Abstract
The role of the RecBCD recombination pathway in PilE antigenic variation in Neisseria gonorrhoeae is contentious and appears to be strain dependent. In this study, N. gonorrhoeae strain MS11 recB mutants were assessed for recombination/repair. MS11 recB mutants were found to be highly susceptible to DNA treatments that caused double-chain breaks and were severely impaired for growth; recB growth suppressor mutants arose at high frequencies. When the recombination/repair capacity of strain MS11 was compared to that of strains FA1090 and P9, innate differences were observed between the strains, with FA1090 and P9 rec+ bacteria presenting pronounced recombination/repair defects. Consequently, MS11 recB mutants present a more robust phenotype than the other strains that were tested. In addition, MS11 recB mutants are also shown to be defective for pilE/pilS recombination. Moreover, pilE/pilS recombination is shown to proceed with gonococci that carry inverted pilE loci. Consequently, a novel RecBCD-mediated double-chain-break repair model for PilE antigenic variation is proposed.
Neisseria gonorrhoeae causes the human mucosal infection gonorrhea. Following sexual transmission of the organism, gonococci bind to columnar epithelial cells utilizing proteins associated with the pilus organelle. The major protein component of the pilus structure is PilE polypeptide. PilE polypeptide undergoes antigenic variation such that the chemical composition of the protein changes and, in doing so, negates an efficacious immune response, thus promoting immune evasion (reviewed in reference 18).
PilE protein is expressed from a single pilE gene (1). However, the genome also contains many truncated, variant, nontranscribed copies of pil sequence which are located in several different loci (pilS) (7). Despite this apparent diversity, the genetic structures of all pil genes are very similar, with variable regions being interspersed with conserved DNA segments, with these conserved regions believed to facilitate recombination between pilE and a pilS gene copy, leading to variant pilE alleles (7). Changes in the pilE gene sequence occur primarily through RecA-mediated gene conversion events (7, 15, 25, 29) where a portion of the pilS gene copy is transferred into the pilE gene, with the resident pilE sequence being ejected or lost from the chromosome (7, 25). Consequently, this results in the expression of a unique PilE polypeptide.
Homologs of many of the Escherichia coli Rec proteins have been identified in N. gonorrhoeae (3, 8, 15, 17). Genetic analysis of pilE gene variation using N. gonorrhoeae rec mutants has proven to be controversial. In N. gonorrhoeae strain MS11, inactivation of the recD gene was shown to cause hyperrecombination at pilE, resulting in progeny with an increased number of nonparental pilus phenotypes (3). Consequently, this led to a proposal that pilE gene variation proceeded via a RecBCD-mediated pathway. However, in studies utilizing a different strain (FA1090), insertion mutations in either recB or recC did not effect gene variation at pilE and caused only a modest recombination/repair deficiency (17). Consequently, these observations indicated a significant difference between the proposed behavior of the N. gonorrhoeae RecBCD complex and the E. coli RecBCD enzyme. To account for these differences, it was proposed that a “RecF-like” pathway accounted for pilE gene variation and recombination/repair in strain FA1090, especially as gonococci possess several homologs to genes that are present in the E. coli RecF pathway, which include recO, recR, recJ, and recQ (8, 17). However, gonococci lack exonuclease I (sbcB and -C) as well as the recF gene, so the relationship of this pathway to the RecBCD pathway is currently unclear. Nonetheless, two studies have demonstrated that this “RecF-like” pathway participates in repairing UV-induced lesions (8, 23).
Although the effects of an insertion mutation in the recB gene of N. gonorrhoeae strain FA1090 have been documented (17), the effects of a null mutation in strain MS11 have yet to be investigated. The purpose of this study is to determine the effect of recB (and recC) mutations on recombination/repair in N. gonorrhoeae strain MS11. From the data presented, an MS11 recB (or recC) null mutation severely impedes repair of double-chain breaks caused by nalidixic acid treatment as well as the repair of DNA alkylation lesions caused by methyl methanesulfate (MMS) treatment. In addition to the robust recombination/repair phenotype exhibited by the MS11 recB mutants, we present evidence that indicates that pilE gene variation proceeds via a RecBCD-mediated pathway in strain MS11. Furthermore, as we are also able to demonstrate that inverting the pilE locus does not impede pilE/pilS recombination, these combined observations allow us to propose a double-chain-break repair model for pilE gene variation in strain MS11.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Neisseria gonorrhoeae strains MS11 (obtained from John Swanson, Rocky Mountain Laboratories, MT), FA1090 (obtained from Fred Sparling, University of North Carolina), and P9 (obtained from John Saunders, University of Liverpool) were used in this study (Table 1). Strain MS11 has been passaged as a laboratory strain for many years; strains FA1090 and P9 were obtained in the mid 1980s and have undergone limited cultivation. During the course of these studies, gonococci were passaged daily on gonococcal (GC) typing medium (GTM) at 37°C in 5% CO2 (24). Occasionally, gonococci were resuspended in GC HEPES medium, which is identical in composition to GTM (24) except that the phosphate salts are replaced by 0.2% HEPES, Na+ salt (Calbiochem, La Jolla, CA), and 0.5% HEPES acid (Calbiochem, La Jolla, CA). Where appropriate, the medium was supplemented with antibiotics: 10 μg/ml of erythromycin or 80 μg/ml of kanamycin (Sigma, St. Louis, MO).
TABLE 1.
