Abstract
Harpins are a subset of type III secretion system (T3SS) substrates found in all phytopathogenic bacteria that utilize a T3SS. Pseudomonas syringae pv. tomato DC3000 was previously reported to produce two harpins, HrpZ1 and HrpW1. DC3000 was shown here to deploy two additional proteins, HopAK1 and HopP1, which have the harpin-like properties of lacking cysteine, eliciting the hypersensitive response (HR) when partially purified and infiltrated into tobacco leaves, and possessing a two-domain structure similar to that of the HrpW1 class of harpins. Unlike the single-domain harpin HrpZ1, the two-domain harpins have C-terminal enzyme-like domains: pectate lyase for HopAK1 and lytic transglycosylase for HopP1. Genetic techniques to recycle antibiotic markers were applied to DC3000 to generate a quadruple harpin gene polymutant. The polymutant was moderately reduced in the elicitation of the HR and translocation of the T3SS effector AvrPto1 fused to a Cya translocation reporter, but the mutant was unaffected in the secretion of AvrPto1-Cya. The DC3000 hrpK1 gene encodes a putative translocator in the HrpF/NopX family and was deleted in combination with the four harpin genes. The hrpK1 quadruple harpin gene polymutant was strongly reduced in HR elicitation, virulence, and translocation of AvrPto1-Cya into plant cells but not in the secretion of representative T3SS substrates in culture. HrpK1, HrpZ1, HrpW1, and HopAK1, but not HopP1, were independently capable of restoring some HR elicitation to the hrpK1 quadruple harpin gene polymutant, which suggests that a consortium of semiredundant translocators from three protein classes cooperate to form the P. syringae T3SS translocon.
The type III secretion system (T3SS) is an important virulence determinant in gram-negative bacterial pathogens of animals and plants (17). In Pseudomonas syringae pv. tomato DC3000, the T3SS, which is encoded by the hrp-hrc (hypersensitive response [HR] and pathogenicity-hr conserved) gene cluster, is required for elicitation of the defense-associated HR in nonhost plants like tobacco and for pathogenicity in host plants like tomato. Both HR elicitation and pathogenicity require the translocation into plant cells of effectors, which are also known as Avr (avirulence) proteins or Hops (Hrp outer proteins) (44).
The T3SS of pathogens must cross three biological membranes, the inner and outer bacterial membranes and the host plasma membrane. Breaching the host plasma membrane is expected to involve a T3SS pore-forming translocon complex (17). In addition, the T3SS of phytopathogens such as P. syringae, Erwinia amylovora, Ralstonia solanacearum, Pantoea spp., and Xanthomonas spp. must cross the plant cell wall, which can be several hundred nanometers in width and is constructed primarily of a meshwork of polysaccharides, with pectic polymers being particularly important in controlling pore size and integrity (59, 66). Because of this additional barrier, the Hrp pilus of phytopathogens is much longer than the T3SS extracellular appendages of animal pathogens, and the translocation process may be more complex (33).
The prototypical translocon complex for T3SS-utilizing animal pathogenic bacteria is possessed by Yersinia spp. and is constructed from the cotranscribed translocator proteins YopB, YopD, and LcrV. These proteins have all been demonstrated to form pores in membranes and to interact with one another (9). The components of the animal translocon complex are completely interdependent; the removal of any of these three proteins abrogates translocation (9, 26). LcrV has been localized to the tip of the secretion needle and is hypothesized to form a platform for YopB and YopD pore assembly (17). Other animal pathogens typically carry homologs of all three proteins, which have been demonstrated to function in a similar manner (17). Loss of the translocon prevents the T3SS from delivering effectors across the plasma membrane and eliminates any translocation-dependent phenotypes, however, it does not block secretion of effectors in culture.
The translocons of phytopathogens appear to be only partially similar to those of animal pathogens. Members of the HrpF/NopX family show limited homology to YopB, but homologs of YopD and LcrV have not been reported in phytopathogens (47). The homologous HrpF of X. campestris pv. vesicatoria and PopF1 of R. solanacearum are typical translocon components in that they are required for effector translocation but not secretion (10, 47, 60). Both proteins are members of the HrpF/NopX family and have predicted transmembrane (TM) domains that are required for function (10, 47, 60). Furthermore, HrpF has been shown to form pores in planar lipid bilayers (10). The P. syringae HrpK1 protein is also a member of the HrpF/NopX family with a predicted TM domain (47, 54). HrpK1 is encoded in the hrp-hrc gene cluster, which suggests a central role in the T3SS. However, hrpK1 mutants have relatively weak and variable T3SS phenotypes, which brings into question the role of HrpK1 as a translocator (47, 54).
Harpins are unique to phytopathogens and represent another class of T3SS-secreted proteins that may contribute to effector translocation. Harpin-like proteins have been identified in all T3SS-utilizing phytopathogens (1, 4, 13, 28, 34, 35, 70). Although these proteins may have little homology, they share several characteristics, such as a low isoelectric point, enrichment for glycine, and a lack of cysteine residues. Importantly, harpins also share the unusual ability to elicit the HR, a defense-related programmed cell death, when applied externally to plant cells as isolated proteins. The biological significance of this activity during infection is unclear. Genetic evidence indicates that effectors rather than harpins are responsible for the HR produced during interactions with nonhosts (2), and isolated harpins elicit the HR in hosts, as well as nonhosts (55). Furthermore, the amount of purified harpin required for exogenous HR elicitation in tobacco has been shown to be 500-fold greater than the amount produced in planta by a concentrated suspension of P. syringae (67). The contribution of harpins to successful (compatible) P. syringae infections is unknown.
P. syringae pv. tomato DC3000 has been shown previously to produce two harpins, HrpZ1 and HrpW1 (13, 55). The hrpZ1 gene is located within the hrp-hrc gene cluster in a polycistronic operon just downstream of the hrpA1 pilin subunit gene. HrpZ1 proteins from various P. syringae pathovars bind lipids and form ion-conducting pores in artificial lipid bilayers (40). HrpZ1Psy 61 was reported to bind to the outermost layers of the plant cell wall but not to tobacco protoplasts (30). Conversely, HrpZ1Pph1448A was reported to bind tobacco protoplasts, as well as tobacco microsomal membrane preparations, at a proposed lipid site (39). Also, HrpZ1Pph1448A binds to a phage-displayed peptide and to a small acidic plant protein from bean with the same binding domain (41).
HrpW1 is encoded in the conserved effector locus, which is contiguous with the hrp-hrc gene cluster. HrpW1 has a two-domain structure with an N-terminal harpin domain and a C-terminal domain that is homologous to a PL3 pectate lyase (18, 19). The pectate lyase domain is capable of binding to calcium pectate beads but has no detectable enzymatic activity (13). Exogenous HR elicitation is associated only with the N-terminal harpin domain (13), but HrpW1 homology is much more strongly conserved in its pectate lyase domain than in its harpin domain (11). HrpZ1 and HrpW1 are both conserved among various P. syringae pathovars and are rapidly expressed at high levels when hrp-hrc genes are induced (21, 43). However, a double knockout of both genes in P. syringae pv. tomato DC3000 produced only a mild reduction in HR elicitation activity and no detectable reduction in virulence (13). It was hypothesized that P. syringae pv. tomato DC3000 might express additional, redundant harpins that mask the phenotype of the double mutation, but without a phenotype, the function of harpins in P. syringae virulence has been difficult to investigate (13).
Here, we demonstrate that P. syringae pv. tomato DC3000 deploys two additional harpins, HopAK1 and HopP1. We adapted techniques to recycle antibiotic markers in P. syringae pv. tomato DC3000, generated a strain carrying mutations in all four harpin genes, and showed that the polymutant has a moderate translocator phenotype. We then combined hrpK1 and harpin gene mutations to produce strains that were strongly deficient in translocation in planta but retained the wild-type ability to secrete T3SS substrates in culture. Lastly, we demonstrate that HrpK1, HrpZ1, HrpW1, and HopAK1, but not HopP1, can independently restore translocation abilities to the polymutant strain. We propose a model for the phytopathogenic T3SS translocon in which a consortium of translocators from three classes, typically with one dominant translocator and several lesser translocators, overlap in their contributions to translocation.
MATERIALS AND METHODS
Bacterial strains.
For the Escherichia coli and P. syringae pv. tomato DC3000 strains and plasmids used in this study, see Table S1 in the supplemental material. Plasmid maintenance and manipulations were typically done with either E. coli DH5α or TOP10, but those involving Invitrogen Gateway manipulations used E. coli DB3.1. Uses of other E. coli strains were as noted.
Media and culture conditions.
P. syringae pv. tomato DC3000 strains were routinely grown in King's B (KB) medium at 25 or 30°C (37). For Hrp induction, P. syringae pv. tomato DC3000 strains were grown in Hrp minimal medium augmented with 0.2% fructose and 0.2% mannitol at 22°C (31). E. coli strains were routinely grown in LB or Terrific broth medium at 37°C. Antibiotics and additives were used at the following final concentrations (in micrograms per milliliter unless otherwise noted): ampicillin, 100; carbenicillin, 100; kanamycin, 50; gentamicin, 10; spectinomycin, 50; streptomycin, 100; rifampin, 50; tetracycline, 20; chloramphenicol, 20; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40; 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc), 20; isopropyl-β-d-thiogalactopyranoside (IPTG), 1 mM.
Construction of transconjugants and transformants.
Triparental and biparental matings were conducted with sterile nitrocellulose squares on LM medium essentially as described previously (22, 27). E. coli HB101/pRK2013 was used routinely as a helper strain in triparental matings. E. coli S17-1 was used as a host donor strain for biparental matings (64). P. syringae pv. tomato DC3000 transconjugants were selected with rifampin and/or ampicillin as chromosomal markers. Transformation of P. syringae pv. tomato DC3000 strains was conducted by electroporation essentially as described by Choi et al. (14). Transformation of E. coli strains was conducted by standard electroporation or chemical competence protocols.
Recombinant DNA techniques.
DNA manipulations and PCR were conducted according to standard protocols (61). Plasmid DNA was purified with the QIAprep Spin Miniprep kit from QIAGEN (Valencia, CA). Genomic DNA was prepared with the Wizard Genomic DNA purification kit from Promega (Madison, WI). DNA gel extraction and DNA enzyme reaction cleanups were conducted with the Gel DNA Recovery kit and Clean-up and Concentrator kit from Zymo Research (Orange, CA). DNA restriction and modification enzymes were from New England BioLabs (Ipswich, MA) and were used according to the manufacturer's recommendations. LR recombination was conducted with LR clonase I or LR clonase II from Invitrogen (Carlsbad, CA) as recommended by the manufacturer. PCR was routinely conducted with Takara Ex Taq Polymerase Premix from Takara Mirus Bio (Otsu, Shiga, Japan). PCR primers were obtained from IDT (Coralville, IA). DNA sequencing was conducted at the Cornell University Biotechnology Resource Center with an Applied Biosystems 3730xl DNA analyzer. Sequences were analyzed with the Vector NTI software package from Invitrogen.
