Abstract
A number of gram-negative bacteria have a quorum-sensing system and produce N-acyl-l-homoserine lactone (AHL) that they use them as a quorum-sensing signal molecule. Pantoea ananatis is reported as a common colonist of wheat heads at ripening and causes center rot of onion. In this study, we demonstrated that P. ananatis SK-1 produced two AHLs, N-hexanoyl-l-homoserine lactone (C6-HSL) and N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL). We cloned the AHL-synthase gene (eanI) and AHL-receptor gene (eanR) and revealed that the deduced amino acid sequence of EanI/EanR showed high identity to those of EsaI/EsaR from P. stewartii. EanR repressed the ean box sequence and the addition of AHLs resulted in derepression of ean box. Inactivation of the chromosomal eanI gene in SK-1 caused disruption of exopolysaccharide (EPS) biosynthesis, biofilm formation, and infection of onion leaves, which were recovered by adding exogenous 3-oxo-C6-HSL. These results demonstrated that the quorum-sensing system involved the biosynthesis of EPS, biofilm formation, and infection of onion leaves in P. ananatis SK-1.
Quorum sensing is one of the cell-cell communication mechanisms depending on cell population density in bacteria (4, 11). In many gram-negative bacteria, several kinds of N-acyl-l-homoserine lactone (AHL) have been identified as signal compounds involved in this mechanism and called autoinducers (4, 11). AHLs are synthesized in bacteria by a member of the LuxI protein family and diffused outside and inside of bacteria. When AHL concentration increases and reaches a threshold due to accumulation of AHL derived from each bacterial cell, AHL receptor protein belonging to the LuxR protein family binds AHL and regulates expression of many genes responsible for bioluminescence, production of pigment or antibiotics, and so on (11). In particular, many gram-negative pathogens control the expression of virulence factors, which include the secretion of extracellular protease, pectinase, and biosurfactant and forming biofilm (4).
Many plant pathogens produce AHLs and regulate their virulence by AHL-mediated quorum sensing (2, 27). Erwinia carotovora, which causes soft rot diseases on many plant species, induces the production of various exoenzymes and plant tissue maceration by AHLs (3). Pantoea stewartii regulates exopolysaccharide (EPS) biosynthesis and pathogenicity in sweet corn by AHLs (28). Erwinia amylovora produces one AHL and regulates EPS biosynthesis, tolerance to hydrogen peroxide, and the development of symptoms on apple leaves (18). Agrobacterium vitis causes necrosis on grape plants and a hypersensitive-like response on tobacco plants by a quorum-sensing system (30). In general, AHL-negative mutants show defects in pathogenicity, so it is expected that disrupting or manipulating quorum-sensing signals inhibits the expression of virulence and infection of host cells. Recently, some AHL-degrading bacteria and enzymes have been reported (7). An AHL lactonase-encoded gene (aiiA) was cloned from Bacillus sp. strain 240B1 (6). The expression of aiiA in transformed E. carotovora significantly attenuates pathogenicity in some crops (6). Expression of aiiA in E. amylovora impairs EPS production and reduces virulence on apple leaves (18). Transgenic plants expressing AHL lactonase exhibited significantly enhanced resistance to E. carotovora infection (5).
P. ananatis is reported as a common colonist of wheat heads at ripening and causes “center rot” disease in onion plants (1, 9, 29). Center rot was first reported on an onion in Georgia in 1997 and has continued to reduce yields and cause postharvest losses (9). Onion leaves infected by P. ananatis are usually collapsed and hang down beside the onion neck (1). For the treatment of center rot, fixed copper materials tank mixed with EBDC fungicides are recommended to suppress infection and spread (1). However, the major virulence factors of P. ananatis are unknown. Recently, it was reported that P. ananatis strains produced AHLs. Yoshida et al. revealed that P. ananatis inhabiting wheat heads produced at least two AHLs, N-hexanoyl-l-homoserine lactone (C6-HSL) and N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL) (29). Pomini et al. reported that P. ananatis (Serrano 1928) produced three AHLs, and the major identified substance is C6-HSL (22). However, these authors did not identify the LuxRI homologs from P. ananatis strains and elucidate the phenotypes controlled by AHL-mediated quorum sensing. We report here the identification of the LuxRI homologs, EanRI, and C6-HSL and 3-oxo-C6-HSL in P. ananatis strain SK-1. We also present evidence for the involvement of the quorum-sensing system in the regulation of EPS biosynthesis, biofilm formation, and the infection of onion leaves.