Bacterial strains utilized in this study
| Strain | Description | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | F ϕlacZ Δ(lacZYA argF)U16 deoR recA1 endA1 hsdR17(rκ− mκ+)phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco BRL, Gaithersburg, MD |
| GY5873 | Hfr argA::Tn9 lacMS286 Φ80dII lacBK1 recB21 | 21 |
| Neisseria gonorrhoeae | ||
| MS11 rec+ | Wild type | J. Swanson |
| P9 rec+ | Wild type | J. Saunders |
| FA1090 rec+ | Wild type | F. Sparling |
| MS11 recB | recB::ermC | This study |
| MS11 recB opaE::recB+ | Wild type recB gene placed in opaE locus | This study |
| MS11 recB sup | Intragenic recB suppressor mutant | This study |
| FA1090 recB | recB::ermC | This study |
| P9 recB | recB::kan | This study |
| MS11 pro | ΔProline residue in recB active site and ermC cassette 3′ of recB gene | This study |
| MS11 inv | 900-bp segment of the pilE locus in an inverse orientation | This study |
| MS11 recC | recC::ermC (or kan) | This study |
| FA1090 recC | recC::ermC (or kan) | This study |
| P9 recC | recC::ermC (or kan) | This study |
| MS11 Sma::Xho | XhoI linker placed in the SmaI site in the Sma/Cla repeat downstream of pilE | 9 |
| FA1090 Sma::Xho | XhoI linker placed in the SmaI site in the Sma/Cla repeat downstream of pilE | This study |
Escherichia coli was grown on Luria-Bertani medium at 37°C, with antibiotics added at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 200 μg/ml; and kanamycin, 40 μg/ml. The lac papillation assay was performed as described previously, using E. coli strain GY5873 (Table 1) and MacConkey lactose medium (21, 30).
DNA manipulations.
All constructs were made in E. coli strain DH5α. N. gonorrhoeae strain MS11 chromosomal DNA was used as a PCR template to construct the MS11 recB mutants by amplifying two segments of the recB gene using primers recB4/B5 and recB9/recB10 (Table 2). The PCR products were then sequentially ligated into the pCRII vector (Invitrogen) with an erythromycin or kanamycin gene cassette inserted in the middle. The recombinant plasmid was used to transform N. gonorrhoeae to drug resistance to create a recB insertional mutant, with the mutation confirmed by PCR in which one of the primers was located within the drug resistance marker (Erm1.5Rev) (Table 2). N. gonorrhoeae recC mutants were constructed following PCR amplification of recC using primer pair RecC2 and RecC4 (Table 2). The resulting 1,834-bp fragment was cloned into the pCRII vector. The insert was sequenced and an erythromycin (or kanamycin) gene cassette being inserted into a unique HincII site. The recombinant plasmid was then used to transform strain MS11 to drug resistance. The mutation was confirmed by PCR in which one of the primer pairs was located within the drug resistance marker (Erm1.5Rev) (Table 2).
TABLE 2.
Oligonucleotides utilized in this study
| Designation | Nucleotide sequence |
|---|---|
| Cys2R | 5′-GCAGGTGACGGCAGGTGC-3′ |
| Erm1.5Rev | 5′-TGTTAGCCAAAGCTTCCAAGC-3′ |
| HVPilS(R) | 5′-CGTTGTCGTTGCCGGCTTTGGT-3′ |
| InterB6 | 5′-GCCTGAAGGTAATAGTGCTGGTGCGCGACGGC-3′ |
| PilRBS | 5′-GGCTTTCCCCTTTCAATTAGGAG-3′ |
| RecC2 | 5′-GATTTGTTCCGCAGCGCACAA-3′ |
| RecC4 | 5′-GGCAACGTATTGATGATTCG-3′ |
| RecB4 | 5′-CCCAAAGCTTGCACTGCACAAATGGCTGCGCGATCAAATC-3′ |
| RecB5 | 5′-CTAGTCTAGAGTGCGGGCAGGAAGGTGTCGCCTTCGT-3′ |
| RecB6 | 5′-CTAGTCTAGAGGTGAGGGTGTGCGTGGTCATA-3′ |
| RecB9 | 5′-CCCTCGAGGCTCAAGATTTTTGGCGGGAACG-3′ |
| RecB10 | 5′-GGTCTAGACCCGAAGTCTGCCAGTTTCTGCAATTC-3′ |
| RecB12 | 5′-AAAAGCTTGGCTGTGCCTGCACGAAATTCTTGAAGA-3′ |
| Tracy1 | 5′-CGATATGGTCTGCCAAGACGACGGCAATATCTGC-3′ |
| Tracy2 | 5′-GCAGATATTGCCGTCGTCTTGGCAGACCATATCG-3′ |
| GC recB1 | 5′-AAACCGCAACCCCGCCGCA-3′ |
| GC recB2 | 5′-GCGTGGGACGCGCAAGATACC-3′ |
| GC recB3 | 5′-CGGCGGCGTTGCCAGCGT-3′ |
| GC recB4 | 5′-GGTGCCGCCGTCCCGAA-3′ |
| probe 245 | 5′-GCCTTTTTGAAGGGTATTCAT-3′ |
Site-specific mutagenesis (Stratagene) was used to delete the codon that specified the proline residue in the recB nuclease active site. A standard PCR was performed using primer pairs RecB12 (upstream primer) and Tracy1 (the deletion primer) (Table 2) with Pfu Taq polymerase (Pfu Ultra; Stratagene) to amplify the active-site DNA. A second PCR was performed using a primer complementary to the first deletion primer (Tracy2) and the downstream primer RecB6 (Table 2). These PCRs were purified, and a third PCR was run, with these template DNAs serving as primers because they are partially complementary. This final PCR product was cloned, and codon deletions were confirmed by DNA sequencing. A drug resistance marker was then inserted in a unique SalI site that is located downstream of the recB transcriptional unit. The codon deletion was crossed into the gonococcal chromosome by transformation. The incorporation of the codon deletion was confirmed by DNA sequencing of selected transformants.
