Abstract
Pseudomonas aeruginosa has served as an important organism in the study of biofilm formation; however, we still lack an understanding of the mechanisms by which this microbe transitions to a surface lifestyle. A recent study of the early stages of biofilm formation implicated the control of flagellar reversals and production of an exopolysaccharide (EPS) as factors in the establishment of a stable association with the substratum and swarming motility. Here we present evidence that SadC (PA4332), an inner membrane-localized diguanylate cyclase, plays a role in controlling these cellular functions. Deletion of the sadC gene results in a strain that is defective in biofilm formation and a hyperswarmer, while multicopy expression of this gene promotes sessility. A ΔsadC mutant was additionally found to be deficient in EPS production and display altered reversal behavior while swimming in high-viscosity medium, two behaviors proposed to influence biofilm formation and swarming motility. Epistasis analysis suggests that the sadC gene is part of a genetic pathway that allows for the concomitant regulation of these aspects of P. aeruginosa surface behavior. We propose that SadC and the phosphodiesterase BifA (S. L. Kuchma et al., J. Bacteriol. 189:8165-8178, 2007), via modulating levels of the signaling molecule cyclic-di-GMP, coregulate swarming motility and biofilm formation as P. aeruginosa transitions from a planktonic to a surface-associated lifestyle.
The gram-negative bacterium Pseudomonas aeruginosa is adept at coordinating individual cells to participate in several surface-associated behaviors. This ability benefits this microbe, presumably through providing protection from environmental insults or by improving access to nutrients (9). Recent work in our laboratory has identified a genetic pathway responsible for modulating two of these surface-associated behaviors: swarming motility and biofilm formation (3).
P. aeruginosa engages in swarming motility, allowing it to move across semisolid surfaces. The flagellum and surfactants, or surface wetting agents, are required for this process in P. aeruginosa (5, 20). The importance of swarming motility is not well understood; however, a recent report has suggested that it may play a role in determining the ultimate structure of biofilms formed by this microbe (48).
Biofilms have long been known as a predominant feature of the lifestyles of many bacteria (8). These microbial communities are common in a wide variety of both environmental and medical settings, where they can be especially problematic since bacteria within a biofilm have a reduced susceptibility to antimicrobial agents (27). The gram-negative organism P. aeruginosa has served as an important model system for understanding the formation of these microbial communities (21).
Biofilm formation is a stepwise process commencing when planktonic cells encounter a surface. Cells enter a transitional state of reversible surface attachment and, if biofilm formation is to proceed, these cells must stabilize this interaction with the substratum. To date, several factors, including the sadB, pelA, and motAB genes (3, 4, 52), have been identified that contribute to the progression toward irreversible attachment.
As the biofilm matures, cells aggregate into microcolonies and larger macrocolonies (19, 43), which are characteristically encased in an extracellular matrix. This material is thought to function in organizing and structuring the bacterial community (2). The P. aeruginosa biofilm matrix is comprised of multiple constituents, including proteinaceous materials (11, 54), membrane vesicles (44), DNA (56), and exopolysaccharides (EPS) (11, 12, 16, 29), although the relative contributions of each of these components to biofilm integrity are not yet fully understood. It is further becoming apparent that some components of the extracellular matrix also function to promote the early steps in biofilm formation (3, 16, 55, 56).
Emerging as an important signaling molecule in the control of aspects of the transition between a motile and a biofilm lifestyle is the intracellular molecule cyclic di-GMP (c-di-GMP). This messenger has been found in multiple systems to regulate motility and extracellular matrix production, two cellular outputs that influence biofilm formation. The existing paradigm in the field is that high concentrations of this molecule correlate with a sessile lifestyle (e.g., biofilm formation and EPS production), while its absence favors motility (e.g., twitching and swarming). The levels of c-di-GMP are enzymatically modulated by diguanylate cyclases (DGCs), proteins containing a GGDEF domain (37), and phosphodiesterases containing either an EAL (7, 49) or HD-GYP domain (42).
Here we report the characterization of the sadC gene, which encodes a membrane-localized diguanylate cyclase that is involved in biofilm formation in P. aeruginosa. We find that mutations in this gene influence both motility and EPS production, both of which may contribute to modulation of biofilm formation and swarming motility. Finally, we present genetic studies to place sadC within the context of the currently proposed pathway for reciprocally regulating biofilm formation and swarming motility.
MATERIALS AND METHODS
Media and chemicals.
P. aeruginosa and Escherichia coli strains were routinely cultured in LB medium supplemented with antibiotics as appropriate. M63 minimal medium supplemented with glucose (0.2%), Casamino Acids (0.5%), and MgSO4 (1 mM) was used as the base for biofilm, Congo red (CR), and swim-reversal measurements as described below. M8 salts (20) with glucose, Casamino Acids, and MgSO4 in the same concentrations as described above were utilized in swarming experiments. A MOPS-based medium (50 mM NaCl, 40 mM MOPS [morpholinepropanesulfonic acid, pH 7.4], 10 g of NH4SO4/liter, 1 mM MgSO4, 30 mM succinate, 0.15 mM K2HPO4) containing limiting amounts of phosphate was used to culture P. aeruginosa for assessment of whole-cell levels of c-di-GMP as described previously (13). Where indicated, arabinose at 0.2% was added to increase expression. Saccharomyces cerevisiae was maintained on yeast extract-peptone-dextrose medium. Synthetic defined medium lacking uracil (Sunrise Science) was used to select for and grow plasmid-harboring yeast.
Bacterial strains, plasmids, and primers.