MATERIALS AND METHODS
Bacterial strains, plasmids, compounds and growth conditions.
Selected bacterial stains and plasmids used in the present study were listed in Table 1. Escherichia coli and Chromobacterium violaceum were grown at 30°C in Luria-Bertani (LB) medium (24). P. ananatis was grown at 30°C in tryptic soy broth (TSB; Becton Dickinson). Solid bacterial media were made by the addition of agar at a final concentration of 1.5%. Antibiotics were added as required at final concentrations of 100 μg of ampicillin/ml, 10 μg of chloramphenicol/ml, 10 μg of colistin/ml, and 10 μg of gentamicin/ml. AHL standards were synthesized by a previously described method (14). pKRP14, which carried gene cassettes imparting resistance to the gentamicin (Gmr), was constructed in the present study. Briefly, a Gmr cassette was amplified from pJN105 by using the following primers, which contained HindIII restriction sites (underlined) at their 5′ ends: 5′-AAGCTTTCGCCTTGCGTATAATATTTGCCC-3′ and 5′-AAGCTTTGACAATTTACCGAACAACTCCGC-3′. PCR fragments were cut out by HindIII digestion and inserted into the HindIII-digested pKRP11 for construction of pKRP14.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Pantoea ananatis | ||
| SK-1 | Natural isolate from Shirakawa river in Kumamoto prefecture (Japan) | This study |
| SK-02I | SK-1 derivative, eanI::Gmr | This study |
| SK-05R | SK-1 derivative, eanR::Gmr | This study |
| Escherichia coli | ||
| DH5α | F−supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Nippon Gene |
| S17-1 λpir | thi pro hsdR hsdM+recA RP4 2Tc::Mu-Km::Tn7 | 25 |
| Chromobacterium violaceum CV026 | ATCC 31532 derivative, cviI::Tn5xylE; Kmr Smr | 17 |
| Plasmids | ||
| pSTV28 | Cloning vector; Cmr | Takara Bio |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pAN01 | 6.0-kb Sau3AI fragment from SK-1 genomic DNA in pSTV28 | This study |
| pAN02 | pGEM-T easy containing eanI-eanR locus | This study |
| pGP704Sac38 | pBR322 derivative with R6K ori, mob RP4, polylinker from M13 tg131 containing sacB; Apr | 19 |
| pGP704EIG | pGP704Sac38 containing eanI::Gmr | This study |
| pGP704ERG | pGP704Sac38 containing eanR::Gmr | This study |
| pJN105 | pBBR1MCS-5 with araC and PBAD regions before the polylinker; Gmr | 20 |
| pJN105ER | pJN105 containing eanR | This study |
| pQF50 | lacZ transcriptional fusion vector; Apr | 8 |
| pQF50ER | pQF50 containing eanR promoter. | This study |
| pKRP11 | Cloning vector; Kmr Apr | 23 |
| pKRP14 | pKRP11 derivative containing the Gmr cassette | This study |
Cmr, chloramphenicol resistance; Smr, streptomycin resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance.
Identification and characterization of AHLs.
AHLs produced by P. ananatis SK-1 were isolated and purified by a previously described method (17). The AHL sample was subjected to analytical and preparative thin-layer chromatography (TLC). TLC analysis was carried out on a C18 reversed-phase TLC plate (Analtech). C6-HSL and 3-oxo-C6-HSL were used for AHL standards. AHL sample and standards were spotted onto a TLC plate and developed with 60% (vol/vol) methanol in water. The air-dried plate was overlaid with LB soft gel (1% agar) with C. violaceum CV026 biosensor (17) and incubated at 30°C. AHL production was also assayed by cross-streaking against CV026 biosensor as the AHL biosensor. Briefly, CV026 was streaked at the center of the LB agar plate. The target bacteria were streaked on the same plate next to CV026 line. Diffusible AHL produced by the target bacteria induces strain CV026 to produce a purple pigment.
Cloning and disruption of chromosomal eanI-eanR locus.