Construction of plasmids for P. syringae pv. tomato DC3000 mutagenesis and expression of AvrPto1-Cya from its native promoter.
Multiple plasmids were constructed and sequenced, including (i) a series of plasmids carrying FRT-flanked cassettes with different antibiotic markers (GenBank accession no. EU24547 to EU24552); (ii) pCPP5301 (GenBank accession no. EU24541), a Gateway destination marker exchange vector with a lacZ marker; (iii) pCPP5264 (GenBank accession no. EU24542), an unstable FLP expression vector; (iv) pCPP5333 (GenBank accession no. EU24543), a Gateway destination suicide vector which is counterselectable with sucrose or the I-SceI meganuclease; (v) pCPP5386 (GenBank accession no. EU24544), an unstable I-SceI meganuclease expression vector; (vi) pCPP5312 (GenBank accession no. EU24545), a native promoter AvrPto1-Cya expression plasmid; and (vii) pCPP5702 (GenBank accession no. EU24546), a native promoter AvrPto1-Cya expression plasmid that constitutively expresses NptII, which is useful as a cytoplasmic marker in secretion assays. For the construction and properties of these plasmids, see Table S1 and the associated information in the supplemental material.
Marker exchange mutagenesis and construction of deletions in P. syringae pv. tomato DC3000.
One-kilobase regions flanking hopAK1 and 1.5-kb regions flanking hopP1 were PCR amplified from P. syringae pv. tomato DC3000 genomic DNA with external XbaI sites and overlapping internal tails with introduced XhoI sites. The flanks were joined by gene splicing by overlap extension PCR and cloned either directly into the XbaI site of pRK415 or TOPO cloned into pENTR/D-TOPO (29). The FRTGmrgus and FRTgusKmr cassettes were PCR amplified from pCPP5212 and pCPP5250, respectively, with primer-introduced terminal XhoI sites. The cassettes were digested with XhoI and cloned between the hopAK1 and hopP1 deletion flanks. Marked entry vector deletion constructs were then LR recombined with pCPP5301. Completed pRK415-based and pCPP5301-based deletion constructs were conjugated by triparental mating into P. syringae pv. tomato DC3000 and derivative strains. To enrich for marker exchange mutants, isolated colonies were grown in KB medium with either half-strength gentamicin (5 μg ml−1) or kanamycin (25 μg ml−1) and were subinoculated 1:1,000 into fresh selective medium once per day for 2 to 6 days. Enrichment cultures were diluted and plated every 2 days onto KB agar augmented with half-strength gentamicin or kanamycin. Cultures carrying pCPP5301-based deletion constructs were additionally augmented with X-Gal for blue-white screening of the lacZ marker. Isolated colonies were patched onto KB containing tetracycline and KB containing half-strength gentamicin or kanamycin to identify colonies that were Tcs but resistant to the mutation marker. In the case of pCPP5301-based deletion constructs, only white colonies were chosen for further analysis, as the blue Lac+ phenotype indicates maintenance of the deletion construct plasmid. Mutations were confirmed by PCR with chromosomal primers outside the deletion construct, as well as sequencing through deletion regions. For strains containing multiple FRT scars, the genomic contexts of all FRT scars in the genome were assessed simultaneously to ensure that neither large-scale deletions nor chromosomal inversions had occurred.
Flp recombination of FRT-flanked cassettes.
To remove the FRT-flanked antibiotic resistance cassettes, strains were conjugated by triparental mating with the Flp-expressing plasmid pCPP5264 and plated onto KB containing tetracycline. Transconjugant colonies were patched on selective and nonselective KB plates to confirm the loss of the FRT-flanked resistances. Plates were also augmented with X-Gluc for cassettes containing gus. Antibiotic-sensitive and Gus− colonies were restreaked to isolation in the absence of tetracycline and patched to confirm curing of pCPP5264. Deletions were screened and confirmed by PCR and sequencing. For strains containing multiple FRT scars, the genomic contexts of all FRT scars were confirmed simultaneously by PCR to check for inversions or large-scale deletions.
Integration-resolution mutagenesis by sacB or SceI counterselection.
Regions flanking hrpK1 (1.1 kb and 1.0 kb) were PCR amplified with internal, primer-introduced XhoI and XmaI sites, respectively. The FRTGmr cassette from pCPP5209 was PCR amplified with terminal, primer-introduced XhoI and XmaI sites. The three PCR fragments were restriction digested with XhoI and XmaI and then ligated with T4 ligase. The ca. 3.2-kb ligation product was gel purified and TOPO cloned into pCR2.1-TOPO. The deletion construct was subcloned from pCR2.1-TOPO by restriction with EcoRI and ligated into the ca. 5.7-kb EcoRI-linearized plasmid pK18mobsacB to create pCPP5645. To create pCPP5336, the unmarked hopP1 pENTR/D-TOPO deletion construct was LR recombined with pCPP5333. E. coli S17-1 was electroporated with either pCPP5336 or pCPP5645. The transformants were used in biparental matings with P. syringae pv. tomato DC3000 and derivative strains. Integration events were selected with half-strength gentamicin for pCPP5336 and with kanamycin and half-strength gentamicin for pCPP5645. CUCPB5405/pCPP5336 integrant colonies were conjugated by triparental mating with pCPP5386 to counterselect the pCPP5336 integration with I-SceI meganuclease. Transconjugants were patched on KB and on KB containing gentamicin to confirm the resolution of the integrated plasmid pCPP5336. Resolved colonies were then streaked to isolation on KB in the absence of tetracycline and patched to confirm the curing of pCPP5386. Sucrose counterselection of pCPP5645 integrant colonies was achieved by suspending one colony in 2 ml of KB and plating 50 μl of the suspension on KB and on KB augmented with 10% sucrose. Sucrose-resistant colonies were patched onto KB containing half-strength gentamicin and onto KB containing half-strength gentamicin and kanamycin to confirm the resolution of the integrated plasmid pCPP5645. Genomic DNA was prepared from pCPP5333- and pCPP5645-resolved colonies. Deletions were screened and confirmed by PCR and sequencing.
Complementation vectors.
To enable complementation tests of P. syringae pv. tomato DC3000 hrpK1 and harpin gene mutations, entry vector clones of hrpK1 and the harpin genes were LR recombined with the PnptII expression broad-host-range vector pBS46. LR recombination of pBS46 with harpin entry clones lacking stop codons creates C-terminal hemagglutinin (HA) epitope fusions. The pBS46 derivatives were transformed into CUCPB5483, and the HR and virulence assays were conducted as described below.
Purification of harpins.
For the purification of soluble harpins, pENTR/SD/D entry clones of the harpin genes lacking stop codons were LR recombined with the pET-DEST42 T7 expression vector. Derivative pET-DEST42 plasmids produce C-terminal fusions with the V5 epitope and His6 affinity tag. An empty-vector derivative of pET-DEST42 was constructed by restriction digestion with BsrGI and T4 ligase recircularization of the ca. 5.7-kb backbone to remove the Gateway cassette. The pET-DEST42 plasmids were transformed into E. coli BL21(DE3). Single colonies of the expression strains were inoculated into 12 ml LB containing carbenicillin and incubated for 12 to 16 h with shaking at 37°C. The starter cultures were used to inoculate 250-ml LB-carbenicillin cultures in fretted, sidearm Erlenmeyer flasks, which were incubated for 3 h with shaking at 30°C to an optical density at 600 nm (OD600) of ca. 0.6. IPTG was added to a final concentration of 1 mM, and the cultures were allowed to incubate for an additional 5 h. Cell pellets were harvested at 4,000 × g for 20 min at 4°C and frozen at −80°C overnight. Pellets were defrosted on ice for 15 min and suspended in 5 ml lysis buffer with final concentrations of 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0 (56). Lysozyme (Sigma, St. Louis, MO) was added to a final concentration of 1 mg ml−1, and the cells were incubated on ice for 30 min. Phenylmethylsulfonyl fluoride protease inhibitor was added to a final concentration of 1 mM, and the suspension was sonicated on ice 12× with a 10-s on-off cycle with a Fisher Scientific (Pittsburgh, PA) 550 sonic Dismembrator and a microtip at power level 4. Cleared lysates were harvested at 10,000 × g for 30 min at 4°C. One milliliter of well-mixed Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) was added to 4 ml of cleared lysate and gently agitated for 1 h at 4°C. The lysate-Ni-NTA agarose mixture was loaded onto an empty disposable polyprene column (QIAGEN), washed twice with 4 ml of 20 mM imidazole wash buffer, and eluted with a 2-ml total volume of 250 mM imidazole elution buffer (56). The elution buffer was replaced by using a Centricon Ultracel YM-30 2-ml centrifugal filter device (Millipore, Billerica, MA), washing three times with equivalent volumes of 5 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 5.5), and suspending in a final volume of 0.5 ml. Purified harpins were stored at 4°C until use. Proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie brilliant blue. Protein concentrations were determined by Bradford assay. Harpin preparations ranged from 0.12 to 2.6 mg protein ml−1 and were diluted 2- to 16-fold for plant reaction assays. Empty-vector preparations were obtained in parallel with each harpin preparation and ranged from 0.02 to 0.18 mg protein ml−1, contained contaminants that ran at 26 and 81 kDa in SDS-PAGE gels, and were always used at full strength in plant tests.
HR assays.
Tobacco plants (Nicotiana tabacum cv. xanthi) were grown under greenhouse conditions until ca. 8 weeks postgermination. Prior to the assay, plants were allowed to acclimate in the laboratory for 2 days at 25°C and low relative humidity with daylight and supplemental light. Primary streaks of P. syringae pv. tomato DC3000 strains were made from isolated colonies onto selective KB plates and grown overnight at room temperature. The plates were then spread with 100 μl sterile KB and incubated overnight at room temperature to produce even bacterial lawns. Cells were scraped from plates with a sterile loop and suspended in 10 mM MgCl2-100 mM sucrose to a final OD600 of 0.3. Dilution series were made from the suspensions with an OD600 of 0.3. Leaf panels of the third- to sixth-oldest leaves were infiltrated with dilutions of the bacterial suspensions by pricking the leaves with a dissecting needle and infiltrating the suspensions with a blunt syringe. Mutant strains were compared with identical dilutions of wild-type P. syringae pv. tomato DC3000 on the same leaf. Purified harpins were diluted in 5 mM MES, pH 5.5, and infiltrated with a blunt syringe as described above, with or without the HR inhibitor lanthanum chloride at a final concentration of 2 mM. Proteins from mock purifications of undiluted empty-vector samples were infiltrated into panels on the same leaves as the negative control. The HR was allowed to develop for at least 24 h in the laboratory at room temperature with supplemental lighting before photography. The red channel was preferentially used in the conversion from RGB to gray scale for each complete figure to enhance visualization of the collapsed and desiccated tissue that characterizes the HR.
