Abstract
Campylobacter jejuni is the leading cause of bacterial gastroenteritis in the developed world. Despite its prevalence, relatively little is known about C. jejuni's precise pathogenesis mechanisms, particularly in comparison to other well-studied enteric organisms such as Escherichia coli and Salmonella spp. Altered expression of phosphate genes in a C. jejuni stringent response mutant, together with known correlations between the stringent response, polyphosphate (poly-P), and virulence in other bacteria, led us to investigate the role of poly-P in C. jejuni stress survival and pathogenesis. All sequenced C. jejuni strains harbor a conserved putative polyphosphate kinase 1 predicted to be principally responsible for poly-P synthesis. We generated a targeted ppk1 deletion mutant (Δppk1) in C. jejuni strain 81-176 and found that Δppk1, as well as the ΔspoT stringent response mutant, exhibited low levels of poly-P at all growth stages. In contrast, wild-type C. jejuni poly-P levels increased significantly as the bacteria transitioned from log to stationary phase. Phenotypic analyses revealed that the Δppk1 mutant was defective for survival during osmotic shock and low-nutrient stress. However, certain phenotypes associated with ppk1 deletion in other bacteria (i.e., motility and oxidative stress) were unaffected in the C. jejuni Δppk1 mutant, which also displayed an unexpected increase in biofilm formation. The C. jejuni Δppk1 mutant was also defective for the virulence-associated phenotype of intraepithelial cell survival in a tissue culture infection model and exhibited a striking, dose-dependent chick colonization defect. These results indicate that poly-P utilization and accumulation contribute significantly to C. jejuni pathogenesis and affect its ability to adapt to specific stresses and stringencies. Furthermore, our study demonstrates that poly-P likely plays both similar and unique roles in C. jejuni compared to its roles in other bacteria and that poly-P metabolism is linked to stringent response mechanisms in C. jejuni.
Campylobacter jejuni is a gram-negative, microaerophilic bacterium belonging to the family Campylobacteraceae (73) and is now considered the leading cause of human gastroenteritis in the developed world (4, 69). C. jejuni lives harmlessly in the intestinal microflora of most mammals and birds, resulting in a commensal relationship (11). However, upon infecting a human host, C. jejuni invades the intestinal mucosa, interrupts intestinal integrity, and causes profuse watery and/or bloody diarrhea (13). Campylobacteriosis has been correlated with other medical sequelae, such as reactive arthritis, hemolytic-uremic syndrome, and inflammatory bowel disease; the most notable complication of infection is Guillain-Barré syndrome, an acute neuromuscular paralysis (34). C. jejuni is typically transmitted to humans via contact with infected animals or through undercooked food, unpasteurized milk, or contaminated water (3, 4, 25).
Despite the prevalence of C. jejuni infection, the molecular mechanisms C. jejuni uses to cause human disease, as well as to adapt to or survive stresses encountered during both in vivo colonization and ex vivo transmission, are not well understood, particularly in comparison to mechanisms used by other well-studied pathogens such as Escherichia coli, Salmonella spp., and Helicobacter pylori. For instance, although C. jejuni is not considered an obligate intracellular pathogen, its ability to invade and survive in intestinal epithelial cells correlates well with virulence (8-10, 16, 22, 41). Despite this, specific bacterial factors contributing to C. jejuni invasion and intracellular survival are poorly characterized. It is likewise perplexing how C. jejuni survives a multitude of environmental stresses given its fastidious laboratory growth requirements. Our current limited understanding of C. jejuni is largely due to its difficult culture conditions, significant interstrain virulence variability, and a historical intractability of C. jejuni to genetic manipulation. Moreover, the recent publication of three complete C. jejuni genome sequences revealed that C. jejuni lacks many virulence characteristics and factors found in other bacterial pathogens, such as pathogenicity islands and certain stress response factors such as RpoS (23, 31, 55). Nonetheless, the high worldwide prevalence of C. jejuni commensal and human infection suggests the presence of factors that allow this fastidious pathogen to navigate a multitude of environments during transmission, colonization, and virulence.
Several lines of evidence led us to hypothesize that polyphosphate (poly-P), unstudied in C. jejuni prior to the work described here, would prove to be a key factor impacting multiple aspects of the C. jejuni pathogenesis cycle. Poly-P is ubiquitous in nature and consists of phosphate residues linked by high-energy phosphoanhydride bonds such as those found in ATP. Analysis of published genome sequences revealed that C. jejuni harbors homologues of several genes predicted to participate in poly-P metabolism, including ppk1, ppk2, and ppx (23, 31, 55). The ppk1 gene encodes a polyphosphate kinase, PPK1, that reversibly forms poly-P from the terminal γ-phosphate of ATP and is responsible for either all or the majority of poly-P formation in several bacteria, including E. coli, H. pylori, and Pseudomonas aeruginosa (1, 43, 70). It was recently shown that some bacteria, including P. aeruginosa and C. jejuni, also harbor a reversible PPK2 enzyme that preferentially synthesizes GTP from poly-P (35, 78). An exopolyphosphatase, encoded by the ppx gene, has been shown to be responsible for the conversion of poly-P into free phosphate residues in E. coli (2).
Poly-P has a multiplicity of functions within bacterial cells. Long chains of poly-P can serve as a phosphate reservoir, a cation chelator (30), a membrane channel for DNA entry (64), a capsular component (71, 72), a pH buffer (36, 57, 58), and likely an ATP substitute (12, 43). Furthermore, in E. coli, poly-P inhibits RNA degradation, promotes translation fidelity, and activates the Lon protease complex that degrades specific ribosomal proteins to meet nutritional requirements during starvation (45).
Important roles for poly-P formation in bacterial pathogenesis have been established in such pathogens as E. coli, P. aeruginosa, Vibrio cholerae, Klebsiella aerogenes, S. enterica serovar Typhimurium, H. pylori, and Shigella flexneri. In these organisms, poly-P formation was shown to be critical for attributes such as motility, quorum sensing, biofilm formation, resistance to oxidative, osmotic, heat, and alkaline stress, and stationary-phase survival (36, 40, 52, 58, 59, 62, 63, 70). Although the importance of poly-P in various bacterial phenotypes has been reported, the precise molecular mechanisms by which poly-P enacts specific functions, as well as the primary and secondary effects of poly-P accumulation, are still not understood in even the best-characterized bacterial species.
Our interest in poly-P was further manifested by its connection to the stringent response (SR). In the SR, RelA and/or SpoT enzymes activated during stress synthesize guanosine tetraphosphate (ppGpp), an alarmone that binds RNA polymerase to alter transcription and allow the organism to cope with the stress condition. We recently showed that the C. jejuni SR is important for several pathogenesis attributes, including survival during the transmission-related stresses of nutrient limitation, aerobiosis, and rifampin exposure (29). The SR was also implicated in virulence: C. jejuni's spoT gene encoding a bifunctional ppGpp synthetase/hydrolase was upregulated in the presence of intestinal epithelial cells, and deletion of spoT resulted in significant epithelial cell invasion and intracellular survival defects (29). These responses occur in the absence of RpoS, which mediates many SR-related phenotypes in other bacteria. Poly-P has been linked to SR mechanisms in E. coli, where SR mutants with diminished ppGpp levels also harbor lower levels of poly-P (44, 60). Furthermore, our microarray analysis of the C. jejuni ΔspoT mutant revealed increased expression of genes regulating inorganic phosphate uptake during stationary phase concurrent with upregulation of genes involved in heat shock (29), suggesting that poly-P may play a role in C. jejuni stress survival.