The MS11 recB gene was cloned from a pBR322 plasmid library that was created using a Sau3A partial digest of gonococcal chromosomal DNA. Plasmids carrying the gonococcal recB gene were identified using Neisseria-specific recB oligonucleotides (Gc recB1 to -4) (Table 2).
The pilE gene was inverted by PCR of specific gene fragments incorporating unique restriction sites, using pVD203 (which carries the pilE gene, the downstream Sma/Cla repeat, and approximately 800 bp upstream of the pilE start codon) (1) and pSX2.7 (which carries the opaE gene, which resides downstream of pilE in the gonococcal chromosome [2]) as templates. The PCR fragments were assembled sequentially through selective restriction digests. The pilE inversion included approximately 500 bp upstream of pilE, the pilE gene itself, and the downstream Sma/Cla repeat. An erythromycin gene cassette was inserted in the SalI site located in the opaE gene. The construct was crossed into the gonococcal chromosome by transformation. The structures of the final construct and of putative transformants were confirmed by DNA sequencing and PCR analysis.
Analysis of N. gonorrhoeae mutants. (i) Exposure to nalidixic acid.
Gonococci were serially diluted in GC HEPES medium (pH 7.4) and were then plated on GTM containing 0.5 μg/ml nalidixic acid and were grown for 2 days. Survival rates were calculated by dividing the total number of colonies growing on the nalidixic acid-containing GTM by the total number of colonies growing on the GTM without antibiotic. The assays were performed concurrently.
(ii) Exposure to MMS.
Gonococci were plated on GTM plates to a confluent density, with paper disks saturated with either 0.01%, 0.05%, 0.1%, 0.2%, or 0.3% freshly diluted MMS (Sigma) solution placed on top of the agar, and the plates were incubated overnight. Zones of inhibition identified the growth-inhibiting concentrations of MMS. The assays were performed concurrently.
(iii) Exposure to UV light.
Gonococci were serially diluted to the appropriate cell density, plated on solid medium, and exposed to various doses of UV radiation (0, 20, 40, 60, and 80 mJ/cm2) using a UV Stratalinker 1800. Survival rates were calculated by dividing the total number of visible colonies on an irradiated plate by the total number of visible colonies on the nonirradiated plate. The assays were performed concurrently.
PilE antigenic variation assays.
Antigenic variation was determined by two methods. The first method, in which conversion of a pilE outside marker is assessed, has been previously described (9). A XhoI linker was inserted into the SmaI site of the Sma/Cla repeat that is located downstream of the pilE locus. Chromosomal DNA was purified from an overnight culture, digested with SmaI, and analyzed by Southern analysis using the pilE-specific probe 245 (Table 2). Conversion of Sma::Xho to the wild-type Sma configuration requires pilE/pilS recombination (9). The second method entailed reverse transcription-PCR (RT-PCR) on purified RNA samples, slightly modifying a previously described procedure (27). RT-PCRs were run at 60°C for 30 min using conserved pilE primer pairs (PilRBS and Cys2R) under conditions outlined by the manufacturer (Boehringer Mannheim). Taq polymerase (2.5 units) was then added, and PCR was performed for 30 cycles. The RT-PCR was then used as a template for a second PCR using pilE primers [PilRBS and HVPilS(R)]. A pilE/pilS1 recombination event is required in order to obtain a product from the second PCR. The products were then analyzed by Southern analysis, using the HVPilS(R) primer as a probe.
RNA isolation.
RNA was isolated for RT-PCR as described previously (10). The purified RNA was then incubated with 2 units of molecular biology grade DNase (United States Biochemical) for 10 min at 37°C. The DNase was inactivated by boiling it for 5 min.
Southern analysis.
Southern blot analyses were performed as described previously, using radiolabeled oligonucleotides as probes (9, 25).
RESULTS
The N. gonorrhoeae RecB protein shares some similarity with its E. coli counterpart (33% identity and 49% similarity). A canonical ATPase domain (16/22 amino acid identity) is located toward the amino-terminal end of the protein, with the nuclease domain being located toward the carboxy-terminal end. A proline residue is located in the center of the nuclease active site (Fig. 1) (28). Given the disparity that has been observed between various gonococcal strains regarding RecBCD involvement in pilE gene variation (3, 17), we explored whether this was due to the presence of the proline residue in the nuclease active site causing a partial recB mutant phenotype in strain FA1090, with the defect in strain MS11 being alleviated through suppressor mutations. Site-directed mutagenesis was used to delete the codon that specified the proline residue in the nuclease active site to determine whether this proline residue influenced the functionality of the protein. No differences in the functionality of the proline-minus RecB protein were observed when these mutants were assessed in either strain MS11 or strain FA1090 (the data obtained with strain MS11 are presented in Fig. 2). The functionality of the MS11 recB gene was further confirmed by cloning recB from an MS11 plasmid library on an approximately 8-kb fragment and then utilizing this clone to complement an E. coli recB21 mutant in a lac papillation assay; extensive lac papillation was apparent in E. coli rec+ and in the E. coli recB21 strain carrying the GC recB complementation plasmid and was absent in the E. coli recB21 strain carrying the vector alone when the various bacteria were grown on solid medium (21, 30).
FIG. 1.