Virulence assays.
Tomato (Solanum lycopersicum cv. Moneymaker) plants were grown under greenhouse conditions until 4 to 5 weeks postgermination. Bacterial lawns were grown as described above. Cells were scraped from plates with a sterile loop and suspended in 10 mM MgCl2-100 mM sucrose to a final OD600 of 0.3. Bacterial suspensions were diluted to ca. 2 × 104 CFU ml−1 in 10 mM MgCl2. The bacterial concentrations of the suspensions were verified by plate count. The suspensions were infiltrated into half leaflets of the third and fourth most recently expanded leaves with a dissecting needle and a blunt syringe. The inoculated tomato plants were incubated at 20 to 25°C with high humidity and a 12-h light cycle. At 3 days postinfection, two 0.8-cm-diameter leaf discs were harvested with a cork borer from each infiltration area. The discs were ground with a sterile mortar and pestle into 0.3 ml 10 mM MgCl2-100 mM sucrose, diluted, and plated to determine the number of CFU per square centimeter.
Translocation assays.
AvrPto1-Cya translocation assays were conducted as described by Schechter et al. (62), with some modifications. Strains carrying AvrPto1-Cya-expressing plasmid pCPP5312 or pCPP5702 were grown as lawns on KB plates and suspended in 10 mM MgCl2-100 mM sucrose to a final OD600 of 0.3 as described above. The suspensions were diluted and infiltrated into the third- or fourth-oldest leaves of mature tobacco plants as described above. All strains and dilutions for an individual experiment were infiltrated into a single tobacco leaf. The plants were incubated for 6 h under laboratory conditions. Two 1-cm-diameter tobacco leaf discs per infiltration area were collected with a cork borer and frozen in liquid N2. For harpin-Cya translocation assays, overnight cultures of strains carrying harpin-Cya-expressing pCPP3234 LR recombinants were suspended in 5 mM MES, pH 5.5, to a final OD600 of 0.3 and induced with 0.1 mM IPTG. The suspensions were infiltrated into the leaves of mature tomato plants as described above and incubated for 7 h in a humid chamber. A 0.63-cm-diameter tomato leaf disc was collected with a cork borer from each infiltration area and frozen in liquid N2. Frozen leaf discs were crushed with a Kontes pestle in a microcentrifuge tube and suspended in 300 μl of 0.1 M HCl. Protein and cyclic AMP (cAMP) concentrations of the suspensions were determined as described previously, by use of the Bradford assay and the Direct Cyclic AMP Correlate EIA kit (Assay Designs, Ann Arbor, MI), respectively (62).
T3SS secretion assays.
Bacterial lawns were grown as described above. Lawns were suspended in 5 ml of Hrp minimal medium with a sterile loop and diluted in 100 ml of Hrp minimal medium in fretted, sidearm Erlenmeyer flasks to a final OD600 of 0.3. The cultures were incubated at 22°C at 200 rpm on a rotary shaker to a final OD600 of 0.5, taking approximately 8 h. The cell and supernatant fractions were separated by centrifugation at 5,200 × g for 15 min at 4°C. The cell pellet from 40 ml of culture was suspended in 1 ml sterile distilled H2O, and 90 μl of suspended cell pellet was added to Laemmli loading buffer to a 1× concentration and saved as the cell fraction. The top 40 ml of supernatant was centrifuged at 21,000 × g for 40 min at 4°C. The top 20 ml of this supernatant was combined with 5 ml of cold trichloroacetic acid and incubated overnight on ice to precipitate the protein. Precipitated protein was harvested by centrifugation at 21,000 × g for 40 min at 4°C. The supernatant was discarded. The pellet was air dried and suspended in 120 μl of 1× Laemmli loading buffer as the supernatant fraction. The cell pellet and supernatant fractions were boiled for 10 min. Samples were subjected to 12% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane with a Hoefer (San Francisco, CA) SemiPhor semidry transfer unit. The blots were probed with either rabbit polyclonal anti-AvrPto1 immunoglobulin G (IgG), rabbit polyclonal anti-HopAA1-1 IgG, rabbit polyclonal anti-NptII IgG (United States Biological, Swampscott, MA), or mouse monoclonal anti-HA.11 (16B12) IgG (Covance Research Products, Princeton, NJ) as primary antibodies. Blots were probed with either anti-rabbit IgG alkaline phosphatase conjugate or anti-mouse IgG alkaline phosphatase conjugate (Sigma) as secondary antibodies. Blots were developed with Tropix CDP-Star chemiluminescent substrate (Applied Biosystems, Foster City, CA) and imaged with Kodak scientific imaging film.
RESULTS
Identification of harpin candidates.
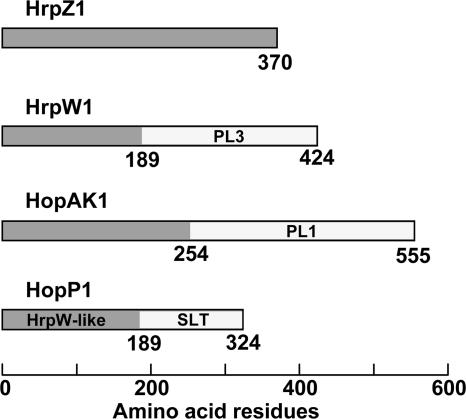
We used a bioinformatic approach to identify novel harpin gene candidates in P. syringae pv. tomato DC3000. The genome of this strain had previously been sequenced and analyzed by microarray and bioinformatic analyses for promoters regulated by the HrpL alternative sigma factor and for downstream open reading frames with N-terminal amino acid patterns predictive of T3SS substrates (8, 21, 38, 63). We screened the open reading frames predicted to encode HrpL-regulated T3SS substrates for those lacking cysteine and found several harpin candidates. hopAK1 (PSPTO4101) and hopP1 (PSPTO2678) were particularly promising. Both genes are rapidly activated by HrpL (21), and they have a two-domain structure similar to that of HrpW1, with C-terminal enzyme-like regions. The domain structures of HrpZ1, HrpW1, HopAK1, and HopP1 are diagrammed in Fig. 1. The enzyme-like domain of HopAK1 is homologous to a PL1 pectate lyase (18, 19). The overall structure of HopAK1 is very similar to HrpW1, although these two proteins are not homologs. HopP1 has an N-terminal region that is homologous to the harpin domain of HrpW1, and its C-terminal enzyme-like region is similar to a soluble lytic transglycosylase. Thus, DC3000 expresses four harpin-like genes.
FIG. 1.
Predicted domains of the P. syringae pv. tomato DC3000 HrpZ1, HrpW1, HopAK1, and HopP1 proteins. Domains were identified by BLASTP and NCBI conserved-domain searches (3, 46). HrpW-like and pectate lyase (PL) domains were further refined by referencing the CAZy database (http://www.cazy.org/index.html) (18, 19) and determining amino acid consensus with AlignX at default settings in the Vector NTI software package. The HrpW1 PL3 (polysaccharide lyase family 3) domain has 43.9% similarity to Bacillus subtilis Pel-15 pectate lyase (GenBank accession no. BAA87892). The HopAK1 PL1 domain (polysaccharide lyase family 1) has 56.1% similarity to Bacillus licheniformis PelB (GenBank accession no. AAU24559). The HopP1 HrpW-like domain has 45.5% similarity to HrpW1 from P. syringae pv. tomato DC3000 (GenBank accession no. AAO54895). The SLT (soluble lytic transglycosylase) domain was previously described (53).
HopP1 is translocated at higher levels than HrpZ1, HrpW1, and HopAK1.
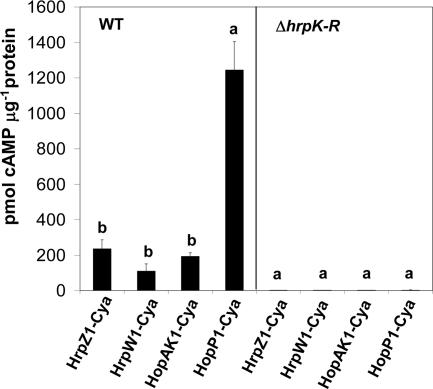
The HrpZ1 and HrpW1 proteins had previously been shown to be secreted in culture by the P. syringae pv. tomato DC3000 T3SS (13), and HopP1 had been shown to be translocated into plant cells by the DC3000 T3SS when expressed from its own promoter as a fusion with the AvrRpt280-255 translocation reporter (12). However, similar HrpW1-AvrRpt280-255 and HopAK1-AvrRpt280-255 fusions were not translocated (12). To determine whether the DC3000 HopAK1 can travel the T3SS pathway and to compare the four harpin candidates for their relative abilities to be translocated when expressed from a common promoter, we constructed plasmid pCPP3234 derivatives that expressed each protein from a vector tac promoter and produced proteins with a C-terminal fusion to the Cya (Bordetella pertussis adenylate cyclase) translocation reporter (62). The strains were infiltrated into the leaves of tomato plants (S. lycopersicum cv. Moneymaker), and cAMP production resulting from calmodulin-dependent Cya activity within plant cells was measured 7 h later (Fig. 2). Translocation levels for HrpZ1-Cya, HrpW1-Cya, and HopAK1-Cya were relatively low and similar to those we previously observed with three substrate components of the Hrp T3SS, HrpF-Cya, HrpJ-Cya, and HrpP-Cya (58). In contrast, HopP1 was translocated much more strongly. Thus, all four of the harpin candidates are capable of being translocated into plant cells by the T3SS when expressed from a heterologous promoter, HopAK1 appears similar to HrpW1 in being poorly translocated when expressed from its seemingly strong native promoter, and HopP1 is distinct from the other three harpin candidates in its strong translocation abilities.
FIG. 2.
The P. syringae pv. tomato DC3000 HrpZ1, HrpW1, HopAK1, and HopP1 proteins are T3SS substrates. Strains expressing harpin-Cya fusions from a Ptac promoter were infiltrated into tomato (S. lycopersicum cv. Moneymaker) leaves at 2 × 108 CFU ml−1. The T3SS-deficient control strain used was the ΔhrpK-R mutant CUCPB5114. A 0.63-cm-diameter leaf disc was harvested at 7 h postinfiltration and processed to determine the number of picomoles of soluble cAMP per microgram of protein. Values represent means and standard deviations of three leaves for each treatment. Means marked with the same letter were not significantly different at the 5% confidence level on the basis of Duncan's multiple-range test. The experiment was repeated four times with similar results.
Partially purified HopP1 and HopAK11-254 can elicit the HR in tobacco leaves.