To test the hypothesis that poly-P plays an important role in C. jejuni stress survival and pathogenesis, the ppk1 gene was disrupted in the highly invasive C. jejuni strain 81-176. Analyses of the Δppk1 mutant strain, which was defective for poly-P accumulation, identified roles for poly-P in numerous aspects of pathogenesis, including the transmission-related stresses of osmotic shock and low-nutrient-stress survival. PPK1 is also now only the third C. jejuni factor found to be involved in prolonged survival of C. jejuni inside human epithelial cells, an important but underexplored aspect of C. jejuni virulence. Our studies also indicate that poly-P metabolism in C. jejuni likely intersects with SR mechanisms, as seen in E. coli. However, contrary to observations in other Δppk1 mutant bacteria, which are generally defective for forming biofilms (18, 63, 67), the C. jejuni Δppk1 mutant exhibited an unexpected increase in biofilm formation. The C. jejuni Δppk1 mutant also displayed a striking, dose-dependent chick colonization defect. These findings identify poly-P as an important new factor involved in C. jejuni transmission, colonization, and pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All studies were performed with the highly invasive Campylobacter jejuni strain 81-176 originally isolated from a diarrheic patient (42). The ΔspoT mutant of this strain was previously described (29). C. jejuni was routinely cultured on Mueller-Hinton (MH; Oxoid, Ltd., Hampshire, England) agar plates, and growth/survival curves were determined in MH broth unless otherwise stated (Oxoid). C. jejuni was always grown in medium supplemented with 10 μg of vancomycin/ml and 5 μg of trimethoprim/ml; chloramphenicol was added at 20 μg/ml when required. All bacteria were enumerated on MH agar plates by serial 10-fold dilutions unless otherwise stated. Plates were incubated in a tri-gas incubator (Heraeus) in 6% O2 and 12% CO2 at 37°C. Broth culture growth and survival curves were determined by using the Oxoid CampyGen system to produce a microaerobic atmosphere of 6% O2 and 12% CO2, and cells were shaken at 200 rpm at 37°C. E. coli strain DH5α was grown on Luria-Bertani agar or broth, with the addition of 30 μg of chloramphenicol/ml as needed, at 37°C under normal atmospheric conditions. Karmali agar was used for growth of C. jejuni for the chick colonization studies.
Construction of C. jejuni 81-176 ppk1 targeted deletion mutant and complemented strains.
A polyphosphate kinase (ppk1) gene with 30.1% identity and 50.7% similarity to the ppk1 gene in E. coli was identified in the C. jejuni 81-176, 11168, and RM1221 genomes using the BLAST features of CampyDB (http://campy.bham.ac.uk [17]). The ppk1 gene was PCR amplified from C. jejuni chromosomal DNA prepared by using the QIAGEN DNeasy kit (QIAGEN, Inc., Valencia, CA), with the primers PPK1-2 Fp (GCAAATATTTACACCAAGAAAAAGAAC) and PPK1-2 Rp (ATCTGCACTCGATATAAAATAATTTGG), yielding a 1,550-bp fragment. The amplified product was cloned into the pGEM-T vector (Promega, Nepean, CA), which is a suicide plasmid in C. jejuni. Two EcoRI sites located within the ppk1 gene were used to remove 1,048 bp of coding sequence. A chloramphenicol acetyltransferase cassette (cat) was excised from pRY109 (77) with EcoRI and ligated into the EcoRI-digested pGEM-ppk1 vector to create the ppk1 knockout construct. Insertion of the cat cassette was confirmed by restriction digestion and sequencing (Nucleic Acid Protein Service Unit, Vancouver, CA). This construct was used to naturally transform C. jejuni 81-176 (74). Recombinants were recovered on MH agar plates supplemented with chloramphenicol. Insertional inactivation of the ppk1 gene via cat cassette insertion was verified by PCR using PPK1-1 Fp (TGCCCTTAGCGTTATAAAAAGTATAAA) and PPK1-1 Rp (AATTTTCGGTCATTTTTGATAGTGTAG) primers that are external to the ppk1 gene and the region originally amplified. Southern analysis also verified a single chromosomal insertion of the cat cassette into the ppk1 gene.
Generation of a reconstituted wild-type strain of C. jejuni, designated ppk1*, was achieved via natural transformation of the Δppk1 mutant with the ppk1 gene in pGEM-T. The naturally transformed Δppk1 mutant was spread onto MH plates, harvested after 2 h, and serial 10-fold dilutions plated on MH plates supplemented with 0.17 M NaCl and incubated for 48 h at 37°C in a tri-gas incubator. Individual colonies were selected and purified on MH agar containing 10 μg of vancomycin/ml and 5 μg of trimethoprim/ml. Colonies were tested for sensitivity to chloramphenicol by plating on MH agar containing 20 μg of chloramphenicol/ml. Colonies representing putative reconstituted wild-type strains were confirmed by using PCR with the PPK1-1 primer set and sequence analysis of 18 bona fide recombinants, represented by ppk1*.
Extraction of poly-P with Glassmilk.
Extraction of poly-P from C. jejuni cells and binding to Glassmilk (qBiogene) was performed essentially as described by Ault-Riche et al. (6). C. jejuni cultures were grown in MH broth to mid-log phase (hereafter, mid-log phase means an optical density at 600 nm [OD600] of ∼0.2 to 0.5) and then diluted to an OD600 of 0.05 to initiate the time course experiment. Cells were harvested after 2, 10, and 24 h by pelleting 1 ml in a table-top centrifuge at 6,000 rpm for 10 min. All pellets were processed directly. To each pellet was added 500 μl of guanidine isothiocyanate (GITC) lysis buffer (4 M GITC, 500 mM Tris-HCl [pH 7.0]) prewarmed at 95°C. Tubes were vortex mixed, incubated for 5 min in a 95°C heating block, and sonicated briefly; a 10-μl sample was removed for total protein estimation. To each poly-P assay tube, 30 μl of 10% sodium dodecyl sulfate, 500 μl of 95% ethanol, and 5 μl of Glassmilk were added. Tubes were vortex mixed and centrifuged briefly to pellet glass, which was then resuspended in 500 μl of ice-cold new wash buffer (5 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM EDTA, 50% ethanol) by vortexing and repelleted; washing was repeated two additional times. The washed pellet was resuspended in 50 μl of 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 20 μg each of DNase and RNase per ml, followed by incubation at 37°C for 10 min. The pellet was washed with 150 μl of 4 M GITC lysis buffer and 150 μl of 95% ethanol and then twice in new wash buffer. Poly-P was eluted from the Glassmilk pellet with 50 μl of 50 mM Tris-HCl (pH 8.0) at 95°C for 2 min, followed by two additional elutions.
Measurement of poly-P levels by a TBO assay.