CLUSTALW alignment of the amino acids that constitute the RecB nuclease active site. Regions of identity are indicated by stars and regions of similarity by dots. The sequences are as follows: N. gonorrhoeae strain MS11, GeneID no. 3282190 (NGON); Neisseria meningitidis strain FAM18, GeneID no. 4675668 (NMEN); Mycobacterium tuberculosis strain CDC151, GeneID no. 925060 (MTB); Escherichia coli strain K-12, GeneID no. 947286 (ECOLI); Haemophilus influenzae strain RdKW20, GeneID no. 950246 (HINF); Borrelia burgdorferi strain B31, GeneID no. 1195485 (BBURG); and Chlamydophila pneumoniae strain J138, GeneID no. 919511 (CPNEU). Also included are the homologous AddA proteins previously identified from Bacillus subtilis strain 168, GeneID no. 939793 (BSUB), and from Treponema pallidum strain Nichols, GeneID no. 2611603 (TPAL) (27).
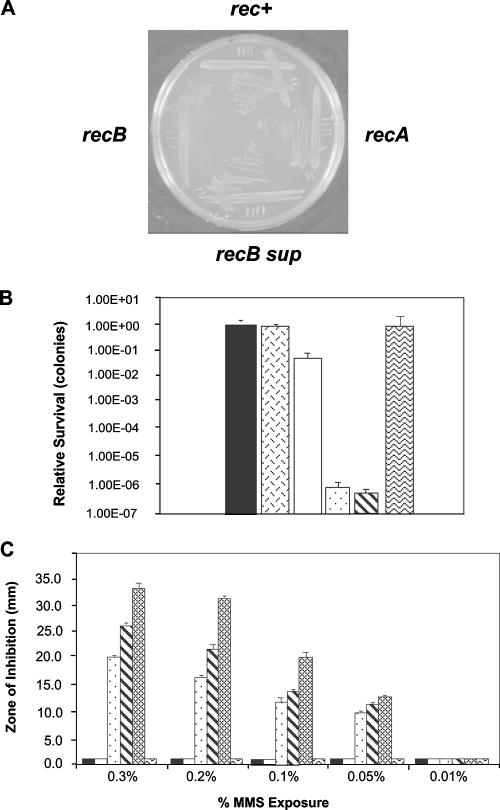
FIG. 2.
Analysis of N. gonorrhoeae strain MS11 recB mutants. (A) Comparison of the growth characteristics of MS11 rec+, recB, recA, and recB suppressor mutants. (B) Exposure to nalidixic acid (0.5 μg/ml). Solid bar, rec+; hexed bar, proline minus mutant; clear bar, recB suppressor; stippled bar, recB; diagonally striped bar, recA; wavy bar, recB opaE::recB+. Error bars indicate standard deviations from the mean (n = 10). (C) Exposure to MMS. Solid bar, rec+; clear bar, recB suppressor; stippled bar, recB; diagonally striped bar, recA; shaded bar, recC; wavy bar, recB opaE::recB+. Data represent two experiments performed in triplicate. Error bars represent standard errors (n = 6).
Analysis of N. gonorrhoeae strain MS11 recB mutants.
Neisseria gonorrhoeae recB mutants yielded small colonies on solid growth medium (Fig. 2A). However, larger colonies appeared (with an estimated frequency of 1 × 10−3 per CFU) upon passaging the small recB mutants on solid medium containing no antibiotic (Fig. 2A). These suppressor mutants arose through excision of the drug resistance marker and a small segment of the recB gene apparently using small direct repeats that flanked the drug resistance cassette in the recB gene (data not shown). Besides identification of these intragenic suppressor mutations, wild-type growth could also be restored through extragenic suppressor mutations that abrogated natural competence for DNA transformation, as observed when pilT and comA mutants were established via DNA transformation of the recB mutants. The MS11 recB mutants were tested for their abilities to undergo recombinational repair of DNA damage. When double-strand breaks were introduced to the chromosome by exposure of the bacteria to nalidixic acid (6, 16), the decrease in the repair capacity of the small-colony N. gonorrhoeae recB mutant was seen to be comparable to that for N. gonorrhoeae recA mutants (Fig. 2B). In contrast, wild-type MS11 and the recB proline-minus mutant displayed little inhibition of growth, while the intragenic recB suppressor mutant showed a modest defect. Complementation of the recB mutation with a recB+ allele restored wild-type growth and repair capabilities. Comparable observations were made following DNA alkylation by MMS treatment (Fig. 2C). Strain MS11 N. gonorrhoeae recC mutants displayed repair phenotypes similar to that of the recB mutant following nalidixic acid (data not shown) or MMS (Fig. 2C) treatment. From these data, we conclude that N. gonorrhoeae strain MS11 uses RecBCD to repair double chain breaks and alkylated DNA.