A defining property of harpins is their ability to elicit HR-like programmed cell death when infiltrated into tobacco leaf intercellular spaces. To test the two new harpin candidates for this ability, we produced HrpZ1, HrpW1, HopAK1, and HopP1 from E. coli cells carrying appropriate derivatives of pET-DEST42, a T7 expression vector that makes C-terminal V5 epitope and His6 tag fusions. HrpZ1, HrpW1, and HopP1 were soluble after overexpression in E. coli and could be readily purified with Ni-NTA agarose (QIAGEN). Full-length HopAK1 was insoluble when overexpressed with this system. To test HopAK1 for the ability to elicit the HR, we subcloned a DNA fragment that encodes residues 1 to 254. This approach was based on the expectation that the harpin domain would be sufficient for HR elicitation, as was shown with HrpW1 (13). The HopAK1 harpin domain was found to be soluble when overexpressed. Comparisons of protein concentrations from mock preparations of the empty-vector strain with harpin and harpin candidate preparations indicated that the harpin preparations were at least 80% pure.
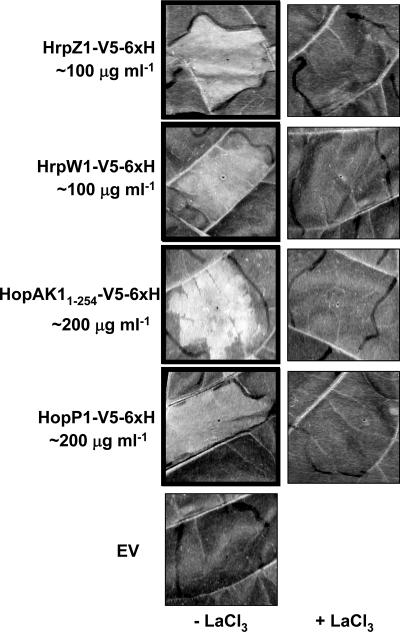
A dilution series of each protein was infiltrated into leaves of tobacco (N. tabacum cv. xanthi) with or without lanthanum chloride, a calcium channel blocker that has been shown to act as an inhibitor of harpin-induced HR (13, 28). Importantly, inhibition by lanthanum chloride indicates that any cell death observed is not the result of pectolytic damage to the plant cell wall and consequent cell lysis. An undiluted empty-vector preparation was always infiltrated into the same leaf as a control for cell death that might be caused by contaminating proteins or high salt concentrations. As expected, both HrpZ1 and HrpW1 elicited a lanthanum chloride-inhibitable HR at about 100 μg ml−1 (Fig. 3), matching previously published concentrations (28, 67). The time required for HR elicitation by HrpZ1 tended to be faster than for HrpW1. HopP1 and HopAK11-254 elicited a lanthanum chloride-inhibitable HR with kinetics similar to that of HrpW1, although they required approximately twice the protein concentration as HrpZ1 and HrpW1. The empty-vector preparations elicited no visible response. Elicitation of the HR by exogenous protein application indicates that HopP1 and HopAK1 possess a defining property of harpins.
FIG. 3.
Partially purified P. syringae pv. tomato DC3000 harpins HrpZ1, HrpW1, HopAK11-254, and HopP1 elicit HR-like tissue collapse in tobacco leaves. V5-His6-tagged recombinant protein preparations were diluted in MES buffer and infiltrated into panels of tobacco (N. tabacum cv. xanthi) leaves with or without 2 mM lanthanum chloride, an inhibitor of HR. The protein concentrations shown represent approximate minimal concentrations that gave full collapse within 24 h postinfiltration. A mock, undiluted, empty-vector (EV) preparation was always tested in parallel. Boxed panels indicate a positive HR. The experiment was repeated twice with similar results.
Construction of harpin gene polymutants of P. syringae pv. tomato DC3000.
To facilitate the investigation of the virulence function of the multiple P. syringae harpins, we constructed a series of polymutants, including one that was mutated in all four harpin genes. Three P. syringae pv. tomato DC3000 mutants that contained ΔhrpZ1, ΩhrpW1, and ΔhrpZ1 ΩhrpW1, respectively, had been constructed previously (13). These mutants are marked with Kmr and Spr Smr, which precludes the further use of these markers in polymutant construction. To remove the remaining harpin genes and maintain antibiotic markers for further rounds of mutation, techniques to recycle antibiotic markers and generate unmarked mutants were applied to P. syringae pv. tomato DC3000. Two genetic strategies were adopted to accomplish this, i.e., FRT-flanked marker exchange mutagenesis and two-step integration-resolution mutagenesis. For a diagram and an explanation of the major steps of the two strategies, see Fig. S1 in the supplemental material. By a combination of these strategies, eight additional harpin gene mutant and polymutant strains were generated (Table 1; see Table S1 in the supplemental material).
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant feature(s) (reference) | Abbreviation used in figures |
|---|---|---|
| P. syringae pv. tomato strains | ||
| DC3000 | Wild type; Rfr Apr (20) | WT |
| CUCPB5113 | DC3000 ΔhrcQ-U::ΩSpr/Smr (5) | ΔhrcQ-U |
| CUCPB5114 | DC3000 ΔhrpK1-R::ΩCmr (23) | ΔhrpK-R |
| CUCPB5481 | DC3000 ΔhrpK1 | ΔK |
| CUCPB5094 | DC3000 ΔhrpZ1::nptII (13) | ΔZ |
| CUCPB5096 | DC3000 hrpW1::ΩSpr/Smr (13) | ΔW |
| CUCPB5405 | DC3000 ΔhopAK1 | ΔAK |
| CUCPB5465 | DC3000 ΔhopP1 | ΔP |
| CUCPB5482 | DC3000 ΔhrpK1 ΔhrpZ1::nptII | ΔK,Z |
| CUCPB5095 | DC3000 ΔhrpZ1::nptII hrpW1::ΩSpr/Smr (13) | ΔZ,W |
| CUCPB5402 | DC3000 ΔhrpZ1::nptII ΔhopAK1 | ΔZ,AK |
| CUCPB5403 | DC3000 hrpW1::ΩSpr/Smr ΔhopAK1 | ΔW,AK |
| CUCPB5446 | DC3000 ΔhopAK1 ΔhopP1 | ΔAK,P |
| CUCPB5396 | DC3000 ΔhrpZ1::nptII hrpW1::ΩSpr/Smr ΔhopAK1 | ΔZ,W,AK |
| CUCPB5394 | DC3000 ΔhrpZ1::nptII hrpW1::ΩSpr/Smr ΔhopP1 | ΔZ,W,P |
| CUCPB5401 | DC3000 ΔhrpZ1::nptII hrpW1::ΩSpr/Smr ΔhopAK1 ΔhopP1 | ΔZ,W,AK,P |
| CUCPB5483 | DC3000 ΔhrpK1 ΔhrpZ1::nptII hrpW1::ΩSpr/Smr ΔhopAK1 ΔhopP1 | ΔK,Z,W,AK,P |
| Plasmids | ||
| pCPP5312 | pBBR Gateway RfB PavrPto1 avrPto1-cya; Gmr | |
| pCPP5702 | pUCP26::ΩKm PavRpto1 avrPto1-cya; Gmr Kmr | |
| pCPP3257 | pCPP3234 hrpZ1-cya; Spr Smr | HrpZ1-Cya |
| pCPP3258 | pCPP3234 hrpW1-cya; Spr Smr | HrpW1-Cya |
| pCPP3255 | pCPP3234 hopAK1-cya; Spr Smr | HopAK1-Cya |
| pCPP3256 | pCPP3234 hopP1-cya; Spr Smr | HopP1-Cya |
| pCPP5749 | pBS46 hrpK1; Gmr | K |
| pCPP5750 | pBS46 hrpZ1-HA; Gmr | Z |
| pCPP5751 | pBS46 hrpW1-HA; Gmr | W |
| pCPP5752 | pBS46 hopAK1-HA; Gmr | AK |
| pCPP5753 | pBS46 hopP1-HA; Gmr | P |
| pCPP5756 | pBS46 hopP1; Gmr | |
| pCPP5643 | pET-DEST42 hrpZ1-V5-His6; Apr | |
| pCPP5644 | pET-DEST42 hrpW1-V5-His6; Apr | |
| pCPP5098 | pET-DEST42 hopP1-V5-His6; Apr | |
| pCPP5760 | pET-DEST42 hopAK11-254-V5-His6; Apr | |
| pCPP3382 | pET-DEST42 ΔGateway cassette; Apr |
For the complete list of strains and plasmids used in this study, see Table S1 in the supplemental material.
Harpin gene polymutants partially reduce the ability of DC3000 to elicit the HR and translocate a model effector in tobacco but do not diminish effector production or secretion.
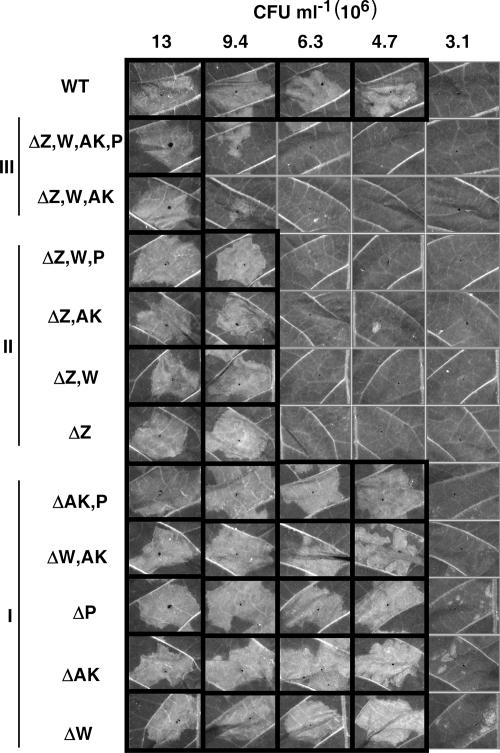
Pilot tests with the P. syringae pv. tomato DC3000 quadruple harpin gene mutant revealed that the mutant was partially reduced in T3SS-dependent plant interaction phenotypes, most notably, HR elicitation. Therefore, an HR threshold assay was designed to document minor variations among the mutants. The strains were infiltrated into panels of tobacco leaves in a tight dilution series over a range of approximately 1 × 107 to 3 × 106 CFU ml−1 (Fig. 4). This range spans the inoculum threshold for elicitation of the macroscopic HR by P. syringae pv. tomato DC3000. The harpin gene mutant strains fell into three HR phenotypic classes on the basis of the results of this assay. Five strains could not be distinguished from the wild type and were designated class I. These included the ΩhrpW1, ΔhopAK1, and ΔhopP1 single mutants and the ΔhopAK1 ΔhopP1 and ΩhrpW1 ΔhopAK1 double mutants. The four strains in class II had a mild but repeatable HR defect. This class included the ΔhrpZ1 single mutant, the ΔhrpZ1 ΩhrpW1 and ΔhrpZ1 ΔhopAK1 double mutants, and the ΔhrpZ1 ΩhrpW1 ΔhopP1 triple mutant. The unifying feature of class II mutants was the deletion of hrpZ1. Class III mutants had a moderate but easily detectable HR defect and included the ΔhrpZ1 ΩhrpW1 ΔhopAK1 triple mutant and the quadruple harpin gene mutant ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1. Interestingly, the ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 quadruple mutant and the ΔhrpZ1 ΩhrpW1 ΔhopAK1 triple mutant did not differ in the strengths of their HR phenotypes (Fig. 4). This suggests that HopP1 does not contribute to effector translocation.