A standard curve was determined by the addition of 100 μl of a known concentration of phosphorus standard (Sigma-Aldrich, Oakville, CA) sample that had been serially diluted (1:10) to 900 μl (64, 32, 16, 8, and 4 nmol of poly-P) of a toluidine blue O (TBO; Sigma-Aldrich, Oakville, CA) dye solution consisting of 6 mg of TBO/liter in 40 mM acetic acid. After the addition to TBO, all samples (standard curve and C. jejuni time course samples) were incubated at room temperature for 15 min. Absorbance at 630-nm and 530-nm levels were assessed spectrophotometrically and used to generate an A530/A630 ratio. Poly-P binding to TBO results in a shift in TBO absorbance from 630 to 530 nm; the 530-nm/630-nm ratio thus reflects the amount of poly-P in a given sample. The levels of poly-P from each experimental sample were then determined by direct comparison to the standard curve. Poly-P levels were expressed in nanomoles of poly-P and per milligram of total cellular protein, as measured via Bradford analysis (Pierce Scientific).
Static biofilm formation.
Biofilm formation was assayed as described previously (54). Briefly, cells were grown microaerobically in MH broth at 37°C to mid-log phase overnight and diluted to an OD600 of 0.20 and 0.02. Aliquots (100 μl) were added to 96-well polyvinyl chloride plates, followed by incubation at 37°C for 48 h. Static biofilm formation as measured by surface-associated bacteria was assessed by adding 25 μl of a 1% crystal violet solution in ethanol, followed by incubation for 15 min at room temperature, and three washes with distilled H2O (54). To quantify biofilm formation, 150 μl of 80% dimethyl sulfoxide (DMSO) was added to each polyvinyl chloride (PVC) microtiter well; then each well was covered and incubated 24 h at room temperature. A 1:10 dilution in 80% DMSO of each well was measured at OD570 to indirectly quantify the amount of biofilm stained by crystal violet.
Nutrient downshift survival.
C. jejuni were grown microaerobically at 37°C in MH broth to mid-log phase overnight, collected by centrifugation at 6,000 rpm for 10 min, and washed twice with minimal essential medium (MEM) or MOPS-MGS with Earle's salts and l-glutamine (Difco) or MOPS-MGS (50). Bacteria were resuspended in MEM and diluted to an OD600 of 0.05. Cultures were placed under microaerobic conditions at 37°C with shaking at 200 rpm. CFU were measured over time by plating on MH agar plates.
Osmotic stress survival.
C. jejuni strains were grown to mid-log phase, serially diluted (1:10), and spotted onto MH agar plates containing 0.17 M NaCl to assess single-colony growth. Survival during osmotic stress was also tested in shaking liquid cultures by growing bacterial strains to mid-log phase in MH and diluting them to an OD600 of 0.05 in MH broth with or without 0.25 M NaCl. CFU were assayed by serial dilution and plating on MH agar.
INT407 cell infection assay for invasion and intracellular survival.
INT407 cells were seeded to semiconfluence (∼1 × 105 cells/well) and confluence (∼5.5 × 105 cells/well) in 24-well plates approximately 16 h prior to infection assays. C. jejuni strains were inoculated at an OD600 of 0.001 into MH broth culture and grown overnight to mid-log phase. Bacteria were pelleted at 6,000 rpm for 10 min, washed twice with MEM, and diluted to an OD600 of 0.002 in MEM. Bacterial suspensions in MEM (1 ml) were used to infect tissue culture cells at multiplicities of infection (MOIs) of ∼100 and ∼20 (∼107 bacteria/ml). All experiments were performed in triplicate. After 3 h of infection in a 5.0% CO2 incubator, bacteria in MEM were removed from the wells, and cells were washed twice with MEM. To all wells, 1 ml of MEM containing 150 μg of gentamicin/ml was added to kill the remaining extracellular bacteria. After 2 h, wells were washed twice with MEM. To assay invasion, 1 ml of distilled H2O was added to some of the wells, and INT407 cells were disrupted by lysis with a 27G syringe. Invaded bacteria were assayed by serial dilution and plating on MH agar. Samples for assaying intracellular survival were covered with MEM with 3% fetal bovine serum and the addition of 10 μg of gentamicin/ml to halt bacterial growth if cell lysis occurred. After 23 to 24 h of incubation, intracellular survival was assayed as described for invasion samples.
Chick colonization assays.
Colonization of 1-day-old chicks was performed essentially as described previously (14). Briefly, broiler chicks were obtained from a local hatchery in Saskatchewan on the day of hatch. Five chicks were euthanized, and their cecal contents cultured for Campylobacter. The remaining birds were randomly assigned into groups of 10 birds and provided with feed and water ad libitum. Birds were cared for in accordance to guidelines of the Canadian Council for Animal Care. Birds were orally challenged with the indicated strain (wild type or Δppk1 mutant) and dose (1.5 × 105, 1.5 × 106, and 1.5 × 107 CFU) of C. jejuni in 0.5 ml of MH broth. Inocula for challenge experiments were produced by harvesting cells grown for 18 h under microaerophilic conditions at 37°C into cold MH broth, diluting them to the indicated concentration in MH broth, and maintaining them on ice until immediately before use. Viable cell counts were determined by plating serial dilutions onto MH agar (Becton Dickinson). Birds were maintained for 7 days after challenge and then euthanized by cervical dislocation. Ceca were aseptically collected for qualitative and quantitative assessment of colonization. Colonization of the birds was measured by culturing the cecal contents, after appropriate dilutions were made in MH broth, on Karmali agar (Bacto) under microaerophilic conditions at 42°C.
RESULTS
The Δppk1 and ΔspoT mutants are defective for poly-P accumulation.
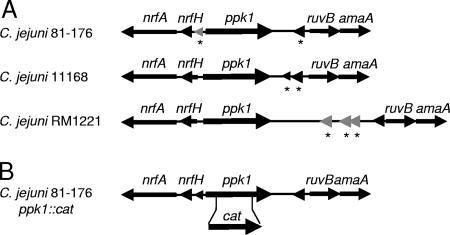
All three published C. jejuni strain sequences contain a highly conserved ppk1 gene (Fig. 1A), encoding a putative PPK1 with significant homology to PPK1 enzymes in other bacteria (23, 31, 55). The predicted C. jejuni amino acid sequences exhibit 45.4, 36.2, and 30.1% identity to PPK1 in H. pylori, P. aeruginosa, and E. coli, respectively. Moreover, two highly conserved histidine residues, required for PPK1 activity in E. coli, are also present at amino acid positions 427 and 580 in the C. jejuni 81-176 strain. In all three strains, ppk1 appears to be a single-gene operon, and microarray data from multiple C. jejuni gene expression experiments with strains 81-176 and 11168 indicate that ppk1 is transcribed independently of neighboring genes (23, 28, 29, 31, 48, 55). To explore the role of poly-P in C. jejuni 81-176, ∼50% of the ppk1 gene was deleted, including the codons for the conserved His residues, and replaced with a chloramphenicol acetyltransferase (cat) cassette conferring chloramphenicol resistance (Fig. 1B). Sequencing and Southern analyses demonstrated a single cat insert in the ppk1 gene (data not shown). To ensure that observed phenotypes were attributed to ppk1, a reconstituted wild-type strain was generated by homologous recombination of a wild-type copy of ppk1 into the Δppk1 strain ppk1::cat locus as described in Materials and Methods. The resulting reconstituted wild-type strain was designated the ppk1* strain.
FIG. 1.
