I. Carroll, T.K.H., J.W., and D.S.S., unpublished observations) is likewise dependent on TAFII250 (Fig. 4). Conversely, if Tat repression depends on its interaction with TAFII250, promoters known to be insensitive to Tat repression should not require TAFII250 for activity. Because the viral promoters/enhancers of SV40 (pSV2) and MuLV are not repressed by Tat101 (11, 12), we examined their dependence on TAFII250 in tsBN462. As predicted, these promoter/enhancers were fully active at the nonpermissive temperature, indicating that functional TAFII250 is not required for their activity (Fig. 4). The HIV LTR is also active in tsBN462 at the nonpermissive temperature (data not shown). Thus, for the promoters we have examined, susceptibility to Tat repression correlates with dependence on TAFII250.
Figure 4.
Promoter sensitivity to TAFII250 correlates with sensitivity to Tat-mediated repression. A series of promoters linked to the reporter CAT with known sensitivities to Tat-mediated repression (as indicated) were transfected into tsBN462 cells. Promoter activity was measured at the permissive (32°C) or restrictive (39°C) temperatures. To allow a direct comparison of the different promoters, activity was normalized to the activity of each at 32°C. Promoters sensitive to Tat repression were 313 (class I), b2 m (β2microglobulin), and pSV3 (basal SV40 promoter); resistant promoters were pSV2 (SV40 enhancer/promoter) and MuLV (murine leukemia virus).
This correlation can be extended to a single promoter whose susceptibility to Tat mediated repression can be modified by upstream enhancer elements. Viral enhancers confer on the SV40 basal promoter resistance to Tat repression (12); although the minimal promoter (pSV3) is repressed by Tat, the extended enhancer/promoter of SV40 (pSV2), containing the 72 bp viral enhancer, is resistant to Tat repression (12). As shown in Fig. 4, these promoters also differ in their dependence on TAFII250: in the tsBN462 cells, the minimal pSV3 promoter is inactive at the nonpermissive temperature whereas the extended pSV2 enhancer/promoter is fully active. These results demonstrate a strong correlation between susceptibility to Tat-mediated repression and dependence on TAFII250 for promoter activity. They further indicate that promoter requirements can be altered or modulated by upstream enhancer elements.
Tat Represses Transcription from TATAA and Inr Promoters.
The interaction of Tat with TAFII250 suggests that Tat mediates repression through the transcription initiation complex. If so, then susceptibility to this repression should not be restricted to a single promoter element or activator. To test this prediction, we examined Tat’s ability to repress basal and activated transcription from a set of synthetic promoter constructs: a TATAA element derived from the AdMLP, an initiator (Inr) element derived from the TdT promoter, and a construct containing both the TATAA and Inr (23). Each synthetic promoter was fused to a tk reporter gene and was flanked by 5′ gal4 binding sites to allow activation by the activators GAL4VP16 or GAL4Sp1. In the presence of two-exon Tat, the activities of all three promoters were reduced, as compared with either a vector control (Table 4) or one-exon Tat (data not shown). Repression was independent of either the basal promoter element or activator. Thus Tat does not target a specific promoter element or activator but, rather, functions through the common transcription initiation complex itself, consistent with its binding to TAFII250.
Table 4.
Tat represses promoter activity of various combinations of promoter elements and activators
| Repression by Tat,
Tat/control
| |||
|---|---|---|---|
| Activator | None | VP16 | Sp1 |
| Promoter | |||
| TATAA/Inr | 0.3 | 0.31 ± 0.09 | 0.38 ± 0.16 |
| TATAA | – | 0.44 ± 0.05 | – |
| Inr | 0.14 | 0.51 ± 0.09 | 0.30 ± 0.04 |
Each of the promoter constructs, fused to a tk reporter gene, was transfected into HeLa cells in the presence or absence of either GaIVP16 or GaISp1 and in the presence or absence of Tat. Promoter activity was determined by primer extension assays of RNA isolated from transfected cells, as described (23). The results are expressed as the ratio of promoter activity in the presence or absence of Tat and are the average of 3–4 independent transfections.
–, no detectable activity.