FIG. 4.
P. syringae pv. tomato DC3000 harpin mutants divide into three classes based on reduced abilities to elicit the HR in tobacco. Strains were infiltrated into panels of tobacco leaves at the indicated concentrations, which represent a ca. 1.5-fold dilution series. HR was scored and photographed ca. 24 h after infiltration. Boxed panels indicate the collapse of at least half of the infiltrated tissue and therefore a positive HR. Phenotypic classes: I, no detectable difference from the wild type; II, mild HR reduction; III, moderate HR reduction. This experiment was repeated six times with similar results.
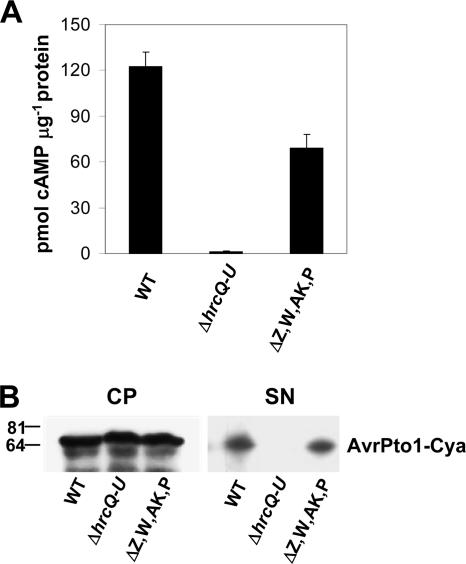
HR elicitation by P. syringae in a nonhost like tobacco is dependent on the translocation of effectors. To directly test the impact of the harpin gene mutations on effector translocation, we examined the abilities of the mutants to translocate AvrPto1-Cya into plant cells. The P. syringae pv. tomato DC3000 quadruple harpin gene mutant, as well as the wild-type and T3SS-deficient ΔhrcQ-U control strains, was transformed with pCPP5312, a vector that expresses the model T3SS effector AvrPto1 fused to Cya and uses the native avrPto1 promoter. The strains were infiltrated into tobacco at 2 × 107 CFU ml−1, close to the HR threshold of the P. syringae pv. tomato DC3000 quadruple harpin gene mutant. The P. syringae pv. tomato DC3000 quadruple harpin gene mutant was shown in repeated experiments to translocate significantly less AvrPto1-Cya than the wild-type strain (Fig. 5A.)
FIG. 5.
A P. syringae pv. tomato DC3000 quadruple harpin gene polymutant is moderately reduced in translocation but not in the secretion of AvrPto1-Cya. (A) AvrPto1-Cya translocation into tobacco. Strains transformed with the avrPto1-cya-expressing plasmid pCPP5312 were infiltrated into panels of tobacco leaves at 2 × 107 CFU ml−1. The T3SS-deficient control strain used was the ΔhrcQ-U mutant CUCPB5113. Two 1-cm-diameter leaf discs were harvested at 6 h postinfiltration and processed to determine the number of picomoles of soluble cAMP per microgram of protein. The values are means and standard deviations of two measurements. The results shown are representative of four experiments, and a paired t test of wild-type versus ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 (ΔZ,W,AK,P) values from the four experiments revealed a significant difference (α < 0.01) between them. (B) AvrPto1-Cya secretion in Hrp-inducing minimal medium. Strains transformed with pCPP5312 were grown in Hrp-inducing minimal medium from an OD600 of 0.3 to an OD600 of 0.5. Cell pellet (CP) and supernatant (SN) fractions were separated by centrifugation. Fractions were analyzed by SDS-PAGE, followed by immunoblotting with polyclonal anti-AvrPto1 antibodies. The assay was repeated four times with similar results. The values on the left are molecular sizes in kilodaltons.
A secretion assay in Hrp minimal medium was conducted with the same AvrPto1-Cya-expressing strains to determine if the quadruple harpin gene mutant possessed a defect in AvrPto1-Cya secretion. The quadruple harpin gene mutant expressed and secreted amounts of AvrPto1-Cya similar to those secreted by wild-type P. syringae pv. tomato DC3000 (Fig. 5B). Overall, these observations indicate that three of the four harpins contribute to effector translocation in planta but none contributes to effector production or secretion in culture.
A P. syringae pv. tomato DC3000 mutant lacking HrpK1 and all four harpins is strongly reduced in its ability to elicit the HR in nonhost tobacco and to grow in host tomato.
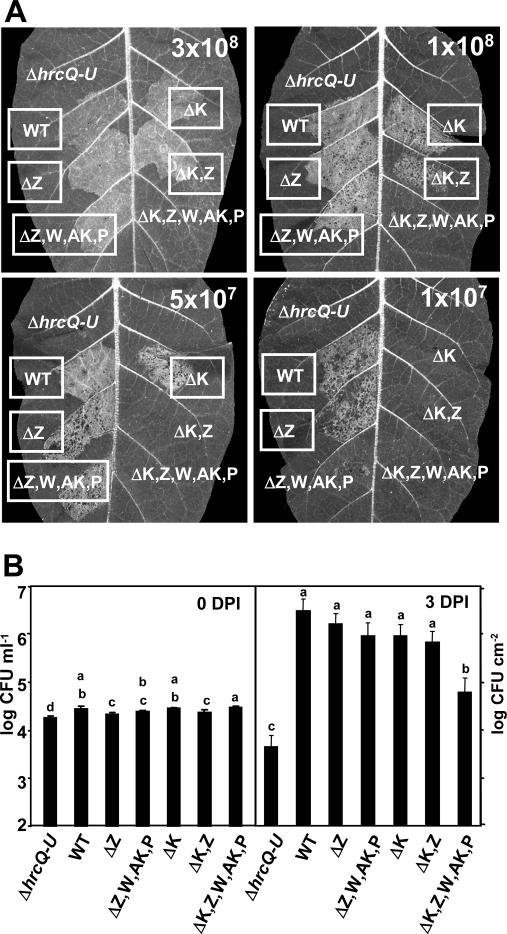
Because HrpK1 had been proposed previously to be a translocator (54), we generated a series of mutants to determine if harpins possess any function that overlaps that of hrpK1. Three hrpK1 mutant strains were generated, a ΔhrpK1 mutant, a ΔhrpK1 ΔhrpZ1 double mutant, and a ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant. The HR phenotype in tobacco leaves of the ΔhrpK1 mutant was very similar to that of the P. syringae pv. tomato DC3000 harpin gene polymutant (Fig. 6A), which is consistent with previous reports of the moderate HR phenotype of a hrpK1 P. syringae pv. tomato DC3000 mutant (54). The ΔhrpK1 ΔhrpZ1 double mutant showed a stronger reduction in HR elicitation ability, and the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant had the strongest phenotype, failing to elicit the HR even at the high concentration of 3 × 108 CFU ml−1 (Fig. 6A). However, the strain did not appear to be a complete null. At 1 × 109 CFU ml−1 elicitation of HR was observed with this strain approximately 50% of the time (data not shown).
FIG. 6.
The combination of mutations in P. syringae pv. tomato DC3000 harpin genes and hrpK1 produces a strong reduction in HR elicitation and virulence. (A) HR elicitation in tobacco. Strains were infiltrated into panels of tobacco leaves at the indicated concentrations. The T3SS-negative control strain was the ΔhrcQ-U mutant CUCPB5113. Leaves were scored for the HR and photographed at ca. 24 h after infiltration. Boxed labels indicate a positive HR. The assay was repeated twice with similar results. (B) Bacterial growth in tomato. Strains were diluted to 2 × 104 CFU ml−1 and infiltrated into half leaflets of tomato plants and incubated at 20 to 25°C with high humidity. Inoculum concentrations were confirmed by plating. Two 0.8-cm-diameter leaf discs were harvested from each infiltration area at 3 days postinfection (DPI). The discs were ground with a sterile mortar and pestle and diluted and plated to determine the number of CFU per square centimeter. Results are means and standard deviations of four infiltration areas per strain. Means marked with the same letter were not significantly different at the 5% confidence level on the basis of Duncan's multiple-range test. The assay was repeated twice with similar results.
The mutant strains also were tested for growth in host tomato and showed similar trends in this assay. Importantly, the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant was the only strain significantly reduced in growth in planta, although the growth of the polymutant was still significantly greater than that of the ΔhrcQ-U control (Fig. 6B).
A DC3000 mutant lacking HrpK1 and all four harpin genes is strongly reduced in translocation but not secretion of type III effectors.
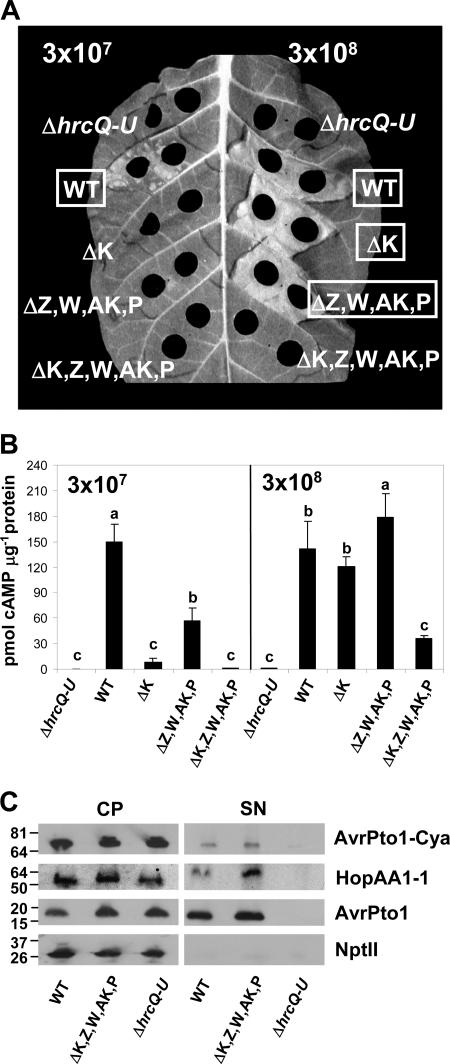
The various hrpK1 and harpin gene mutants were transformed with pCPP5702, a vector that expresses AvrPto1-Cya from its native promoter and also constitutively expresses NptII, a useful cytoplasmic marker for secretion assays. These strains were infiltrated into tobacco leaves at two inoculum levels and then simultaneously assayed for HR elicitation and AvrPto1-Cya translocation (Fig. 7A and B). The HR phenotypes were consistent with those observed in Fig. 6A, although the HR was generally weaker when AvrPto1-Cya was produced (Fig. 7A). At the lower inoculum level of 3 × 107 CFU ml−1, all three mutant strains failed to elicit an HR and were significantly reduced in AvrPto1-Cya translocation (Fig. 7B). Importantly, the translocation deficiency of the ΔhrpK1 mutant and the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant was not significantly different from that of the ΔhrcQ-U control (Fig. 7B). At the higher concentration of 3 × 108 CFU ml−1, only the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant failed to elicit an HR (Fig. 7A). Furthermore, only this mutant showed a significant reduction in the translocation of AvrPto1-Cya, and this reduction was not significantly different from that of the ΔhrcQ-U mutant. In interpreting the residual level of translocation by the polymutant, it is important to note that the Cya assay approaches saturation for the wild type at this high inoculum level (see Fig. S2 in the supplemental material), and therefore the relative translocation by the mutant is lower than the figure suggests.