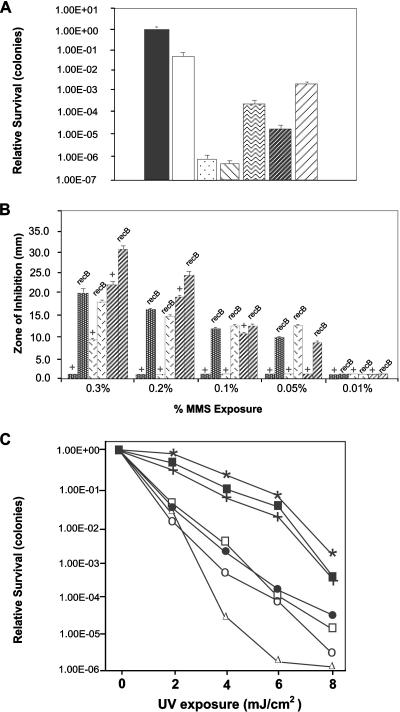
The preceding results obtained with strain MS11 differ from those found in a different strain background (FA1090) where only a modest defect in recombinational repair was observed (17). Consequently, we constructed an N. gonorrhoeae strain FA1090 recB mutant, as well as a recB mutant in a European isolate (N. gonorrhoeae strain P9), and repeated the assays. As can be seen in Fig. 3A and B, with these other strains, the effect of a recB mutation is less severe than that observed with strain MS11, with these data essentially confirming the previously published conclusions for strain FA1090 (14, 17). Comparable results were also found using FA1090 and P9 recC mutants (data not shown). However, what became apparent was that the wild-type repair capacities of strains FA1090 and P9 were significantly impaired compared to the wild-type MS11 repair capacity (e.g., repair of nalidixic acid-induced double chain breaks shows a difference of approximately 3 orders of magnitude) (Fig. 3A). Therefore, these observations indicate that for strains FA1090 and P9, wild-type bacteria appear deficient for recombinational repair, with the result that a recB mutation has little impact on the repair phenotype. When a recombinant FA1090 strain in which the MS11 recB allele replaced the FA1090 recB gene was constructed, the repair capability remained that of the FA1090 wild type, indicating that the recombinational repair phenotype presented by the FA1090 wild-type strain probably reflects genetic differences outside the recB locus. In contrast to these observations, no difference was observed between the three strains for the repair of UV-induced lesions (Neisseria possesses the uvr repair pathway); the wild-type strains grouped together, as did the various recB mutants (Fig. 3C).
FIG. 3.
Comparing the effects of DNA damage-causing reagents between N. gonorrhoeae strains MS11, FA1090, and P9. (A) Exposure to nalidixic acid (0.5 μg/ml). Solid bar, MS11 rec+; clear bar, MS11 recB suppressor; stippled bar, MS11 recB; diagonally striped bar, MS11 recA; squiggly bar, FA1090 rec+; dark diagonal bar, FA1090 recB; light diagonal bar, FA1090 recBMS11. Strain P9 rec+ and recB mutants were unable to tolerate nalidixic acid exposure at this concentration. Error bars indicate standard deviations from the mean (n = 10). (B) Exposure to MMS. Dark stippled bars, strain MS11; hexed bars, strain FA1090; diagonal bars, strain P9. The + symbol reflects rec+ bacteria. Data represent two experiments performed in triplicate. Error bars represent standard errors (n = 6). (C) Exposure to UV irradiation. MS 11 rec+ (filled squares), FA 1090 rec+ (stars), P9 rec+ (crosses), MS11 recB (open circles), FA 1090 recB (closed circles), P9 recB (open squares), and MS 11 recA (open triangles). Data represent two experiments performed in triplicate. Standard error bars are omitted for clarity.
Effect of an N. gonorrhoeae strain MS11 recB mutation on pilE gene variation.
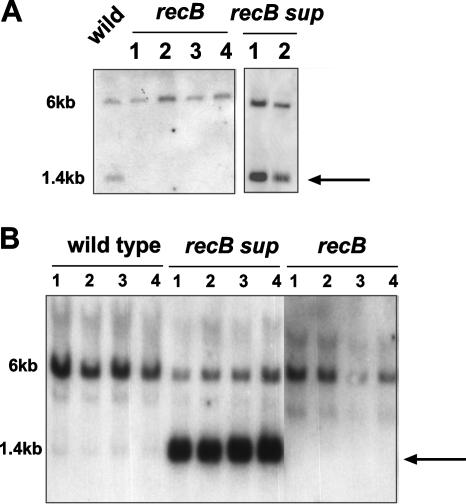
Two previously published qualitative assays were used to test whether an MS11 recB mutation influenced pilE gene variation: (i) an RT-PCR assay in which the selection of primers demands a pilE/pilS recombination event (Fig. 4A) (27) and (ii) a marker conversion assay that has also been shown to be dependent on pilE/pilS recombination (Fig. 5) (9). The RT-PCR assessment of the recB effect is shown in Fig. 4B. Following standardization of the reaction with respect to the amount of RNA that was used for RT as well as of the amount of the template that was used for the second PCR amplification (Fig. 4B), it is evident that a recB mutation reduces the extent of pilE/pilS recombination compared to that for either the wild type or the intragenic recB suppressor strain (Fig. 4B). As would be predicted, no signal was obtained when RNA was isolated and tested from an MS11 recA mutant. These observations were then confirmed in the coconversion assay whose results are presented in Fig. 5A. In this assay, a DNA linker is placed downstream of the pilE locus. Following pilE/pilS recombination, the linker can be either converted back to the wild-type configuration or not, due to the presence of homology downstream of pilE and the various pilS loci. As shown in Fig. 5A, the introduction of the recB mutation abrogates conversion of the marker back to the wild-type configuration, whereas with rec+ bacteria and the intragenic recB suppressor, conversion of the marker from the mutated state to the wild-type configuration is apparent. This conversion assay was also used to test pilE/pilS recombination with strain FA1090. In Fig. 5B, FA1090 rec+ bacteria show very little conversion of the pilE outside the marker, and what little that appears to occur is abrogated in the recB mutants. However, when recB growth suppressors were isolated, considerable conversion of the outside marker was demonstrable. Coconversion results comparable to those displayed by strain FA1090 were also evident with strain P9 (data not shown).
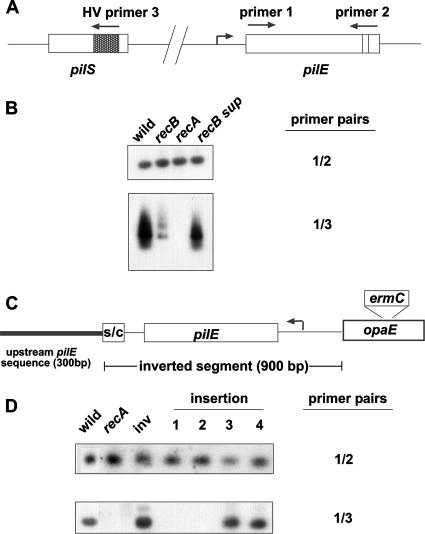
FIG. 4.