DISCUSSION
The present observations significantly extend our original model of Tat as a bifunctional protein with separable and distinct domains mediating transactivation and repression (11, 15). We report that Tat interacts with the TAFII250 component of TFIID, resulting in both repression of transcription and inhibition of the TAFII250 HAT activity. Tat has been shown to interact with a variety of other components of the preinitiation complex (including TBP, TAFII55, RNA polymerase II) as well as multiple other cellular factors (17–20, 34). Tat also interacts with a TFIIH-associated kinase, resulting in enhanced kinase activity (35). Of interest, all of the interactions with cellular proteins that result in promoter activation occur with the N-terminal activation domain of Tat. In contrast, the interaction of Tat with TAFII250 was observed by using the C-terminal repression domain. Based on the present studies, we now propose that these structural domains interact with distinct sets of proteins. The transactivation domain of Tat interacts with transcription factors to enhance their activities—acetylation, phosphorylation—thereby augmenting transcription. The C-terminal repression domain interacts with TAFII250 to reduce transcription. Together, these factors modulate levels of repression and activation.
Transcription initiation depends on the recruitment of general transcription factors to the promoter. Among these is the general transcription factor TFIID, which nucleates the transcription initiation complex (36). TFIID contains the TBP in association with the TBP-associated factors (TAFs). TAFII250, the largest component of TFIID, recently has been shown to possess HAT activity (27). The notion that HAT activity is important in initiating transcription is strengthened by the finding that at least two other transcriptional coactivators, p300 and CBP, are also histone acetyltransferases (37).
The observation that the site of interaction of HIV Tat with TAFII250 is coincident with the HAT domain and results in the inhibition of HAT activity raises the possibility that Tat repression of class I transcription may be mediated through its inhibition of HAT activity. It has been speculated that TAFII250 acetylates nucleosome core histones, relaxing chromatin folding and facilitating transcription from chromatin templates (27, 28). However, as shown here, Tat inhibits in vitro transcription from naked DNA. Thus, either Tat’s inhibition of TAFII250 HAT activity is unrelated to its repression of transcription or the substrates of the TAFII250 HAT activity are not limited to nucleosomal histones. Consistent with the possibility that protein acetylation functions as a regulatory mechanism is the recent report that the proto-oncogene p53 is acetylated, which results in increased sequence specific DNA binding (38). Recently, a Tat-interacting cellular protein, Tip60, was isolated (39). Although its function is not known, Tip60 contains a HAT domain (16). The fact that Tat interacts with both TAFII250 and Tip60—two otherwise unrelated proteins—suggests that the HAT activity may be an important target for Tat. Future studies should be directed at distinguishing these possibilities.
HIV-1, which infects CD4+ T cells and monocytes, is able to avoid immune surveillance to establish a persistent infection that ultimately leads to a profound immunodeficiency (1, 14, 33). One mechanism by which HIV-1 may avoid elimination by the immune system is through its down-regulation of MHC class I expression (11, 13). At least three viral proteins are known to affect levels of class I. The viral proteins Nef and Vpu both reduce cell surface expression of class I heavy chain molecules whereas Tat represses class I gene transcription (13, 40–42). The present studies demonstrate that Tat directly affects levels of transcription through its selective interactions with components of the transcription initiation complex. Its interaction with TAFII250 through the second exon domain represses the class I promoter. We propose that this repression, together with the effects of Nef and Vpu, leads to reduced surface levels of class I on HIV infected cells. This reduced expression provides a mechanism for the virus to avoid immune surveillance.
Acknowledgments
The authors very gratefully acknowledge Steve Smale and Bob Tjian for many helpful discussions and for providing the model promoter and hTAFII250 constructs, respectively. We thank J. Gnarra, S. Kirshner, V. Ravichandran, I. Carroll, and C. Cornell for many helpful discussions and Drs. T. Saito, S. Miyatake and S. Elledge for making available cDNA libraries. We also appreciate the critical reading of the manuscript by Drs. D. Levens, L. Glimcher, A. Singer, A. Weissman, F. Kashanchi, and J. Brady. A.C. was supported by the Howard Hughes Medical Institute.
ABBREVIATIONS
- LTR
long terminal repeat
- MHC
major histocompatibility
- TBP
TATAA binding protein
- HAT
histone acetyl transferase
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF022178).
Present address: Department of Biochemistry and Medicine, Louisiana State University Medical Center, 1901 Perdido Street, Suite 3205, New Orleans, LA 70112.