All bacterial strains, plasmids, and primers are listed in Table 1. Primers were obtained from Integrated DNA Technologies.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant genotype or sequence (5′-3′)a | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PA14 | WT | 38 |
| sadC mutant | PA14 ΔsadC | This study |
| pelA mutant | PA14 ΔpelA | 11 |
| sadB mutant | PA14 sadB::Tn5B21; Tcr | 4 |
| bifA mutant | PA14 ΔbifA | 22 |
| bifA sadC mutant | PA14 ΔbifA ΔsadC | This study |
| Plasmids | ||
| pMQ30 | Suicide vector; Gmr Sac+URA3 CEN6/ARSH4 | 47 |
| pKO4332 | PA4332 knockout construct in pMQ30 | This study |
| pMQ80 | Cloning vector, GmrURA3 PBADaraC, GFP+ | 47 |
| pMQ72 | Cloning vector, GmrURA3 PBADaraC | 47 |
| psadC | C-terminal HA-tagged sadC under the control of the PBAD promoter in pMQ80; GmrURA3 | This study |
| psadCHis | C-terminal 10-His-tagged sadC under the control of the PBAD promoter in pMQ80; GmrURA3 | This study |
| pD/C637 | C-terminal 10-His-tagged PleD*-SadC chimera (pleD bases 1 to 896, sadC bases 637 to 1125) under the control of the PBAD promoter in pMQ80; GmrURA3 | This study |
| pD/C700 | C-terminal 10-His-tagged PleD*-SadC chimera (pleD bases 1 to 932, sadC bases 700 to 1125) under the control of the PBAD promoter in pMQ80; GmrURA3 | This study |
| pUCP18 | Cloning vector, Cbr Apr | 46 |
| pNC5 (pSadB+) | sadB in pUCP18; Cbr Apr | 4 |
| pRP89 | C-terminal His6-tagged PleD* | 37 |
| Primers | ||
| ko4332inF | CCACGCTTGGCACGGTAGAGCGCCTGGTCGGCCCGCTGCAAGGCCAGGCCGAGCAGGTAAGTG | |
| ko4332inR | CCGGTTCGGCATGGCCGCAGGCACTTACCTGCTCGGCCTGGCCTTGCAGCGGGCCGAC | |
| ko4332outF | CCAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGTCAGGCGTGGGGCAGGAAC | |
| ko4332outR | CCATGATTACGAATTCGAGCTCGGTACCCGGGGATCCTCGTCGCCGGGAACGATG | |
| 4332mq80F | GCTTTTTATCGCAACTCTCTACTGTTTCTCCATAACCGTTCTTCAGGCGGGTAATTCGAATG | |
| mq80-4332HAF | CTTGCATGCCTGCAGACTAGTTTATGCATAATCAGGAACATCATAAGGATA | |
| mq80-4332HAR | CCTGGGAGGTCACCAGTGCCTATCCTTATGATG | |
| 4332hahisF | AAGCTTGCATGCCTGCAGACTAGTTTAGTGGTGGTGATGATGGTGGTGATGATGGTGGCC | |
| 4332hahisR | GCCACCTGGGAGGTCACCAGTGCCGGCCACCATCATCACCACCATCATCACCACCACTAA | |
| pleD1R | GAGCTCGGTACCCGGGGAAGGAGATATACATATGAGCGCCCGGATCCTCGTCGTC | |
| pleD869-C637F | GCGGTTGAACAGCCCGGTGAGTTCGTCGGTGACGGCCAGCTCCAGCGAGTG | |
| pleD932-C700F | GTGCTGCCGGTTCGGCAGCAGGTCTTCGACCAGCGAGTCGAGCTGACC |
Gmr, gentamicin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Cbr, carbenicillin resistance; Kanr, kanamycin resistance. HA, hemagglutinin.
Molecular techniques.
Plasmids constructed during the course of the present study were prepared by using homologous recombination in Saccharomyces cerevisiae (47). Constructs were then electroporated into Escherichia coli for confirmation by colony PCR or sequencing. Restriction enzymes were obtained from New England Biolabs unless otherwise noted. A deletion was introduced into the sadC gene by using an allelic replacement strategy (45) with the plasmid pKO4332. Expression vectors were constructed by replacing the green fluorescent protein open reading frame in pMQ80 with PCR products containing the sadC gene and desired modifications. These were then transferred to Pseudomonas by electroporation (6).
For quantitative real-time PCR (qRT-PCR), P. aeruginosa in static planktonic cultures or on plates was grown and harvested as previously described (3). Preparation of RNA and cDNA and analysis by qRT-PCR were performed according to published methods (23).
Biofilm and attachment assays.
Static biofilm assays were performed in 96-well microtiter plates as previously described (31), followed by incubation at 37°C for 8 h unless otherwise indicated. To monitor the initial stages of biofilm formation, bacteria from a stationary-phase culture were diluted 1:100 in M63 minimal medium supplemented with glucose (0.2%), Casamino Acids (0.5%), and MgSO4 (1 mM). A 500-μl portion of this solution was pipetted into a sterile 24-well plate, followed by incubation at 37°C for 30 min for initial attachment or 1 h to assay reversible attachment (3). The medium was then gently replaced, and phase-contrast images of the surface-associated bacteria were captured at 60 frames/min at ×1,400 magnification using OpenLab software. The images were converted into QuickTime movies and analyzed as reported (15).
Motility assays.
Swarm assays were conducted as previously described on 0.5% agar plates (53). The area of the plate surface covered by the swarming bacteria was calculated by using ImageJ software (National Institutes of Health) as previously described (3). Directional reversals during swimming were quantified as described by Caiazza et al. (3) using M63 glucose-Casamino Acids medium as for biofilm assays. Ficoll was added to 15% to provide the desired viscosity in the medium. M63 glucose-Casamino Acids medium was solidified with 0.3% agar for swimming motility plates (35). Twitching motility assays were performed in LB medium solidified with 1.5% agar (34).
Assessment of EPS production.
M63 salt solution was supplemented as for biofilm assays with the addition of CR (40 μg/ml) and Coomassie brilliant blue (20 μg/ml). For plate-based assay, the medium was solidified with 1% agar (11). Wells and plates were inoculated from overnight cultures grown in LB broth with antibiotics as appropriate, followed by incubation at 37°C for approximately 24 h or as indicated. CR plates were further incubated for 1 or 2 days at room temperature to improve color development.
Protein localization and detection.
Strains were grown in LB medium supplemented with gentamicin (60 μg/ml) and arabinose (0.2%). Cells were harvested and resuspended in lysis buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 5% glycerol, and a complete EDTA-free protease inhibitor cocktail tablet (Roche). Lysis was carried out with the addition of 300 μg of lysozyme/ml, followed by incubation at 4°C for 2 h, and samples were subsequently sonicated to fragment the DNA. Unbroken cells were pelleted by centrifugation at 13,000 × g. Cellular fractionations were performed based on the method of Nunn and Lory (33), as modified by Hinsa and O'Toole (15). Fractions were normalized based on protein concentration and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a gradient (4 to 15%) gel, followed by Western blotting. Proteins were detected by using antibodies that recognize the His epitope (QIAGEN), SadB (4), and SecY (15) to distinguish the SadC protein, and cytoplasmic and inner-membrane fractionation controls, respectively.
Protein purification.