RT-PCR analysis assessing pilE/pilS recombination. (A) Schematic showing the relative locations of the oligonucleotide primers used in the assay. (B) Southern hybridizations of RT-PCR products using MS11 RNAs prepared from the wild type, recB mutants, recA mutants, and the recB intragenic growth suppressor. The blots were probed with primer 2 (top) or primer 3 (bottom). (C) Schematic showing the inverted pilE chromosomal context (not drawn to scale). S/C represents the Sma/Cla repeat located downstream of pilE. (D) Southern hybridizations of RT-PCR products performed on MS11 RNAs prepared from various MS11 strains. The inverted pilE locus is designated inv. The insertion (no. 1 to 4) mutants carry an erythromycin gene cassette at positions −192 (insertion 1), −221 (insertion 2), −336 (insertion 3), and −743 (insertion 4) relative to the ATG start codon. The blots were probed with primer 2 (top) or primer 3 (bottom).
FIG. 5.
Coconversion assay assessing pilE/pilS recombination. (A) XhoI linker DNA is located in the SmaI site in the Sma/Cla repeat downstream of the pilE locus. Following restriction of chromosomal DNA with SmaI, the XhoI linker can either be converted back to a wild-type configuration (1.4 kb; arrow) or remain in the mutated state (6 kb) if pilE/pilS recombination extends beyond the pilE locus. (A) Analysis of MS11 pilE variant 6 (9) and its recB derivatives. (B) Analysis of FA1090 and its recB derivatives. recB sup are intragenic growth suppressors derived from the cognate recB population. Each blot was probed with the pilE-specific probe 245. In each panel, lanes 1, 2, 3, and 4 represent independent mutants.
Effect of inverting the pilE locus on pilE gene variation.
A model that has been proposed for pilE/pilS recombination involves duplicating the pilE locus following a pilE/pilS recombination event, which then leads to the excision of a pilE::pilS fusion on a closed-circular piece of DNA. The closed circle then recombines with pilE, leading to pilE gene variation event (reviewed in references 18 and 26). In this model, in order for the closed circle to be efficiently excised from the chromosome, the duplicated pilE genes need to be in direct orientation. Consequently, if this mechanism actually operates in the gonococcus, inverting pilE should either totally abrogate or severely curtail pilE/pilS recombination. However, if RecBCD operates during pilE gene variation in strain MS11, this should occur irrespective of pilE locus orientation (22). The pilE locus was inverted on the MS11 chromosome (Fig. 4C), and the effect of this inversion on pilE/pilS interactions was assessed by RT-PCR. The blot presented in Fig. 4D shows that an inverted pilE locus recombines with pilS as efficiently as is observed with the pilE locus in direct orientation. The blot also confirms previously published observations (12) that show that large nonhomologous insertions placed immediately upstream of the pilE promoter abrogate pilE/pilS recombination (Fig. 4D, insertions 1 and 2).
DISCUSSION
From the data presented, we conclude that the RecBCD recombination pathway in N. gonorrhoeae strain MS11 plays a pronounced role in recombinational repair and pilE gene variation. This contrasts with previously reported observations with a different gonococcal strain, where the data indicated that the gonococcal RecBCD recombination pathway was noticeably different from the equivalent pathway in E. coli (17). Initially, we examined whether the presence of a proline residue in the RecB nuclease active site may have caused some disruption of RecB functionality, possibly accounting for the previously reported observations. However, we found that the presence or absence of this residue had no effect on enzyme function in the gonococcus. Subsequently, we were able to demonstrate that the MS11 recB gene was fully capable of complementing an E. coli recB21 mutant in a lac recombination assay.
MS11 recB null mutants were severely deficient in the repair of nalidixic acid-induced chromosomal breaks as well as deficient in the repair of alkylated DNA lesions caused by MMS treatment. Accompanying this repair deficiency was a severe growth defect. Therefore, MS11 recB mutants appeared to present a more pronounced phenotype than the corresponding FA1090 recB mutants and, as such, behave very much like their E. coli counterparts. In addition, MS11 recB growth suppressors were readily obtainable, which is commonly seen with E. coli recB mutants. Two types of growth suppressors were identified in the MS11 recB cultures: (i) intragenic suppressors that arose through the deletion of the antibiotic marker plus a small segment of the recB gene and (ii) extragenic suppressors, with two suppressor mutations mapping to genes involved in DNA transformation (pilT and comA). In the latter suppressors, the growth defect was alleviated. However, the repair defect was retained (data not shown). A possible explanation for the observed discrepancy between the two phenotypes is that the differences reflect ATP usage between the various mutants, as uptake of DNA during DNA transformation requires extensive ATP hydrolysis, as does repair of broken chromosomes. Therefore, by eliminating DNA transformation, higher cellular-ATP levels now become available, leading to an enhanced growth phenotype.