C. jejuni ppk1 and generation of a single insert Δppk1 disruption strain. (A) Genomic location of the ppk1 gene is conserved among the sequenced and annotated strains C. jejuni 81-176, C. jejuni 11168 and C. jejuni RM1221. MUMer alignment was performed by using the CampyDB gene viewer (17). Hypothetical proteins are represented by an asterisk (*), and genes with no predicted orthologues are colored gray. (B) The approximate site of the cat-marked insertion-deletion mutation generated in C. jejuni 81-176 ppk1 is shown. The resultant mutant strain was designated Δppk1.
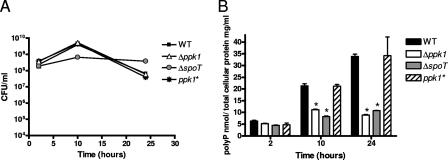
Growth rates (Fig. 2A) and poly-P levels (Fig. 2B) for wild-type C. jejuni 81-176, the Δppk1 mutant, the ΔspoT mutant, and the ppk1* strain were assessed in shaking MH broth cultures harvested at various growth stages. Poly-P was extracted after cell lysis by binding to Glassmilk and measured by using the metachromatic dye TBO as described in Materials and Methods. Poly-P levels in C. jejuni samples are expressed as nanomoles of poly-P and normalized to milligrams of total cellular protein.
FIG. 2.
Poly-P accumulates in wild-type and ppk1* strains at later growth stages but remains at low levels in Δppk1 and ΔspoT strains. Wild-type (WT) C. jejuni 81-176, Δppk1 mutant, the complemented strain ppk1*, and the ΔspoT mutant were grown microaerobically in shaking broth culture to early log phase and diluted to an OD600 of 0.05. Cultures (1 ml) were harvested at 2, 10, and 24 h and assayed for CFU/ml (A) and poly-P levels versus total cellular protein (B). Triplicate samples were harvested and assayed for each time point. Statistical significance (P < 0.05) is represented by an asterisk.
We found that wild-type, Δppk1, and ppk1* strains exhibited identical growth profiles under normal laboratory culture conditions by both OD600 (not shown) and CFU/ml (Fig. 2A) analyses, whereas the ΔspoT mutant, as previously reported (29), exhibited some growth differences from the wild type. In wild-type C. jejuni, the poly-P levels were significantly higher than those of the Δppk1 and ΔspoT mutants, with levels peaking in stationary phase at 33.8 nmol of poly-P/mg of total protein (Fig. 2B). The Δppk1 mutant exhibited significantly lower levels of poly-P at both 10 and 24 h (P < 0.05), with the largest difference (∼3.8-fold) observed at the 24-h stationary-phase time point. The ΔspoT mutant exhibited poly-P levels that were also significantly lower than those of the wild type at both 10 and 24 h (P < 0.05), with the largest difference (∼3.2-fold) again observed in the stationary phase. The complemented ppk1* strain displayed levels of poly-P similar to that of the wild type at all time points assayed. Together, these data confirm that the ppk1 gene is important for poly-P synthesis in C. jejuni and demonstrate a connection between the SR and poly-P metabolism.
Poly-P is critical for C. jejuni survival during nutritional downshift.
Initial tests of the C. jejuni Δppk1 mutant based on known Δppk1 mutant defects in other bacteria (i.e., motility and oxidative shock survival) revealed that the C. jejuni Δppk1 mutant was indistinguishable from the wild type for a number of predicted phenotypes and stress resistance characteristics (see Table S1 in the supplemental material).
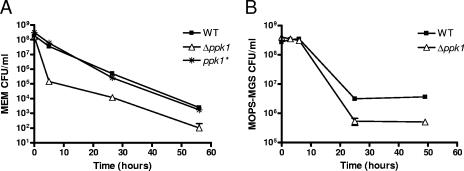
In contrast, poly-P was found to be important for the ability of C. jejuni to survive low-nutrient stress. To test this, wild-type, Δppk1, and ppk1* strains were shifted from rich MH broth to nutrient-poor MEM and assayed for CFU/ml over a 55-h time course (Fig. 3A). The Δppk1 mutant exhibited survival defects during low-nutrient stress compared to the wild type at all of the time points assayed. The most pronounced effect of nutrient downshift was seen after 5 h, at which time the culturability of the mutant dropped >250-fold relative to the wild-type and ppk1* strains. Survivability was also assessed in another limited-nutrient medium, MOPS-MGS buffered medium (50) without phosphate (Fig. 3B). As expected, the Δppk1 mutant showed a considerable defect in survivability at later time points assayed; however, the differences were not as pronounced as those in MEM. These data demonstrate that the Δppk1 mutant exhibits nutrient downshift tolerance defects in two different minimal media.
FIG. 3.
Poly-P is important for C. jejuni survival during nutritional downshift. Wild type (WT), Δppk1, and ppk1* strains were grown in MH broth to mid-log phase. Cells were subjected to nutritional downshift by centrifugation, followed by resuspension to an OD600 of 0.05 in minimum essential medium (A) or MOPS-MGS buffered medium (B).
Poly-P accumulation is required for C. jejuni to grow and survive during osmotic shock.
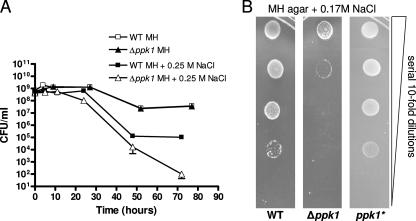
To test survival under osmotic stress, strains were grown to mid-log phase and shifted to MH broth with or without NaCl added to 0.25 M, and the CFU/ml were enumerated at the time points indicated (Fig. 4A). During osmotic shock, both the wild-type and the Δppk1 mutant strains ceased growth at 5 h. After 24 h, Δppk1 CFU/ml were >10-fold lower than wild-type CFU/ml. By 72 h, the Δppk1 mutant CFU/ml were >1,000-fold lower than the wild-type levels. Wild-type and Δppk1 grew identically in MH broth without the addition of salt (Fig. 4A); levels less than 0.25 M NaCl did not inhibit the survival of wild-type or Δppk1 strains (data not shown).
FIG. 4.
Poly-P is required for C. jejuni osmotic stress survival. (A) C. jejuni 81-176 wild-type and Δppk1 strains were grown to mid-log phase and shifted to MH broth with or without 0.25 M NaCl. Growth and/or survivability was monitored by CFU/ml plate counts. (B) Wild-type, Δppk1, and ppk1* strains were grown to mid-log phase in MH broth and serially diluted (1:10) from 2 × 105 to 2 × 101 CFU/ml and spotted onto 0.17 M NaCl MH agar plates. Growth was assessed after 48 h under microaerobic conditions.
The ability of single bacteria to grow into colonies during continuous osmotic stress was tested by growing wild-type, Δppk1, and ppk1* strains to mid-log phase in MH broth and then serially diluting and spotting the bacteria onto MH plates containing 0.17 M NaCl. Under these conditions, the Δppk1 mutant was significantly defective for growth compared to wild-type and ppk1* strains (Fig. 4B); all strains grew identically on MH agar without added salt (data not shown).
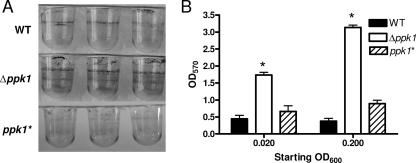
The Δppk1 mutant displays enhanced static biofilm formation.