Chromosomal DNA of SK-1 was extracted to construct genomic library by the standard protocol (24). DNA was digested partially with Sau3AI, and the fragments were inserted into the BamHI site of cloning vector, pSTV28. The genomic library of SK-1 was transformed into E. coli DH5α, and we checked the AHL-producing ability by cross-streaking with a CV026 biosensor. One of the AHL-producing plasmids, pAN01, was sequenced by using BigDye terminator version 3.1 and an ABI Prism 3100 genetic analyzer (Applied Biosystems). The eanI/eanR locus on the chromosomal DNA of SK-1 was amplified by PCR using the primers 5′-GTAAAATCAGTACAGGATAGCCGTGAGGGC-3′ and 5′-TAAAGGAGGACAATCAGGTGTGGGAAAGCG-3′ and cloned into pGEM-T Easy cloning vector for construction of pAN02. To disrupt the eanI gene, pAN02 was digested with HindIII and inserted the 900-bp Gmr cassette from HindIII-digested pKRP14. The eanI::Gmr region was cut out by EcoRI digestion and inserted into the MunI site of pGP704Sac38 for construction of pGP704EIG. To disrupt the eanR gene, pAN02 was digested with BglII, and the Gmr cassette was inserted from BamHI-digested pKRP14. The eanR::Gmr region was cut out by EcoRI digestion and inserted into the MunI site of pGP704Sac38 for construction of pGP704ERG. Disruption of chromosomal eanI and eanR in strain SK-1 was performed by bacterial conjugation and homologous recombination (19). Conjugation was conducted between SK-1 and E. coli S17-1 λpir carrying pGP704EIG or pGP704ERG. The chromosomal disruption of eanI and eanR was checked by PCR using the same primers, and the insertion mutants of eanI and eanR were designated SK-02I and SK-05R, respectively.
Promoter assay.
The putative promoter region of eanR was amplified by PCR using the primers 5′-TAAAGGAGGACAATCAGGTGTGGGAAAGCG-3′ and 5′-GTTTAAAGGCGGTAAGGATAACCGGATCGG-3′ and cloned into pGEM-T Easy. The putative promoter region was cut out by SphI and SalI digestion and cloned into the SphI and SalI sites of vector pQF50 for the construction of pQF50ER. To construct the EanR expression plasmid, the promoter-less eanR gene was amplified by PCR using the primers 5′-ATCGTTAAGTAAAAGAAGCAGCATGGAGCC-3′ and 5′-TACTCAAACGGTCCGGATGGCAAATCAGCG-3′ and cloned into pGEM-T Easy. The promoter-less eanR was cut out by EcoRI digestion and ligated with EcoRI-digested pJN105 for the construction of pJN105ER. Both pQF50ER and pJN105ER were transformed into E. coli DH5α for the promoter assay. The DH5α reporter strain was grown for 15 h and inoculated into 4 ml of fresh LB medium (1% inoculum). AHLs were added into the subculture at a final concentration of 10 μM. Arabinose was used for the induction of expression of EanR at a concentration of 0.4%. After incubation for 20 h, β-galactosidase activity was measured by using a Galacto-Light Plus kit (Tropix) as described previously (16). The results are given in units of β-galactosidase activity relative to the optical density at 600 nm.
Detection of EPS biosynthesis.
EPS biosynthesis was evaluated by a previously described method (28). P. ananatis strains were streaked onto the TSB agar plate. After incubation for 24 h at 30°C, colonies producing EPS have a mucoid appearance, whereas those deficient in EPS have a nonmucoid phenotype. Experiments were repeated at least three times.
Biofilm formation assay.
Biofilm formation was determined by the previously described method with slight modification (15). The full-grown cultures of P. ananatis strains were diluted 100-fold in the TSB medium, and 200 μl of the dilution was added to each well of a 96-well polypropylene microtiter plate (Corning, Inc.). After incubation at 30°C for 20 h, 25 μl of a 1% crystal violet solution was added to each well. The plates were incubated at room temperature for 15 min and rinsed with distilled water. The crystal violet was dissolved in 200 μl of 95% ethanol, and biofilm formation was analyzed at 570 nm by using a Spectra Max 250 spectrophotometer (Molecular Devices).
Plant infection assays.
Pathogenicity of P. ananatis strains was determined on onion leaves as described previously with slight modifications (10). Briefly, a sterile needle was dipped into the bacterial colonies on TSB agar plates grown for 24 h. The needle was then inserted under the epidermis of a leaf. P. ananatis strains were inoculated at two sites per leaf. Inoculated leaves were incubated at room temperature and observed for the development of symptoms. All infection assays contained at least two leaves per treatment, and experiments were performed at least twice.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene and the eanI/eanR locus from the strain SK-1 have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AB304809 and AB304810, respectively.