In the three gonococcal strains that were assessed in this study, recB mutants for each strain showed diminished repair/recombination capacities compared to the wild type. However, the magnitudes of the defect varied considerably among strains and appeared to reflect the innate repair capacity of the wild type rec+ strain, with strain MS11 apparently presenting a full repair capacity. This was evident from the relatively high sensitivities to MMS of P9 rec+ and FA1090 rec+. Similarly, FA1090 rec+ was relatively sensitive to nalidixic acid whereas strain P9 rec+ was unable to tolerate these nalidixic acid levels. Consequently, when these innate differences in recombination/repair are taken into account, MS11 recB mutants present a more robust phenotype than that observed for the other two strains, which begs the question as to whether wild-type strain FA1090 and wild-type strain P9 are fully functional with respect to recombination/repair. As the functionality of the FA1090 RecBCD protein was not addressed in the previously reported study (17), it is not known whether the observed recombination/repair deficiency of the FA1090 rec+ strain is due to a defect in the RecBCD enzyme. However, if the FA1090 RecBCD pathway is defective in wild-type bacteria, this may explain why that particular strain relies on an alternative “RecF-like” pathway for recombination/repair (11). Interestingly, in meningococcal clinical isolates, epidemic Neisseria meningitidis clones that present recB mutant phenotypes due to small deletions located at the 5′ end of the gene have recently been identified (21). Furthermore, these strains were shown to undergo increases in pilE gene variation. Indeed, considerable variation in DNA repair capacity apparently is manifest by clinical meningococcal strains (4, 5). Therefore, the pathogenic Neisseria strains may simply utilize whatever active recombination proteins are at their disposal.
The major conclusion of this study is the finding that the RecBCD recombination pathway mediates PilE antigenic variation in N. gonorrhoeae strain MS11 in contrast to previously reported observations that proposed that a “RecF-like” pathway mediated pilE gene variation (11, 17). From the two assays presented in this paper (as well as a colony morphological analysis not presented), MS11 recB mutants appear to be constrained with respect to pilE/pilS recombination. In the more sensitive RT-PCR assay, the apparent low-level signal obtained from the recB RNA sample may reflect suppressor activation of an alternative pathway, such as the “RecF-like” pathway. This then leads to the intriguing possibility that if strain FA1090 is deficient in recombination/repair and does not show decreased levels of PilE antigenic variation in recB mutants, then perhaps the identification of a “RecF-like” pathway is due to suppressor activation countering this recombination/repair defect. The blot presented in Fig. 5B supports this contention.
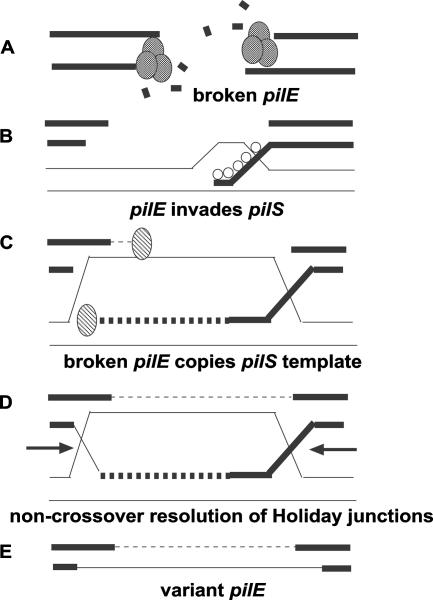
MS11 recB mutants show reduced levels of pilE/pilS recombination, augmenting previous studies that have implicated the RecBCD recombination pathway in pilE gene variation. Previously published work has shown that MS11 recD mutants presented a hyperrecombination phenotype when pilE gene variation was assessed by a colony morphological assay in conjunction with DNA sequencing (3). In addition, the RecBCD pathway was shown to influence pilE/pilS templated deletion formation across the pilE locus, especially l-pilin-to-pilus+ transitions (9). Given these considerations, coupled with the observation that pilE/pilS recombination can proceed with an inverted pilE locus, a parsimonious double-chain-break repair model that utilizes the RecBCD enzyme can now be proposed for pilE gene variation occurring by a gene conversion mechanism in N. gonorrhoeae strain MS11 (Fig. 6). In this model, a break occurs at the pilE locus, each end of the broken chromosome is resected using the RecBCD enzyme, the single-chain DNAs then search for homology (i.e., pilS) using RecA protein, pilE/pilS pairing occurs, and pilS DNA then serves as a template to repair the break at pilE, which then leads to pilE gene variation and an unchanged pilS gene copy. Therefore, we propose that in recombination/repair of proficient gonococci, pilE gene variation proceeds via a double-chain-break repair model that utilizes the RecBCD enzyme, whereas in those strains where the RecBCD pathway is compromised, a half-crossing-over model for pilE gene variation remains valid (13), as inverting the pilE locus should not prevent the formation of the initial recombination intermediate.
FIG. 6.
Double-chain-break repair model for pilE gene variation. The model is based on the yeast mating-type switching model in yeast (20). (A) The pilE locus is broken, and the ends are acted upon by the RecBCD nuclease yielding 3′ overhangs. (B) The single chain overhangs bind RecA protein, which then seeks homology and invades a homologous pilS gene copy. (C) Following invasion of the pilS gene copy, the 3′ end is extended by DNA polymerase, using the pilS gene copy as a template. (D) Following extension, the Holiday junctions are resolved (arrows). (E) Creation of a variant pilE gene consisting of novel pilS sequence that was obtained during the DNA polymerase extension.
The proposed model is very similar to the mating-type switching model for Saccharomyces cerevisiae (20). The model proposes that pilE is actively broken, which may simply be through the action of cellular nucleases. In the RT-PCR assay, pilE promoter insertions (Fig. 4D) that disrupted pilE/pilS recombination were identified (12). The insertions that disrupted pilE/pilS recombination lie adjacent to the integration host factor binding site which is located in the pilE promoter (10). Consequently, as integration host factor bends the gonococcal promoter (10) and that DNA bending has been implicated in chromosome breaks (19), the effect of the promoter insertions on pilE/pilS recombination may be due to prevention of pilE locus bending, causing pilE to remain intact. In the proposed model for pilE/pilS recombination, no role for potential Chi site activation is stated, as it is unknown whether Chi activates the gonococcal RecBCD enzyme. A BLAST search of the gonococcal chromosome using the Chi sequence (5′-GCTGGTGG-3′) reveals approximately 140 Chi sites occurring on average in 9-kb intervals (the predicted number of Chi sites randomly occurring in the gonococcal genome is approximately 40). This is a smaller number than that observed in E. coli, where Chi is found approximately every 3 kb. However, because the gonococcal recB, recC, and recD genes are unlinked on the chromosome, assessment of potential Chi stimulation is difficult to demonstrate experimentally.