Biofilm formation contributes to bacterial virulence, colonization, environmental survival, and antibiotic resistance. Poly-P has been shown to be essential for biofilm formation in several bacteria, including P. aeruginosa (63). The role of poly-P in C. jejuni biofilm formation was assayed by growing standing broth cultures of C. jejuni wild-type, Δppk1, and ppk1* strains in PVC plates from starting OD600 levels of 0.02 and 0.20. Biofilm formation at the air-liquid interface and on the sides of the PVC wells were assayed by staining the wells with a crystal violet solution; in this assay, increased crystal violet staining correlates with increased biofilm production (54; M. M. McLennan et al., unpublished data). After 2 days, wild-type C. jejuni formed a faint biofilm at the air-liquid interface for both starting doses, while the Δppk1 mutant exhibited statistically significant (P < 0.05) ∼3-fold and ∼6-fold increases in biofilm formation at the air-liquid interface and on the bottom and sides of the wells compared to wild-type at the lower and higher inoculating doses, respectively (Fig. 5). The Δppk1 mutant also exhibited a dose-dependent biofilm phenotype: the higher inoculating dose of the Δppk1 mutant yielded a statistically significant (P < 0.05) increase in biofilm formation compared to the lower inoculating dose of the Δppk1 mutant, whereas a similar phenomenon was not observed for the wild type (Fig. 5B). Consistent with our other observations, the ppk1* strain exhibited wild-type biofilm formation levels (Fig. 5).
FIG. 5.
The Δppk1 mutant exhibits increased static biofilm formation. Microtiter PVC plates were inoculated with C. jejuni in MH broth at OD600s of 0.02 and 0.20. At 48 h, biofilms were qualitatively observed by crystal violet staining. (A) Representative wells from the OD600 0.02 inoculation are shown for wild-type, Δppk1, and ppk1* strains. (B) Biofilm formation was quantified by the addition of 80% DMSO to each crystal violet-stained well, followed by OD570 absorbance measurements of solubilized crystal violet. Statistical significance (P < 0.05) is represented by an asterisk.
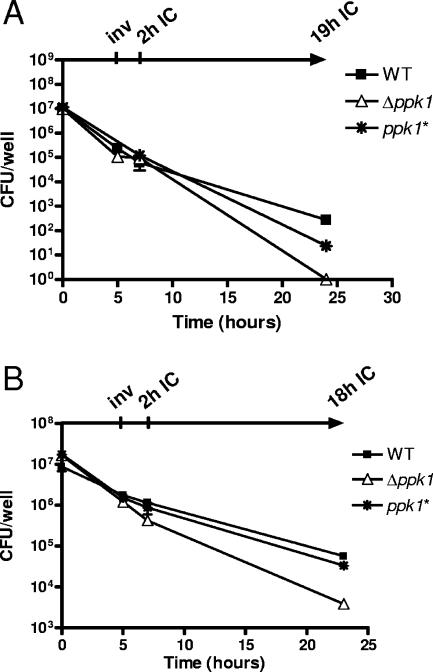
The Δppk1 mutant is defective for prolonged intracellular survival.
To assay the virulence-associated phenotypes of invasion and intracellular survival, wild-type, Δppk1, and ppk1* strains were allowed to infect INT407 cells at an MOI of ∼20 or ∼100 for 3 h. Gentamicin was added to all wells to kill extracellular bacteria, after which cells were assayed for invasion, as well as for both short-term and long-term intracellular (IC) survival, at 2 and 18 h (MOI, ∼20) or at 2 and 19 h (MOI, ∼100) after invasion, respectively. The wild type and the Δppk1 mutant exhibited similar invasion profiles (Fig. 6A, B). However, the Δppk1 mutant was reproducibly defective for longer-term IC survival (18 and 19 h), exhibiting a statistically significant (P < 0.05) >100-fold defect compared to the wild-type IC survival profile. The complemented ppk1* strain survival was similar to that of the wild type. All strains survived equally well in MEM at the 3-h time point, showed identical gentamicin susceptibilities, and were fully resistant to distilled H2O-syringe lysis (data not shown).
FIG. 6.
The Δppk1 mutant is defective for long-term intracellular survival in an epithelial cell model of infection. Wild-type (WT), Δppk1, and ppk1* strains were grown to mid-log phase in MH broth. At the zero time point, semiconfluent or confluent monolayers of INT407 cells were inoculated with bacteria at MOIs of ∼100 (A) and ∼20 (B). After 3 h, the cells were washed, and gentamicin was added at 150 μg/ml to all wells for 2 h to kill extracellular bacteria. Gentamicin was washed from the cells at 5 h, and invaded intracellular bacteria were harvested and plated for enumeration (inv). To all remaining wells, fresh media containing 10 μg of gentamicin/ml and 3% fetal bovine serum were added. After an additional 2 h (2h IC) or 18 or 19 h (18h or 19h IC) of incubation, cells were washed, and surviving intracellular bacteria were harvested. For all time points, experiments were performed in triplicate, and error bars are shown.
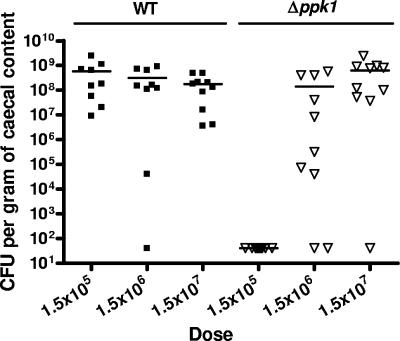
The Δppk1 mutant exhibits a dose-dependent chick colonization defect.
To assess the role of poly-P in commensal colonization, 1-day-old chicks were infected with wild-type and Δppk1 strains at increasing inoculation levels (1.5 × 105, 1.5 × 106, and 1.5 × 107 CFU/infection). Chicks were sacrificed after 7 days, and cecal contents were assayed for viable C. jejuni. Wild-type C. jejuni colonized chicks at a minimum average of 1.79 × 108 CFU/g of cecal content at all inoculating doses, with nearly all infected chicks colonized to high levels (Fig. 7). In contrast, the Δppk1 mutant colonization levels were dependent upon the inoculating dose. Strikingly, at an inoculating dose of 1.5 × 105 CFU, no Δppk1 bacteria were recovered from any chick infected. The intermediate dose of the Δppk1 mutant (1.5 × 106 CFU/0.5 ml) resulted in colonization of 8 of 10 chicks. The highest dose of the Δppk1 mutant (1.5 × 107 CFU/0.5 ml) yielded colonization of all chicks, at levels similar to that for wild-type C. jejuni, with a mean concentration of 6.79 × 108 CFU per g of cecal content.
FIG. 7.
The Δppk1 mutant exhibits a dose-dependent defect for chick cecal colonization. Chicks were challenged orally with C. jejuni 81-176 wild type (▪) and the Δppk1 mutant (▿), using doses of 1.5 × 105, 1.5 × 106, and 1.5 × 107 CFU in 0.5 ml of broth. After 7 days postinfection, chicks were sacrificed, and bacterial colonization of ceca was determined by plating on Karmali agar. Levels of colonization at specific doses are expressed as CFU/g of cecal content. The detection limited was 40 CFU. Each data symbol represents CFU recovered from an individual chick. The geometric mean of bacterial concentration recovered is represented by a bar for each dosage.