References
- 1. Cullen B. FASEB J. 1991;5:2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 2.Haseltine W A. FASEB J. 1991;5:2349–2360. doi: 10.1096/fasebj.5.10.1829694. [DOI] [PubMed] [Google Scholar]
- 3.Sastry K, Raghava H, Pandita R, Tatpal K, Aggarwal B. J Biol Chem. 1990;265:20091–20093. [PubMed] [Google Scholar]
- 4.Scala G, Ruocco M, Ambrosino C, Mallardo M, Giordano V, Baldasarre F, Dragonetti E, Quinto I, Venuta S. J Exp Med. 1994;179:961–971. doi: 10.1084/jem.179.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tansey W P, Ruppert S, Tjian R, Herr W. Genes Dev. 1994;8:2756–2769. doi: 10.1101/gad.8.22.2756. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnav Y, Wong-Staal F. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]
- 7.Westendorp M O, Shatrov V A, Schulze-Osthoff K, Rainer F, Kraft M, Los M, Krammer P H, Droge W, Lehmann V. EMBO J. 1995;14:546–554. doi: 10.1002/j.1460-2075.1995.tb07030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pocsik E, Higuchi M, Aggarwal B. Lymphokine Cytokine Res. 1992;11:317–325. [PubMed] [Google Scholar]
- 9.Purvis S F, Georges D, Williams T, Lederman M. Cell Immunol. 1992;144:32–42. doi: 10.1016/0008-8749(92)90223-c. [DOI] [PubMed] [Google Scholar]
- 10.Flores S, Marecki J, Harper K, Bose S, Nelson S, McCord J. Proc Natl Acad Sci USA. 1993;90:7632–7636. doi: 10.1073/pnas.90.16.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howcroft T K, Strebel K, Martin M, Singer D S. Science. 1993;260:1320–1322. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 12.Howcroft T K, Palmer L A, Brown J, Rellahan B, Kashanchi F, Brady J N, Singer D S. Immunity. 1995;3:127–138. doi: 10.1016/1074-7613(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 13.Scheppler J, Nicholson J, Swan D, Ahmed-Ansari A, McDougal J S. J Immunol. 1989;143:2858–2866. [PubMed] [Google Scholar]
- 14.Cullen B. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 15.Brown J, Howcroft T K, Singer D S. J Acquired Immune Defic Syndr. 1998;17:9–16. doi: 10.1097/00042560-199801010-00002. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto Y, Horikoshi M. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 17.Kashanshi R, Piras G, Radonovich M F, Duvall J, Fattaey A, Chiang C-H, Roeder R G, Brady J N. Nature (London) 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 18.Chiang C, Roeder R. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 19.Cujec T, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin M. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeang K-T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima T, Sekiguchi T, Sunamoto H, Yura K, Tomoda S, Go M, Kere J, Schlessinger D, Nishimoto T. Gene. 1994;141:193. doi: 10.1016/0378-1119(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 23.Emami K, Navarre W, Smale S. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durfee T, Becherer K, Chen P, Yeh S, Yang Y, Kilburn A, Lee W, Elledge S. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 25.Ohno H, Stewart J, Fournier M, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Mizzen C, Yang X, Kokubo T, Brownell J, Bannister A, Owen-Hughes T, Workman J, Wang L, Berger S, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 28.Brownell J, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth S, Allis C D. Cell. 1196;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 30.Dikstein R, Ruppert S, Tjian R. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 31.Moqtaderi Z, Bai Y, Poon D, Weil P, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki-Yagawa, Guermah M, Roeder R. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang E, Tjian R. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 34.Rice A P, Carlotti F. J Virol. 1990;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Martinez L, Mavankal G, Neveu J, Lane W, Ivanov I, Gaynor R. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 37.Ogryzko V, Schiltz L, Russanova V, Howard B, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 38.Gu W, Roeder R. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 39.Kamine J, Elangovan B, Subranmanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 40.Kerkau T, Schmitt-Landgraf R, Schimpl A, Wecker E. AIDS Res Hum Retroviruses. 1989;5:613–620. doi: 10.1089/aid.1989.5.613. [DOI] [PubMed] [Google Scholar]
- 41.Kerkau T, Bacik I, Bennink J, Yewdell J, Hunig T, Schimpl A, Schubert U. J Exp Med. 1997;185:1295–1305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz O, Marechal V, LeGall S, Lemonnier F, Heard J. Nature Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]