PleD*-His was purified essentially as previously described by using a 5-ml HisTrap FF column (GE Healthcare) (37). To purify the His-tagged SadC protein or His-tagged PleD-SadC chimeras, an overnight culture of wild-type (WT) PA14 carrying the appropriate plasmid was diluted into 1 liter of LB medium with gentamicin and grown until the optical density at 600 nm (OD600) reached 0.4. Arabinose was added to 0.2%, and cells were incubated in a shaking flask at 37°C to an OD600 of ∼1.0. Cells were harvested and resuspended in binding buffer (20 mM sodium phosphate [pH 7.4], 0.5 M NaCl, 20 mM imidazole) to which an EDTA-free protease inhibitor (Roche) had been added. Triton X-100 (1%) and lysozyme were also included for the full-length His-tagged SadC construct. The PleD-SadC chimera-containing strains were lysed by passage through a French pressure cell, while the cells containing the full-length sadC construct were enzymatically lysed on a rocker at 4°C for 2 to 4 h. Sonication was used to decrease the viscosity of the lysate prior to clearing by centrifugation at 13,000 × g for 30 min. The supernatant was passed through a 0.22-μm-pore-size filter for clarification and then loaded onto a 5-ml HisTrapFF column (GE Healthcare). Purification was carried out according to the manufacturer's instructions. Fractions containing SadC were pooled, concentrated, and dialyzed as reported elsewhere (32).
Diguanylate cyclase assays.
c-di-GMP was radiolabeled in vivo and separated from cellular nucleotides by thin-layer chromatography (TLC) on polyethyleneimine cellulose plates (Selecto Scientific) as described by Hickman et al. (13). Plates were exposed to a phosphor screen and analyzed as previously described (32).
In vitro enzyme assays were conducted essentially as reported by Paul et al. (37) and modified by Monds et al. (32) with incubation times of 2 and 24 h at room temperature. Reactions were stopped with addition of 10 μl of 0.5 M EDTA and an equal volume of running buffer (1:1.5 saturated NH4SO4 and 1.5 M KH2PO4 [pH 3.6]). Reaction products were resolved and analyzed as previously described (32).
RESULTS
A sadC (PA4332) mutant is defective in biofilm formation.
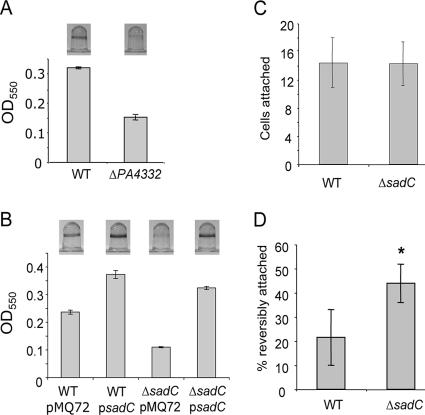
A strain containing a transposon mutation in the PA4332 gene was first identified in a screen for biofilm-defective mutants (34). To confirm a role for the PA4332 gene in biofilm formation, we constructed a strain in which a large portion of the predicted open reading frame of the PA4332 gene was deleted. The ΔPA4332 mutant was then tested for its ability to form a biofilm under static conditions as reported (36). When compared to WT P. aeruginosa PA14, the ΔPA4332 mutant showed a defect in colonization of the surface after an 8-h incubation (Fig. 1A). This difference becomes less pronounced, although it is still evident, at 24 h.
FIG. 1.
The sadC gene (PA4332) is involved in the transition to irreversible attachment during biofilm formation. (A) Biofilm formation of the WT and ΔPA4332 mutant in a microtiter plate assay. Image of CV-stained wells (top) and quantification of staining by OD550 readings of the ethanol-solubilized dye (bottom). The graph depicts averages of four replicates with the standard deviation indicated by error bars. (B) Biofilm assay as described above comparing the addition of a vector control (pMQ72) or a sadC-containing plasmid (psadC) to either the WT or ΔsadC mutant strains. (C) Static attachment assay in minimal medium containing glucose and Casamino Acids. Surface-attached bacteria were quantified from 10 fields of view using phase-contrast microscopy. Average numbers of attached cells per field of view are shown. (D) Reversible attachment under static conditions. Bacteria associated with the substratum were observed and scored as to whether they were reversibly or irreversibly attached defined on the basis of whether movement was observed. The average percentage of reversibly attached cells is represented comparing the WT to the ΔsadC mutant (n = 10 fields of view; *, P = 0.00069).
In further support of a role for the PA4332 gene in biofilm formation, when a multicopy plasmid carrying this gene was constructed and used to transform the WT and ΔPA4332 mutant strains, biofilm formation was stimulated in both of these backgrounds (Fig. 1B). In light of these data we have elected to rename the PA4332 open reading frame sadC (for surface attachment defective) to reflect its role in biofilm formation and relationship to previously described genes that when mutated yield similar phenotypes.
The fact that the ΔsadC mutant was defective in the microtiter plate assay suggested that this mutation might disrupt early steps in biofilm formation. Accordingly, we further probed the role of the sadC gene in initial attachment and subsequent reversible or irreversible attachment phenotypes through microscopic analysis of these behaviors under similar static conditions.
When planktonic bacteria first attach to a surface, the association is a relatively unstable polar attachment event designated reversible attachment (43). This initial stage of biofilm formation was monitored by inoculating individual compartments of a 24-well plate and quantifying the attached bacteria after a 30-min incubation. We found that the ΔsadC mutant was present on the substratum at levels indistinguishable from those of the WT, suggesting that it is not an inability to initiate attachment that leads to the observed defect in biofilm formation (Fig. 1C).
Because of its early biofilm defect, we also determined the role of sadC in the transition from reversible to irreversible attachment. Microscopic observation of cell-surface association was performed as previously described (3). Bacteria harboring a deletion in sadC are approximately twice as likely to be reversibly attached to the surface as the WT (P = 0.0007, Fig. 1D). These data indicate that the ΔsadC mutant is defective in forming a stable association with the substratum compared to its parental strain.
The sadC mutant displays an increased swarming phenotype.
Published studies conducted in our laboratory (3, 4, 52) found that strains with mutations in the sadB, pelA, and motAB genes display a defect in the transition from reversible to irreversible attachment during biofilm development. Interestingly, these mutants have also been shown to share another trait: they are hyperswarmers. In light of these recent findings, we sought to determine whether the ΔsadC mutant also had an altered swarming phenotype.