DISCUSSION
The poly-P molecule has been hypothesized as playing multiple diverse roles in the bacterial cell, including acting as a phosphate reservoir, an energy source, a regulatory molecule, a pH modulator, a metal chelator, and a structural component of transport channels (12, 43). Given C. jejuni's relatively small genome (23, 31, 55), a molecule such as poly-P would be predicted to impact numerous aspects of C. jejuni biology, including those related to pathogenesis. In the present study, we have found that, similar to other bacteria, wild-type C. jejuni exhibited a significant, ppk1-dependent increase in poly-P levels as the bacteria transitioned from exponential to stationary phase (Fig. 2). As discussed below, we have also found that poly-P accumulation in C. jejuni is important for numerous transmission-, colonization-, and virulence-related phenotypes. Although certain C. jejuni Δppk1 phenotypes are consistent with those in other bacteria, interesting differences are also noted that may reflect genome dissimilarities and/or species-specific stresses. Finally, phenotype comparisons and poly-P assays suggest that poly-P and the SR likely intersect in C. jejuni; however, the two responses also impact distinct functions. To our knowledge, this is the first study demonstrating the importance of poly-P in C. jejuni biology and pathogenesis.
Our data demonstrate that poly-P is important for the transmission-related phenotypes of low-nutrient-stress survival, osmotic stress survival, and biofilm formation. C. jejuni's ability to survive in nutritionally poor environments is particularly critical during conditions such as waterborne transmission, which despite the organism's fastidious laboratory culture requirements, is a major source of larger-scale C. jejuni outbreaks (5, 25, 66). Our low-nutrient-stress observations (Fig. 3) are consistent with studies of ppk1 mutants in E. coli and other organisms, which also exhibit reduced survivability during starvation (40, 50, 70, 71). A mechanistic model to explain this has been developed from work in E. coli, where in wild-type bacteria a nutrient downshift causes an immediate upsurge in poly-P, which in turn complexes with and activates the ATP-dependent Lon protease to selectively degrade free ribosomal proteins, liberating amino acids to meet the nutritional requirements of the cell (44). C. jejuni harbors a putative but currently uncharacterized Lon protease (23, 31, 55); a similar link between poly-P and Lon in C. jejuni might explain the decreased survivability of the Δppk1 mutant under nutrient deprivation conditions.
Resistance to high osmolarity is important for survival during food processing, in certain aquatic environments, and in fecal matter. Although a role for the heat shock and lipo-oligosaccharide gene htrB in osmotic shock survival has been proposed (56), little else is known about this phenomenon in C. jejuni. Our data indicate that C. jejuni requires poly-P for both growth and survival during osmotic stress, most acutely (i) when the organism must grow from isolated single bacteria into colonies (Fig. 4B) and (ii) during later growth stages in broth culture (Fig. 4A), where poly-P levels were shown to rise dramatically in wild-type but not the Δppk1 mutant (Fig. 2B). A number of enteric pathogens lacking ppk1 are also less tolerant to osmotic stress than the parental wild-type strains; these include E. coli, Salmonella spp., and V. cholerae (36, 49, 59). Although poly-P has been hypothesized to participate in osmotic shock protection by virtue of its polyanion structure (59), specific molecular mechanisms have not yet been elucidated.
In contrast to the osmotic-shock and low-nutrient data, and in marked contrast to poly-P mutants in other bacteria, the C. jejuni Δppk1 mutant exhibited enhanced biofilm development versus the wild type (Fig. 5), with no obvious differences in planktonic growth rate (Fig. 2A). The role of poly-P in biofilm formation was first studied in P. aeruginosa, where a Δppk1 mutant was defective for motility, biofilm maturation, and quorum sensing (63). Poly-P is likewise required by V. cholerae, Bacillus cereus, P. aeruginosa, and Porphyromonas gingivalis for motility and biofilm formation (18, 24, 61, 67). Biofilm formation is thought to protect bacteria from adverse environmental conditions and is considered an important virulence factor. Environmental biofilms have also been proposed as a likely mechanism by which C. jejuni survives hostile environments and overcomes its fastidious survival requirements, thereby contributing significantly to its worldwide prevalence (21, 37). We recently found that the C. jejuni SR mutant also exhibits increased biofilm formation (McLennan et al., unpublished). As with Δppk1, this is contrary to observations in other bacteria, where the loss of the SR typically leads to decreased biofilm formation. An ensuing hypothesis is that in C. jejuni, both the SR mutant and the Δppk1 mutant may be constantly stressed, resulting in activation of alternative stress response pathways that may be distinct from those found in gammaproteobacteria (see the discussion of rpoS below) and which in turn accelerate conversion to a protective biofilm state.
We have also identified clear roles for poly-P in both virulence- and colonization-related attributes of C. jejuni. Intraepithelial cell survival is thought to be important for C. jejuni immune and chemotherapeutic evasion, in addition to damage, relapse, and persistence in the human host (19, 20, 38, 65). Although it represents an important virulence phenotype, very little is known about this aspect of C. jejuni pathogenesis. PPK1 is now the third C. jejuni factor, apart from SpoT and the ferrous iron Fe2+ transporter FeoB (51), shown to be required for extended intracellular survival in epithelial cells (Fig. 6). Previous cell biology-based work suggested that C. jejuni resides in a vacuole or vacuole-like compartment after cell internalization (32, 33), a hypothesis corroborated by a recent report describing a role for NOD1 in the innate immune response to C. jejuni cell infection (79). A requirement for SpoT, FeoB, and PPK1 in intracellular survival also supports this notion. Vacuoles are typically low-nutrient, low-iron environments (53). The SR is induced in such an environment, which would also be expected to require FeoB for reduced iron uptake. Poly-P's importance in low-nutrient survivability is consistent with this hypothesis and likely provides a mechanistic explanation for the Δppk1 intracellular survival defect. An S. enterica serovar Typhimurium Δppk1 mutant also displayed an invasion and long-term intracellular survival defect in HEp-2 epithelial cells (40). The only other study investigating a role for poly-P in Campylobacter spp. showed that a C. coli UA585 Δppk1 mutant was as sensitive to macrophage killing as the wild type and that PPK1 was not involved in protection against oxygen radicals in macrophage cells (75). However, poly-P was shown to be important for macrophage survival of S. enterica serovar Typhimurium (40).