Acknowledgments
We thank Frank Stahl for his comments on the manuscript and Ichizo Kobayashi and Frank Stahl for fruitful discussions of the data.
Funding was provided through the Biology Department and the Plant Molecular Biology Center at Northern Illinois University.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Bergstrom, S., K. Robbins, J. M. Koomey, and J. Swanson. 1986. Piliation control mechanisms in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 83:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat, K., C. P. Gibbs, O. Barrera, S. G. Morrison, F. Jahnig, A. Stern, E. M. Kupsch, T. F. Meyer, and J. Swanson. 1991. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5:1889-1901. [DOI] [PubMed] [Google Scholar]
- 3.Chaussee, M. S., J. Wilson, and S. A. Hill. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145:389-400. [DOI] [PubMed] [Google Scholar]
- 4.Davidsen, T., E. K. Amundsen, E. A. Rodland, and T. Tonjum. 2007. DNA repair profiles of disease-associated isolates of Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 49:243-251. [DOI] [PubMed] [Google Scholar]
- 5.Davidsen, T., H. K. Tuven, M. Bjørås, E. A. Rødland, and T. Tønjum. 2007. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J. Bacteriol. 189:5728-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas, R., and T. F. Meyer. 1986. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44:107-115. [DOI] [PubMed] [Google Scholar]
- 8.Hill, S. A. 2000. Neisseria gonorrhoeae recJ mutants show defects in recombinational-repair of alkylated bases and UV-induced pyrimdine dimers. Mol. Gen. Genet. 264:268-275. [DOI] [PubMed] [Google Scholar]
- 9.Hill, S. A., and C. C. R. Grant. 2002. Recombination-error and deletion formation in Neisseria gonorrhoeae: a role for RecJ in pilEL deletions. Mol. Genet. Genomics 266:962-972. [DOI] [PubMed] [Google Scholar]
- 10.Hill, S. A., D. S. Samuels, J. H. Carlson, J. Wilson, D. Hogan, L. Lubke, and R. J. Belland. 1997. Integration host factor is a transcription cofactor of pilE in Neisseria gonorrhoeae. Mol. Microbiol. 23:649-656. [DOI] [PubMed] [Google Scholar]
- 11.Howell-Adams, B., and H. S. Seifert. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146-1158. [DOI] [PubMed] [Google Scholar]
- 12.Kline, K. A., A. K. Criss, A. Wallace, and H. S. Seifert. 2007. Transposon mutagenesis identifies sites upstream of the Neisseria gonorrhoeae pilE gene that modulate pilin antigenic variation. J. Bacteriol. 189:3462-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kline, K. A., E. V. Sechman, E. P. Skaar, and H. S. Seifert. 2003. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R's of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 50:3-13. [DOI] [PubMed] [Google Scholar]
- 14.Kline, K. A., and H. S. Seifert. 2005. Mutation of the priA gene of Neisseria gonorrhoeae affects DNA transformation and DNA repair. J. Bacteriol. 187:5347-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDaniel, L. S., L. H. Rogers, and W. E. Hill. 1978. Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J. Bacteriol. 134:1195-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, T. F., and S. A. Hill. 2003. Genetic variation in the pathogenic Neisseria spp., p. 142-164. In A. Craig and A. Scherf (ed.), Antigenic variation. Academic Press, London, United Kingdom.
- 19.Moitoso de Vargas, L., S. Kim, and A. Landy. 1989. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science 244:1457-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvatore, P., C. Bucci, C. Pagliarulo, M. Tredici, R. Colicchio, G. Cantalupo, M. Bardaro, L. Del Giudice, D. R. Massardo, A. Lavitola, C. B. Bruni, and P. Alifano. 2002. Phenotypes of a naturally defective recB allele in Neisseria meningitidis clinical isolates. Infect. Immun. 70:4185-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segall, A. M., and J. R. Roth. 1994. Approaches to half-tetrad analysis in bacteria: recombination between repeated, inverse-order chromosomal sequences. Genetics 136:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skaar, E. P., M. P. Lazio, and H. S. Seifert. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J. Bacteriol. 184:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson, J. 1982. Colony opacity and protein II compositions of gonococci. Infect. Immun. 37:359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson, J., S. Berstrom, K. Robbins, O. Barrera, D. Corwin, and J. M. Koomey. 1986. Gene conversion involving the pilin structural gene correlates with pilus+ to pilus− changes in Neisseria gonorrhoeae. Cell 47:267-276. [DOI] [PubMed] [Google Scholar]
- 26.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wainwright, L. A., K. H. Pritchard, and H. S. Seifert. 1994. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol. Microbiol. 13:75-87. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J., R. Chen, and D. Julin. 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275:507-513. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Q. Y., D. DeRyckere, P. Lauer, and M. Koomey. 1992. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc. Natl. Acad. Sci. USA 89:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zieg, J., and S. R. Kushner. 1977. Analysis of genetic recombination between two partially deleted lactose operons of Escherichia coli K-12. J. Bacteriol. 131:123-132.24. [DOI] [PMC free article] [PubMed] [Google Scholar]