RESULTS AND DISCUSSION
P. ananatis SK-1 produces AHLs.
P. ananatis SK-1 was isolated from the Shirakawa River in Kumamoto Prefecture (Japan). The nucleotide sequence of the 16S rRNA fragment from SK-1 showed 99.9% identity with that of P. ananatis strain LMG 20103 (accession no. AF364847). Morphological and biochemical test results were evaluated according to Bergey's Manual of Determinative Bacteriology (12). These test results also suggested that strain SK-1 belonged to P. ananatis (data not shown). We screened for the AHL production of SK-1 by cross-streaking against C. violaceum CV026 as the AHL biosensor. Since SK-1 stimulated violacein production of the CV026 biosensor, it revealed that SK-1 had the ability to produce AHLs (Fig. 1A). We also checked the structure of produced AHLs by the TLC analysis. AHLs were extracted with dichloromethane and fractionated by TLC. TLC-overlaid CV026 biosensor revealed two AHL spots in the culture supernatants of SK-1. After comparison with AHL standards, these spots corresponded with C6-HSL and 3-oxo-C6-HSL (Fig. 1B). In a previous report, P. ananatis inhabiting wheat heads produces C6-HSL and 3-oxo-C6-HSL (29). It was noteworthy that a wide range of P. ananatis strains generally produced C6-HSL and 3-oxo-C6-HSL.
FIG. 1.
Identification and characterization of AHLs produced by P. ananatis SK-1. (A) Cross-streaks of SK-1 and its mutants against C. violaceum CV026. Diffusible AHL production by SK-1 and SK-05R induced CV026 biosensor to produce a violacein. (B) TLC analysis of AHLs produced by strain SK-1. AHLs were visualized as pigments produced by CV026 biosensor and identified after comparison with the AHL standards C6-HSL and 3-oxo-C6-HSL.
Cloning and characterization of the luxI and luxR homologs eanI and eanR.
For cloning the AHL synthase gene, we constructed a genomic library of SK-1 based on pSTV28 cloning vector. Approximately 2,000 transformants were screened for the presence of a luxI homolog by toothpicking colonies and cross-streaking them against the CV026 biosensor. One clone was able to induce the production of violacein in CV026. Plasmid DNA extracted from the positive clone contained 6.0-kb DNA fragment and was designated pAN01. The nucleotide sequence of the pAN01 revealed the presence of luxI and luxR homologs. These luxI and luxR homologs were designated eanI (for E. ananatis luxI) and eanR (for E. ananatis luxR), respectively. The putative gene product of eanI (EanI) encoded a 210-amino-acid protein and showed 91.9% identity with EsaI from P. stewartii and 44.3% identity with CarI from E. carotovora (Fig. 2A). The previous report indicated that P. stewartii produced 3-oxo-C6-HSL as a sole AHL compound (28). Although EsaI from P. stewartii showed high homology with EanI, EsaI did not produce C6-HSL. EanI and EsaI shared well-known conserved amino acid residues that have been demonstrated to be important to AHL synthase (data not shown) (21). A comparative study of EanI and EsaI could not explain the reason for the structural difference of AHLs produced EanI and EsaI. The putative gene product of eanR (EanR) encoded 249 amino acids and showed 95.2% identity with EsaR from P. stewartii and 32.8% identity with CarR from E. carotovora (Fig. 2B). The eanR promoter region contained putative −10 and −35 sequences, and a 20-bp imperfect inverted repeat spanned the −10 region (Fig. 3A). This inverted repeat sequence can be observed at the promoter region of luxR homolog in P. stewartii (28) and Serratia marcescens (13). These inverted repeats were very similar to the lux box consensus sequence (Fig. 3B). Thus, the inverted repeat sequence in the eanR promoter region is thought to represent the binding site of LuxR homolog.
FIG. 2.
Multiple alignments of EanI (A) and EanR (B). Gray and black shading indicates similar and identical amino acids, respectively. Sequence alignment was performed with CLUSTAL W (http://www.ddbj.nig.ac.jp/search/clustalw-j.html) and presented with Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The sequences used in the alignments with EanI were EsaI from P. stewartii (accession no. L32183) and CarI from E. carotovora (X74299). The sequences used in the alignments with EanR were EsaR from P. stewartii (L32184) and CarR from E. carotovora (AF041840).