The importance of poly-P accumulation in vivo has been shown in various pathogens, including Salmonella spp. and P. aeruginosa, and for colonization of certain strains of H. pylori (7, 49, 63, 70). Chickens are a natural zoonotic reservoir for C. jejuni, and contamination of commercial broiler flocks is thought to account for the majority of human C. jejuni infections (47). Interestingly, the C. jejuni Δppk1 mutant exhibited a dose-dependent colonization defect in chicks: no chicks were colonized to any detectable level at a ∼105 CFU inoculum of Δppk1 mutant, whereas the same dose of wild type colonized to 107 to 109 CFU/g of cecal content (Fig. 7). Although 105 CFU is the lowest inoculating dose used in our studies, wild-type C. jejuni 81-176 colonizes chicks well at doses as low as 103 CFU (48); thus, this is a significant colonization defect for a fully motile C. jejuni mutant (see Table S1 in the supplemental material). The Δppk1 mutant hyperbiofilm formation phenotype also appears to be dose dependent (Fig. 5). One hypothesis to explain the dose-dependent colonization for the Δppk1 mutant is that at low doses, the mutant is primarily planktonic and significantly more susceptible than the wild type to in vivo stresses. As the dose increases, biofilm formation is enhanced in the mutant versus the wild type, protecting the Δppk1 mutant during the initial (or later) stages of colonization. Recent reports indicate that several bacteria, including P. aeruginosa and H. pylori, form biofilms during infection (15, 26). Although such studies have not yet been conducted for C. jejuni, it is of note that we have found that the C. jejuni SR mutant, which also exhibits certain planktonic sensitivities yet forms highly exaggerated biofilms, is fully colonization competent in both chicks and mice (29; McLennan et al., unpublished). Future in vivo studies to address this directly should yield insight into an in vivo role for C. jejuni biofilms.
As described above, poly-P clearly affects certain conserved phenotypes in most bacteria studied, while other phenotypes are much more species specific. Specific differences between C. jejuni and other organisms may occur via a number of genetic and genomic mechanisms. For instance, in most bacteria, and especially other enteric pathogens, poly-P-deficient mutants are unable to express rpoS, and this has been proposed as the reason for certain ppk1 mutant phenotypes (24, 39, 49, 68). C. jejuni lacks rpoS, which may account for some of the surprising phenotypic differences between the C. jejuni Δppk1 mutant (Fig. 2 to 7 and see Table S1 in the supplemental material) and Δppk1 mutants in E. coli, Salmonella spp., and P. aeruginosa. Alternatively, a double deletion of ppk1 and ppk2 may be necessary to completely deplete or severely limit poly-P in C. jejuni. However, a single ppk1 deletion sufficiently induced motility defects in both P. aeruginosa and V. cholerae, each of which harbor both ppk1 and ppk2 (24, 52, 61), while the C. jejuni Δppk1 single mutant was fully motile. C. jejuni ppk1 gene regulation may also differ from that of other organisms. In E. coli, ppk1 and the ppx exopolyphosphatase gene are in an operon; thus, the levels of PPK1 and PPX are also transcriptionally coregulated. In contrast, ppk1 and ppx in C. jejuni are not found in an operon and thus may not be transcriptionally linked. There is also conflicting evidence as to whether the phosphate (pho) regulon regulates ppk1 expression in various bacteria (27). C. jejuni was recently shown to harbor a PhoSR two-component signal transduction system which, like PhoBR in E. coli, controls numerous phosphate acquisition genes via binding to promoter pho box regions (76). The C. jejuni ppk1 gene does not appear to be under the molecular control of the pho regulon, since neither a “traditional” pho box nor the recently identified PhoSR consensus binding sequence is found upstream of the ppk1 gene, and ppk1 was not reported as downregulated in a ΔphoR mutant (76).
Finally, previous work in E. coli has identified functional and regulatory links between the SR and poly-P accumulation (6, 44, 46, 60). In that organism, ppGpp inhibits poly-P hydrolysis by blocking the activity of PPX; in E. coli SR mutants lacking ppGpp, PPX remains active, resulting in diminished levels of poly-P (44). Consistent with this, and despite the above-mentioned lack of conserved ppx/ppk operon structure in C. jejuni, we also observed diminished levels of poly-P in the C. jejuni ΔspoT mutant (Fig. 2), suggesting that this mechanism of poly-P regulation may be conserved between C. jejuni and E. coli. Our phenotype data indicate that certain C. jejuni ΔspoT and Δppk1 defects are similar (see above), while others are not (i.e., invasion and aerobic survival defects for ΔspoT but not Δppk1, osmotic shock and commensal colonization defects for Δppk1 but not ΔspoT), indicating that these two stress responses provide both overlapping and complementary cellular functions. Further elucidation of how these multifunctional factors intersect as well as the precise nature of poly-P regulation in C. jejuni will provide many interesting avenues for future study and may serve as unique models for other bacteria as well.
In summary, the present study demonstrates the importance of poly-P in C. jejuni transmission, colonization, and infection of host cells and has established that poly-P likely interacts with SR mechanisms in C. jejuni. The phenotypic differences between the C. jejuni Δppk1 mutant and Δppk1 mutants in other organisms suggest that this work may also provide a model for further exploring novel, RpoS-independent roles for poly-P in C. jejuni and other bacterial species. Finally, the present study will serve as a platform for numerous future studies exploring the up- and downstream molecular events surrounding poly-P accumulation in C. jejuni, which should in turn lend significant additional insight into mechanisms allowing C. jejuni to remain such a prevalent human pathogen.
Supplementary Material
Acknowledgments
We thank all members of the Gaynor laboratory for scientific advice and helpful discussion. In particular, we thank Emilisa Frirdich, Sarah Svensson, and Meghan McLennan for critically reviewing the manuscript.
E.C.G. is supported by a Canada Research Chair award, the Michael Smith Foundation for Health Research, and a Burroughs Wellcome Fund Career Development Award in the Biomedical Sciences. This study and H.L.C. were funded by Canadian Institutes of Health Research operating grant MOP-68981 to E.C.G.