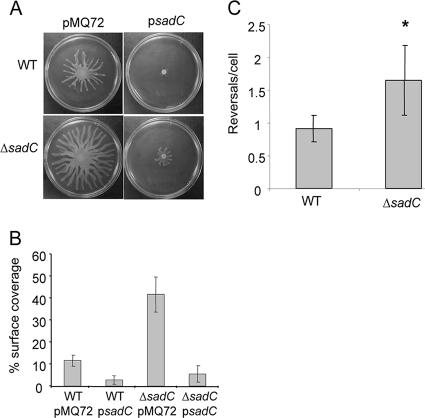
We tested swarming motility as previously described (5) by inoculating strains on 0.5% agar and incubating the swarm agar plates for 16 h. The ΔsadC mutant formed swarms that were substantially larger than those of the WT strain (Fig. 2A and B). The ΔsadC mutant was indistinguishable from WT for swimming and twitching motilities (data not shown).
FIG. 2.
The ΔsadC mutant displays hypermotility phenotypes. (A) Swarming motility assays on 0.5% agar plates. Overnight cultures of the WT or ΔsadC strains carrying a vector control (pMQ72) or a sadC-containing plasmid (psadC) were spotted onto swarm agar and incubated at 37°C for 16 h. (B) Average percentage of the plate surface occupied by the respective swarms. Replicate swarm plates (n = 5) for each test strain were grown under identical conditions and then photographed for calculation of the surface area coverage. Averages for the WT (WT/pMQ72) and ΔsadC (ΔsadC/pMQ72) strains carrying vector controls differ significantly (P = 0.001). In both cases, addition of a multicopy sadC-containing plasmid (WT/psadC, ΔsadC/psadC) results in a significant decrease in respective swarm coverage at this time point (P = 0.0088, P = 0.0004). (C) Swim reversals in 15% Ficoll-containing medium quantified from six separate fields of view. The ΔsadC mutant cells on average are observed to reverse more frequently than the parental strain (*, P = 0.01).
Given that the loss of SadC function results in increased swarming motility, we hypothesized that increased SadC activity might result in suppression of swarming motility. To test this hypothesis, we examined strains in which the sadC gene was introduced on a multicopy plasmid for their ability to swarm. In either the WT or ΔsadC mutant backgrounds, expression of the sadC gene on a high-copy plasmid significantly decreases the extent of swarming (Fig. 2A and B).
Mutations in either the sadB, pilJ, or motAB genes were also previously shown to affect the reversal rates of single cells under conditions of high viscosity (3, 53), conditions that are likely analogous to those encountered during swarming motility (53). We counted the number of direction reversals during swimming in the presence of 15% Ficoll and determined that the ΔsadC mutant exhibits a ∼50% increase in reversals/cell compared to the WT (P = 0.01, Fig. 2C). Taken together, mutating the sadC gene results in phenotypes similar to those observed for the previously reported sadB, pilJ, and motAB mutants, that is, decreased biofilm formation, increased swarming motility, and increased flagellar reversals in high-viscosity medium.
The sadC gene enhances CR staining in a pel-dependent manner.
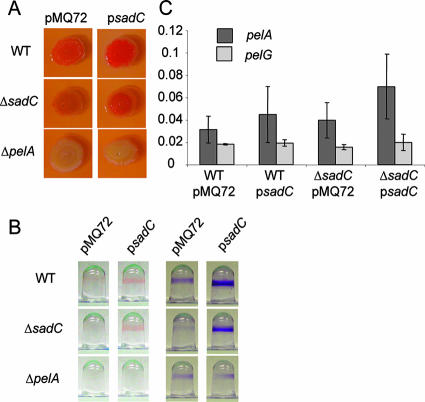
In addition to the aforementioned suite of phenotypes, it was also demonstrated that mutations in the sadB or pilJ genes led to decreased adsorption of the dye CR. CR adsorption has been shown to positively correlate with the presence of EPS in a number of bacterial species including P. aeruginosa (12, 26), Salmonella enterica serovar Typhimurium, and Escherichia coli (59). Furthermore, we noted that when strains maintaining a multicopy plasmid carrying the sadC gene were spotted onto an agar plate, they developed a wrinkled surface, a phenotype frequently associated with overproduction of EPS (13, 18, 58, 59). We therefore decided to assess whether SadC plays a role in the production of the polysaccharide component of the biofilm extracellular matrix. Strains were inoculated onto CR-containing plates and incubated as described previously (3). We observed that a strain harboring a deletion in the sadC gene absorbed less dye than the WT (Fig. 3A), a finding consistent with a diminution in EPS production in this strain. Strikingly, strains in which sadC is expressed from a multicopy plasmid show greatly enhanced amounts of dye bound (Fig. 3A, right panels). These results are consistent with a role for the sadC gene in enhancing production of a CR-binding component of the extracellular matrix.
FIG. 3.
sadC influences production of the Pel EPS. (A) CR binding assay for EPS production. Aliquots of WT, ΔsadC, and ΔpelA strains carrying a vector control (pMQ72) or a plasmid expressing the sadC gene (psadC) were spotted onto CR plates. Plates were incubated for 24 h at 37°C, followed by 24 h at room temperature. (B) CR binding in biofilms. Representative wells showing attached cells grown in the presence of CR (left) or post-stained with crystal violet (right) after a 24-h incubation at 37°C. (C) qRT-PCR analysis of gene expression from agar-grown strains. The picograms of input cDNA is plotted on the y axis. There is no statistically significant difference comparing expression between the strains for the pelA (P > 0.09) or pelG gene (P > 0.3).
The pelA-G gene cluster contains genes whose products are predicted to be the structural proteins directly involved in polysaccharide production based on sequence analysis. The pel locus is required for the production of a glucose-rich polysaccharide that adsorbs the dye CR and is involved in maintaining the integrity of multicellular structures, including biofilms and pellicles (11), leading us to speculate that the elevation in CR binding observed in response to the sadC gene in high copy is dependent on this locus. To test this hypothesis, the sadC gene was expressed in multicopy in a strain harboring a deletion in the pelA gene. In this background, overexpression of sadC was unable to enhance CR staining (Fig. 3A), suggesting that the pel locus is indeed required for this phenotype.
We next assessed whether the difference in EPS production observed on agar plates was also discernible in the context of biofilm formation. By adding CR to our usual microtiter dish biofilm assay, we found that the presence of a sadC-containing plasmid enhanced CR staining at the air-liquid interface, the site of biofilm formation in this assay (Fig. 3B). This is true either in the WT or in the ΔsadC mutant backgrounds, although there is slightly less total crystal violet (CV) staining in the ΔsadC+psadC strain compared to WT+psadC (Fig. 3B). Although the ΔpelA mutant forms some biofilm in this situation, CV staining is no longer stimulated by addition of psadC, nor is there any CR staining visible with this mutant (left panel), a finding consistent with the lack of EPS production.