FIG. 3.
(A) Nucleotide sequence analysis of the upstream region of eanR. Putative −35 and −10 promoter sequences, the ribosome binding site (RB), and the translation initiation site are shown in boldface type. The inverted repeat exhibiting similarity to lux box was underlined. (B) Nucleotide sequence comparison of consensus lux box sequences (13) and the upstream region of luxR homolog in P. ananatis SK-1, P. stewartii DC283, and S. marcescens SS-1. Conserved sequences are shown in boldface type.
EanR acts as a negative regulator.
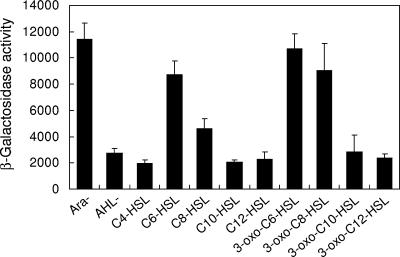
To analyze the function of EanR for the activation of the lux box-like sequence (ean box), the upstream region of eanR was amplified by PCR and cloned upstream of the promoterless lacZ gene of reporter plasmid pQF50. We monitored the promoter activity of the ean box in E. coli DH5α carrying the EanR expression plasmid pJN105ER and the ean box-lacZ plasmid pQF50ER. The expression of eanR on the pJN105ER was controlled by the PBAD promoter. The ean box promoter was strongly activated without arabinose but only at a very low level with 0.4% arabinose (Fig. 4). This result suggested that the activation of the ean box sequence was negatively regulated by EanR. To confirm whether the addition of AHLs results in derepression of ean box, various kinds of AHLs were added at a concentration of 10 μM. The ean box promoter was activated by C6-HSL and C8-HSL, and their 3-oxo-substitution (Fig. 4). 3-Oxo-substituted AHLs demonstrated higher activity than 3-oxo-unsubstituted AHLs (Fig. 4). In the case of P. stewartii, EsaR also worked as a negative regulator of AHL-mediated quorum sensing (28). EsaR binds the lux box-like sequence in the absence of AHL and blocks the transcriptional activity of RNA polymerase (26). In addition, the ability of EsaR to bind to its DNA recognition site is antagonized by the presence of 3-oxo-C6-HSL (26). Our data suggested that EanR might behave in a fashion similar to its closest homolog, EsaR, and was likely to bind the ean box promoter in the absence of AHLs.
FIG. 4.
Induction of the ean box-lacZ transcriptional fusion in E. coli DH5α by different AHL compounds. E. coli DH5α carrying pJN105ER and pQF50ER was grown in the LB medium with various synthetic AHLs. After 15 h of incubation, the β-galactosidase activity was measured. Arabinose was used for the induction of expression of EanR at a concentration of 0.4%. Ara− indicates activity without arabinose and AHL. AHL− indicates the result with 0.4% arabinose and without AHL. The results were reproduced in three repeated experiments, and error bars indicate standard deviations.
AHL synthesis requires eanI but not eanR.
To determine whether eanI and eanR are required for AHL synthesis, we disrupted the genomic eanI and eanR genes in SK-1. The Gmr gene was inserted to the HindIII or BglII site of the eanI/eanR locus for construction of eanI and eanR mutants, respectively. When cross-streaked against CV026 biosensor, the eanR mutant SK-05R showed obvious AHL-producing activity, as well as the SK-1 parent strain, but the eanI mutant SK-02I did not produce any AHLs (Fig. 1A). This result demonstrated that the eanI gene was necessary for the production of both C6-HSL and 3-oxo-C6-HSL, and the expression of eanI was not regulated by EanR. In P. stewartii, esaI is expressed constitutively and not regulated by EsaR (28). The lux box-like sequence was absent in the upstream region of eanI, as well as esaI (data not shown). It was assumed that eanI was also expressed constitutively.
EPS biosynthesis and biofilm formation require eanI or AHLs.