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, K., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 2.Akiyama, M., E. Crooke, and A. Kornberg. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 267:22556-22561. [PubMed] [Google Scholar]
- 3.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 4.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni: an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auld, H., D. MacIver, and J. Klaassen. 2004. Heavy rainfall and waterborne disease outbreaks: the Walkerton example. J. Toxicol. Environ. Health A 67:1879-1887. [DOI] [PubMed] [Google Scholar]
- 6.Ault-Riche, D., C. D. Fraley, C. M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayraud, S., B. Janvier, L. Salaun, and J. L. Fauchere. 2003. Modification in the ppk gene of Helicobacter pylori during single and multiple experimental murine infections. Infect. Immun. 71:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babakhani, F. K., G. A. Bradley, and L. A. Joens. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 61:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 11.Beery, J. T., M. B. Hugdahl, and M. P. Doyle. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, M. R., and A. Kornberg. 2004. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 101:16085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butzler, J. P., and M. B. Skirrow. 1979. Campylobacter enteritis. Acta Paediatr. Belg. 32:89-94. [PubMed] [Google Scholar]
- 14.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 15.Carron, M. A., V. R. Tran, C. Sugawa, and J. M. Coticchia. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J. Gastrointest. Surg. 10:712-717. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho, A. C., G. M. Ruiz-Palacios, P. Ramos-Cervantes, L. E. Cervantes, X. Jiang, and L. K. Pickering. 2001. Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 39:1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri, R. R., and M. J. Pallen. 2006. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res. 34:D335-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, W., R. J. Palmer, and H. K. Kuramitsu. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect. Immun. 70:4708-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day, W. A., Jr., J. L. Sajecki, T. M. Pitts, and L. A. Joens. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Melo, M. A., G. Gabbiani, and J. C. Pechere. 1989. Cellular events and intracellular survival of Campylobacter jejuni during infection of HEp-2 cells. Infect. Immun. 57:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykes, G. A., B. Sampathkumar, and D. R. Korber. 2003. Planktonic or biofilm growth affects survival, hydrophobicity, and protein expression patterns of a pathogenic Campylobacter jejuni strain. Int. J. Food Microbiol. 89:1-10. [DOI] [PubMed] [Google Scholar]
- 22.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 23.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraley, C. D., M. H. Rashid, S. S. Lee, R. Gottschalk, J. Harrison, P. J. Wood, M. R. Brown, and A. Kornberg. 2007. A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc. Natl. Acad. Sci. USA 104:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 26.Garcia-Medina, R., W. M. Dunne, P. K. Singh, and S. L. Brody. 2005. Pseudomonas aeruginosa acquires biofilm-like properties within airway epithelial cells. Infect. Immun. 73:8298-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavigan, J. A., L. M. Marshall, and A. D. Dobson. 1999. Regulation of polyphosphate kinase gene expression in Acinetobacter baumannii 252. Microbiology 145(Pt. 10):2931-2937. [DOI] [PubMed] [Google Scholar]
- 28.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 30.Harold, F. M. 1966. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol. Rev. 30:772-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, L., J. P. McDaniel, and D. J. Kopecko. 2006. Signal transduction events involved in human epithelial cell invasion by Campylobacter jejuni 81-176. Microb. Pathog. 40:91-100. [DOI] [PubMed] [Google Scholar]
- 34.Hughes, R. 2004. Campylobacter jejuni in Guillain-Barre syndrome. Lancet Neurol. 3:644. [DOI] [PubMed] [Google Scholar]
- 35.Ishige, K., H. Zhang, and A. Kornberg. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 99:16684-16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jahid, I. K., A. J. Silva, and J. A. Benitez. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalmokoff, M., P. Lanthier, T. L. Tremblay, M. Foss, P. C. Lau, G. Sanders, J. Austin, J. Kelly, and C. M. Szymanski. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiehlbauch, J. A., R. A. Albach, L. L. Baum, and K. P. Chang. 1985. Phagocytosis of Campylobacter jejuni and its intracellular survival in mononuclear phagocytes. Infect. Immun. 48:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, H. J., K. Y. Yang, B. H. Cho, K. Y. Kim, M. C. Lee, Y. H. Kim, A. J. Anderson, and Y. C. Kim. 2007. Transcript accumulation from the rpoS gene encoding a stationary-phase sigma factor in Pseudomonas chlororaphis strain O6 is regulated by the polyphosphate kinase gene. Curr. Microbiol. 54:219-223. [DOI] [PubMed] [Google Scholar]
- 40.Kim, K. S., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. USA 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konkel, M. E., M. D. Corwin, L. A. Joens, and W. Cieplak. 1992. Factors that influence the interaction of Campylobacter jejuni with cultured mammalian cells. J. Med. Microbiol. 37:30-37. [DOI] [PubMed] [Google Scholar]
- 42.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 43.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 44.Kuroda, A., H. Murphy, M. Cashel, and A. Kornberg. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240-21243. [DOI] [PubMed] [Google Scholar]
- 45.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in Escherichia coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 46.Kuroda, A., S. Tanaka, T. Ikeda, J. Kato, N. Takiguchi, and H. Ohtake. 1999. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:14264-14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, M. D., and D. G. Newell. 2006. Campylobacter in poultry: filling an ecological niche. Avian Dis. 50:1-9. [DOI] [PubMed] [Google Scholar]
- 48.MacKichan, J. K., E. C. Gaynor, C. Chang, S. Cawthraw, D. G. Newell, J. F. Miller, and S. Falkow. 2004. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol. Microbiol. 54:1269-1286. [DOI] [PubMed] [Google Scholar]
- 49.McMeechan, A., M. A. Lovell, T. A. Cogan, K. L. Marston, T. J. Humphrey, and P. A. Barrow. 2007. Inactivation of ppk differentially affects virulence and disrupts ATP homeostasis in Salmonella enterica serovars Typhimurium and Gallinarum. Res. Microbiol. 158:79-85. [DOI] [PubMed] [Google Scholar]
- 50.Mendrygal, K. E., and J. E. Gonzalez. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogawa, N., C. M. Tzeng, C. D. Fraley, and A. Kornberg. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Riordan, M., and D. A. Portnoy. 2002. The host cytosol: front-line or home front? Trends Microbiol. 10:361-364. [DOI] [PubMed] [Google Scholar]
- 54.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 55.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 56.Phongsisay, V., V. N. Perera, and B. N. Fry. 2007. Expression of the htrB gene is essential for responsiveness of Salmonella typhimurium and Campylobacter jejuni to harsh environments. Microbiology 153:254-262. [DOI] [PubMed] [Google Scholar]
- 57.Pick, U., and M. Weiss. 1991. Polyphosphate hydrolysis within acidic vacuoles in response to amine-induced alkaline stress in the halotolerant alga Dunaliella salina. Plant Physiol. 97:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price-Carter, M., T. G. Fazzio, E. I. Vallbona, and J. R. Roth. 2005. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J. Bacteriol. 187:3088-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao, N. N., S. Liu, and A. Kornberg. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reusch, R. N., and H. L. Sadoff. 1988. Putative structure and functions of a poly-β-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc. Natl. Acad. Sci. USA 85:4176-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russell, R. G., M. O'Donnoghue, D. C. Blake, Jr., J. Zulty, and L. J. DeTolla. 1993. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168:210-215. [DOI] [PubMed] [Google Scholar]
- 66.Schuster, C. J., A. G. Ellis, W. J. Robertson, D. F. Charron, J. J. Aramini, B. J. Marshall, and D. T. Medeiros. 2005. Infectious disease outbreaks related to drinking water in Canada, 1974-2001. Can. J. Public Health 96:254-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi, X., N. N. Rao, and A. Kornberg. 2004. Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc. Natl. Acad. Sci. USA 101:17061-17065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiba, T., K. Tsutsumi, H. Yano, Y. Ihara, A. Kameda, K. Tanaka, H. Takahashi, M. Munekata, N. N. Rao, and A. Kornberg. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. USA 94:11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter, and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 70.Tan, S., C. D. Fraley, M. Zhang, D. Dailidiene, A. Kornberg, and D. E. Berg. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 187:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tinsley, C. R., and E. C. Gotschlich. 1995. Cloning and characterization of the meningococcal polyphosphate kinase gene: production of polyphosphate synthesis mutants. Infect. Immun. 63:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tinsley, C. R., B. N. Manjula, and E. C. Gotschlich. 1993. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect. Immun. 61:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandamme, P. 2000. Microbiology of Campylobacter infections: taxonomy of the family Campylobacteraceae, p. 3-26. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 74.van Vliet, A. H., M. L. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wassenaar, T. M., M. Engelskirchen, S. Park, and A. Lastovica. 1997. Differential uptake and killing potential of Campylobacter jejuni by human peripheral monocytes/macrophages. Med. Microbiol. Immunol. 186:139-144. [DOI] [PubMed] [Google Scholar]
- 76.Wosten, M. M., C. T. Parker, A. van Mourik, M. R. Guilhabert, L. van Dijk, and J. P. van Putten. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278-291. [DOI] [PubMed] [Google Scholar]
- 77.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zilbauer, M., N. Dorrell, A. Elmi, K. J. Lindley, S. Schuller, H. E. Jones, N. J. Klein, G. Nunez, B. W. Wren, and M. Bajaj-Elliott. 2007. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol. 9:2404-2416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.