FIG. 7.
A P. syringae pv. tomato DC3000 polymutant lacking four harpin genes and hrpK1 is strongly reduced in HR elicitation and in translocation but not secretion of test effectors. (A) HR elicitation in tobacco. Strains transformed with pCPP5702, a plasmid that expresses AvrPto1-Cya from its native promoter and NptII from a constitutive promoter, were infiltrated into tobacco leaves at the indicated concentrations. The T3SS-negative control strain used was the ΔhrcQ-U mutant CUCPB5113. HR was scored and photographed at ca. 24 h after infiltration. Boxed labels indicate a positive HR. The assay was repeated twice with similar results. (B) AvrPto1-Cya translocation into tobacco. Two 1-cm-diameter leaf discs were harvested at 6 h postinfiltration from the leaf panels shown in panel A and processed to determine the number of picomoles of soluble cAMP per microgram of protein. Values are means and standard deviations of three measurements. Means marked with the same letter are not significantly different at the 5% confidence level on the basis of Duncan's multiple-range test. The assay was repeated twice with similar results. (C) Secretion of T3SS substrates in Hrp minimal medium. P. syringae pv. tomato DC3000 and derivatives carrying pCPP5702 were grown in Hrp minimal medium from an OD600 of 0.3 to an OD600 of 0.5. Cell pellet (CP) and supernatant (SN) fractions were separated by centrifugation. Fractions were analyzed by SDS-PAGE, followed by immunoblotting with polyclonal antibodies against AvrPto1, HopAA1-1, and NptII as indicated. Plasmid-expressed AvrPto1-Cya was probed with polyclonal antibodies against AvrPto1. The assay was repeated three times with similar results. The values on the left are molecular sizes in kilodaltons.
To further define the T3SS deficiency in the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant, we examined the ability of the mutant to secrete in culture the native effectors AvrPto1 and HopAA1-1 and also plasmid-expressed AvrPto1-Cya. We observed no reduction in the ability of the mutant to secrete AvrPto1, HopAA1-1, or AvrPto1-Cya relative to that of the wild type. Thus, both the production and secretion of AvrPto1-Cya from the plasmid vector and the production and secretion of native HopAA1-1 and AvrPto1 were indistinguishable from the levels observed with the wild-type strain (Fig. 7C). Thus, the ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant has T3SS phenotypes typical of translocator mutants in that it can secrete but not translocate effectors.
HrpK1 and three of the four harpins can independently restore HR elicitation to a DC3000 mutant lacking HrpK1 and all four harpins.
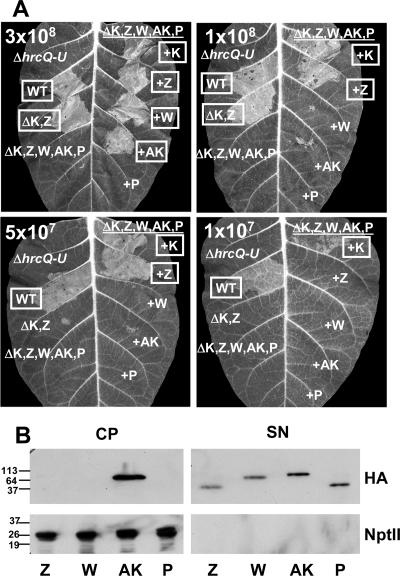
The ΔhrpK1 ΔhrpZ1 ΩhrpW1 ΔhopAK1 ΔhopP1 polymutant was transformed with complementing vectors expressing HrpK1 or harpins with C-terminal HA epitope tags. The transformed strains were assessed for their relative abilities to elicit the HR (Fig. 8A). HrpK1, HrpZ1, HrpW1, and HopAK1 could each independently restore some level of HR elicitation above that of the polymutant. HrpK1 produced the strongest complementation, followed by HrpZ1. HrpW1 and HopAK1 were equivalently moderate in their abilities to restore HR elicitation. This ranking, based on the strength of HR complementation, is similar to the ranking based on the relative reduction in HR observed with the various mutants (Fig. 4 and 6). HopP1 showed no ability to restore the HR, although it was secreted at levels similar to those of the other three harpins (Fig. 8B). Expression of HopP1 lacking the HA epitope tag did not alter the phenotype (data not shown). These observations with HopP1 are consistent with the failure of a hopP1 deletion to alter the HR phenotype of a ΔhrpZ1 ΩhrpW1 ΔhopAK1 triple mutant and argue against a role for HopP1 as a translocator. Interestingly, in repeated tests for secretion, HopAK was different from the other harpins in showing a strong signal in the cell pellet. However, as expected, all four harpins were secreted in culture.
FIG. 8.
HrpK1, HrpZ1, HrpW1, and HopAK1, but not HopP1, are independently capable of restoring HR elicitation on tobacco to the P. syringae pv. tomato DC3000 hrpK1 quadruple harpin gene polymutant. (A) HR complementation on tobacco. The P. syringae pv. tomato DC3000 hrpK1 quadruple harpin gene polymutant was transformed with plasmids expressing either HrpK1 or one of the four harpins with a C-terminal HA epitope tag. Strains were infiltrated into panels of tobacco leaves at the indicated concentrations. The left sides of the leaves were infiltrated with control strains and mutants. The right sides of the leaves were infiltrated with the P. syringae pv. tomato DC3000 hrpK1 quadruple harpin gene polymutant transformed with complementing vectors. The T3SS-negative control strain used was the ΔhrcQ-U mutant CUCPB5113. HR was scored and photographed at ca. 24 h postinfiltration. Boxed labels indicate a positive HR. The assay was repeated three times with similar results. (B) Harpin secretion in Hrp minimal medium. The P. syringae pv. tomato DC3000 hrpK1 quadruple harpin gene polymutant strains transformed with complementing vectors were grown in Hrp minimal medium from an OD600 of 0.3 to an OD600 of ca. 0.5. Cell pellet (CP) and supernatant (SN) fractions were separated by centrifugation. Fractions were analyzed by SDS-PAGE, followed by immunoblotting with monoclonal anti-HA antibodies and polyclonal anti-NptII antibodies as a lysis control. The assay was repeated twice with similar results. The values on the left are molecular sizes in kilodaltons.
DISCUSSION
We have found that P. syringae pv. tomato DC3000 produces four proteins with key characteristics of harpins and that three of these proteins functionally overlap HrpK1 in promoting the translocation of T3SS effectors into plant cells. None of these five proteins is essential for effector translocation, but a mutant lacking all of them has the translocon-deficient phenotype of being fully able to secrete effectors into the cultural milieu but poorly able to translocate effectors into host cells. These observations provide new insights into the nature of harpins and the translocation process in P. syringae, which we will discuss below.
The complete repertoire of harpins deployed by P. syringae pv. tomato DC3000.
Our conclusion that DC3000 deploys four harpins is based on an additional iteration of the bioinformatic/experimental process that yielded the complete Hop/Avr/effector repertoire of P. syringae pv. tomato DC3000 (12, 43, 63, 68). Specifically, we identified all of the DC3000 proteins that are actively deployed by the DC3000 T3SS, lack cysteine, and can elicit the HR when partially purified and infiltrated into tobacco leaf intercellular spaces. Importantly, the two new harpins found, HopAK1 and HopP1, have the same two-domain structure of HrpW1, and the C-terminal enzyme-like domains predicted substrates in the walls of the host and pathogen, respectively (thus, outside of the host cytoplasm). Because of HrpW-like structure, HopAK1 and HopP1 had previously been classified as “candidate T3SS helpers” (43).
In assessing the completeness of the harpin repertoire, it is important to note that DC3000 harbors genes that encode three other proteins with a HrpW-like structure (HopAH1, HopAH2-1, and HopAH2-2) and four apparent effectors that lack cysteine (HopI1, HopM1, HopAA1-1, and HopAQ1), as well as a cysteine-lacking homolog of the HrpA1 pilin (HopY1). The HopAH family proteins possess C-terminal cellulase-glycosyl hydrolase family 5 domains and can be translocated with C-terminal Cya fusions when expressed from the tac promoter in pCPP3234 (63). However, their genes are not preceded by Hrp promoters and they are not expressed under Hrp-inducing conditions (63). Thus, these harpin candidates appear to be inactive in DC3000. In contrast, the cysteine-lacking apparent effectors are expressed, but HopI1, HopM1, and HopAA1-1 are typical of effectors, not harpins, in being translocated when expressed from their native promoter (12) and in having biological activity inside plant cells (32, 42, 50). HopAQ1 is unlike P. syringae harpins in being very small (8.4 kDa) and having a low glycine content (4.8%) and high isoelectric point (11.0). We do not know why harpins characteristically lack cysteine, but our analysis of the DC3000 Hop repertoire supports the predictive value of this feature as a step in the comprehensive identification of harpin candidates in phytopathogen genomes.
Harpins have long been hypothesized to be involved in the translocation of effectors (40, 47-49, 54, 57). Our evidence that P. syringae harpins contribute to the translocation of effectors, and therefore function in the apoplast, is consistent with two important properties of these proteins. The first is the ability of harpins from various phytopathogens to elicit the HR from outside, rather than inside, plant cells (4, 13, 24, 28, 36, 67, 70). Although the relationship between this activity (based on experiments with purified harpin proteins and harpin genes expressed in planta) and harpin functions in natural infections is unclear, these observations nevertheless establish that harpins are distinct from effectors in having biological activity in the apoplast. Also pointing to activity in the apoplast is the increasing evidence that harpins are typically secreted more strongly but translocated less efficiently than effectors. For example, HrpW1 and HopAK1 are not translocated as fusions with AvrRpt280-255 when expressed from their native promoter (12). Similarly, a proteomic analysis of proteins secreted by P. syringae pv. tomato DC3000 in Hrp minimal medium by mass spectrometry and tandem mass spectrometry combined searches and data obtained on an AB4700 tandem time of flight instrument revealed all four harpins and HrpK1 to be secreted (K. Lee, S. Cartinhour, and D. Schneider, personal communication; www.leelab.org/). However, this same analysis found only three effectors in the medium, i.e., AvrPto1, HopM1, and HopAM1. It is noteworthy that all three of these effectors are strongly expressed (21) and hopAM1 is duplicated in the DC3000 genome (43). On the basis of all of these observations, we conclude that the repertoire of active harpins in DC3000 is complete with HrpZ1, HrpW1, HopAK1, and HopP1.