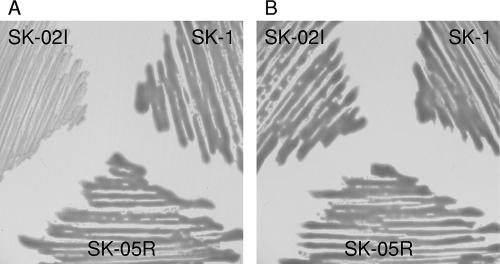
In a previous report on P. stewartii, EPS biosynthesis was regulated by the esaI gene or 3-oxo-C6-HSL (28). Thus, we investigated the ability to produce EPS in SK-1 and its mutants. We tested the abilities of these strains to stimulate EPS biosynthesis on TSB agar plates. After incubation for 24 h, SK-1 and SK-05R displayed a mucoid phenotype resulting from the production of EPS, but SK-02I did not (Fig. 5A). When the TSB agar plate was supplemented with 10 μM 3-oxo-C6-HSL, the colonies of SK-02I became mucoid, as well as those of other strains (Fig. 5B). These results demonstrated that AHLs produced by eanI gene induced the production of EPS in SK-1. Pathogenesis in P. stewartii correlates with the ability to produce EPS, and the production of the EPS requires 3-oxo-C6-HSL (28). Thus, it is possible that P. ananatis produces EPS as a major virulence factor under quorum-sensing control.
FIG. 5.
Production of EPS by wild-type and mutant strains of P. ananatis SK-1. Strains were streaked onto the TSB agar plates without (A) or with (B) 10 μM 3-oxo-C6-HSL. Mucoid and slimy colony morphology indicated EPS production after incubation at 30°C for 24 h.
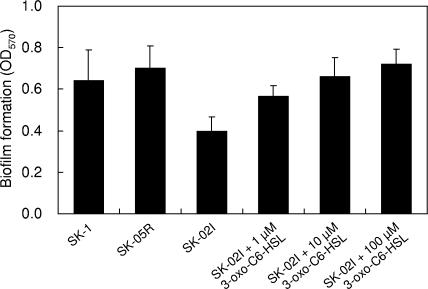
We also tested biofilm formation on a polypropylene plastic surface. Although SK-1 and SK-05R formed a certain amount of biomass that adhered to the polypropylene, biofilm formation of SK-02I was reduced to ca. 60% of the parental level (Fig. 6). When 3-oxo-C6-HSL was added into each well, the biofilm formation of SK-02I was increased in a dose-dependent manner (Fig. 6). This behavior implied that biofilm formation of SK-1 was influenced by AHL-mediated quorum sensing. In the case of P. stewartii, a nonvirulent mutant lacking the esaI gene adheres strongly to surfaces, and QS mutants lacking the EsaR repressor attach poorly to surfaces (15). Interestingly, P. ananatis and P. stewartii showed opposite behavior in terms of biofilm-forming ability.
FIG. 6.
Quantification of bacteria in biofilm formed on polypropylene plastic by wild-type and mutant strains of P. ananatis SK-1. Biofilms were allowed to form in a 96-well polypropylene microtiter dish, stained with crystal violet, and estimated by analysis at 570-nm absorbance. Six wells of each sample were used for measuring biofilm formation, and error bars indicate standard deviations.
AHLs contribute to symptom expression in onion leaves.
We conducted pathogenicity tests on onion leaves. SK-1 and its mutants were inoculated into onion leaves, and the development of the necrotic symptoms was monitored. After 3 days of incubation at room temperature, the onion leaves infected by SK-1 and SK-05R were collapsed, hanging down beside the inoculation site, and displayed typical symptoms of center rot. SK-1 and SK-05R induced necrotic symptoms at the inoculation site, but SK-02I did not (Fig. 7). In order to confirm whether the exogenous 3-oxo-C6-HSL induces the virulence of SK-02I, 100 nmol of 3-oxo-C6-HSL was spotted onto the leaves, and the needle-dipped SK-02I was inserted at the same site. As a result, SK-02I-exposed 3-oxo-C6-HSL induced necrotic symptoms as well as SK-1 and SK-05R (Fig. 7). Treatment of inoculated leaves with 3-oxo-C6-HSL did not elicit any detectable symptoms (Fig. 7). These results demonstrated that the pathogenicity of P. ananatis was regulated by the AHL-mediated quorum-sensing system.
FIG. 7.
Virulence assay of wild-type and mutant strains of P. ananatis SK-1. Bacterial strains were injected into onion leaves at two sites per leaf. 3-oxo-C6-HSL was spotted onto the leaves at 100 nmol per spot. Inoculated onion leaves were incubated at room temperature for 3 days and monitored for the development of the necrotic symptoms.