SadC does not alter pelA/G mRNA levels.
The experiments described above suggest a role for SadC in regulating the production of the pel-dependent polysaccharide matrix. In order to further probe this relationship, qRT-PCR was used to gauge whether transcriptional upregulation of the pelA-G locus might be involved in the apparent alteration in exopolysaccharide production observed in strains either lacking or carrying extra copies of the sadC gene.
We assessed transcript levels from the first and last genes in the pel locus: pelA, which shares similarity to endo-α-1,4-polygalactosaminidase and oligogalacturonide lyase (11), and pelG, a putative member of the PST family of proteins, which are generally involved in the translocation of glycolipid precursors out of the cytoplasm to position them for further processing (55). Mutations in either of these genes cause a loss of CR binding (11) and decreased biofilm formation in nonpiliated strains (55), and the ΔpelA strain has also been shown to be defective for biofilm formation (3, 11, 22). Transcript levels from the pelA and pelG genes were unchanged (P = 0.09) by the deletion or overexpression of sadC compared to the vector control when grown on agar plates (Fig. 3C) or as a static planktonic culture (data not shown). These data suggest that the alterations in pel-dependent CR binding in these strains are not due to changes in pelA or pelG mRNA levels.
The SadC protein is localized to the cytoplasmic membrane.
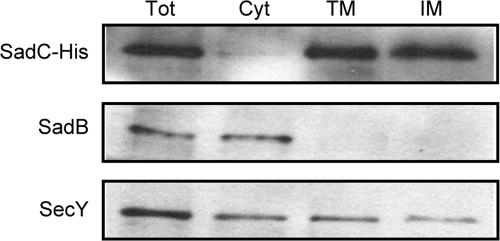
The TMpred prediction program calculates that the sadC open reading frame encodes an integral inner membrane protein with five transmembrane domains (http://www.ch.embnet.org/software/TMPRED_form.html). In order to verify this localization prediction, we performed a cellular fractionation of a strain expressing a C-terminally His-tagged SadC protein to isolate the outer and inner membranes, as well as the cytoplasmic compartment. These fractions were probed for appropriate localization controls, as well as an anti-His antibody to recognize the SadC-His protein.
We found that SadC-His can be detected in the inner membrane but not the cytoplasmic fraction, in agreement with the computer prediction (Fig. 4). Antibodies recognizing the SadB and SecY proteins reacted with bands of the expected sizes predominantly in the cytoplasm and inner membrane, respectively (1, 4, 14). We did, however, see the SecY antibody reacting with a protein in the cytoplasm as well. Since there is no cytoplasmic contamination from the SadC-His protein, the recognition of a cytoplasmic band by the SecY antibody may be due to cross-reactivity of the antibody rather than to substantial contamination of the cytoplasmic fraction with the inner membrane.
FIG. 4.
SadC is localized to the cytoplasmic membrane. Subcellular protein localization by Western blotting of fractions separated by differential centrifugation. Shown are the total cell lysate (Tot) and the cytoplasmic (Cyt), total-membrane (TM), and inner-membrane (IM) fractions.
The GGDEF domain of SadC has diguanylate cyclase activity.
The Pseudomonas genome project annotates the sadC gene as containing a putative GGDEF domain (http://www.pseudomonas.com). The amino acid sequence of the sadC GGDEF domain shares 49% identity to the consensus GGDEF domain according to an NCBI conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Furthermore, the SadC GGDEF domain exhibits nearly perfect conservation of the eponymous residues except for a glutamate in place of the aspartate. Interestingly, this GGEEF motif is found in all of the P. aeruginosa PA14 proteins that contain a GGDEF domain but not an EAL domain (24) and is found in proteins such as WspR that have been demonstrated to possess DGC activity in vivo (13, 24) and in vitro (28).
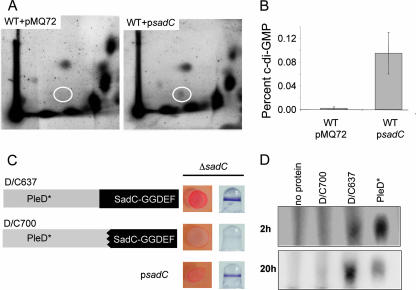
Due to the presence of a putative GGDEF domain, we next sought to determine whether the SadC protein possessed DGC activity. To begin to evaluate SadC's function, we first examined whether the expression of SadC could affect the cellular pools of c-di-GMP produced in vivo. Briefly, radiolabeled nucleotides were extracted from cells either harboring a vector control or a plasmid carrying the sadC gene and separated by two-dimensional TLC (Fig. 5A). Expression of the sadC gene from a multicopy plasmid coincides with an approximately 10-fold increase in the amount of c-di-GMP detected compared to the vector control (Fig. 5A and B).
FIG. 5.
The GGDEF domain of SadC has diguanylate cyclase activity. (A) Resolution of labeled extracts by two-dimensional TLC from WT carrying a vector control (WT/pMQ72) or the sadC gene on a multicopy plasmid (WT/psadC) grown in the presence of [32P]orthophosphate. Shown is a representative image from an experiment performed with three independent replicates. c-di-GMP is indicated in the white oval. (B) The percent c-di-GMP (normalized to the total 32P label) present in the WT/psadC strain is ∼10-fold greater than that of the WT/pMQ72 strain (*, P < 0.05). (C) Diagram of PleD/SadC chimeric proteins and respective phenotypes on CR plates and in biofilm assays. Both chimeric proteins are translational fusions comprised of the PleD* N terminus and the SadC C terminus, the net effect being to replace all or most of the GGDEF domain of PleD* with that of SadC. The D/C637 construct encodes the entire SadC GGDEF domain while the D/C700 construct lacks the first 21 amino acids of the cognate SadC GGDEF domain (left). Plasmids containing the pD/C637 or pD/C700 construct expressed in the ΔsadC background were spotted onto CR plates and assayed for biofilm formation (right). Arabinose to 0.2% was added to both assays. (D) In vitro diguanylate cyclase activity assay. Purified D/C700 or D/C637 proteins were incubated with [32P]GTP under diguanylate cyclase reaction conditions. Reaction products at 2 and 20 h are shown resolved by TLC with concomitantly run no-protein and PleD* 1:100 dilution control reactions.