The atypical properties of HopP1 as a harpin.
HopP1 possesses the primary characteristics of a harpin, but it differs from other harpins in several aspects. HopP1 is unique to P. syringae pv. tomato DC3000, whereas HrpZ1, HrpW1, and HopAK1 are widely distributed among P. syringae pathovars. The predicted LT domain of HopP1 implies a potential interaction with peptidoglycan in the bacterial periplasm rather than any extracellular activity. Also, HopP1 behaves like an effector in that it is translocated at a much higher level than other harpins, although it also appears to be abundantly secreted in culture. The deletion of hopP1 does not affect the T3SS-dependent phenotypes of the harpin gene polymutants, and expression in trans of HopP1 cannot restore HR elicitation to the P. syringae pv. tomato DC3000 ΔhrpK1 quadruple harpin gene polymutant, which argues against a role for HopP1 in effector translocation. Lastly, HopP1 has been demonstrated recently to have an overlap in function with HrpH (PSPTO1378), the T3SS-associated specialized lytic transglycosylase (53). All of this implies that the function of HopP1 differs from that of the other harpins which appear to have an overlap in function with HrpK1.
Evaluation of the role of HrpK1 as a translocator.
Because our complementation experiments with the ΔhrpK1 quadruple harpin gene polymutant suggest that HrpK1 and three of the harpins have overlapping functions, it is important to assess the evidence that HrpK1 is a translocator. An overriding problem in this regard is that P. syringae hrpK1 mutants of various pathovars have been reported to have only variably reduced or conditional phenotypes (6, 7, 16, 45, 54). A recent report by Petnicki-Ocwieja et al. (54) showed that P. syringae pv. tomato DC3000 HrpK1 is itself secreted and capable of presenting C-terminally fused Cya or AvrRpt281-255 to the plant cell cytoplasm. Furthermore, a hrpK1 mutant was partially reduced in the translocation-dependent phenotypes of virulence and HR elicitation. These phenotypes could not be restored by expression of hrpK1 in the plant, further suggesting that extracellular HrpK1 is functioning in the T3SS pathway, rather than as an effector itself. Finally, deletion of a TM domain in the C terminus, which is characteristic of YopB and HrpF/NopX family translocators, abolished these HrpK1 abilities. Here, we have extended this evidence by showing directly that HrpK1 is needed for robust translocation but not secretion of T3SS effectors, and we have generated a strong translocation-deficient phenotype with a polymutant and showed that HrpK1 can restore T3SS translocation function to the polymutant. Although there is currently no direct evidence of membrane pore-forming ability by HrpK1, the congruence of structure and phenotype with better-studied members of the HrpF/NopX family strongly suggests that HrpK1 is a translocator.
The distribution and relative roles of harpins and other classes of T3SS translocators in phytopathogens.
The P. syringae pv. tomato DC3000 translocators fall into three classes, a HrpF/NopX class (HrpK1), a one-domain harpin class (HrpZ1), and a two-domain harpin class (HrpW1 and HopAK1). (Our use of the term domain refers to obvious structural differences in the two classes of harpins and does not preclude the potential for multiple functional or folding domains in these proteins.) As summarized in Table 2, diverse other phytopathogens that have sequenced genomes and well-studied T3SSs also possess representatives of each of these three classes of putative translocators. However, these pathogens differ in the relative importance of each class in translocation-dependent phenotypes and in gene amplification within each class. The HrpF/NopX class is illustrative. HrpF appears to be the dominant, essential translocator in X. campestris pv. vesicatoria, although the XopA single-domain harpin was also implicated in translocation, based on a mutant showing partial reduction in virulence and HR elicitation without an effect on T3SS secretion (49). In contrast, HrpK, the only member of this class in E. amylovora, appears to make little to no contribution to translocation; instead, HrpN, a single-domain harpin, appears to be the dominant translocator because hrpN mutants are strongly reduced in both HR and virulence but not in T3SS secretion (34, 51, 70).
TABLE 2.
T3SS translocators of model phytopathogens
| Translocator class, enzyme-like domaina | Protein (reference[s])
|
|||
|---|---|---|---|---|
| Hrp1 T3SS
|
Hrp2 T3SS
|
|||
| P. syringae pv. tomato DC3000 | E. amylovora Ea273/321 | X. campestris pv. vesicatoria 85-10 | R. solanacearum GMI1000 | |
| HrpF/NopX family, NAb | HrpK1**c (54, this study) | HrpK° (52) | HrpFd**** (10, 60) | PopF1*** (47), PopF2* (47) |
| One-domain harpins, NA | HrpZ1* (40, this study) | HrpN*** (48) | XopA* (49) | PopA1° (57) |
| Two-domain harpins | ||||
| PL3 | HrpW1* (this study) | HrpW° (24) | —e | PopW |
| PL1 | HopAK1* (this study) | Eop2 | ||
| SLT | HopP1° (this study) | |||
| CEL | (HopAH1, HopAH2-1, HopAH2-2)f | |||
PL3, pectate lyase 3; PL1, pectate lyase 1; SLT, soluble lytic transglycosylase; CEL, cellulase (18, 19, 46).
NA, not applicable.
Demonstrated translocator function is indicated by symbols that show phenotypic strength as follows: ****, translocation null; ***, strong reduction/almost null; **, moderate reduction; *, mild reduction/almost wild type; °, indistinguishable from wild type; no symbol, uncharacterized.
Proteins in bold font have been shown to form pores in lipid bilayers.
—, Uncharacterized HrpW1 homologs are present in X. campestris pv. campestris and X. axonopodis pv. citri.
The R. solanacearum translocator repertoire provides further contrasts. Strain GMI1000 encodes two HrpF/NopX family members, PopF1 and PopF2. Only the deletion of popF1 produces a translocator phenotype characterized by a strong decrease in HR, virulence, and effector translocation without the loss of effector secretion. This implies that PopF1 is the dominant translocator. However, popF2 expression in trans was shown to complement popF1 phenotypes, indicating that PopF2 is functionally redundant to PopF1. Another interesting observation is that R. solanacearum UW551 codes for PopF1 and a different HrpF/NopX family member, PopX. It has been proposed, on this basis, that the translocation strategies of Ralstonia may be diverse (47). R. solanacearum GMI1000 also expresses the single-domain harpin PopA, which has been shown to form ion-conducting pores in artificial and biological membranes. However, popA mutants have no detectable phenotype (4, 57). It would be interesting to determine if PopA overexpression could complement any of the ΔpopF1 phenotypes. Lastly, R. solanacearum GMI1000 also encodes a HrpW1 homolog, PopW, that has not been characterized.
The comparison of the P. syringae pv. tomato DC3000 translocator repertoire with other phytopathogen repertoires in Table 2 reveals that two-domain PL1 harpins appear to be unique to Hrp1 T3SSs and that there is a striking amplification of two-domain harpin genes in P. syringae pv. tomato DC3000. This amplification includes hopP1, which is unique to DC3000 among the sequenced P. syringae strains, and the hopAH family, which appears to be unexpressed in DC3000. However, the P. syringae pv. syringae B728A hopAH1 gene is preceded by a Hrp promoter (25, 43) and the protein can be translocated into plant cells as a full-length fusion with the AvrRpt2101-255 reporter, suggesting that HopAH1 is deployed by B728a (69). Variation in P. syringae translocator repertoires can also extend to the single-domain harpin class, as indicated by the failure of P. syringae pv. tabaci to produce functional HrpZ (65). In summary, effector translocation by Hrp2 T3SSs is strongly dependent on HopF/NopX translocators, whereas effector translocation by P. syringae pv. tomato DC3000 involves a consortium of interchangeable translocators from all three classes.
Translocator repertoire expansion in phytopathogens in the context of plant barriers to the T3SS.
What has driven the amplification of translocator genes in phytopathogens? One explanation is that translocation is a two-step process and that harpins are important in the first step of crossing the plant cell wall, whereas HrpF/NopX family members are important in the second step of crossing the plant plasma membrane. Unfortunately, we currently have no way to experimentally distinguish these putative steps in the overall translocation process. Observations supporting a two-step model include that HrpW1 and HopAK1 have pectate lyase domains, which predicts interaction with plant cell wall components, whereas HrpK1 is a member of a well-studied family of translocators predicted to interact with the plasma membrane rather than the cell wall. The two-step translocation model implies that the HrpA pilus is poorly able to breach the cell wall without factors like the two-domain harpins, which would be secreted by the growing pilus. Thus, the need to service this first step in translocation would at least partially explain the amplification of translocator-like factors.
An alternative model is that the HrpA pilus has an intrinsic ability to breach the plant cell wall and that harpins and the HrpF/NopX family proteins function as a consortium in TM translocation. We favor the translocator consortium model for the following reasons. First, the weak phenotype of the ΩhrpW1 ΔhopAK1 double mutant does not support a role for these proteins in an essential first step in translocation. Second, these proteins can at least partially restore translocator function to a polymutant lacking all known translocators. Third, the N-terminal region of two-domain harpins is biologically similar to that of HrpZ1, which is known to form pores in membranes. Finally, it is noteworthy that the type IV secretion system of Agrobacterium tumefaciens can translocate proteins and a nucleoprotein complex through plant cell walls via a similarly sized pilus without the benefit of any harpins (15).
In the translocator consortium model, the pectate lyase domains in the two-domain harpins could function as scaffolding factors supporting translocon assembly, which is analogous to the role of LcrV as a scaffold for the assembly of the YopB/D translocator pore in Yersinia. Our data suggest that such a scaffold is not essential in P. syringae and that all components of the consortium have at least some ability to serve as independent translocators. Future testing of the ability of the two-domain harpins to interact with membranes will be important in evaluating this model.
Plant defense surveillance is thought to have driven the expansion of effector repertoires in phytopathogens, and such surveillance may similarly explain the amplification and diversity of translocators. For instance, tobacco responds with strong defenses to purified harpins, and P. syringae pv. tabaci, a compatible tobacco pathogen, may have diminished this defense through loss of a functional hrpZ gene (65). Similarly, the single-domain harpin and proposed translocator, XopA, of X. campestris pv. vesicatoria is truncated at a conserved alpha helix essential for exogenous HR elicitation by its homologs (35). The use of a variable consortium of translocators may benefit phytopathogens by keeping the concentration of individual translocators below the recognition thresholds of host surveillance systems and by enabling individual translocators that have come under host surveillance to be lost from the consortium without loss of overall translocon function.