In summary, our work is the first report that the quorum-sensing system involves the biosynthesis of EPS, biofilm formation, and infection of onion leaves in P. ananatis. We also show that P. ananatis and P. stewartii had very similar quorum-sensing systems. The virulence factors of P. ananatis have not been elucidated clearly. More study of quorum sensing in P. ananatis may contribute to the detection of novel virulence factors and the treatment of infected plants.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Boyhan, G., D. Granberry, and T. Kelley. 2001. Onion production guide. Cooperative Extension Service, University of Georgia, Tifton, GA.
- 2.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 61:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 6.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y. H., and L. H. Zhang. 2005. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43:101-109. [PubMed] [Google Scholar]
- 8.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitaitis, R. D., R. R. Walcott, M. L. Wells, J. C. Diaz Perez, and F. H. Sanders. 2003. Transmission of Pantoea ananatis, causal agent of center rot of onion, by tobacco thrips, Frankliniella fusca. Plant Dis. 87:675-678. [DOI] [PubMed] [Google Scholar]
- 10.Goszczynska, T., S. N. Venter, and T. A. Coutinho. 2006. PA 20, a semi-selective medium for isolation and enumeration of Pantoea ananatis. Microbiol. Methods 64:225-231. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg, E. P. 1997. Quorum sensing in gram-negative bacteria. ASM News 63:371-377. [Google Scholar]
- 12.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. The Williams & Wilkins Co., Baltimore, MD.
- 13.Horng, Y. T., S. C. Deng, M. Daykin, P. C. Soo, J. R. Wei, K. T. Luh, S. W. Ho, S. Swift, H. C. Lai, and P. Williams. 2002. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45:1655-1671. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, T., K. Kajiyama, T. Kita, N. Takiguchi, A. Kuroda, J. Kato, and H. Ohtake. 2001. The synthesis of optically pure enantiomers of N-acyl-homoserine lactone autoinducers and their analogues. Chem. Lett. 30:314-315. [Google Scholar]
- 15.Koutsoudis, M. D., D. Tsaltas, T. D. Minogue, and S. B. von Bodman. 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 103:5983-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. H., Y. Lequette, and E. P. Greenberg. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602-609. [DOI] [PubMed] [Google Scholar]
- 17.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycrof, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 18.Molina, L., F. Rezzonico, G. Défago, and B. Duffy. 2005. Autoinduction in Erwinia amylovora: evidence of an acyl-homoserine lactone signal in the fire blight pathogen. J. Bacteriol. 187:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morohoshi, T., T. Inaba, N. Kato, K. Kanai, and T. Ikeda. 2004. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J. Biosci. Bioeng. 98:276-281. [DOI] [PubMed] [Google Scholar]
- 20.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 21.Parsek, M. R., A. L. Schaefer, and E. P. Greenberg. 1997. Analysis of random and site-directed mutations in rhII, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 26:301-310. [DOI] [PubMed] [Google Scholar]
- 22.Pomini, A. M., W. L. Araújo, and A. J. Marsaioli. 2006. Structural elucidation and biological activity of acyl-homoserine lactones from the phytopathogen Pantoea ananatis Serrano 1928. J. Chem. Ecol. 32:1769-1778. [DOI] [PubMed] [Google Scholar]
- 23.Reece, K. S., and G. J. Phillips. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141-142. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Simon, R., J. Quandt, and W. Klipp. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 26.von Bodman, S. B., J. K. Ball, M. A. Faini, C. M. Herrera, T. D. Minogue, M. L. Urbanowski, and A. M. Stevens. 2003. The quorum sensing negative regulators EsaR and ExpREcc, homologues within the LuxR family, retain the ability to function as activators of transcription. J. Bacteriol. 185:7001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Bodman, S. B., W. D. Bauer, and E. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 28.von Bodman, S. B., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida, S., L. L. Kinkel, H. Shinohara, N. Numajiri, S. Hiradate, M. Koitabashi, K. Suyama, H. Negishi, and S. Tsushima. 2006. Production of quorum-sensing-related signal molecules by epiphytic bacteria inhabiting wheat heads. Can. J. Microbiol. 52:411-418. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, D., H. Zhang, S. Carle, G. Hao, M. R. Holden, and T. J. Burr. 2003. A luxR homolog, aviR, in Agrobacterium vitis is associated with induction of necrosis on grape and a hypersensitive response on tobacco. Mol. Plant-Microbe Interact. 16:650-658. [DOI] [PubMed] [Google Scholar]