To more directly assess the activity of SadC, we utilized an in vitro DGC assay. Purified full-length SadC-His was found to promote synthesis of c-di-GMP in vitro, but a 24-h incubation period was required to observe c-di-GMP, and we did not observe robust accumulation of this end product (data not shown). Furthermore, purification of full-length SadC-His was low yield, and we were not confident that the structure of the protein was maintained during the purification process. We next attempted to express a His-tagged protein containing only the GGDEF domain of SadC which lacks the entire N terminus of the protein, including all predicted membrane domains. We were unable to detect this protein by Western blotting, suggesting that the GGDEF domain of SadC is not stable in isolation. We next constructed chimeric proteins that fused the N terminus of PleD* to the GGDEF domain-containing C terminus of SadC. The D/C637 construct maintains the entire GGDEF domain of SadC, while the D/C700 construct is lacking the first 21 amino acids of the cognate GGDEF domain (Fig. 5C).
To begin to examine the functionality of these constructs, both were expressed in the WT or ΔsadC mutant backgrounds. Expression of D/C637 in the ΔsadC mutant background resulted in an increase in CR binding and biofilm formation compared to the D/C700 construct (Fig. 5C). Similar results were obtained when expressing the D/C637 or D/C700 constructs in a WT strain (data not shown). These data provide evidence that the D/C637 protein may indeed contain an active GGDEF domain. In cellular fractionation experiments we found that for both of these constructs, while some of the protein is cytoplasmic as expected, a portion of the D/C637 and D/C700 proteins appears to be retained in the membrane fraction (data not shown).
We further explored the diguanylate cyclase activity of these constructs in vitro. The D/C637 and D/C700 chimeras were purified and assayed under conditions that support DGC activity of a PleD* control. We found that the D/C637 but not the D/C700 chimera supported detectable accumulation of c-di-GMP, as assessed by migration of this product by TLC compared to the PleD* control (Fig. 5D). The accumulation of c-di-GMP in this reaction is detectable at 2 h (top) and is more pronounced after a longer incubation period (lower panel). These data suggest that the GGDEF domain of SadC is capable of catalyzing the synthesis of c-di-GMP. Taken together with the in vivo labeling studies and CR data, this finding suggests that SadC is indeed a functioning DGC.
sadC interacts genetically with other biofilm-related genes.
In exploring the phenotypes of the sadC mutant, it rapidly became obvious that the profile of phenotypes displayed was also shared by other mutants that had been studied in the laboratory, most notably in the sadB and pilJ genes. The sadB and pilJ genes were previously suggested to be in the same genetic pathway (3). We were impelled to consider whether these genes might be operating in a genetic pathway with the sadC gene, and thus we performed epistasis studies to explore this question.
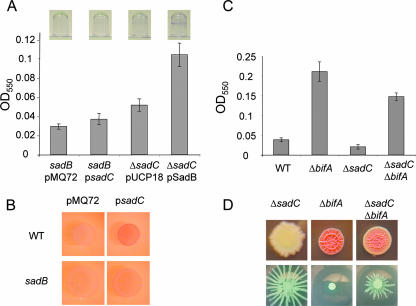
We first investigated the relationship between the sadB and sadC genes. Multicopy sadC was unable to stimulate biofilm formation in the sadB mutant at early time points (Fig. 6A), a very different outcome than what we observed when the same plasmid is placed in a WT or ΔsadC mutant background (compare with Fig. 1B). Overexpression of the sadB gene in a ΔsadC mutant background was able to increase biofilm formation in this strain, completely rescuing the defect in biofilm formation (Fig. 6A). Furthermore, in a sadB mutant background, the expression of the sadC gene was prevented from enhancing CR binding at early time points (Fig. 6B), although a similar amount of CR adsorption was eventually attained. Taken together, these experiments suggest that SadB is important for SadC function, although such robust overexpression of a DGC may also affect accessory c-di-GMP responsive systems within the cell.
FIG. 6.
sadC is in a genetic pathway with other genes that influence biofilm formation, swarming motility, and EPS production. (A) Biofilm formation was assessed for the strains indicated after 4 h. Images of representative wells (top) are shown with averaged OD readings from dye solubilized from four replicate wells (bottom). (B) CR binding of the WT and sadB strains carrying a vector control or a sadC-containing multicopy plasmid. CR plates were incubated for 24 h at 37°C. (C) Biofilm formation by the indicated strains was assayed after 8 h at 37°C. (D) The indicated strains were assayed for CR binding (top) and swarming motility (bottom). CR plates were incubated for 24 h at 37°C, followed by 24 h at room temperature, while swarm plates were imaged after a 16-h incubation at 37°C.
We next investigated the interaction between SadC and the recently identified BifA, a c-di-GMP phosphodiesterase involved in biofilm formation and swarming motility that is described in a companion manuscript (22). The ΔbifA mutant accumulates c-di-GMP and is a hyperbiofilm former. Mutating sadC in the ΔbifA background reduced the biofilm formation of this strain, although not completely to the level of the WT (Fig. 6C). A similar trend was observed in respect to the CR binding and swarming phenotypes (Fig. 6D). In both cases, the phenotypes of the double mutants were intermediate between that of the ΔbifA mutant and the WT. In combination, these data suggest that while BifA acts downstream of SadC, there are likely to be other DGCs contributing to the supply of c-di-GMP influencing these phenotypes.
Taken together, these data and previously published work (3) suggest the following ordered genetic pathway: sadC, bifA, sadB, pelA/pilJ. In addition, work presented by Kuchma et al. demonstrated that the sadB gene is required for the phenotypes of the ΔbifA mutant, thus providing further evidence for the proposed genetic model (Fig. 7).
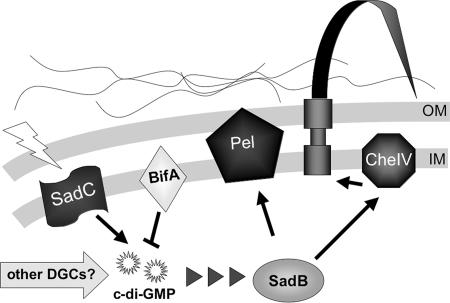
FIG. 7.