Supplementary Material
Acknowledgments
This work was supported by NSF grant MCB-0544066.
We thank Kent Loeffler for photography, Stephen Lory for pEXGmGW, and Bryan Swingle for pBS46. We also thank Kelvin Lee, Samuel Cartinhour, and David Schneider for making P. syringae pv. tomato DC3000 proteomic data available.
Footnotes
Published ahead of print on 14 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahmad, M., D. R. Majerczak, S. Pike, M. E. Hoyos, A. Novacky, and D. L. Coplin. 2001. Biological activity of harpin produced by Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 14:1223-1234. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., D. W. Bauer, T. M. Milos, and A. Collmer. 1996. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19:715-728. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badel, J. L., R. Shimizu, H. S. Oh, and A. Collmer. 2006. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant-Microbe Interact. 19:99-111. [DOI] [PubMed] [Google Scholar]
- 6.Bozso, Z., P. G. Ott, M. L. Kecskes, and Z. Klement. 1999. Effect of heat and cycloheximide treatment of tobacco on the ability of Pseudomonas syringae pv. syringae 61 hrp/hrmA mutants to cause HR. Physiol. Mol. Plant Pathol. 55:215-223. [Google Scholar]
- 7.Bozso, Z., P. G. Ott, A. Szatmari, A. Czelleng, G. Varga, E. Besenyei, E. Sardi, E. Banyai, and Z. Klement. 2005. Early detection of bacterium-induced basal resistance in tobacco leaves with diaminobenzidine and dichlorofluorescein diacetate. J. Phytopathol. 153:596-607. [Google Scholar]
- 8.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büttner, D., and U. Bonas. 2002. Port of entry—the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 10.Büttner, D., D. Nennstiel, B. Klusener, and U. Bonas. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, J. H., A. K. Goel, S. R. Grant, and J. L. Dangl. 2004. Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr. Opin. Microbiol. 7:11-18. [DOI] [PubMed] [Google Scholar]
- 12.Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu, S. Gombar, S. R. Grant, F. M. Ausubel, and J. L. Dangl. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charkowski, A. O., J. R. Alfano, G. Preston, J. Yuan, S. Y. He, and A. Collmer. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 15.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collmer, A., J. L. Badel, A. O. Charkowski, W. L. Deng, D. E. Fouts, A. R. Ramos, A. H. Rehm, D. M. Anderson, O. Schneewind, K. van Dijk, and J. R. Alfano. 2000. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 97:8770-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 18.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 19.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.
- 20.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira, A. O., C. R. Myers, J. S. Gordon, G. B. Martin, M. Vencato, A. Collmer, M. D. Wehling, J. R. Alfano, G. Moreno-Hagelsieb, W. F. Lamboy, G. DeClerck, D. J. Schneider, and S. W. Cartinhour. 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis clement, and identifies novel coregulated genes. Mol. Plant-Microbe Interact. 19:1167-1179. [DOI] [PubMed] [Google Scholar]
- 22.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouts, D. E., J. L. Badel, A. R. Ramos, R. A. Rapp, and A. Collmer. 2003. A Pseudomonas syringae pv. tomato DC3000 Hrp (type III secretion) deletion mutant expressing the Hrp system of bean pathogen P. syringae pv. syringae 61 retains normal host specificity for tomato. Mol. Plant-Microbe Interact. 16:43-52. [DOI] [PubMed] [Google Scholar]
- 24.Gaudriault, S., M. N. Brisset, and M. A. Barny. 1998. HrpW of Erwinia amylovora, a new Hrp-secreted protein. FEBS Lett. 428:224-228. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg, J. T., and B. A. Vinatzer. 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6:20-28. [DOI] [PubMed] [Google Scholar]
- 26.Håkansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 28.He, S. Y., H. C. Huang, and A. Collmer. 1993. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73:1255-1266. [DOI] [PubMed] [Google Scholar]
- 29.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 30.Hoyos, M. E., C. W. Stanley, S. Y. He, S. Pike, X. A. Pu, and A. Novacky. 1996. The interaction of harpinPss, with plant cell walls. Mol. Plant-Microbe Interact. 9:608-616. [Google Scholar]
- 31.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 32.Jelenska, J., N. Yao, B. A. Vinatzer, C. M. Wright, J. L. Brodsky, and J. T. Greenberg. 2007. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin, Q., R. Thilmony, J. Zwiesler-Vollick, and S. Y. He. 2003. Type III protein secretion in Pseudomonas syringae. Microbes Infect. 5:301-310. [DOI] [PubMed] [Google Scholar]
- 34.Kim, J. F., and S. V. Beer. 1998. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, J. G., E. Jeon, J. Oh, J. S. Moon, and I. Hwang. 2004. Mutational analysis of Xanthomonas harpin HpaG identifies a key functional region that elicits the hypersensitive response in nonhost plants. J. Bacteriol. 186:6239-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J.-G., B. K. Park, C.-H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 38.Lan, L., X. Deng, J. Zhou, and X. Tang. 2006. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS. Mol. Plant-Microbe Interact. 19:976-987. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J., D. F. Klessig, and T. Nurnberger. 2001. A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J., B. Klusener, G. Tsiamis, C. Stevens, C. Neyt, A. P. Tampakaki, N. J. Panopoulos, J. Noller, E. W. Weiler, G. R. Cornelis, J. W. Mansfield, and T. Nurnberger. 2001. HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. USA 98:289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, C. M., M. Haapalainen, J. Lee, T. Nurnberger, M. Romantschuk, and S. Taira. 2005. Harpin of Pseudomonas syringae pv. phaseolicola harbors a protein binding site. Mol. Plant-Microbe Interact. 18:60-66. [DOI] [PubMed] [Google Scholar]
- 42.Li, X., H. Lin, W. Zhang, Y. Zou, J. Zhang, X. Tang, and J. M. Zhou. 2005. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102:12990-12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindeberg, M., S. Cartinhour, C. R. Myers, L. M. Schechter, D. J. Schneider, and A. Collmer. 2006. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant-Microbe Interact. 19:1151-1158. [DOI] [PubMed] [Google Scholar]
- 44.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 45.Mansfield, J., C. Jenner, R. Hockenhull, M. A. Bennett, and R. Stewart. 1994. Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol. Plant-Microbe Interact. 7:726-739. [DOI] [PubMed] [Google Scholar]
- 46.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer, D., S. Cunnac, M. Gueneron, C. Declercq, F. Van Gijsegem, E. Lauber, C. Boucher, and M. Arlat. 2006. PopF1 and PopF2, two proteins secreted by the type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 188:4903-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nissinen, R. M., A. J. Ytterberg, A. J. Bogdanove, K. J. Van Wijk, and S. V. Beer. 2007. Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8:55-67. [DOI] [PubMed] [Google Scholar]
- 49.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the Hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nomura, K., S. Debroy, Y. H. Lee, N. Pumplin, J. Jones, and S. Y. He. 2006. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313:220-223. [DOI] [PubMed] [Google Scholar]
- 51.Oh, C. S., and S. V. Beer. 2005. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253:185-192. [DOI] [PubMed] [Google Scholar]
- 52.Oh, C. S., J. F. Kim, and S. V. Beer. 2005. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6:125-138. [DOI] [PubMed] [Google Scholar]
- 53.Oh, H.-S., B. H. Kvitko, J. E. Morello, and A. Collmer. 2007. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effectors into plant cells. J. Bacteriol. 189:8277-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petnicki-Ocwieja, T., K. van Dijk, and J. R. Alfano. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preston, G., H. C. Huang, S. Y. He, and A. Collmer. 1995. The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol. Plant-Microbe Interact. 8:717-732. [DOI] [PubMed] [Google Scholar]
- 56.QIAGEN. 2001. The QIAexpressionist: a handbook for high-level expression and purification of 6xHis-tagged proteins, 5th ed. QIAGEN, Valencia, CA.
- 57.Racapé, J., L. Belbahri, S. Engelhardt, B. Lacombe, J. Lee, J. Lochman, A. Marais, M. Nicole, T. Nurnberger, F. Parlange, S. Puverel, and H. Keller. 2005. Ca-dependent lipid binding and membrane integration of PopA, a harpin-like elicitor of the hypersensitive response in tobacco. Mol. Microbiol. 58:1406-1420. [DOI] [PubMed] [Google Scholar]
- 58.Ramos, A. R., J. E. Morello, S. Ravindran, W. L. Deng, H. C. Huang, and A. Collmer. 2007. Identification of Pseudomonas syringae pv. syringae 61 type III secretion system Hrp proteins that can travel the type III pathway and contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189:5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose, J. K. C. 2003. The plant cell wall. Blackwell Press Ltd., Oxford, United Kingdom.
- 60.Rossier, O., G. Van den Ackerveken, and U. Bonas. 2000. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38:828-838. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schechter, L. M., M. Vencato, K. L. Jordan, S. E. Schneider, D. J. Schneider, and A. Collmer. 2006. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant-Microbe Interact. 19:1180-1192. [DOI] [PubMed] [Google Scholar]
- 64.Simon, R., U. Priefer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 65.Taguchi, F., R. Tanaka, S. Kinoshita, Y. Ichinose, Y. Imura, S. Andi, K. Toyoda, T. Shiraishi, and T. Yamada. 2001. Harpinpsta from Pseudomonas syringae pv. tabaci is defective and deficient in its expression and HR-inducing activity. J. Gen. Plant Pathol. 67:116-123. [Google Scholar]
- 66.Tampakaki, A. P., V. E. Fadouloglou, A. D. Gazi, N. J. Panopoulos, and M. Kokkinidis. 2004. Conserved features of type III secretion. Cell. Microbiol. 6:805-816. [DOI] [PubMed] [Google Scholar]
- 67.Tampakaki, A. P., and N. J. Panopoulos. 2000. Elicitation of hypersensitive cell death by extracellularly targeted HrpZPsph produced in planta. Mol. Plant-Microbe Interact. 13:1366-1374. [DOI] [PubMed] [Google Scholar]
- 68.Vinatzer, B. A., J. Jelenska, and J. T. Greenberg. 2005. Bioinformatics correctly identifies many type III secretion substrates in the plant pathogen Pseudomonas syringae and the biocontrol isolate P. fluorescens SBW25. Mol. Plant-Microbe Interact. 18:877-888. [DOI] [PubMed] [Google Scholar]
- 69.Vinatzer, B. A., G. M. Teitzel, M. W. Lee, J. Jelenska, S. Hotton, K. Fairfax, J. Jenrette, and J. T. Greenberg. 2006. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62:26-44. [DOI] [PubMed] [Google Scholar]
- 70.Wei, Z. M., R. J. Laby, C. H. Zumoff, D. W. Bauer, S. Y. He, A. Collmer, and S. V. Beer. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257:85-88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.