Model for the role of SadC in modulating biofilm formation, swarming motility, and related phenotypes. A diagram of the putative roles of SadC and other factors in this pathway involved in the inverse regulation of biofilm formation and swarming motility. SadC, presumably upon receipt of the proper environmental signals (such as nutritional cues or contact with an appropriate surface), transmits this information to the cytoplasm by modulating the production of c-di-GMP. This signal can be further regulated by the activity of the BifA protein, a c-di-GMP phosphodiesterase. We hypothesize that the status of this c-di-GMP pool, which may also be impacted by the action of additional DGCs, acts as a signal to the downstream constituents of this pathway. SadB is predicted to play a role in the transmission of this signal to the Pel machinery and components of the CheIV chemotaxis-like cluster. We propose that when there are relatively high c-di-GMP levels, biofilm formation is promoted at the expense of swarming motility. This is consistent with the observed increase in EPS production and decrease in swarming motility. Based on its previously reported function as an allosteric effector of EPS production, we suggest a similar role for c-di-GMP-based regulation of Pel production. If the c-di-GMP pool is low, this state is associated with decreased biofilm formation and increased swarming motility. In the case of modulating flagellar function, PilJ or potentially other components of the Che IV chemotaxis cluster may be involved. How c-di-GMP communicates with the CheIV cluster is not known, but it may parallel regulation of the pel locus.
DISCUSSION
We present here evidence that the sadC gene, encoding an inner-membrane diguanylate cyclase, plays an integral role in modulating two surface-associated behaviors of P. aeruginosa PA14: swarming motility and biofilm formation. As the body of literature regarding these DGC enzymes and their counterpart phosphodiesterases has developed, it has been noted that their activities are often associated with control of motility and biofilm formation (39). How is it that this c-di-GMP signal functions to regulate these two complex multicellular behaviors? Previous work in this laboratory identified a genetic pathway, which includes the sadB gene (3), that functions in regulation of flagellar function and production of the exopolysaccharide matrix. We present evidence that sadC functions upstream of this sadB-dependent pathway and is likely to be important for the induction of this conduit to regulate the surface-associated lifestyles of P. aeruginosa.
Prior to and during the course of the present study, our laboratory identified several genes that appear to impact both swarming and biofilm formation. The similarity of these phenotypes, and subsequent epistasis analyses, suggested the possibility that these factors may operate in a common genetic pathway to regulate both biofilm formation and swarming motility (3, 4, 22). We used a genetic approach in order to better understand these findings and to identify additional components of the pathway regulating these surface-associated behaviors. This led us to identify the sadC gene, which when disrupted leads to alterations in both biofilm formation and swarming motility. Specifically, the observed decrease in biofilm formation and increase in swarming motility in the ΔsadC mutant led us to contemplate the potential interrelation of these two aspects of P. aeruginosa biology. In thinking about this question, we first felt it essential to further dissect these two complex behaviors by assessing other contributing phenotypes. We ascertained that, relative to the WT, the ΔsadC mutant produced a reduced amount of the Pel EPS matrix and underwent an abnormally high number of flagellar reversals during incubation in a high-viscosity medium. Could these two phenotypes assist in our understanding of the decreased biofilm formation and hyperswarming of the ΔsadC mutant?
EPS production has previously been shown to play a role in both early and late stages of biofilm development (11, 16, 26, 55), but might it also act as a negative effector of swarming motility? One could easily imagine that bacteria producing an extracellular meshwork of polysaccharides might be hampered in their movements. In support of this idea, a mutation in the pelA gene, required for the manufacture of a glucose-rich polysaccharide, leads to elevated swarming in this organism (3). Thus, we can envision a role for the Pel polysaccharide matrix in both promoting biofilm formation and hindering swarming motility, although this idea requires further experimental verification.
Likewise, we contemplated the consequences of increased flagellar reversal rates in either swarming motility or biofilm formation. At this point, we can only speculate as to the role (if any) of changing flagellar reversal rates in these two surface behaviors. It has been suggested that in E. coli the modulation of flagellar reversals is indeed important in surface attachment (30), although the precise role of reversals is not yet understood. In an examination of motility in semisolid agar, Wolfe and Berg found a general trend that a greater reversal frequency correlated with an overall greater swarm rate. Those authors observed that direction changes prevent the bacteria from becoming “trapped” in the agar matrix (57). In the case of the Sadc mutant, we postulate that direction reversals help to “unstick” the bacteria from the surface, allowing for maximal forward progress (57). Perhaps the increased reversal behavior that promotes swarming motility hampers the ability of the microbe to make a stable commitment to a surface.
Whatever the mechanism, a clear correlation is emerging between the ability to control flagellar reversals and the surface-associated behavior adopted by P. aeruginosa. For example, mutations in the sadC, sadB, pilJ, and motAB genes all yield hyperswarming strains that are biofilm defective at the transition to irreversible attachment; all of these mutants also show increased flagellar reversal rates in high-viscosity medium (3, 4, 52). In contrast, in the accompanying manuscript, we show that a mutation in the BifA phosphodiesterase suppresses swarming motility, even in the absence of the Pel polysaccharide, and shows a 50% decrease in flagellar reversals compared to the WT (22). A further link between control of flagellar function and biofilm formation is presented in a recent report from our group, showing that cells harboring mutations in one of the flagellar stators, which show no defect in swimming motility, still result in reduced biofilm formation (52). Taken together, our data suggest that control of flagellar function, and in particular the rate of reversals, is key for the control of swarming motility and biofilm formation.
The presence of a GGDEF domain in the sadC gene product immediately leads us to reason that SadC might play a role in signaling rather than as part of the machinery directly functioning in flagellar function or EPS production. Proteins containing this motif have been shown to catalyze the synthesis of c-di-GMP, a molecule that was first identified for its role in allosterically regulating cellulose synthesis in Gluconacetobacter xylinus (formerly Acetobacter xylinum) (40, 41). Mutations in genes whose products synthesize or degrade this nucleotide are often associated with alterations in motility- and sessility-related phenotypes such as biofilm formation (25, 32, 50, 51), twitching motility (17), and production of exopolysaccharides (25). The presence of a GGDEF domain is consistent with a model in which SadC is part of a relay system responsible for transmission of an as-yet-unidentified environmental signal to the control of flagellar and polysaccharide-producing machinery of the cell via modulation of c-di-GMP levels.
To test the above hypothesis, we examined the ability of SadC to impact production of the signaling molecule c-di-GMP. Overexpression of SadC results in an increase in the total intracellular levels of c-di-GMP (Fig. 5A). In an in vitro DGC assay, we also found that a chimera protein containing the GGDEF domain of SadC is able to generate c-di-GMP compared to a second chimera missing the first 21 amino acids of the cognate GGDEF domain of SadC. Taken together, with the fact that sadC encodes an intact GGDEF domain, this is strong evidence that the SadC protein possesses the catalytic activity required to synthesize c-di-GMP. A previous study has also explored the diguanylate cyclase activity of SadC in the context of surveying all of the P. aeruginosa PA14 GGDEF or EAL domain-containing proteins. Kulasakara et al. (24) failed to observe production of c-di-GMP in an extract prepared from a strain carrying a plasmid expressing the sadC gene. Given the low levels of c-di-GMP we observe, this disparity may simply be attributable to differential detection limits of the respective methods utilized.
How do the sadC gene and its product fit into the known factors involved in early biofilm formation? We have shown that the ΔsadC mutant is blocked in the transition from reversible to irreversible attachment during the early events in establishing a surface community. Through a series of epistasis experiments, we have placed sadC in the context of other genes that impact this step in the biofilm pathway. The sadB mutant is also defective in irreversible attachment and displays a hyperswarming phenotype. In a sadB mutant background, adding sadC in multicopy no longer enhances CR adsorption to the same degree as that observed in the WT. Likewise, there is a decreased stimulation of biofilm formation in the sadB mutant compared to what we normally observe in response to overexpression of the sadC gene. These data suggest that sadB is genetically downstream of sadC and that its product perhaps functions in transmission of the c-di-GMP signal to downstream effectors. Although a role for sadB in biofilm formation has been previously established (4), its biochemical activity has remained elusive. In addition, SadB has been shown previously to act upstream of pelA, which is required for production of the Pel polysaccharide, and pilJ, a member of the CheIV chemotaxis cluster, which is consistent with its role in controlling flagellar reversals (3). Deletion of the motAB genes, which comprise one of the two P. aeruginosa flagellar stators, also causes a defect in irreversible attachment, hyperswarming, and an increase in swim reversals, suggesting that these genes may be additional downstream candidates of this pathway (10, 52, 53). Alternatively, it may be that some of the components involved in the early stages of biofilm formation are instead functioning in convergent or parallel genetic pathways. Future studies will be required to obtain biochemical data to explain the underpinnings of the phenotypic relationships observed thus far.
In an accompanying manuscript, we show that BifA (22), which contains an inactive GGDEF domain and an EAL domain that functions as a phosphodiesterase, is capable of hydrolyzing c-di-GMP and that deletion of the bifA gene leads to an accumulation of this nucleotide within the cell, a hyperbiofilm phenotype, and complete loss of swarming motility (22). Deletion of the sadC gene in the ΔbifA mutant results in partial restoration of swarming motility, a decrease in CR binding, and decreased biofilm formation. We interpret these data to mean that SadC contributes some but not all of the c-di-GMP to this system under these conditions. Therefore, we can further infer that c-di-GMP from at least one other DCG is also converging on BifA. A possible candidate for this other DGC is the WspR protein which is required for increased CR binding and attachment in a WspF mutant (13). Finally, it has been shown that the sadB gene is required for the phenotypes of the ΔbifA mutant, supporting the conclusion that these genes are all in the same genetic cascade (22).
Based on the data presented here and in other recent studies, we can construct a model of the pathway by which SadC may be regulating aspects of motility and biofilm formation in P. aeruginosa (Fig. 7). We propose that the diguanylate cyclase SadC produces c-di-GMP in response to some as-yet-unknown signal such as contact with a surface or changes in medium viscosity. The amount of this signaling molecule accumulating can be attenuated by the phosphodiesterase BifA, providing a second control point whereby c-di-GMP pools are regulated. This c-di-GMP signal is then transmitted, perhaps via SadB based on our genetic studies, to the pel genes and/or members of the CheIV chemotaxis-like cluster, resulting in control of exopolysaccharide production and flagellar function. The mechanisms linking c-di-GMP pools to the control of EPS production and flagellar function are still under investigation since there are no known c-di-GMP binding proteins present in the genes we have examined. One tempting candidate for allosteric regulation of the Pel polysaccharide is PelD protein, which contains a putative GAF domain. In other systems GAF domains have been shown to bind the cyclic nucleotide cyclic AMP or cyclic GMP. Although studies of SadC and other members of this pathway (3, 22) are providing us with insight into the cellular functions coregulating biofilm formation and swarming motility as cells transition from a planktonic to a surface-associated lifestyle, there remains much to be elucidated in our understanding of this pathway.
Acknowledgments
This study was supported by NIH training grant T32 GM08704 predoctoral fellowship to J.H.M. and grants AI51360 and P20-RR018787 from the Institutional Development Award (IDeA) to G.A.O.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1985. The SecY membrane component of the bacterial protein export machinery: analysis by new electrophoretic methods for integral membrane proteins. EMBO 4:3351-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 3.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caiazza, N. C., R. M. Shanks, and G. A. O'Toole. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K.-H., A. Kumar, and H. P. Schweizer. 2005. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 7.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829-30837. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle, T. B., A. C. Hawkins, and L. L. McCarter. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 15.Hinsa, S. M., and G. A. O'Toole. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375-1383. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739-749. [DOI] [PubMed] [Google Scholar]
- 19.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 20.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolter, R., and E. P. Greenberg. 2006. The superficial life of microbes. Nature 441:300-302. [DOI] [PubMed] [Google Scholar]
- 22.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 26.Ma, L., K. D. Jackson, R. M. Landry, M. R. Parsek, and D. J. Wozniak. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 28.Malone, J. G., R. Williams, M. Christen, U. Jenal, A. J. Spiers, and P. B. Rainey. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153:980-994. [DOI] [PubMed] [Google Scholar]
- 29.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClaine, J. W., and R. M. Ford. 2002. Reversal of flagellar rotation is important in initial attachment of Escherichia coli to glass in a dynamic system with high- and low-ionic-strength buffers. Appl. Environ. Microbiol. 68:1280-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merritt, J. H., D. E. Kadouri, and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.2. In Current protocols in microbiology. J. Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed]
- 32.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:659-679. [DOI] [PubMed] [Google Scholar]
- 33.Nunn, D., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175:4375-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 37.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 39.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. c-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 40.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose synthesis in Acetobacter xylinum: a unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101-117. [Google Scholar]
- 41.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 103:109-112. [DOI] [PubMed] [Google Scholar]
- 47.Shanks, R. M. Q., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. A yeast-based molecular tool kit for manipulation of gram-negative bacterial genes. Appl. Environ. Microbiol. 72:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shrout, J., D. Chopp, C. Just, M. Hentzer, M. Givskov, and M. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 49.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toutain, C. M., N. C. Caiazza, M. E. Zegans, and G. A. O'Toole. 2007. Roles for the flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res. Microbiol. 158:471-477. [DOI] [PubMed] [Google Scholar]
- 53.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasseur, P., I. Vallet-Gely, C. Soscia, S. Genin, and A. Filloux. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985-997. [DOI] [PubMed] [Google Scholar]
- 56.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 57.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]